Abstract
Clostridium perfringens is the causative pathogen of avian necrotic enteritis. Lactobacillus spp. are well-characterized probiotics with anti-microbial and immune-modulatory activities. In the present study, we investigated the effects of L. acidophilus and L. fermentum on the growth, α-toxin production and inflammatory responses of C. perfringens. In in vitro culture experiments, both lactobacilli inhibited the growth of C. perfringens (P < 0.01), accompanied with a decrease in pH (P < 0.01). Supernatants from lactobacilli cultures also suppressed the growth of C. perfringens during 24 h of incubation (P < 0.01), but this inhibitory effect disappeared after 48 h. Both lactobacilli decreased the α-toxin production of C. perfringens (P < 0.01) without influencing its biomass, and even degraded the established α-toxin (P < 0.01). Lower environmental pH reduced the α-toxin production as well (P < 0.01). Preincubation with L. acidophilus decreased the attachment of C. perfringens to cells (P < 0.01) with the cell cytotoxicity being unaffected. Both lactobacilli pretreatment reduced the up-regulation of proinflammatory factors, peptidoglycan (PGN) receptors and nuclear factor kappa B (NF-κB) p65 in C. perfringens-challenged chicken intestinal epithelial cells (P < 0.05). In conclusion, L. acidophilus and L. fermentum inhibited the pathological effects of C. perfringens in vitro conditions.
Keywords: Lactobacillus, Clostridium perfringens, α-toxin, chicken, intestinal epithelial cell, inflammation
Introduction
Clostridium perfringens is a robust Gram-positive anaerobic spore-forming and rod-shaped bacterium, and able to produce up to 18 toxins and extracellular enzymes, causing diseases, such as necrotic enteritis (NE) in poultry, equine colitis, food poisoning, and gas gangrene (Songer and Meer, 1996; Revitt-Mills et al., 2015). C. perfringens strains are classified into five toxinotypes (A, B, C, D, and E) based on the production of four major toxins (α, β, ε, and ι) (Petit et al., 1999). Type A strains are the most widespread in the intestines of warm-blooded animals and in the environment, mainly responsible for avian NE (Songer, 1996). In 2015, the costs of NE came close to USD six billion (Wade and Keyburn, 2015).
The α-toxin is produced by all five types of C. perfringens and belongs to a family of bacterial zinc-metallo phospholipase C enzymes. Although α-toxin is not the essential virulence factor to cause NE (Keyburn et al., 2006), it might still be important in the pathogenesis. It was reported that α-toxin was detected in the intestines of broiler chickens inoculated with a α-toxin mutant strain of C. perfringens and there was a direct relationship between intestinal lesion severity and amount of α-toxin determined in the gut contents and mucosa (R = 0.89–0.99) (Coursodon et al., 2010). Furthermore, α-toxin is a key mediator of human gas gangrene and bovine necro-haemorrhagic enteritis (Bryant et al., 2000; Goossens et al., 2017). It could result in myonecrosis, hemorrhage and neutrophil infiltration in human and animals.
Microscopic examination of early stages of NE shows strong intestinal inflammation to C. perfringens infection (Timbermont et al., 2011). The lamina propria of intestine is hyperaemic and infiltrated with numerous inflammatory cells, mainly heterophilic granulocytes. Moreover, the gene expression of cytokines, such as interferon (IFN)-γ, interleukin (IL)-1β, and IL-10 was significantly up-regulated in the intestine of NE chickens (Collier et al., 2008; Park et al., 2008). Using in vitro primary culture of chicken intestinal epithelial cells, we previously demonstrated that both C. perfringens and α-toxin challenge induced intense cytokine expression by activating the nuclear factor kappa B (NF-κB) signaling pathway (Guo et al., 2015). Furthermore, the cell receptors, Toll-like receptor (TLR) 2 and nucleotide-binding oligomerization domain (NOD) 1, might play a role in the recognition of peptidoglycan (PGN) component of C. perfringens (Lu et al., 2009; Guo et al., 2015).
Lactobacillus spp. are members of the normal intestinal flora and used as probiotics to prevent the growth and colonization of C. perfringens in NE model of poultry (La Ragione et al., 2004; Rahimi et al., 2011). It was reported that orally administration of Lactobacillus fermentum strain 1.2029 reduced the inflammatory damage of NE in chickens by up-regulating IL-10 mRNA levels and down-regulating the mRNA expression of IFN- γ and TLR2 (Cao et al., 2012). In a co-infection model with Eimeria spp. and C. perfringens, broiler chickens fed L. johnsonii had lower NE lesion scores and improved feed efficiency (Geier et al., 2010). In vitro co-culture system, L. fermentum strain 104R silenced C. perfringens β2 toxin production by decreasing cpb2 mRNA, which was most likely a result of the decline in environmental pH (Allaart et al., 2011). However, the effect of Lactobacillus spp. on the α-toxin production and inflammation of C. perfringens as well as the underlying mechanisms are largely unknown. The purpose of this study was to investigate the effects of Lactobacillus acidophilus and Lactobacillus fermentum on the growth and α-toxin production of C. perfingens as well as the inflammatory responses induced by this pathogen in the intestinal epithelial cells of chicken embryos.
Materials and methods
Materials
All experimental procedures used were approved by the China Agricultural University Animal Care and Use Committee. Bacterium culture media and agars were obtained from Beijing Land Bridge Technology Co. Ltd. (Beijing, China). Cell culture media, buffers and fetal bovine serum (FBS) were purchased from Gibco (Carlsbad, CA, USA). Epidermal growth factor (EGF) was purchased from BD Biosciences (San Jose, CA, USA). Bacterium and cell culture plastics were obtained from Corning Life Science (Tewksbury, MA, USA). All other chemicals, unless specified, were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Bacterial strains and culture conditions
Lactobacillus acidophilus (CGMCC No. 1.1878) and L. fermentum (CGMCC No. 1.2029) were obtained from China General Microbiological Culture Collection Center (CGMCC, Beijing, China), and cultured in Mann-Rogosa-Sharpe (MRS) broth. The chicken C. perfringens type A field strain (CVCC No. 2030) was obtained from China Veterinary Culture Collection Center (CVCC, Beijing, China) and cultured in cooked meat medium. Both lactobacilli and C. perfringens were incubated at 37°C under anaerobic conditions. All bacterial experiments were performed with MRS broth, since this provided the best overall growth environment for all species (Allaart et al., 2011). For cell experiments, overnight-incubated bacterial cultures were centrifuged at 4,000 × g for 10 min, washed twice by sterile phosphate buffered saline (PBS, pH 7.4) and resuspended in cell culture medium. Bacterial concentration was estimated by measuring the absorbance at 600 nm and by relating the optical density to colony forming units (CFU, data not shown).
