Abstract
Circadian rhythms are fundamental biological systems in most organisms. Epidemiological and animal studies have demonstrated that disruption of circadian rhythms is linked to tumor progression and mammalian tumorigenesis. However, the clinical significance of in situ clock gene expression in precancerous and cancerous colorectal lesions remains unknown. The present study aimed to investigate mRNA transcript levels of circadian clock genes within human colorectal cancer and adenoma tissue sections. Using in situ hybridization, the expression of key clock genes, including period circadian protein homolog (Per) 1 and 2, cryptochrome 1 (Cry1), circadian locomoter output cycles protein kaput (Clock), brain and muscle ARNT-like protein 1 (Bmal1) and casein kinase 1ε (CK1ε) were retrospectively examined in 51 cases of colorectal carcinoma and 10 cases of adenoma. The expression of clock genes was almost undetectable in the majority of adenomas, whereas positive expression of clock genes was observed in 27–47% of carcinomas. Notably, positive Per1, Per2 and Clock staining in colorectal carcinomas were each significantly associated with a larger tumor size (P=0.012, P=0.011 and P=0.009, respectively). Tumors with positive Per2 and Clock expression tended to exhibit deeper depth of invasion and were generally more advanced than tumors that did not express these genes (P=0.052 and P=0.064, respectively). However, no statistically significant association was observed between clock gene expression and clinicopathological variables, including histopathological differentiation, lymph node metastasis, depth of invasion or disease stage, although Per2-positive tumors tended to be associated with poorer overall survival (P=0.060). The results of the current study suggest that dysregulated expression of clock genes may be important in human colorectal tumorigenesis.
Keywords: circadian rhythm, colorectal cancer, clock gene, in situ hybridization
Introduction
A number of biochemical, physiological and behavioral processes have demonstrated that an internal time-keeping mechanism, referred to as the biological clock, regulates circadian rhythms. The master circadian clock coordinates peripheral clocks elsewhere in the body and is located in the suprachiasmatic nuclei (SCN) within the anterior hypothalamus (1). The core oscillator driving this clock is intergrated by an auto-regulatory transcription-(post) translation-based feedback loop, which is comprised of genes related to the circadian rhythm (1,2).
Epidemiological studies have suggested that disruption of the circadian clock may increase cancer risk in humans (3–5). In particular, it has been observed that shift workers have an increased risk of developing malignancies, including breast, endometrial, prostate and colorectal cancer, due to their disrupted circadian cycles (5–9). Fu et al (10) previously demonstrated that a period circadian protein homolog 2 (Per2) mutation induced upregulation of c-Myc and downregulation of p53 transcription in mice; furthermore, the incidence of spontaneous and radiation-induced lymphoma increased, as did lymphoma-associated mortality. Other in vivo studies have identified an association between alterations of the circadian rhythm and tumorigenesis (2,9,11). In a number of types of human solid cancer, including breast, endometrial and colorectal cancer, the dysregulated expression of circadian genes has been investigated by immunohistochemistry and/or reverse transcription quantitative polymerase chain reaction (RT-qPCR) (9,12–14).
The aim of the present study was to investigate the clinical significance of the mRNA expression of clock genes in human colorectal carcinoma and adenoma tissues, using in situ hybridization.
Patients and methods
Patients and tumor samples
A total of 51 patients (32 males and 19 females) with colorectal carcinoma, and 10 patients with colorectal adenoma were examined. All patients underwent endoscopic or surgical resection to completely remove tumors in the Department of Organ Regulatory Surgery, Fukushima Medical University Hospital (Fukushima, Japan) between April 1999 and July 2005. In several tissue specimens, the surrounding normal mucosa was also examined. None of the patients had received prior chemotherapy or irradiation or had experienced any other form of cancer. The clinicopathological characteristics of the 51 patients with colorectal cancer investigated in this study are summarized in Table I.
Table I.
