Abstract
Individuals with autism spectrum disorder (ASD) have altered gut microbiota, which appears to regulate ASD symptoms via gut microbiota-brain interactions. Rapid assessment of gut microbiota profiles in ASD individuals in varying physiological contexts is important to understanding the role of the microbiota in regulating ASD symptoms. Microbiomes secrete extracellular membrane vesicles (EVs) to communicate with host cells and secreted EVs are widely distributed throughout the body including the blood and urine. In the present study, we investigated whether bacteria-derived EVs in urine are useful for the metagenome analysis of microbiota in ASD individuals. To address this, bacterial DNA was isolated from bacteria-derived EVs in the urine of ASD individuals. Subsequent metagenome analysis indicated markedly altered microbiota profiles at the levels of the phylum, class, order, family, and genus in ASD individuals relative to control subjects. Microbiota identified from urine EVs included gut microbiota reported in previous studies and their up- and down-regulation in ASD individuals were partially consistent with microbiota profiles previously assessed from ASD fecal samples. However, overall microbiota profiles identified in the present study represented a distinctive microbiota landscape for ASD. Particularly, the occupancy of g_Pseudomonas, g_Sphingomonas, g_Agrobacterium, g_Achromobacter, and g_Roseateles decreased in ASD, whereas g_Streptococcus, g_Akkermansia, g_Rhodococcus, and g_Halomonas increased. These results demonstrate distinctively altered gut microbiota profiles in ASD, and validate the utilization of urine EVs for the rapid assessment of microbiota in ASD.
Keywords: Autism spectrum disorder, gut microbiota, Extracellular membrane vesicles, Bacteria-derived EVs, urine marker
INTRODUCTION
Autism spectrum disorder (ASD) is a group of neurodevelopmental disabilities characterized by two domains of core symptoms, persistent social deficits and restricted repetitive patterns of behavior [1]. Most individuals with ASD suffer from various behavioral and physical symptoms, including abnormal preferences regarding specific foods and problems in the digestive system [2,3]. Approximately 70~80% of ASD subjects have food selectivity and restricted food interests due to the texture, smell, or color of specific foods, and food intolerance [3,4]. The limited food intake behavior in ASD subjects leads to health problems including nutrition imbalance and gastrointestinal (GI) symptoms, such as diarrhea and constipation [2,5,6]. Furthermore, studies of a positive correlation between GI symptoms and ASD have been reported; the c-Met promoter variant rs1858830 is associated with ASD and GI symptoms, and the serum level of hepatocyte growth factor (HGF) that binding to the c-Met receptor, is correlated with severity of GI symptom in ASD subjects [7,8,9].
Several lines of evidence indicate that ASD patients have altered microbiota composition in the gut compared to healthy subjects [10,11,12,13,14,15,16,17,18,19,20,21]. The occupancy of the phyla Firmicutes, Fusobacteria, Verrucomicrobia, and Actinobacteria was decreased, whereas Bacteroidetes and Proteobacteria were increased in ASD groups [13,21]. More specifically, in ASD, the genera Bifidobacterum and Akkermansia were found to be decreased in ASD, while Lactobacillus was increased [17,18]. Furthermore, the treatment with a probiotics mix containing Streptococcus (thermophiles), Bifidobacterium breve, B. longum, B. infantis, and Lactobacillus acidophilus, L. plantarum, L. paracasei, and L. delbrueckii (subsp. Bulgaricus) or the transplant of fecal microbiota from healthy subjects to ASD individuals increased overall bacterial diversity and the abundance of Bifidobacterium, Prevotella, and Desulfovibrio among other taxa, and alleviated GI symptoms and ASD core symptoms [22,23]. To date, all available microbiota composition in ASD were mostly assessed from fecal samples [11,13,14,17,18,19,20,21] or directly from the cecum and ileum [15,16]. The fact that some microbiota commonly change in independent studies, and others are not consistently reported (e.g., [13,15,19]), increases the possibility of highly complex dynamics in bodily microbiota composition in ASD individuals under different physiological contexts.
Gram-negative bacteria secrete extracellular membrane vesicles (EVs), also called nanovesicles, to communicate with host cells [24], and are detected in stools, and also in urine and blood serum [25,26,27]. EVs secreted by gram-negative bacteria contain DNA, RNA, proteases, phospholipases, adhesins, toxins, and immunomodulatory compounds. Bacteria-derived EVs are associated with cytotoxicity, bacterial attachment, intercellular DNA transfer, and invasion [24,28,29]. Gram-positive bacteria also produce EVs, which contain peptidoglycan, lipoteichoic acid, virulence proteins, DNA and RNA [30,31]. When bacterial EVs were intraperitoneally injected in mice, EVs were rapidly distributed throughout the body with accumulation in the liver, lung, spleen, and kidney within 3 h [27]. Bacteria-derived EVs in the blood and urine represent the major constituents of microbes in the body, namely the gut microbiota [25,26], and indicate the microbiota that are metabolically or pathologically active [25,27].
In the present study, we investigated bodily microbiota represented by bacterial EVs in the urine of ASD individuals. The results of the present study identify markedly altered microbiota profiles in ASD relative to non-ASD healthy controls and suggest that bacterial EVs in urine can be served as a useful tool for the evaluation of microbiota composition in ASD.
MATERIALS AND METHODS
Subjects and urine sample preparation
Individuals who were enrolled at the Ewha Special Education Research Institute (Seoul, Republic of Korea) or Ewha Womans University MokDong Hospital (Seoul, Republic of Korea) were diagnosed according to the DSM-5 diagnostic criteria by a child and adolescent psychiatrist followed by characterization using the Korean Childhood Autism Rating Scale (K-CARS) as described previously [32]. The K-CARS is a well-established scale for the diagnosis of ASD with good agreement with the DSM-5 diagnostic criteria [33]. This questionnaire contained 15 items, each with 4 symptom scales, and all individual scores on each of the questions were summed to obtain the total score. When the total score was higher than 30 points, the subject was classified as autistic. Individuals who had any associated additional psychiatric and neurological diagnoses, or individuals who were on any antipsychotic medications were excluded from the present study.
Among the characterized ASD individuals, 18 male and 2 female ASD individuals (22.4+/−4.9 years) (Table 1) were joined to this study and their urine was collected during the day. The collected urine samples were frozen and stored at −20℃ until use. Age-matched normal healthy subjects (24 males and 4 females, 21.1+/−9.5 years) (Table 1) were selected from the Inje University Haeundae Paik Hospital (IRB No. 1297992-2015-064) and Seoul National University Hospital Healthcare System Gangnam Center (IRB No. 1502-034-647). The control subjects were not related to ASD and had no clinical findings suggestive of gastrointestinal problems or neuropsychiatric disorders. The control subjects of this study had not taken antibiotics, probiotics or prebiotics in the 3 months prior to the sample collection.