Lactobacilli and C. perfringens co-culture experiments
The procedures for bacterial co-culture experiments used in the current study were previously described by Jiang et al. (2014) with some modifications. Strains of lactobacilli and C. perfringens were cultured individually under anaerobic conditions in MRS broth and cooked meat medium, respectively, overnight to reach a stationary phase. Both cultures were centrifuged at 4,000 × g for 10 min. The pellet was then washed once with PBS and resuspended in fresh MRS broth with a concentration of 1 × 107 CFU/mL. Thirty microliters of L. acidophilus and L. fermentum suspensions were individually inoculated into 3 mL of fresh MRS broth additionally containing 30 μL of C. perfringens suspension. Therefore, the ratio of Lactobacilli to C. perfringens was 1:1 in a 3.06 mL co-culture system. Another 30 μL suspensions of L. acidophilus, L. fermentum, and C. perfringens were individually added to the 3.03 mL of fresh MRS broth. This experiment was performed in triplicate. Cultures were anaerobically incubated at 37°C for 20 h. Following incubation, CFU of lactobacilli and C. perfringens were determined on MRS agar plates and tryptose-sulfite-cycloserine (TSC) agar plates, respectively. Meanwhile, the pH of cultures was measured.
Agar well diffusion assay and broth culture inhibition assay
The preparation of cell-free supernatant from lactobacilli cultures and agar well diffusion assay as well as broth culture inhibition assay was performed as previously described (Schoster et al., 2013), with some modifications. Briefly, lactobacilli suspensions were adjusted to an optical density of 0.220 at 600 nm and 1 mL of this suspension was inoculated into 9 mL of MRS broth. After 24 and 48 h of incubation, the lactobacilli cultures were centrifuged at 4,000 × g for 10 min and the supernatant was collected, filter-sterilized using a 0.22 μm membrane syringe filter and divided into two aliquots, one of which was neutralized to pH 6.20 using NaOH. Both aliquots were checked for absence of viable bacteria by plating on MRS agar and then frozen at −80°C for further use.
Overnight culture of C. perfringens was diluted 10-fold up to 10−2 to achieve confluent growth. When the autoclaved TSC medium cooled down to about 55°C, 100 μL of bacterial dilution were mixed well with 20 mL TSC medium in a 10-cm plastic plate. Five 9-mm wells were made in each agar plate and 100 μL of lactobacilli supernatants were added to the four outer wells whereas sterile MRS broth was put into the central well (control well). When the effect of lactobacilli supernatants without neutralization was evaluated, MRS broth adjusted to pH 4.20 (using HCl) was added to the control well. When using neutralized supernatants, MRS broth at pH 6.20 was used instead. The presence of an inhibition zone >1 mm was assessed visually following 16 h of anaerobic incubation at 37°C. Each assay was performed in duplicate.
Fifty microliters of C. perfringens suspension were added to 5 mL of fresh MRS broth and 1 mL of lactobacilli supernatants harvested after 48 h of incubation. MRS broth at pH 6.20 and pH 4.20 was used as controls for the supernatant with and without pH neutralization, respectively. Growth was measured spectrophotometrically at 0, 24 and 48 h after anaerobic incubation at 37°C. The differences of OD600 measurements between 0 h of incubation and 24 or 48 h of incubation were calculated. Each assay was performed in triplicate.
The effect of lactobacilli on C. perfringens α-toxin production
Fifteen milliliters of C. perfringens cultures in a stationary phase were centrifuged at 4,000 × g for 10 min and washed once with PBS and resuspended in the same volume of fresh MRS broth. This suspension was divided into three equal parts. Two of them were supplemented with the bacterial pellets of L. acidophilus and L. fermentum, respectively. The suspension without lactobacilli pellets was used as the control. After 4 h of anaerobic incubation at 37°C, the numbers of CFU of C. perfringens were determined on TSC agar plates and the pH was measured. The cultures were centrifuged at 4,000 × g for 10 min, and α-toxin level in the supernatants was analyzed.
As lactobacilli could greatly decrease the pH of cultures, we examined α-toxin production at normal and lower pHs. Pellets from stationary culture of C. perfringens were resuspended in MRS broth set at pH 5.20 or pH 6.20 using HCl and incubated anaerobically at 37°C for 2 or 4 h. After each time period, the numbers of CFU of C. perfringens were determined and the pH of each culture was measured. The supernatants of cultures were collected for α-toxin assay. All experiments were performed in triplicate.
C. perfringens α-toxin degradation by lactobacilli
Pellets from stationary culture of C. perfringens were resuspended in the same volume of fresh MRS broth and incubated anaerobically at 37°C for 4 h. The supernatant of this culture were collected by centrifugation (4,000 × g, 10 min) and divided into three equal parts. Pellets of L. acidophilus and L. fermentum cultures were individually resuspended in two parts of this supernatant and incubated anaerobically at 37°C for 2 or 4 h. The culture without lactobacilli pellets was used as the control. At each time point, the pH of each culture was measured and the supernatants of cultures were sampled for α-toxin assay. This experiment was performed in triplicate.
Elisa assay of C. perfringens α-toxin
The content of C. perfringens α-toxin in the supernatants of cultures was determined using antigenic ELISA kit (Bio-X Diagnostics, Rochefort, Belgium), according to the manufacturers' instructions. Each sample and the control antigen (supplied by the kit) were tested in both positive and negative wells. The data were expressed as a percentage of delta optical density of each sample (between positive and negative wells) to that of the control antigen.
Primary culture of chicken intestinal epithelial cells
The methods of isolating primary intestinal epithelial cells from chicken embryos were previously described in detail by Guo et al. (2015). Isolated cells were grown in a 1:1 mixture of Dulbecco's modified Eagles medium and Ham's F12 medium (DMEM/F12) supplemented with 2.5% FBS, 100 U/mL penicillin, 100 μg/mL of streptomycin, 20 ng/mL of EGF, 100 μg/mL of heparin sodium salt, 5 μg/mL of insulin. Cells were maintained in a humidified environment with an atmosphere of 5% CO2 at 37°C.
Cell adhesion and cytotoxicity assay
The assay of bacterial adhesion to cells was performed as previously described (Martin and Smyth, 2010) with some modification. The isolated cells were seeded in 12-well cell culture plates at a density of 1.0 × 106 cells/mL with a total volume of 1 mL. After 48 h of incubation, cells were gently washed with pre-warmed PBS. One milliliter of DMEM/F12 medium containing L. acidophilus or L. fermentum with multiplicity of infection (MOI) at 10 was added to wells. Cells without lactobacilli were used as the control. Following 3 h of anaerobic incubation, a few microliters of resuspended C. perfringens was inoculated into each well (MOI = 1) and incubated at 37°C under anaerobic condition for 1 h. All experiments were performed in triplicate. After incubation, the cells were washed twice with PBS and 200 μL of 0.05% trypsin was added to each well and incubated for 15 min at 37°C. An additional 800 μL of PBS was added to each well and mixed gently by pipetting. The number of C. perfringens released per well was determined by plating serial 10-fold dilutions of the cell suspension onto TSC plates. The percentage of adhering bacteria was calculated by dividing the number of bacteria recovered from wells with the total number of bacteria added to the wells. Meanwhile, the remaining cell suspension were centrifuged at 4,000 × g for 10 min and the supernatant was collected for cytotoxicity quantification. Lactate dehydrogenase (LDH), an enzyme present in the cytoplasm of cells, is quickly resealed upon damage of the plasma membrane. The LDH production was determined using commercially available kit (Nanjing Jiancheng Biological Product, Nanjing, Jiangsu Province, China) following the manufacturer's protocol.