Associations between clock gene expression and clinicopathological variables.
| Per1 | Per2 | Cry1 | Clock | Bmal1 | CKIε | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | |||||||
| Variables | 51 | 27 | 24 | P-value | 25 | 26 | P-value | 27 | 24 | P-value | 30 | 21 | P-value | 31 | 20 | P-value | 37 | 14 | P-value |
| Age | 0.242 | 0.048 | 0.820 | 0.931 | 0.423 | 0.727 | |||||||||||||
| Mean | 65.1 | 63.7 | 67.6 | 68.9 | 62.2 | 65.1 | 65.9 | 65.6 | 65.4 | 65.3 | 65.9 | 65.8 | 64.6 | ||||||
| Range | 33–84 | 33–81 | 43–84 | 49–84 | 33–81 | 33–84 | 41–84 | 41–81 | 33–84 | 41–84 | 33–84 | 33–84 | 46–80 | ||||||
| Gender | 1 | 1 | 0.772 | 0.563 | 0.774 | 0.333 | |||||||||||||
| Male | 32 | 17 | 15 | 16 | 16 | 16 | 16 | 20 | 12 | 20 | 12 | 25 | 7 | ||||||
| Female | 19 | 10 | 9 | 9 | 10 | 11 | 8 | 10 | 9 | 11 | 8 | 12 | 7 | ||||||
| Tumor size (mm) | 0.012a | 0.011a | 0.164 | 0.009a | 0.082a | 0.342 | |||||||||||||
| <50 | 29 | 20 | 9 | 19 | 10 | 18 | 11 | 22 | 7 | 21 | 8 | 23 | 6 | ||||||
| ≥50 | 22 | 7 | 15 | 6 | 16 | 9 | 13 | 8 | 14 | 10 | 12 | 14 | 8 | ||||||
| Tumor location | 0.782 | 1 | 0.577 | 0.578 | 0.567 | 0.363 | |||||||||||||
| Colon | 24 | 12 | 12 | 12 | 12 | 14 | 10 | 13 | 11 | 16 | 8 | 19 | 5 | ||||||
| Rectum | 27 | 15 | 12 | 13 | 14 | 13 | 14 | 17 | 10 | 15 | 12 | 18 | 9 | ||||||
| Histological differentiation | 0.636 | 0.972 | 0.991 | 0.754 | 0.422 | 0.212 | |||||||||||||
| Well | 34 | 17 | 17 | 17 | 17 | 18 | 16 | 20 | 14 | 22 | 12 | 27 | 7 | ||||||
| Moderately | 13 | 7 | 6 | 6 | 7 | 7 | 6 | 7 | 6 | 6 | 7 | 7 | 6 | ||||||
| Poorly | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| Mucinous | 4 | 3 | 1 | 2 | 2 | 2 | 2 | 3 | 1 | 3 | 1 | 3 | 1 | ||||||
| Depth of invasion | 0.482 | 0.103 | 0.978 | 0.303 | 0.580 | 0.240 | |||||||||||||
| pT1 | 5 | 3 | 2 | 4 | 1 | 3 | 2 | 4 | 1 | 2 | 3 | 2 | 3 | ||||||
| pT2 | 7 | 4 | 3 | 5 | 2 | 4 | 3 | 5 | 2 | 4 | 3 | 6 | 1 | ||||||
| pT3 | 31 | 14 | 17 | 11 | 20 | 16 | 15 | 15 | 16 | 21 | 10 | 22 | 9 | ||||||
| pT4 | 8 | 6 | 2 | 5 | 3 | 4 | 4 | 6 | 2 | 4 | 4 | 7 | 1 | ||||||
| Depth of invasion | 0.749 | 0.052 | 0.749 | 0.315 | 0.502 | 0.715 | |||||||||||||
| pT1-2 | 12 | 7 | 5 | 9 | 3 | 7 | 5 | 9 | 3 | 6 | 6 | 8 | 4 | ||||||
| pT3-4 | 39 | 20 | 19 | 16 | 23 | 20 | 19 | 21 | 18 | 25 | 14 | 29 | 10 | ||||||
| Lymph node metastasis | 1 | 1 | 0.264 | 1 | 1 | 0.363 | |||||||||||||
| Absent | 27 | 14 | 13 | 13 | 14 | 12 | 15 | 16 | 11 | 16 | 11 | 18 | 9 | ||||||
| Present | 24 | 13 | 11 | 12 | 12 | 15 | 9 | 14 | 10 | 15 | 9 | 19 | 5 | ||||||
| Stage | 0.723 | 0.240 | 0.506 | 0.180 | 0.987 | 0.363 | |||||||||||||
| I | 9 | 6 | 3 | 6 | 3 | 5 | 4 | 8 | 1 | 5 | 4 | 6 | 3 | ||||||
| II | 16 | 7 | 9 | 6 | 10 | 6 | 10 | 7 | 9 | 10 | 6 | 10 | 6 | ||||||
| III | 21 | 11 | 10 | 12 | 9 | 13 | 8 | 12 | 9 | 13 | 8 | 18 | 3 | ||||||
| IV | 5 | 3 | 2 | 1 | 4 | 3 | 2 | 3 | 2 | 3 | 2 | 3 | 2 | ||||||
| Stage | 0.473 | 0.291 | 1 | 0.064 | 0.724 | 0.692 | |||||||||||||
| I | 9 | 6 | 3 | 6 | 3 | 5 | 4 | 8 | 1 | 5 | 4 | 6 | 3 | ||||||