Table 1. The number, age, and sex of control and ASD subjects.
| Control | ASD | p-value | |
|---|---|---|---|
| Age (years) | 21.1±9.5 | 22.4±4.9 | 0.57 |
| N | 28 | 20 | - |
| Sex (Male/Female) | 24/4 | 18/2 | 0.66 |
The experimental protocol of human subjects was reviewed and approved by the Institutional Review Board of Ewha Womans University Hospital (IRB No. 2015-08-005-002). All eligible participants had been told about the purpose, procedures, risks and benefits of the present study and informed consent was obtained from all ASD subjects.
Isolation of bacteria-derived EVs and DNA extraction from human urine samples
Bacteria EVs were isolated from the urine of ASD individuals following the procedure described previously [25,26]. Briefly, each urine sample was centrifuged at 10,000 × g for 10 min at 4℃. The supernatant was taken and passed through a 0.22-µm membrane filter to eliminate foreign particles. Isolated EVs were dissolved in 100 µl PBS, and quantified on the basis of protein.
Bacterial DNA extraction from prepared EVs was performed as described previously [25,26]. Briefly, isolated EVs (1 µg by protein, each sample) were boiled at 100℃ for 40 min, centrifuged at 13,000 g for 30 min, and the supernatants were collected. Collected samples were then subjected to bacterial DNA extraction using a DNA extraction kit (PowerSoil DNA Isolation Kit, MO BIO, USA) following the manufacturer's instructions, Isolated DNA was quantified by using the QIAxpert system (QIAGEN, Germany).
PCR amplification, library construction, and sequencing of 16S rRNA gene variable regions
Prepared bacterial DNA was used for PCR amplification of the V3-V4 hypervariable regions of the 16S ribosomal RNA genes using the primer set of 16S_V3_F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) and 16S_V4_R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′). The PCR products were used for the construction of 16S rDNA gene libraries following the MiSeq System guidelines (Illumina Inc., San Diego, CA, USA). The 16S rRNA gene libraries for each sample were quantified using QIAxpert (QIAGEN, Germany), pooled at the equimolar ratio, and used for pyrosequencing with the the MiSeq System (Illumina, USA) according to manufacturer's recommendations.
Taxonomic assignments by 16S rRNA genes sequence reads
Obtained raw pyrosequencing reads were filtered on the basis of the barcode and primer sequences using MiSeq Control Software version 1.1.1 (Illumina, USA). Sequence reads were taxonomically assigned using the MDx-Pro ver.1 profiling program (MD Healthcare Inc., Seoul, Korea). Briefly, the quality of sequence reads was retained by controlling an average PHRED score higher than 20 and read length of more than 300 bp. Operational taxonomic units (OTUs) were clustered using CD-HIT sequence clustering algorithms and were assigned using UCLUST [34] and QIIME [35] on the basis of the GreenGenes 8.15.13 16S rRNA sequence database [36]. Based on the sequence similarities, taxonomic assignments were achieved at the following levels: genus, >94% similarity; family, >90% similarity; order, >85% similarity; class, >80% similarity; and phylum, >75% similarity. In cases where clustering was not possible at the genus level due to a lack of sequence information at the database or redundant sequences, the taxon was named based on the higher-level taxonomy with parentheses.
Visualization and principal component analysis (PCA)
Data were normalized to have a mean of 0 and a standard deviation of 1 by linear normalization. PCA and two-dimensional scatter plots with axis of the first and second principal component were calculated and drawn using Matlab 2011a.
Statistical analysis
Two-sample comparisons were performed using Student's t-test. Data clustering of pyrosequencing reads were compared using the χ2 test or t-test, while the comparisons between phylum compositions were tested by Fisher's exact test using GraphPad Prism 6 (San Diego, CA, USA). All data are presented as the mean±SEM, and a statistical difference was accepted at the 5% level.
RESULTS
Metagenome analysis of bodily microbiota in ASD individuals using bacteria-derived EVs in urine
Bacteria-derived EVs were isolated from the urine of 20 ASD individuals and 28 normal healthy subjects. The average age of the control and ASD subjects was 21.1+/−9.5 years and 22.4+/−4.9 years, respectively (Table 1). ASD subjects showed mild impairment of social interaction and stereotypies. The average K-CARS values of these ASD individuals was in the range between 31.5 and 33.5 and IQ values were in the range between 65 and 86. The control group was composed of healthy volunteers who had no medical problems including those related to ASD.
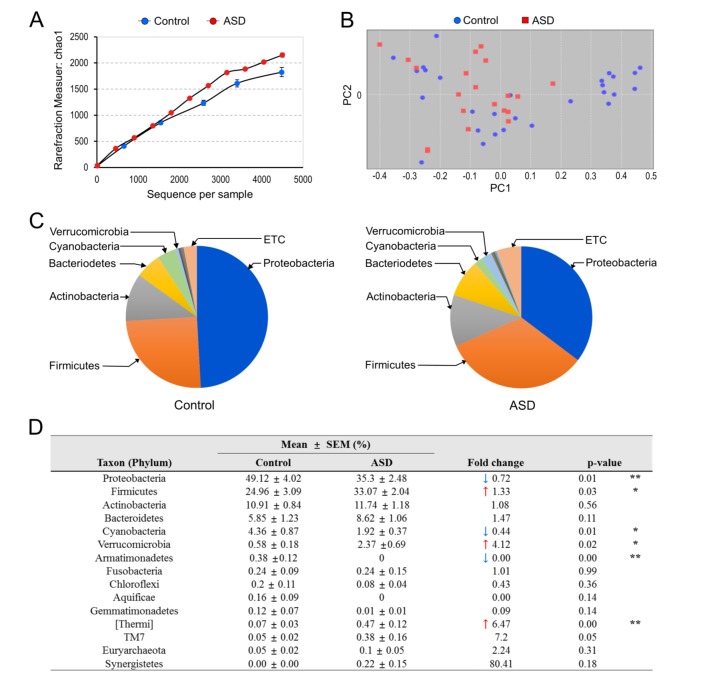
After the extraction of bacterial genomic DNA from the isolated EVs, variable regions of the 16S rRNA genes were amplified by PCR, and the libraries were constructed, as described previously [25,26]. Subsequent DNA sequencing analyses led us to identify over 2,000 operational taxonomic units (OTUs) for ASD and normal individuals. There was no significant difference in the alpha diversity between the two groups (Fig. 1A). Among the identified OTUs, we assigned 30 OTUs at the phylum level, 75 OTUs at the class level, 141 OTUs at the order level, 279 OTUs at the family level, and 619 OTUs at the genus level. Among these OTUs, we primarily focused on OTUs that occupied more than 0.1% of the identified taxons in the following analyses.