Inflammatory responses
Cells were seeded in 6-well cell culture plates at a concentration of 1.0 × 106 cells/mL (2 mL per well). At 90% confluence, cells were pretreated with either L. acidophilus or L. fermentum at MOI of 1 for 2 h, and then stimulated with and without C. perfringens (MOI = 0.1) for 6 h. The DMEM/F12 medium and C. perfringens infection alone served as the negative and positive controls, respectively. Experiments were run in triplicate. After stimulation, the supernatant of cell culture medium was collected for the assay of LDH activity and cells were lysed for RNA extraction.
RNA isolation and quantitative real-time PCR
The methods of RNA extraction and quantitative real-time PCR were described in our previous publication (Guo et al., 2015). Total RNA of the cells was extracted using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. The concentration and purity of total RNA was determined by measuring its optical density at 260 and 280 nm, and the RNA integrity was assessed via agarose gel electrophoresis. One microgram of total RNA was reverse transcribed with PrimeScriptTM RT Master Mix (Perfect Real Time) (TakaRa, Dalian, Liaoning Province, China). All cDNA preparations were stored frozen at −20°C until further use. The qRT-PCR analysis was performed with the 7,500 fluorescence detection system (Applied Biosystems, Foster City, CA) using the SYBR® Premix Ex TaqTM (TaKaRa). The primer pairs for the amplification of chicken IL-6, IL-8, lipopolysaccharide-induced tumor necrosis factor-alpha factor (LITAF), inducible nitric oxide synthase (iNOS), IL-1β, NOD1, TLR2.2, TLR4, NF-κB p65, and β-actin cDNA fragments, used as an endogenous reference gene, were used as listed in Table 1. To confirm amplification specificity, the PCR products from each primer pair were subjected to a melting analysis and subsequent agarose gel electrophoresis. Samples were run in triplicate in 96-well plates. Relative gene expression data were analyzed using the 2−ΔΔCt method (Heid et al., 1996).
Table 1.
Primers used for quantitative real-time PCR.
| Gene name | Accession number | Forward sequence (5′-3′) | Reverse sequence (5′-3′) |
|---|---|---|---|
| β-actin | L08165 | GAGAAATTGTGCGTGACATCA | CCTGAACCTCTCATTGCCA |
| IL-6 | AJ309540 | CAAGGTGACGGAGGAGGAC | TGGCGAGGAGGGATTTCT |
| IL-8 | AJ009800 | ATGAACGGCAAGCTTGGAGCTG | TCCAAGCACACCTCTCTTCCATCC |
| iNOS | U46504 | CAGCTGATTGGGTGTGGAT | TTTCTTTGGCCTACGGGTC |
| LITAF | NM_204267 | GAGCGTTGACTTGGCTGTC | AAGCAACAACCAGCTATGCAC |
| IL-1β | NM_204524 | ACTGGGCATCAAGGGCTA | GGTAGAAGATGAAGCGGGTC |
| TLR2.2 | NM_001161650 | GGGGCTCACAGGCAAAATC | AGCAGGGTTCTCAGGTTCACA |
| NOD1 | JX465487 | AGCACTGTCCATCCTCTGTCC | TGAGGGTTGGTAAAGGTCTGCT |
| NF-κB p65 | NM_205129 | GTGTGAAGAAACGGGAACTG | GGCACGGTTGTCATAGATGG |
IL, interleukin; iNOS, inducible nitric oxide synthase; LITAF, lipopolysaccharide induced TNF factor; TLR, toll-like receptor; NOD, nucleotide-binding oligomerization domain; NF-κB, nuclear factor kappa B.
Statistical analysis
All data were analyzed with SPSS Version 17.0 (SPSS Inc., Chicago, IL). For the data of inflammatory responses, the comparison between negative and positive controls was conducted using an independent-samples t-test. The other data were subjected to one-way analysis of variance (ANOVA) and Tukey test was used for multiple comparisons between treatments. Significance was accepted at P < 0.05 and results were reported as means and standard error (SE).
Results
Lactobacilli and their supernatants inhibit the growth of C. perfringens
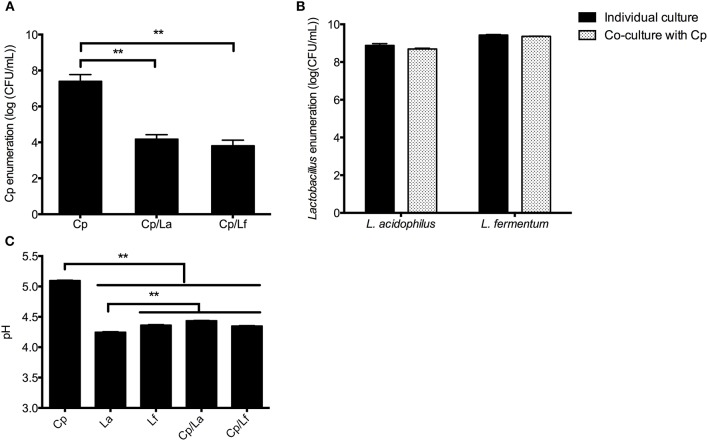
Compared with culture alone, co-culture of C. perfringens with either L. acidophilus or L. fermentum decreased the number of C. perfringens after 20 h of incubation (P < 0.01; Figure 1A), but did not significantly affect the enumeration of both lactobacilli (Figure 1B). Unsurprisingly, lactobacilli growth, alone or together with C. perfringens, reduced the pHs of cultures in comparison with individual C. perfringens culture (P < 0.01; Figure 1C).
Figure 1.
Bacterial enumeration and pH measurement in the co-culture experiment. Lactobacilli and C. perfringens were individually or together inoculated into the MRS broth with equal CFU and cultured at 37°C for 20 h. The enumeration of C. perfringens (A) and lactobacilli (B) as well as pH values of cultures (C) were determined at the end of incubation. Data are presented as mean ± standard error (n = 3). **P < 0.01. La, L. acidophilus; Lf, L. fermentum; Cp, C. perfringens.
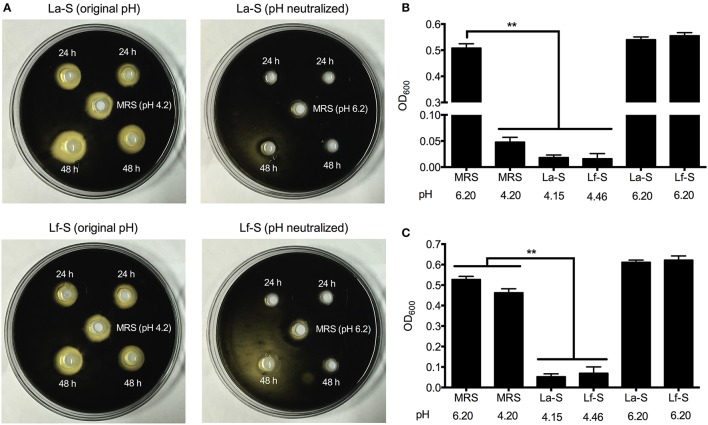
The data of agar well diffusion assay showed that lactobacilli supernatants from 24 to 48 h-cultures exhibited inhibition zones to the growth of C. perfringens (Figure 2A). However, when the pHs of supernatants were neutralized, the inhibition zones disappeared. Furthermore, the MRS broth with pH adjusted to 4.20 also exerted inhibitory effect on C. perfringens growth. Consistently, addition of both lactobacilli supernatants and MRS broth (pH 4.20) to C. perfringens cultures significantly decreased the OD600 values (P < 0.01), which reflected the number of C. perfringens, following 24 h of incubation (Figure 2B). As the incubation time reached 48 h, the lactobacilli supernatants still suppressed the OD600 values of cultures compared with the control (MRS broth at pH 6.20) (P < 0.01), but no significant differences were observed between the control and MRS broth at pH 4.20 (Figure 2C).
Figure 2.
Inhibition of C. perfringens by cell-free supernatants with and without pH neutralization, obtained from L. acidophilus and L. fermentum. Agar well diffusion assay was performed and the inhibition zone was assessed visually (A). Lactobacilli supernatants were collected after 24 or 48 h of incubation. Central wells were filled with sterile MRS broth adjusted to pH 4.2 or standard MRS broth. The colonies of C. perfringens were black in the TSC agar. The growth of C. perfringens in fresh MRS broth supplemented with lactobacilli supernatants was measured by OD600 after 24 (B) and 48 (C) hours of incubation. Data are presented as mean ± standard error (n = 3). **P < 0.01. La-S, supernatants from L. acidophilus cultures; Lf-S, supernatants from L. fermentum cultures; MRS, Mann-Rogosa-Sharpe medium.
Lactobacilli repress the α-toxin production of C. perfringens
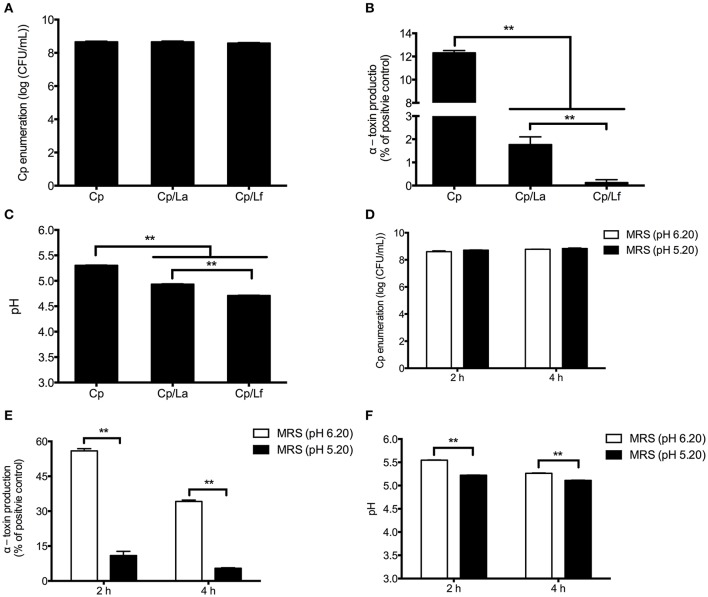
Following 4 h of incubation, L. acidophilus and L. fermentum did not significantly influence the number of C. perfringens in the co-culture systems (Figure 3A), but decreased the α-toxin secretion compared with C. perfringens cultured alone (P < 0.01; Figure 3B). What's more, L. fermentum showed a greater inhibitory effect on α-toxin production than L. acidophilus. The pHs of cultures exhibited similar changes as α-toxin production. Both lactobacilli reduced the pHs of co-cultures and L. fermentum induced a further decrease of pH compared with L. acidophilus (P < 0.01; Figure 3C).
Figure 3.
The effect of lactobacilli and pH on the number and α-toxin production of C. perfringens as well as pH of cultures. C. perfringens enumeration (A), α-toxin production (B) and pH values (C) were determined after 4-h co-culture of C. perfringens and lactobacilli. C. perfringens was grown in the MRS broth with normal (6.20) and lower (5.20) pHs for 2 and 4 h. The CFU of C. perfringens (D), α-toxin secretion (E) and pH (F) were measured at each time point. Data are presented as mean ± standard error (n = 3). **P < 0.01. La, L. acidophilus; Lf, L. fermentum; Cp, C. perfringens; MRS, Mann-Rogosa-Sharpe medium.
As shown in Figure 3D, the number of C. perfringens was not significantly affected by MRS broth at a lower pH (5.20) compared with normal pH (6.20) after 2 and 4 h of culture. However, the α-toxin secretion was impaired by the MRS broth with pH 5.20 following incubation (P < 0.01; Figure 3E). As expected, the MRS broth at pH 5.20 maintained a lower level of pH along the treatment (P < 0.01; Figure 3F).
Lactobacilli degrade established C. perfringens α-toxin
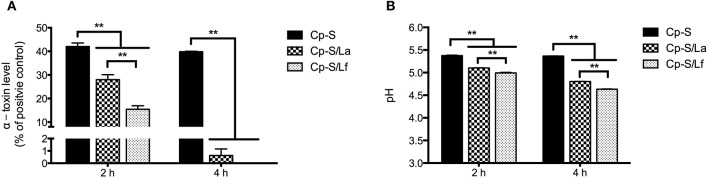
As shown in Figure 4A, the addition of L. acidophilus and L. fermentum to the supernatants from C. perfringens cultures significantly reduced the levels of α-toxin after 2 and 4 h of incubation (P < 0.01). And L. fermentum resulted in a greater reduction of α-toxin compared to L. acidophilus at 2 h of treatment. During incubation, both lactobacilli significantly decreased the pH of C. perfringens supernatants in contrast to the control (P < 0.01) and lower pH was detected in the L. fermentum-treated group in comparison with L. acidophilus treatment (P < 0.01; Figure 4B).
Figure 4.
The degradation of C. perfringens α-toxin by lactobacilli and associated changes of culture pH. Either L. acidophilus or L. fermentum were added into the supernatants from C. perfringens cultures and incubated for 2 or 4 h. The α-toxin levels were detected by ELISA assay (A) and pH values were measured (B). Data are presented as mean ± standard error (n = 3). **P < 0.01. La, L. acidophilus; Lf, L. fermentum; Cp-S, supernatants from C. perfringens cultures.
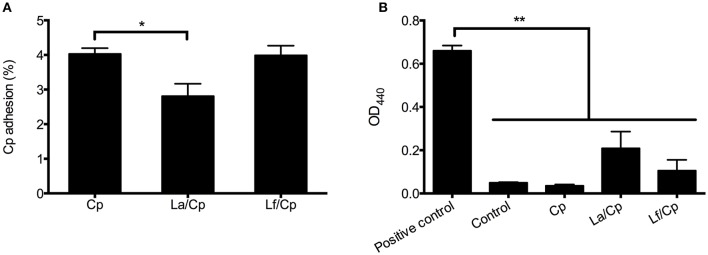
The effect of lactobacilli on the adhesion of C. perfringens to cells
Compared with C. perfringens infection alone, L. acidophilus pretreatment significantly decreased the percentage of C. perfringens adhering to chicken intestinal epithelial cells (P < 0.05), but L. fermentum did not show inhibitory effect on the C. perfringens adhesion (Figure 5A). The levels of LDH released from cells were not significantly changed after 1 h of incubation with C. perfringens alone or together with 3 h of lactobacilli pretreatments (Figure 5B). The LDH levels in the bacterial treatments were also not significantly different from that in the control (cell medium alone) (P > 0.05), but lower than that in the positive control, in which all cells were lysed by 0.5% of Triton X-100 (P < 0.01).
Figure 5.