Fig. 1. The diversity and percent composition of microbiota at the phylum level in control vs. ASD subjects. (A) Rarefication curves representing the mean OTUs over the identified sequences of variable regions of 16S rRNA gene in control (blue) and ASD (red) subjects. Data are the mean +/− SEM (n=5, each). (B) Principal component analysis of microbiota diversity based on the weighted UniFrac distance and Bray-Curtis dissimilarity. Data were normalized to have a mean of 0 and a standard deviation of 1. Control (blue) and ASD (red). (C) Overall composition of microbiota at the phylum level in control (blue) and ASD (red) subjects. Those with occupancy 0.1% or higher in control and/or ASD subjects are presented. (D) The percent composition of microbiota at the phylum level in control and ASD subjects. ↑ and ↓ denote an increase and decrease in the percent composition, respectively. Data are the mean +/− SEM (n=5, each). * and ** denote the differences between the indicated groups at p<0.05 and p<0.01, respectively (Student's t-test).
Altered microbiota profiles between ASD individuals and healthy subjects
Sequence readings of EVs-based 16S rDNA indicated that the top five members of the phyla p_Proteobacteria, p_Firmicutes, p_Actinobacteria, p_Bacteroidetes, and p_Cyanobacteria comprised 95.2% of the identified OTUs in healthy subjects, whereas these members covered 90.65% of the total OTUs in ASD individuals, suggesting that ASD individuals have altered phyla composition. More specifically, the occupancy of p_Proteobacteria decreased from 49.12 to 35.30%, p_Cyanobacteria decreased from 4.36 to 1.92%, and p_Armatimonadetes decreased from 0.38 to 0.00% in ASD individuals (Fig. 1B~D). In contrast, the occupancy of p_Firmicutes increased from 24.96 to 33.07% and p_Verrucomicrobia increased from 0.58 to 2.37% in ASD.
The microbiota whose occupancy decreased or increased in ASD individuals were further analyzed at the class, order and family levels (Table 2, Supplemental Fig. S1). The decrease of f_Sphingomonadaceae and f_Rhizobiaceae accounted for the major decrease in p_Proteobacteria in ASD. In p_Cyanobacteria, o_Streptophyta decreased from 3.8 to 1.68%. In p_Verrucomicrobia, f_Verrucomicrobiaceae was the major increase (0.52 to 2.35%). In p_Firmicutes, f_Streptococcaceae, f_Clostridiaceae, an unclassified member of o_Clostridiales and f_Eubacteriaceae increased from 3.43 to 8.09% (Table 2).
Table 2. The percent composition of microbiota at the class, order, and family taxonomic levels in control and ASD subjects.
| Class | Order | Family | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Taxon | Mean (%) | Fold change | p-value | Taxon | Mean (%) | Fold change | p-value | Taxon | Mean (%) | Fold change | p-value | |||
| Control | ASD | Control | ASD | Control | ASD | |||||||||
| Gammaproteobacteria | 23.51 | 23.47 | 1 | 0.99 | Oceanospirillales | 0.21 | 2.62 | 12.68 | 0.02* | Halomonadaceae | 0.2 | 2.61 | ↑ 13.16 | 0.02* |
| Alphaproteobacteria | 15.29 | 4.84 | 0.32 | 0** | Sphingomonadales | 6.29 | 1.98 | 0.31 | 0** | Sphingomonadaceae | 6.27 | 1.98 | ↓ 0.32 | 0** |
| Rhizobiales | 5.69 | 1.05 | 0.19 | 0** | Rhizobiaceae | 4.58 | 0.18 | ↓ 0.04 | 0** | |||||
| Bradyrhizobiaceae | 0.46 | 0.08 | ↓ 0.17 | 0.01* | ||||||||||
| Rickettsiales | 0.94 | 0.1 | 0.11 | 0.04* | mitochondria | 0.84 | 0.1 | ↓ 0.12 | 0.02** | |||||
| Betaproteobacteria | 10.09 | 6.26 | 0.62 | 0.1 | Burkholderiales | 9.5 | 5.73 | 0.6 | 0.11 | Comamonadaceae | 2.55 | 4.37 | ↑ 1.71 | 0.01* |
| Alcaligenaceae | 2.53 | 0.12 | ↓ 0.05 | 0** | ||||||||||
| Deltaproteobacteria | 0.2 | 0.64 | 3.23 | 0.01* | Desulfovibrionales | 0.06 | 0.53 | 9.66 | 0** | Desulfovibrionaceae | 0.06 | 0.53 | ↑ 9.66 | 0** |
| Bacilli | 12.84 | 17.55 | 1.37 | 0.07 | Lactobacillales | 7.92 | 11.94 | 1.51 | 0.07 | Streptococcaceae | 1.93 | 4.88 | ↑ 2.52 | 0.04* |
| Clostridia | 11.63 | 15.4 | 1.32 | 0.18 | Clostridiales | 11.59 | 15.38 | 1.33 | 0.18 | Unclassified | 1.06 | 1.87 | ↑ 1.75 | 0.04* |
| Clostridiaceae | 0.44 | 1.19 | ↑ 2.70 | 0.02* | ||||||||||
| Eubacteriaceae | 0 | 0.15 | ↑ 146.23 | 0.04* | ||||||||||
| Actinobacteria | 9.9 | 11.09 | 1.12 | 0.38 | Actinomycetales | 7.84 | 10.29 | 1.31 | 0.09 | Nocardiaceae | 0.4 | 1.56 | ↑ 3.91 | 0.02* |
| Bifidobacteriales | 2.06 | 0.8 | 0.39 | 0.03* | Bifidobacteriaceae | 2.06 | 0.8 | ↓ 0.39 | 0.03* | |||||
| Bacteroidia | 5.05 | 7.28 | 1.44 | 0.21 | Bacteroidales | 5.05 | 7.28 | 1.44 | 0.21 | S24-7 | 0.84 | 2.02 | ↑ 2.40 | 0.04* |
| Flavobacteriia | 0.56 | 1.19 | 2.11 | 0.03* | Flavobacteriales | 0.56 | 1.19 | 2.11 | 0.03* | [Weeksellaceae] | 0.43 | 1.12 | ↑ 2.63 | 0.01* |
| Chloroplast | 3.81 | 1.76 | 0.46 | 0.03* | Streptophyta | 3.8 | 1.68 | 0.44 | 0.03* | Unclassified | 3.8 | 1.68 | ↓ 0.44 | 0.03* |
| Verrucomicrobiae | 0.52 | 2.35 | 4.49 | 0.02* | Verrucomicrobiales | 0.52 | 2.35 | 4.49 | 0.02* | Verrucomicrobiaceae | 0.52 | 2.35 | ↑ 4.49 | 0.02* |
| [Fimbriimonadia] | 0.38 | 0 | 0 | 0** | [Fimbriimonadales] | 0.38 | 0 | 0 | 0** | [Fimbriimonadaceae] | 0.38 | 0 | ↓ 0.00 | 0** |
| Deinococci | 0.07 | 0.47 | 6.47 | 0** | Thermales | 0.03 | 0.21 | 6.3 | 0.02* | Thermaceae | 0.03 | 0.21 | ↓ 6.30 | 0.02* |
| TM7-3 | 0.05 | 0.37 | 6.96 | 0.05* | ||||||||||
Microbiota at the family level whose occupancy was significantly different in ASD subjects are presented with associated higher taxonomy levels. Those occupying 0.1% or higher in either normal healthy or ASD subjects were included. ↑ and ↓ denote an increase and decrease in the percent composition, respectively. Data are the mean percentages. * and ** denote significant differences between the indicated groups at p<0.05 and p<0.01, respectively (Student's t-test).