The effect of lactobacilli on the adhesion of C. perfringens to cells and the cytotoxicity induced by C. perfringens. Cells were pretreated with L. acidophilus or L. fermentum with multiplicity of infection (MOI) at 10 for 3 h and then infected with C. perfringens (MOI = 1) for 1 h. The percentage of C. perfringens adhered to cells were calculated (A) and LDH levels in the cell culture medium were detected (B). Data are presented as mean ± standard error (n = 3). *P < 0.05; **P < 0.01. La, L. acidophilus; Lf, L. fermentum; Cp, C. perfringens.
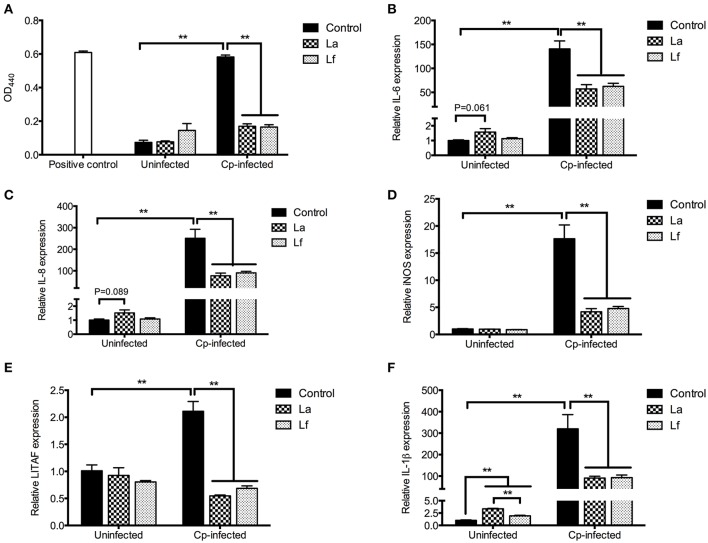
The anti-inflammatory effect of lactobacilli in the C. perfringens-infected cells
The data of LDH level and cytokine mRNA expression were shown in Figure 6. Compared with the uninfected control, C. perfringens infection at MOI of 0.1 for 6 h significantly increased the LDH level released from cells (P < 0.01), which indicated the induction of cytotoxicity by infection. However, pre-incubation of L. acidophilus and L. fermentum at MOI of 1 for 2 h reduced LDH levels in the infected cells (P < 0.01). Consistently, the relative mRNA expression of IL-6, IL-8, iNOS, LITAF and IL-1β was up-regulated by the C. perfringens challenge (P < 0.01), but down-regulated in challenged cells by pretreatment with Lactobacilli (P < 0.01). In the uninfected cells, both lactobacilli significantly increased the IL-1β mRNA level (P < 0.01), and L. acidophilus pre-incubation tended to elevate the mRNA expression of IL-6 (P = 0.061) and IL-8 (P = 0.089).
Figure 6.
The cytotoxicity and cytokine mRNA expression in the C. perfringens-infected cells pretreated with lactobacilli. Cells were pre-incubated with L. acidophilus or L. fermentum (MOI = 1) for 2 h and then stimulated with C. perfringens (MOI = 0.1) for 6 h. Following infection, LDH released from cells to culture medium were detected (A). The mRNA expression of IL-6 (B), IL-8 (C), iNOS (D), LITAF (E) and IL-1β (F) were determined by qRT-PCR analysis. Data are presented as mean ± standard error (n = 3). **P < 0.01. La, L. acidophilus; Lf, L. fermentum; Cp, C. perfringens; IL, interleukin; iNOS, inducible nitric oxide synthase; LITAF, lipopolysaccharide induced TNF factor.
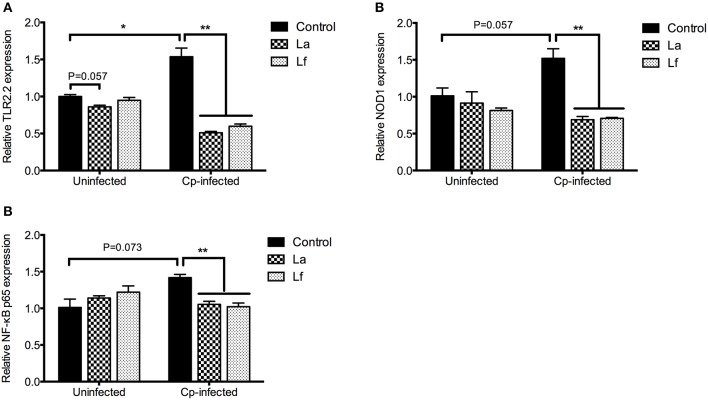
To investigate the possible mechanism mediating the modulation of cytokine expression by lactobacilli pretreatment, the mRNA expression of receptors (TLR2.2 and NOD1) and NF-κB p65 was determined and the data were presented in Figure 7. The C. perfringens infection significantly up-regulated the TLR2.2 mRNA expression (P < 0.05) and tended to increase the mRNA levels of NOD1 (P = 0.057) and NF-κB p65 (P = 0.073). However, the relative expression of TLR2.2, NOD1, and NF-κB p65 in the infected cells was reduced by the preincubation of lactobacilli (P < 0.01). There was a tendency that L. acidophilus decreased TLR2.2 mRNA expression in the uninfected cells (P = 0.057).
Figure 7.
Quantification of TLR2.2, NOD1 and NF-κB p65 transcripts in the cells. Cells were pre-incubated with L. acidophilus or L. fermentum (MOI = 1) for 2 h and then stimulated with C. perfringens (MOI = 0.1) for 6 h. Following infection, the mRNA expression of TLR2.2 (A), NOD1 (B) and NF-κB p65 (C) were analyzed by qRT-PCR. Data are presented as mean ± standard error (n = 3). *P < 0.05, **P < 0.01. La, L. acidophilus; Lf, L. fermentum; Cp, C. perfringens; TLR, toll-like receptor; NOD, nucleotide-binding oligomerization domain; NF-κB, nuclear factor kappa B.
Discussion
Probiotics are one of the alternative strategies to feed antibiotics for prevention against infectious diseases. The beneficial roles of probiotics include production of antimicrobials, alteration of the intestinal microbiota, modulation of mucosal immunity and barrier function (Bermudez-Brito et al., 2012). Lactobacillus spp. are the most frequently used lactic acid bacteria as probiotic agents. They also showed species- and strain-specific characteristics in conferring probiotic benefits (Zheng et al., 2013; Valeriano et al., 2017). In the present study, we investigated the inhibitory effects of L. acidophilus and L. fermentum on the growth and α-toxin production of C. perfringens. Using primary intestinal epithelial cells of chicken embryos, we further examined the modulation of these lactobacilli on inflammatory signaling pathways in C. perfringens-infected cells.
It was demonstrated that probiotics outcompeted pathogens for nutrients and space by secreting inhibitory substances, such as organic acid, hydrogen peroxide and bacteriocins (Salminen et al., 2010; Messaoudi et al., 2013). We observed that L. acidophilus and L. fermentum greatly repressed the growth of C. perfringens, accompanied with the decrease of pH. Actually, C. perfringens was a pH sensitive bacterium. The lowest pH supporting the growth of C. perfringens type A isolates carrying chromosomal or plasmid-borne entertoxin genes was 5.1 ± 0.1 or 5.2 ± 0.1, respectively (Li and McClane, 2006). This was consistent with our finding that the terminal pH in the 20 h-fermentation of C. perfringens was 5.1.
To further demonstrate whether pH played an important role in inhibiting the growth of C. perfringens, we performed the agar well diffusion assay and broth culture inhibition assay. The pH-neutralized supernatants of lactobacilli cultures could not suppress the pathogen growth, which indicated that lower pH was the main factor. However, supernatants from lactobacilli cultures rather than MRS broth at pH 4.20 maintained inhibitory effects on the proliferation of C. perfringens until 48 h. This suggested that pH is not the only contributor and lactobacilli might secret antimicrobials, such as bacteriocins against C. perfringens. It was reported that both L. acidophilus and L. fermentum could produce antibacterial substances displaying a wide inhibitory spectrum including Gram-negative and Gram-positive pathogenic strains (Coconnier et al., 1997; Pascual et al., 2008).
The intestinal microflora forms a delicate ecosystem, in which the different bacterial species may regulate each other's growth and gene expression (Allaart et al., 2011). This regulation is achieved through the quorum sensing (QS) system, where inter- and intra-species communication occurs via released signal molecules (Bassler, 1999). It is clarified that the transcription of plc, the gene encoding the α-toxin, is modulated by the accessory gene regulator (agr) system (Ohtani et al., 2009; Ohtani and Shimizu, 2015). The autoinducing peptide encoded by agr is secreted into the environment and acts as a QS signal molecular resulting in α-toxin production. Recent studies demonstrated that Lactobacillus spp. or their secretions interfered the agr QS system mediated virulence gene expression of pathogens, such as Staphylococcus aureus (Li et al., 2011) and C. difficile (Yun et al., 2014). Therefore, we speculated that both L. acidophilus and L. fermentum reduced α-toxin production likely by quenching the similar system.
However, it could not be neglected that the environmental pH during lactobacilli fermentation also played a role on the toxin secretion of C. perfringens, which was evidenced by Allaart et al. (2011). Thus, we measured the α-toxin production in the absence of lactobacilli cells at different pHs (5.20 and 6.20). The lower environmental pH significantly decreased the production of α-toxin, suggesting that the acid secreted by lactobacilli had a clear inhibitory effect on C. perfringens α-toxin production during coculturing. Furthermore, lactobacilli exhibited the ability of degrading established α-toxin, which might be due to the acid production. Actually, early in 1964, Pivnick et al. reported that the production and stability of C. perfringens toxins were influenced by the pH of growth medium (Pivnick et al., 1964). The optimum pH for synthesis of α-toxin is 6.7, and α-toxin appears unstable, evidenced by the rapid synthesis and degradation. In the current study, the α-toxin levels varied greatly in different experiments, indicating the instability of α-toxin. Whether pH affected α-toxin production at the transcriptional or translational level needs further investigation.
The ability to bind to various components of the host extracellular matrix, including collagen, is a trait often observed in enteric pathogens (Wade et al., 2015). The adherent abilities of C. perfringens strains are crucial for the pathogenesis of avian NE. It was demonstrated that the ability of avian strains of C. perfringens binding to specific collagen types correlated with their virulence (Wade et al., 2015) and the putative adhesion-encoding gene cnaA involved in the adherence (Wade et al., 2016). To prevent the pathogenic infection, probiotic bacterial strains should be able to compete with pathogens to occupy their potential binding sites in the gut (Štyriak et al., 2003; Schillinger et al., 2005). Probiotics usually exhibit high affinity to mucus by specific adhesins and adhesion is one of the important criteria for the selection of probiotic strains (Ouwehand et al., 2001). It was demonstrated that certain strains of lactic acid bacteria significantly reduced the adhesion of C. perfringens to immobilized mucus isolated from canine jejunal chyme in vitro (Rinkinen et al., 2003).
In the present work, when L. acidophilus adhered to the intestinal epithelial cells in advance and with a dose 10 times that of C. perfringens, the adhesion of C. perfringens was inhibited by 30% and the cell cytotoxicity was not significantly affected. On the one hand, L. acidophilus might gain an advantage in the space competition for binding sites. The bacterial size of L. acidophilus (>1,000 nm) (Nagy et al., 2016) was greater than that of L. fermentum (500–1,000 nm) (Sintubin et al., 2009), which was could be directly observed using microscopy (data not shown). This might be the one reason why L. fermentum did not show the similar inhibitory effect on the adhesive ability of C. perfringens. On the other hand, the expression of cnaA in C. perfringens was also regulated by agr QS system (Ohtani and Shimizu, 2015), which might be obstructed by L. acidophilus or its metabolites. Overall, L. acidophilus decreased the adhesion of C. perfringens to chicken intestinal epithelial cells, which might weaken its virulence.
Lactobacillus spp. plays an important role in the regulation of intestinal homeostasis and immunity (van Baarlen et al., 2013; Valeriano et al., 2017). The anti-inflammatory effects of Lactobacillus spp. were largely reported in pathogen-infected broiler chickens (Cao et al., 2012; Wang et al., 2017). In the primary culture of chicken intestinal epithelial cells, we found that pre-incubation with L. acidophilus or L. fermentum decreased the C. perfringens-induced up-regulation of pro-inflammatory factors (IL-6, IL-8, iNOS, LITAF, IL-1β). The alleviation of inflammation by lactobacilli was accordance with their preventative effects on cell cytotoxicity caused by C. perfringens infection.
As a Gram-positive bacterium, the main cell wall component of C. perfringens is PGN. It could be recognized by TLR2.2 or NOD1 in chicken intestinal epithelial cells and thereby activates the NF-κB signaling pathway. Consequently, the inflammatory responses were induced (Lu et al., 2009; Guo et al., 2015). Lactobacillus spp. are Gram-positive bacteria as well, but a host cell response to a microorganism being probiotic or pathogenic depends on the combination of the distinct microorganism-associated molecular patterns (MAMPs) that can interact with the various pattern recognition receptors (PRRs) and associated co-receptors that fine-tune signaling, as well as the presence of other microbial effector molecules, such as toxins (Lebeer et al., 2010). Numerous studies demonstrated that Lactobacillus spp. reduced inflammation through the inactivation of NF-κB signaling pathway in intestinal epithelial cells challenged by infectious agents including pathogens (Yang et al., 2012; Borthakur et al., 2013; Chen et al., 2013). The present study suggested that L. acidophilus and L. fermentum not only decreased the transcription of NF-κB p65, but also suppressed the mRNA expression of TLR2.2 and NOD1, which could be induced by C. perfringens and play a vital role in the signaling transaction. Therefore, the reduction of pro-inflammatory factor expression was not unexpected.
Conclusion
In in vitro co-culture system, both L. acidophilus and L. fermentum inhibited the growth and α-toxin production of C. perfringens mainly by lowering the environmental pH. In primary chicken intestinal cells, pre-treatment with both probiotics alleviated the C. perfringens-induced inflammation via down-regulation of TLR2.2 and NOD1 mRNA expression as well as inactivation of NF-κB p65. Furthermore, L. acidophilus suppressed the adhesion of C. perfringens to the intestinal epithelial cells. L. acidophilus and L. fermentum showed great potential to weaken the virulence of C. perfringens during the pathogeneses of NE in broiler chickens.
Author contributions
SG, DL, and YG conceived and designed the experiments. SG, BZ, and ZL performed the experiments. SG and YL analyzed the experimental data. SG, YG, and BD wrote this paper. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was financially supported by China Agriculture Research System (CARS-42-G13).