The members of the genus occupied by more than 0.1% in either control or ASD individuals are summarized in Table 3 and Supplemental Table 1. Overall, 14 members at the genus level were down-regulated in ASD and their total occupancy in ASD dropped from 34.77 to 14.06%. On the contrary, 17 genus members were up-regulated in ASD and their total occupancy in ASD increased from 6.47 to 22.58%.
Table 3. The percent composition of microbiota at the genus level in control and ASD subjects.
| Class | Order | Family | Taxon | Mean±SEM (%) | Fold change | p-value | |
|---|---|---|---|---|---|---|---|
| Control | ASD | ||||||
| Gammaproteobacteria | Oceanospirillales | Halomonadaceae | Halomonas | 0.12±0.06 | 1.72±0.51 | ↑ 14.61 | 0.01* |
| Pseudomonadales | Pseudomonadaceae | Pseudomonas | 7.48±0.86 | 5.10±0.6 | ↓ 0.68 | 0.03* | |
| Enterobacteriales | Enterobacteriaceae | Erwinia | 0.26±0.1 | 0.64±0.15 | ↑ 2.41 | 0.04* | |
| Citrobacter | 0.66±0.24 | 0.08±0.05 | ↓ 0.12 | 0.02* | |||
| Unclassified | 8.85±1.01 | 6.08±0.58 | ↓ 0.69 | 0.02* | |||
| Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | 4.17±0.83 | 0.71±0.2 | ↓ 0.17 | 0.00** |
| Rhizobiales | Rhizobiaceae | Agrobacterium | 3.83±0.96 | 0.11±0.05 | ↓ 0.03 | 0.00** | |
| Unclassified 1 | 0.63±0.17 | 0.07±0.03 | ↓ 0.11 | 0.00** | |||
| Unclassified 2 | 0.11±0.03 | 0.00±0.00 | ↓ 0.00 | 0.00* | |||
| Bradyrhizobiaceae | Unclassified | 0.24±0.07 | 0.05±0.02 | ↓ 0.21 | 0.02* | ||
| Rickettsiales | Mitochondria | Unclassified | 0.84±0.3 | 0.1±0.06 | ↓ 0.12 | 0.02* | |
| Betaproteobacteria | Rhodobacterales | Rhodobacteraceae | Rhodobacter | 0.01±0.01 | 0.23±0.08 | ↑ 20.34 | 0.02* |
| Burkholderiales | Comamonadaceae | Roseateles | 1.12±0.35 | 0.02±0.01 | ↓ 0.02 | 0.00** | |
| Delftia | 0.22±0.08 | 0.01±0.00 | ↓ 0.05 | 0.02* | |||
| Comamonas | 0.08±0.05 | 0.36±0.12 | ↑ 4.48 | 0.04* | |||
| Unclassified | 0.92±0.15 | 3.79±0.49 | ↑ 4.14 | 0.00** | |||
| Alcaligenaceae | Achromobacter | 2.42±0.75 | 0.05±0.03 | ↓ 0.02 | 0.00** | ||
| Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Desulfovibrio | 0.04±0.02 | 0.48±0.14 | ↑ 10.88 | 0.00* |
| Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | 1.58±0.26 | 4.77±1.28 | ↑ 3.03 | 0.02* |
| Bacillales | Staphylococcaceae | Jeotgalicoccus | 0.03±0.02 | 0.50±0.11 | ↑ 14.70 | 0.00* | |
| Clostridia | Clostridiales | Unclassified | Unclassified | 1.06±0.26 | 1.87±0.26 | ↑ 1.75 | 0.04* |
| Clostridiaceae | Unclassified | 0.27±0.08 | 0.70±0.18 | ↑ 2.58 | 0.04* | ||
| Ruminococcaceae | Oscillospira | 0.10±0.04 | 0.47±0.14 | ↑ 4.53 | 0.02* | ||
| Actinobacteria | Actinomycetales | Nocardiaceae | Rhodococcus | 0.40±0.13 | 1.56±0.43 | ↑ 3.91 | 0.02* |
| Micrococcaceae | Kocuria | 0.06±0.05 | 0.30±0.08 | ↑ 4.75 | 0.01* | ||
| Bacteroidia | Bacteroidales | S24-7 | Unclassified | 0.84±0.36 | 2.02±0.44 | ↑ 2.40 | 0.04* |
| Flavobacteriia | Flavobacteriales | [Weeksellaceae | Cloacibacterium | 0.13±0.06 | 0.62±0.2 | ↑ 4.92 | 0.03* |
| Chloroplast | Streptophyta | Unclassified | Unclassified | 3.80±0.84 | 1.68±0.38 | ↓ 0.44 | 0.03* |
| Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobiaceae | Akkermansia | 0.52±0.18 | 2.35±0.68 | ↑ 4.52 | 0.02* |
| [Fimbriimonadia] | [Fimbriimonadales] | [Fimbriimonadaceae] | Fimbriimonas | 0.38±0.12 | 0.00±0.00 | ↓ 0.00 | 0.00** |
| Deinococci | Thermales | Thermaceae | Thermus | 0.03±0.02 | 0.21±0.07 | ↑ 6.30 | 0.02* |
Microbiota at the genus level whose occupancy was significantly different in ASD subjects are presented with associated higher taxonomy levels. Microbiota with occupancy 0.1% or higher in either normal healthy or ASD subjects were considered. ↑ and ↓ denote an increase and decrease in the percent composition, respectively.