References
- Allaart J. G., van Asten A. J. A. M., Vernooij J. C. M., Gröne A. (2011). Effect of Lactobacillus fermentum on beta2 toxin production by Clostridium perfringens. Appl. Environ. Microbiol. 77, 4406–4411. 10.1128/AEM.03002-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B. L. (1999). How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2, 582–587. 10.1016/S1369-5274(99)00025-9 [DOI] [PubMed] [Google Scholar]
- Bermudez-Brito M., Plaza-Díaz J., Mu-oz-Quezada S., Gómez-Llorente C., Gil A. (2012). Probiotic mechanisms of action. Ann. Nutr. Metab. 61, 160–174. 10.1159/000342079 [DOI] [PubMed] [Google Scholar]
- Borthakur A., Bhattacharyya S., Kumar A., Anbazhagan A. N., Tobacman J. K., Dudeja P. K. (2013). Lactobacillus acidophilus alleviates platelet-activating factor-induced inflammatory responses in human intestinal epithelial cells. PLoS ONE 8:e75664. 10.1371/journal.pone.0075664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant A. E., Chen R. Y., Nagata Y., Wang Y., Lee C. H., Finegold S., et al. (2000). Clostridial gas gangrene. I. Cellular and molecular mechanisms of microvascular dysfunction induced by exotoxins of Clostridium perfringens. J. Inft. Dis. 182, 799–807. 10.1086/315756 [DOI] [PubMed] [Google Scholar]
- Cao L., Yang X. J., Li Z. J., Sun F. F., Wu X. H., Yao J. H. (2012). Reduced lesions in chickens with Clostridium perfringens-induced necrotic enteritis by Lactobacillus fermentum 1.20291. Poult. Sci. 91, 3065–7301. 10.3382/ps.2012-02548 [DOI] [PubMed] [Google Scholar]
- Chen K., Liang N., Luo X., Zhang T. C. (2013). Lactobacillus acidophilus strain suppresses the transcription of proinflammatory-related factors in human HT-29 cells. J. Microbiol. Biotechnol. 23, 64–68. 10.4014/jmb.1208.04067 [DOI] [PubMed] [Google Scholar]
- Coconnier M. H., Liévin V., Bernet-Camard M. F., Hudault S., Servin A. L. (1997). Antibacterial effect of the adhering human Lactobacillus acidophilus strain LB. Antimicrob. Agents Chemother. 41, 1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier C. T., Hofacre C. L., Payne A. M., Anderson D. B., Kaiser P., Mackie R. I., et al. (2008). Coccidia-induced mucogenesis promotes the onset of necrotic enteritis by supporting Clostridium perfringens growth. Vet. Immunol. Immunopathol. 122, 104–115. 10.1016/j.vetimm.2007.10.014 [DOI] [PubMed] [Google Scholar]
- Coursodon C. F., Trinh H. T., Mallozzi M., Vedantam G., Glock R. D., Songer J. G. (2010). Clostridium perfringens alpha toxin is produced in the intestines of broiler chicks inoculated with an alpha toxin mutant. Anaerobe 16, 614–617. 10.1016/j.anaerobe.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Geier M. S., Mikkelsen L. L., Torok V. A., Allison G. E., Olnood C. G., Boulianne M., et al. (2010). Comparison of alternatives to in-feed antimicrobials for the prevention of clinical necrotic enteritis. J. Appl. Microbiol. 109, 1329–1338. 10.1111/j.1365-2672.2010.04758.x [DOI] [PubMed] [Google Scholar]
- Goossens E., Valgaeren B. R., Pardon B., Haesebrouck F., Ducatelle R., Deprez P. R., et al. (2017). Rethinking the role of alpha toxin in Clostridium perfringens-associated enteric diseases: a review on bovine necro-haemorrhagic enteritis. Vet. Res. 48:9. 10.1186/s13567-017-0413-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Li C., Liu D., Guo Y. (2015). Inflammatory responses to a Clostridium perfringens type a strain and α-toxin in primary intestinal epithelial cells of chicken embryos. Avian Pathol. 44, 81–91. 10.1080/03079457.2015.1005573 [DOI] [PubMed] [Google Scholar]
- Heid C. A., Stevens J., Livak K. J., Williams P. M. (1996). Real time quantitative PCR. Genome Res. 6, 986–994. 10.1101/gr.6.10.986 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Kong Q., Roland K. L., Wolf A., Curtiss R., III. (2014). Multiple effects of Escherichia coli Nissle 1917 on growth, biofilm formation, and inflammation cytokines profile of Clostridium perfringens type A strain CP4. Pathog. Dis. 70, 390–400. 10.1111/2049-632X.12153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyburn A. L., Sheedy S. A., Ford M. E., Williamson M. M., Awad M. M., Rood J. I., et al. (2006). Alpha-toxin of Clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Infect. Immun. 74, 6496–6500. 10.1128/IAI.00806-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Ragione R. M., Narbad A., Gasson M. J., Woodward M. J. (2004). In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett. Appl. Microbiol. 38, 197–205. 10.1111/j.1472-765X.2004.01474.x [DOI] [PubMed] [Google Scholar]
- Lebeer S., Vanderleyden J., De Keersmaecker S. C. J. (2010). Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8, 171–184. 10.1038/nrmicro2297 [DOI] [PubMed] [Google Scholar]
- Li J., McClane B. A. (2006). Comparative effects of osmotic, sodium nitrite-induced, and pH-induced stress on growth and survival of Clostridium perfringens type A isolates carrying chromosomal or plasmid-borne enterotoxin genes. Appl. Environ. Microbiol. 72, 7620–7625. 10.1128/AEM.01911-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang W., Xu S. X., Magarvey N. A., McCormick J. K. (2011). Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc. Natl. Acad. Sci. U.S.A. 108, 3360–3365. 10.1073/pnas.1017431108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Sarson A. J., Gong J., Zhou H. J., Zhu W. Y., Kang Z. M., et al. (2009). Expression profiles of genes in Toll-like receptor-mediated signaling of broilers infected with Clostridium perfringens. Clin. Vaccine Immunol. 16, 1639–1647. 10.1128/CVI.00254-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T. G., Smyth J. A. (2010). The ability of disease and non-disease producing strains of Clostridium perfringens from chickens to adhere to extracellular matrix molecules and Caco-2 cells. Anaerobe 16, 533–539 10.1016/j.anaerobe.2010.07.003 [DOI] [PubMed] [Google Scholar]
- Messaoudi S., Manai M., Kergourlay G., Prevost H., Connil N., Chobert J. M., et al. (2013). Lactobacillus salivarius: bacteriocin and probiotic activity. Food Microbiol. 36, 296–304. 10.1016/j.fm.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Nagy G., Pinczes G., Pinter G., Pocsi I., Prokisch J., Banfalvi G. (2016). In situ electron microscopy of lactomicroselenium particles in probiotic bacteria. Int. J. Mol. Sci. 17:E1047. 10.3390/ijms17071047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K., Shimizu T. (2015). Regulation of toxin gene expression in Clostridium perfringens. Res. Microbiol. 166, 280–289. 10.1016/j.resmic.2014.09.010 [DOI] [PubMed] [Google Scholar]