Data are the mean percentage±SEM. * and ** denote significant differences between the indicated groups at p<0.05 and p<0.01, respectively (Student's t-test).
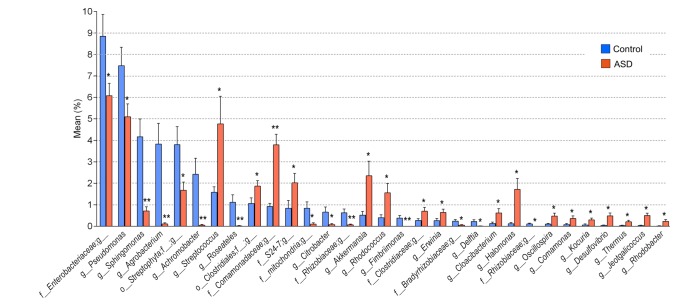
More specifically, an unclassified member of f_Enterobacteriaceae decreased from 8.85 to 6.08%, g_Pseudomonas decreased from 7.48 to 5.10%, g_Sphingomonas decreased from 4.17 to 0.71%, g_Agrobacterium decreased from 3.83 to 0.11%, an unclassified member of o_Streptophyta decreased from 3.80 to 1.68%, g_Achromobacter decreased from 2.42 to 0.05%, g_Roseateles decreased from 1.12 to 0.02%, and an unclassified member of f_mitochondria decreased from 0.84 to 0.10% (Table 3, Fig. 2).
Fig. 2. Genus members that were downregulated or upregulated in ASD subjects. The percent composition of microbiota whose occupancy was significantly changed at the genus level among those occupying 0.1% or higher in either control (blue) or ASD (red) subjects. Data are presented as the mean percentage±SEM. * and ** denote the differences between the indicated groups at p<0.05 and p<0.01, respectively (Student's t-test).
On the other hand, g_Streptococcus increased from 1.58 to 4.77%, an unclassified member of o_Clostridiales increased from 1.06 to 1.87%, an unclassified member of f_Comamonadaceae increased from 0.92 to 3.79%, an unclassified member of f_S24-7 increased from 0.84 to 2.02%, g_Akkermansia increased from 0.52 to 2.35%, g_Rhodococcus increased from 0.40% to 1.56%, and g_Halomonas increased from 0.12 to 1.72% (Table 3, Fig. 2).
DISCUSSION
Metagenome analysis of bacterial EVs in urine identifies altered microbiota profiles in ASD
In the present study, we demonstrated that bacteria-derived EVs in urine were useful for the rapid assessment of microbiota profiles in ASD. The metagenome analysis of urine EVs indicated that p_Verrucomicrobia (0.58 to 2.37%, p=0.02) and p_Firmicutes (24.96 to 33.07%, p=0.03) increased in ASD, whereas p_Cyanobacteria (4.36 to 1.92%, p=0.01) and p_Proteobacterium (49.12 to 35.3%, p=0.01) decreased. There was no significant change in p_Bacteroidetes (5.85 to 8.62%, p=0.11) and p_Actinobacterium (10.91 to 11.74%, p=0.56). The altered microbiota compositions identified from urine EVs of ASD were partially consistent with microbiota compositions assessed from fecal samples reported in recent studies. The analyses of fecal microbiota compositions in previous studies reported that p_Actinobacteria, p_Verrucomicrobia and p_Cyanobacteria decreased or tended to decrease in ASD, but there were conflicting results for p_Firmicutes, p_Bacteroidetes and p_Proteobacteria (Table 4) [13,15,16,19].
Table 4. Summary and comparison of microbiota characterized in the present study with those identified from fecal samples in previous studies.
| Taxons | Mean (%) | Fold change | p-value | Literatures | ||
|---|---|---|---|---|---|---|
| Control | ASD | |||||
| Phylum | Proteobacteria | 49.12 | 35.30 | ↓ 0.72 | 0.01** | ↓ [19]; ↑ [13] |
| Firmicutes | 24.96 | 33.07 | ↑ 1.33 | 0.03* | ↓ [13]; ↑ [15] | |
| Actinobacteria | 10.91 | 11.74 | ↑ 1.08 | 0.56 | ↓ [13] | |
| Bacteroidetes | 5.85 | 8.62 | ↑ 1.47 | 0.11 | ↑ [13]; ↓ [15]; ↓ [19] | |
| Verrucomicrobia | 0.58 | 2.37 | ↑ 4.12 | 0.02* | ↓ [19] | |
| Class | Betaproteobacteria | 10.09 | 6.26 | ↓ 0.62 | 0.10 | ↑ [15] |
| Order | Clostridiales | 11.59 | 15.38 | ↑ 1.33 | 0.18 | ↑ [15] |
| Family | Ruminococcaceae | 5.46 | 6.11 | ↑ 1.12 | 0.68 | ↑ [15] |
| Lachnospiraceae | 3.12 | 2.57 | ↓ 0.82 | 0.54 | ↑ [15] | |
| Corynebacterium | 2.60 | 3.38 | ↑ 1.3 | 0.34 | ↑ [11] | |
| Alcaligenaceae | 2.53 | 0.12 | ↓ 0.05 | 0.00** | ↑ [16] | |
| Pseudomonas | 7.48 | 5.10 | ↓ 0.68 | 0.03* | ↑ [21] | |
| Lactobacillus | 2.56 | 5.45 | ↑ 2.13 | 0.08 | ↑ [18]; ↑ [11] | |
| Bacteroides | 2.48 | 2.93 | ↑ 1.18 | 0.65 | ↑ [13]; ↑ [21] | |
| Staphylococcus | 2.23 | 2.57 | ↑ 1.15 | 0.66 | ↓ [21] | |
| Faecalibacterium | 2.14 | 2.05 | ↓ 0.96 | 0.90 | ↑ [20] | |
| Bifidobacterium | 1.90 | 0.80 | ↓ 0.42 | 0.06 | ↓ [13]; ↓ [18]; ↓ [21]; ↓ [17] | |
| Streptococcus | 1.58 | 4.77 | ↑ 3.03 | 0.02* | ↓ [13]; ↓ [21] | |
| Akkermansia | 0.52 | 2.35 | ↑ 4.52 | 0.02* | ↓ [19]; ↓ [17] | |
| Blautia | 0.47 | 0.28 | ↓ 0.59 | 0.38 | ↓ [20] | |
| Enterococcus | 0.40 | 0.78 | ↑ 1.92 | 0.06 | ↑ [21]; ↓ [18] | |
| Collinsella | 0.37 | 0.11 | ↓ 0.29 | 0.16 | ↓ [13]; ↑ [11] | |
| Veillonella | 0.37 | 0.50 | ↑ 1.34 | 0.43 | ↓ [11] | |
| Lactococcus | 0.35 | 0.11 | ↓ 0.3 | 0.16 | ↓ [13]; ↓ [21] | |
| [Ruminococcus] | 0.34 | 0.15 | ↓ 0.45 | 0.16 | ↓ [13] | |
| Coprococcus | 0.28 | 0.27 | ↓ 0.94 | 0.90 | ↓ [19] | |
| Leuconostoc | 0.27 | 0.01 | ↓ 0.02 | 0.14 | ↓ [13] | |
| Dialister | 0.23 | 1.39 | ↑ 6.16 | 0.18 | ↓ [13]; ↓ [11] | |
| Parabacteroides | 0.21 | 0.18 | ↓ 0.83 | 0.76 | ↓ [11]; ↑ [13] | |
| Weissella | 0.21 | 0.07 | ↓ 0.31 | 0.33 | ↓ [13] | |
| Turicibacter | 0.19 | 0.02 | ↓ 0.12 | 0.07 | ↓ [13] | |
| Dorea | 0.17 | 0.10 | ↓ 0.6 | 0.44 | ↑ [11] | |
| Clostridium | 0.11 | 0.31 | ↑ 2.72 | 0.09 | ↑ [21]; ↓ [13] | |
| [Prevotella] | 0.09 | 0.12 | ↑ 1.34 | 0.74 | ↑ [21]; ↓ [19] | |
| Desulfovibrio | 0.04 | 0.48 | ↑ 10.88 | 0.00** | ↑ [13]; ↓ [19] | |
| Genus | 0.03 | 0.37 | ↑ 11.53 | 0.08 | ↑ [21] | |
The microbiota whose percent composition were significantly different in ASD subjects as characterized in the present study were compared with those identified from fecal samples in previous studies. ↑ and ↓ denote an increase and decrease in the percent composition, respectively. * and ** denote significant differences between the indicated groups at p<0.05 and p<0.01, respectively (Student's t-test).
In p_Firmicutes, g_Streptococcus (1.58 to 4.77%, p=0.02), g_Jeotgalicoccus (0.03 to 0.5%, p<0.01), g_Oscillospira (0.1 to 0.47%, p=0.02), and an unclassified member of f_Clostridiaceae (0.27 to 0.7%, p=0.04) significantly increased in ASD by more than two-fold. The genera g_Akkermansia occupied the greatest proportion of the increased phylum p_Verrucomicrobia (0.52 to 2.35%, p=0.02), whereas in p_Proteobacterium, 12 genera decreased and 5 genera increased in ASD. The genera g_Pseudomonas, g_Citrobacter, an unclassified member of f_Enterobacteriaceae, g_Sphingomonas, g_Agrobacterium, unclassified members of f_Rhizobiaceae, an unclassified member of f_Bradyrhizobiaceae, and an unclassified member of f_mitochondria, g_Roseateles, g_Delftia and g_Achromobacter were decreased (total 30.58 to 12.37%), whereas g_Halomonas, g_Erwinia, g_Rhodobacter, g_Comamonas, and an unclassified member of f_Comamonadaceae were increased (total 1.39 to 6.74%).
At the genus level, the present study identified g_Sphingomonas, g_Agrobacterium, an unclassified member of o_Streptophyta, g_Achromobacter, and g_Roseateles as being decreased in ASD by more than two-fold among the total OTUs having occupancy 0.1% or higher in either healthy control or ASD subjects, and g_Streptococcus, an unclassified member of f_Comamonadaceae, an unclassified member of f_S24-7, g_Akkermansia, g_Rhodococcus, and g_Halomonas were increased in ASD. Particularly, g_Desulfovibrio increased in ASD (0.04 to 0.48%, p<0.01), g_Lactobacillus tended to increase (2.56 to 5.45%, p=0.08), g_Bifidobacterium tended to decrease (1.9 to 0.8%, p=0.06), and g_Turicibacter tended to decrease (0.19 to 0.02%, p=0.07) in ASD, whereas g_Enterococcus (0.4 to 0.78%, p=0.06), g_Enterobacter (0.03 to 0.37, p=0.08), and g_Clostridium (0.11 to 0.31%, p=0.09) tended to increase in ASD. The changes in these genus members are broadly consistent with the results of previous reports assessed for fecal microbiota compositions (Table 4) [11,13,17,18,21].
The EV levels of g_Oscillospira, unclassified members of f_Clostridiaceae and f_Eubacteriaceae, and an unclassified member of o_Clostridiales were increased in ASD subjects. Previous studies have reported that several species of c_Clostridia produced 4-ethylphenyl sulfate (4-EPS) and p-cresol, which were found at high concentrations in the urine of ASD children. Administration of 4-EPS in healthy mice produced myelination deficits in the prefrontal cortex and sociability defects [37,38,39]. It was also reported that c_Clostridia produced propionic acid (PPA) and its related short-chain fatty acids (SCFAs) as fermentation products, and PPA infusions in rats induced ASD-linked neurochemical and behavioral changes [40]. These results suggest that bacteria-derived metabolites induce neurochemical and structural changes and shape behavioural abnormalities.
Oral treatment with Bifidobacteria fragilis ameliorated ASD-related gastrointestinal deficits and associated behavioural abnormalities behavioral abnormalities in the poly (I:C)-injection model [41]. Bifidobacteria infantis attenuated pro-inflammatory immune responses and production of serotonergic precursor, tryptophan, and has potential anti-depressant properties [42,43]. Considering these results, ASD groups with decreased EV levels of g_Bifidobacterium might have benefits by probiotic treatment with g_Bifidobacterium.