- Ohtani K., Yuan Y., Hassan S., Wang R., Wang Y., Shimizu T. (2009). Virulence gene regulation by the agr system in Clostridium perfringens. J. Bacteriol. 191, 3919–3927. 10.1128/JB.01455-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwehand A. C., Tuomola E. M., Tölkkö S., Salminen S. (2001). Assessment of adhesion properties of novel probiotic strains to human intestinal mucus. Int. J. Food Microbiol. 64, 119–126. 10.1016/S0168-1605(00)00440-2 [DOI] [PubMed] [Google Scholar]
- Park S. S., Lillehoj H. S., Allen P. C., Park D. W., FitzCoy S., Bautista D. A., et al. (2008). Immunopathology and cytokine responses in broiler chickens coinfected with Eimeria maxima and Clostridium perfringens with the use of an animal model of necrotic enteritis. Avian Dis. 52, 14–22. 10.1637/7997-041707-Reg [DOI] [PubMed] [Google Scholar]
- Pascual L. M., Daniele M. B., Giordano W., Pájaro M. C., Barberis I. L. (2008). Purification and partial characterization of novel bacteriocin L23 produced by Lactobacillus fermentum L23. Curr. Microbiol. 56, 397–402. 10.1007/s00284-007-9094-4 [DOI] [PubMed] [Google Scholar]
- Petit L., Gibert M., Popoff M. R. (1999). Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7, 104–110. 10.1016/S0966-842X(98)01430-9 [DOI] [PubMed] [Google Scholar]
- Pivnick H., Habeeb A. F. S. A., Gorenstein B., Stuart P. F., Hauschild A. H. W. (1964). Effect of pH on toxinogenesis by Clostridium perfringens type C. Can. J. Microbiol. 10, 329–344. 10.1139/m64-045 [DOI] [PubMed] [Google Scholar]
- Rahimi S., Kathariou S., Grimes J. L., Siletzky R. M. (2011). Effect of direct-fed microbials on performance and Clostridium perfringens colonization of turkey poults. Poult. Sci. 90, 2656–2662. 10.3382/ps.2011-01342 [DOI] [PubMed] [Google Scholar]
- Revitt-Mills S. A., Rood J. I., Adams V. (2015). Clostridium perfringens extracellular toxins and enzymes: 20 and counting. Microbiol. Aust. 36, 114–117. 10.1071/MA15039 [DOI] [Google Scholar]
- Rinkinen M., Jalava K., Westermarck E., Salminen S., Ouwehand A. C. (2003). Interaction between probiotic lactic acid bacteria and canine enteric pathogens: a risk factor for intestinal Enterococcus faecium colonization? Vet. Microbiol. 92, 111–119. 10.1016/S0378-1135(02)00356-5 [DOI] [PubMed] [Google Scholar]
- Salminen S., Nybom S., Meriluoto J., Collado M. C., Vesterlund S., El-Nezami H. (2010). Interaction of probiotics and pathogens-benefits to human health? Curr. Opin. Biotechnol. 21, 157–167. 10.1016/j.copbio.2010.03.016 [DOI] [PubMed] [Google Scholar]
- Schillinger U., Guigas C., Holzapfel W. H. (2005). In vitro adherence and other properties of lactobacilli used in probiotic yoghurt-like products. Int. Dairy J. 15, 1289–1297. 10.1016/j.idairyj.2004.12.008 [DOI] [Google Scholar]
- Schoster A., Kokotovic B., Permin A., Pedersen P. D., Dal Bello F., Guardabassi L. (2013). In vitro inhibition of Clostridium difficile and Clostridium perfringens by commercial probiotic strains. Anaerobe 20, 36–41. 10.1016/j.anaerobe.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Sintubin L., De Windt W., Dick J., Mast J., van der Ha D., Verstraete W., et al. (2009). Lactic acid bacteria as reducing and capping agent for the fast and efficient production of silver nanoparticles. Appl. Microbiol. Biotechnol. 84, 741–749. 10.1007/s00253-009-2032-6 [DOI] [PubMed] [Google Scholar]
- Songer J. G. (1996). Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9, 216–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songer J. G., Meer R. R. (1996). Genotyping of Clostridium perfringens by polymerase chain reaction is a useful adjunct to diagnosis of clostridial enteric disease in animals. Anaerobe 2, 197–203. 10.1006/anae.1996.0027 [DOI] [Google Scholar]
- Štyriak I., Nemcová R., Chang Y. H., Ljungh A. (2003). Binding of extracllular matrix molecules by probiotic bacteria. Lett. Appl. Microbiol. 37, 329–333. 10.1046/j.1472-765X.2003.01402.x [DOI] [PubMed] [Google Scholar]
- Timbermont L., Haesebrouck F., Ducatelle R., Immerseel F. V. (2011). Necrotic enteritis in broilers: an updated review on the pathogenesis. Avian Pathol. 40, 341–347. 10.1080/03079457.2011.590967 [DOI] [PubMed] [Google Scholar]
- Valeriano V. D. V., Balolong M. P., Kang D. K. (2017). Probiotic roles of Lactobacillus sp. In swine: insights from gut microbiota. J. Appl. Microbiol. 122, 554–567. 10.1111/jam.13364 [DOI] [PubMed] [Google Scholar]
- van Baarlen P., Wells J. M., Kleerebezem M. (2013). Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. 34, 208–215. 10.1016/j.it.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Wade B., Keyburn A. (2015). The true cost of necrotic enteritis. World Poult. 31, 16–17. [Google Scholar]
- Wade B., Keyburn A. L., Haring V., Ford M., Rood J. I., Moore R. J. (2016). The adherent abilities of Clostridium perfringens strains are critical for the pathogenesis of avian necrotic enteritis. Vet. Microbiol. 197, 53–61. 10.1016/j.vetmic.2016.10.028 [DOI] [PubMed] [Google Scholar]
- Wade B., Keyburn A. L., Seemann T., Rood J. I., Moore R. J. (2015). Binding of Clostridium perfringens to collagen correlates with the ability to cause necrotic enteritis in chickens. Vet. Microbiol. 180, 299–303. 10.1016/j.vetmic.2015.09.019 [DOI] [PubMed] [Google Scholar]
- Wang S., Peng Q., Jia H. M., Zeng X. F., Zhu J. L., Hou C. L., et al. (2017). Prevention of Escherichia coli infection in broiler chickens with Lactobacillus plantarum B1. Poult. Sci. 96, 2576–2586. 10.3382/ps/pex061 [DOI] [PubMed] [Google Scholar]
- Yang Y. J., Chuang C. C., Yang H. B., Lu C. C., Sheu B. S. (2012). Lactobacillus acidophilus ameliorates H. pylori-induced gastric inflammation by inactivating the Smad7 and NFκB pathways. BMC Microbiol. 12:38. 10.1186/1471-2180-12-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B., Oh S., Griffiths M. W. (2014). Lactobacillus acidophilus modulates the virulence of Clostridium difficile. J. Dairy Sci. 97, 4745–4758. 10.3168/jds.2014-7921 [DOI] [PubMed] [Google Scholar]
- Zheng X., Lyu L., Mei Z. (2013). Lactobacillus-containing probiotic supplementation increases Helicobacter pylori eradication rate: evidence from a meta-analysis. Rev. Esp. Enferm. Dig. 105, 445–453. 10.4321/S1130-01082013000800002 [DOI] [PubMed] [Google Scholar]