Bacterial EVs in urine are useful for rapid assessment of bodily microbiota profiles in ASD
The microbiota profile assessed from urine EVs might reflect a large part of the gut microbiota. Nonetheless, we do believe that the microbiota profile assessed from urine EVs is not likely a simple alternative for microbiota profile assessed from stool. Possible sources for metagenome analysis of bodily microbiota may include stool bacteria, stool EVs, gut (ex, stomach and/or specific regions of the small and large intestines) bacteria, respiratory exhale EVs, oral/nasal bacteria and EVs, urinary system bacteria and EVs, and blood EVs. Generally speaking, microbiota in stool represents the intestinal compartment, whereas microbiota in urine or blood reflects the whole body including the intestinal compartments, oral system, respiratory system, and urinary system. Nonetheless, among the body parts, the gut is the major source of bodily microbiota. It was reported that the metabolites of intestinal microbiota activities, including phenyllactate, p-cresol sulfate, concentrations, and serotonin in urine, plasma, and stool of mouse pups undernourished by timed separation from lactating dams, then resumed ad libitum nursing, were different from each other, although they had some correlations [44,45]. Similar to metabolite profiles of intestinal microbiota activities, available information suggests that metagenome analysis assessed from these sources might be closely related, but represent some distinct landscapes. For an example, metagenome analysis of bacteria and bacteria-derived EV in stool of inflammatory bowel disease model mice indicated that the EV composition in stool was more drastically altered compared to that of bacterial composition in stool [25]. Considering that bacteria-derived EV indicates the metabolically or pathologically activated microbiota [25,27], urine EV may be more representative of the host's microbiota activities than stool bacteria.
To the best of our knowledge, this is the first report characterizing microbiota in ASD individuals on the basis of urine EVs. Compared to blood and feces, urine is easily obtained in large volume and is readily available via a non-invasive method. Considering the general difficulties in repeated sampling microbiota sources from ASD individuals, particularly low functioning individuals with ASD or toddlers with ASD, using urine as a sample source would be a great advantage for rapid and repeated assessments of microbiota changes under varying physiological contexts compared to the use of blood and feces. Comparative analyses of EV profiles from urine, blood and stool of ASD individuals will be valuable. Also it will be worth to understand EV profiles of ASD with diverse factors including age, sex, familial history, genetics, and ethnics.
Overall, the present study assessed urine EVs from individuals with mildly autistic subjects. We believe that further systematic and unbiased analyses of male and female subjects with broad ASD spectrums are necessary. This study focused on young adult subjects. Considering that ASD should be diagnosed in young children as early as 1.5~3 yr of age, this analysis should be expanded to toddlers and infants.
ACKNOWLEDGEMENTS
This study was supported by an intramural research promotion grant from Ewha Womans University School of Medicine.
SUPPLEMENTARY MATERIALS
The diversity of microbiota at the class, order, family, and genus levels in control vs. ASD subjects. (A~D) Principal component analysis (PCA) of microbiota diversity at the class (A), order (B), family (C), and genus (D) levels based on the weighted UniFrac distance and Bray-Curtis dissimilarity. Data were normalized to have a mean of 0 and a standard deviation of 1. Control (blue) and ASD (red).
The percent composition of microbiota at the genus level in control and ASD subjects
References
- 1.American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-5. 5th ed. Washington, D.C.: American Psychiatric Association; 2013. [Google Scholar]
- 2.Kral TV, Eriksen WT, Souders MC, Pinto-Martin JA. Eating behaviors, diet quality, and gastrointestinal symptoms in children with autism spectrum disorders: a brief review. J Pediatr Nurs. 2013;28:548–556. doi: 10.1016/j.pedn.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Marí-Bauset S, Zazpe I, Mari-Sanchis A, Llopis-González A, Morales-Suárez-Varela M. Food selectivity in autism spectrum disorders: a systematic review. J Child Neurol. 2014;29:1554–1561. doi: 10.1177/0883073813498821. [DOI] [PubMed] [Google Scholar]
- 4.Bryant-Waugh R, Markham L, Kreipe RE, Walsh BT. Feeding and eating disorders in childhood. Int J Eat Disord. 2010;43:98–111. doi: 10.1002/eat.20795. [DOI] [PubMed] [Google Scholar]
- 5.Hyman SL, Stewart PA, Schmidt B, Cain U, Lemcke N, Foley JT, Peck R, Clemons T, Reynolds A, Johnson C, Handen B, James SJ, Courtney PM, Molloy C, Ng PK. Nutrient intake from food in children with autism. Pediatrics. 2012;130(Suppl 2):S145–S153. doi: 10.1542/peds.2012-0900L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McElhanon BO, McCracken C, Karpen S, Sharp WG. Gastrointestinal symptoms in autism spectrum disorder: a meta-analysis. Pediatrics. 2014;133:872–883. doi: 10.1542/peds.2013-3995. [DOI] [PubMed] [Google Scholar]
- 7.Campbell DB, Buie TM, Winter H, Bauman M, Sutcliffe JS, Perrin JM, Levitt P. Distinct genetic risk based on association of MET in families with co-occurring autism and gastrointestinal conditions. Pediatrics. 2009;123:1018–1024. doi: 10.1542/peds.2008-0819. [DOI] [PubMed] [Google Scholar]
- 8.Russo AJ, Krigsman A, Jepson B, Wakefield A. Decreased serum hepatocyte growth factor (HGF) in autistic children with severe gastrointestinal disease. Biomark Insights. 2009;4:181–190. doi: 10.4137/bmi.s3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiao EY. Gastrointestinal issues in autism spectrum disorder. Harv Rev Psychiatry. 2014;22:104–111. doi: 10.1097/HRP.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 10.Mulle JG, Sharp WG, Cubells JF. The gut microbiome: a new frontier in autism research. Curr Psychiatry Rep. 2013;15:337. doi: 10.1007/s11920-012-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, Jousson O, Leoncini S, Renzi D, Calabrò A, De Filippo C. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017;5:24. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenfeld CS. Microbiome disturbances and autism spectrum disorders. Drug Metab Dispos. 2015;43:1557–1571. doi: 10.1124/dmd.115.063826. [DOI] [PubMed] [Google Scholar]
- 13.Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, Youn E, Summanen PH, Granpeesheh D, Dixon D, Liu M, Molitoris DR, Green JA., 3rd Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Tomova A, Husarova V, Lakatosova S, Bakos J, Vlkova B, Babinska K, Ostatnikova D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol Behav. 2015;138:179–187. doi: 10.1016/j.physbeh.2014.10.033. [DOI] [PubMed] [Google Scholar]
- 15.Williams BL, Hornig M, Buie T, Bauman ML, Cho Paik M, Wick I, Bennett A, Jabado O, Hirschberg DL, Lipkin WI. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 2011;6:e24585. doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams BL, Hornig M, Parekh T, Lipkin WI. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio. 2012;3:e00261–e00211. doi: 10.1128/mBio.00261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microbiol. 2011;77:6718–6721. doi: 10.1128/AEM.05212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism--comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011;11:22. doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, Krajmalnik-Brown R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8:e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue R, Sakaue Y, Sawai C, Sawai T, Ozeki M, Romero-Pérez GA, Tsukahara T. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci Biotechnol Biochem. 2016;80:2450–2458. doi: 10.1080/09168451.2016.1222267. [DOI] [PubMed] [Google Scholar]
- 21.De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, Cristofori F, Guerzoni ME, Gobbetti M, Francavilla R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8:e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santocchi E, Guiducci L, Fulceri F, Billeci L, Buzzigoli E, Apicella F, Calderoni S, Grossi E, Morales MA, Muratori F. Gut to brain interaction in Autism Spectrum Disorders: a randomized controlled trial on the role of probiotics on clinical, biochemical and neurophysiological parameters. BMC Psychiatry. 2016;16:183. doi: 10.1186/s12888-016-0887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, Khoruts A, Geis E, Maldonado J, McDonough-Means S, Pollard EL, Roux S, Sadowsky MJ, Lipson KS, Sullivan MB, Caporaso JG, Krajmalnik-Brown R. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 2017;5:10. doi: 10.1186/s40168-016-0225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 25.Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ, Gho YS, Kim JG, Kim YK. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One. 2013;8:e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo JY, Rho M, You YA, Kwon EJ, Kim MH, Kym S, Jee YK, Kim YK, Kim YJ. 16S rRNA gene-based metagenomic analysis reveals differences in bacteria-derived extracellular vesicles in the urine of pregnant and non-pregnant women. Exp Mol Med. 2016;48:e208. doi: 10.1038/emm.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jang SC, Kim SR, Yoon YJ, Park KS, Kim JH, Lee J, Kim OY, Choi EJ, Kim DK, Choi DS, Kim YK, Park J, Di Vizio D, Gho YS. In vivo kinetic biodistribution of nano-sized outer membrane vesicles derived from bacteria. Small. 2015;11:456–461. doi: 10.1002/smll.201401803. [DOI] [PubMed] [Google Scholar]
- 28.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maldonado R, Wei R, Kachlany SC, Kazi M, Balashova NV. Cytotoxic effects of Kingella kingae outer membrane vesicles on human cells. Microb Pathog. 2011;51:22–30. doi: 10.1016/j.micpath.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S, Kim SH, Desiderio DM, Kim YK, Kim KP, Gho YS. Grampositive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics. 2009;9:5425–5436. doi: 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- 31.Brown L, Kessler A, Cabezas-Sanchez P, Luque-Garcia JL, Casadevall A. Extracellular vesicles produced by the Gram-positive bacterium Bacillus subtilis are disrupted by the lipopeptide surfactin. Mol Microbiol. 2014;93:183–198. doi: 10.1111/mmi.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shim MS, Kim YH. Standardization study for the Korean version of childhood autism rating scale: reliability, validity and cut-off score. Korean J Clin Psychol. 1998;17:1–15. [Google Scholar]
- 33.Park EY, Kim J. Factor structure of the Childhood Autism Rating Scale as per DSM-5. Pediatr Int. 2016;58:139–145. doi: 10.1111/ped.12770. [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 35.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altieri L, Neri C, Sacco R, Curatolo P, Benvenuto A, Muratori F, Santocchi E, Bravaccio C, Lenti C, Saccani M, Rigardetto R, Gandione M, Urbani A, Persico AM. Urinary p-cresol is elevated in small children with severe autism spectrum disorder. Biomarkers. 2011;16:252–260. doi: 10.3109/1354750X.2010.548010. [DOI] [PubMed] [Google Scholar]
- 38.Gabriele S, Sacco R, Cerullo S, Neri C, Urbani A, Tripi G, Malvy J, Barthelemy C, Bonnet-Brihault F, Persico AM. Urinary p-cresol is elevated in young French children with autism spectrum disorder: a replication study. Biomarkers. 2014;19:463–470. doi: 10.3109/1354750X.2014.936911. [DOI] [PubMed] [Google Scholar]
- 39.Gacias M, Gaspari S, Santos PM, Tamburini S, Andrade M, Zhang F, Shen N, Tolstikov V, Kiebish MA, Dupree JL, Zachariou V, Clemente JC, Casaccia P. Microbiota-driven transcriptional changes in prefrontal cortex override genetic differences in social behavior. Elife. 2016;5:e13442. doi: 10.7554/eLife.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macfabe DF. Short-chain fatty acid fermentation products of the gut microbiome: implications in autism spectrum disorders. Microb Ecol Health Dis. 2012;23:19260. doi: 10.3402/mehd.v23i0.19260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desbonnet L, Garrett L, Clarke G, Bienenstock J, Dinan TG. The probiotic Bifidobacteria infantis: an assessment of potential antidepressant properties in the rat. J Psychiatr Res. 2008;43:164–174. doi: 10.1016/j.jpsychires.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 43.Desbonnet L, Garrett L, Clarke G, Kiely B, Cryan JF, Dinan TG. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience. 2010;170:1179–1188. doi: 10.1016/j.neuroscience.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Preidis GA, Ajami NJ, Wong MC, Bessard BC, Conner ME, Petrosino JF. Microbial-derived metabolites reflect an altered intestinal microbiota during catch-up growth in undernourished neonatal mice. J Nutr. 2016;146:940–948. doi: 10.3945/jn.115.229179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Armstrong CW, McGregor NR, Lewis DP, Butt HL, Gooley PR. The association of fecal microbiota and fecal, blood serum and urine metabolites in myalgic encephalomyelitis/chronic fatigue syndrome. Metabolomics. 2017;13:8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The diversity of microbiota at the class, order, family, and genus levels in control vs. ASD subjects. (A~D) Principal component analysis (PCA) of microbiota diversity at the class (A), order (B), family (C), and genus (D) levels based on the weighted UniFrac distance and Bray-Curtis dissimilarity. Data were normalized to have a mean of 0 and a standard deviation of 1. Control (blue) and ASD (red).
The percent composition of microbiota at the genus level in control and ASD subjects