| II–IV | 42 | 21 | 21 | 19 | 23 | 22 | 20 | 22 | 20 | 26 | 16 | 31 | 11 | ||||||
P<0.05, indicates a significant difference. P, P-value; Per, period circadian protein homolog; Cry1, cryptochrome 1; Clock, circadian locomoter output cycles protein kaput; Bmal1, brain and muscle ARNT-like protein 1; CK1ε, casein kinase 1ε.
All tissue samples were embedded in optimal cutting temperature (OCT) compound (Sakura Finetek USA, Inc., Torrance, CA, USA) and immediately stored at −8°C. Tumors were histopathologically classified as well-differentiated, moderately differentiated, poorly differentiated or mucinous adenocarcinomas (15), and tumor size was defined as the largest diameter of the tumor. Histopathological diagnoses were performed at the Department of Pathology, Fukushima Medical University Hospital following standard procedures. Informed consent was obtained from each patient and the Fukushima Medical University Committee approved the protocol of the present study.
In situ hybridization
Digoxigenin (DIG)-UTP labeled cRNA probes were used to evaluate the mRNA expression of clock genes. The DIG-labeled cRNA probes were synthesized using a DIG RNA Labeling kit (Roche Diagnostics, Basel, Switzerland), and were labeled with SP6 or T7 RNA polymerase in the presence of DIG-UTP. Sections 5-µm thick were formed from the embedded tissue specimens, sufficiently dried with cold air and fixed in 4% paraformaldehyde diluted with phosphate-buffered saline for 30 min. Sections were hybridized overnight at 42°C in hybridization buffer containing 1 µg/ml of DIG-labeled probe. The DIG-labeled probes were diluted to 1 µg/ml with hybridization buffer (Nippon Gene Co., Ltd., Tokyo, Japan) and dropped to the sections, which were incubated at 42°C for 16–20 h to hybridize with each probe. Following hybridization, the sections were washed in 2 × standard citrate buffered saline and 0.2 × saline sodium citrate buffer at 5°C for 20 min and treated with 1% blocking solution at room temperature for 30 min, using the DIG Nucleic Acid Detection Kit (Roche Diagnostics). The sections were subsequently incubated at room temperature for 30 min with alkaline phosphatase labeled anti-digoxigenin antibody (Roche) diluted with a blocking solution (1:5,000). Color reaction was conducted using NTB/BCIP at 4°C for 12 h. As a negative control, the serial section was hybridized with a sense probe. Sections were simultaneously evaluated by two investigators. The tumor cells were classified into 4 groups based on intensity of staining (none, weak, moderate or strong) indicating levels of gene expression within the cells.
Statistical analysis
Differences between groups were evaluated by the χ2 test, Fisher's exact test, Student's t test or the Mann-Whitney U test. Cumulative survival was estimated by the Kaplan-Meier method and differences were analyzed by the log-rank test. All statistical analyses were two-sided and P<0.05 was considered to indicate a statistically significant difference.
Results
Potential involvement of clock genes in colorectal tumor progression
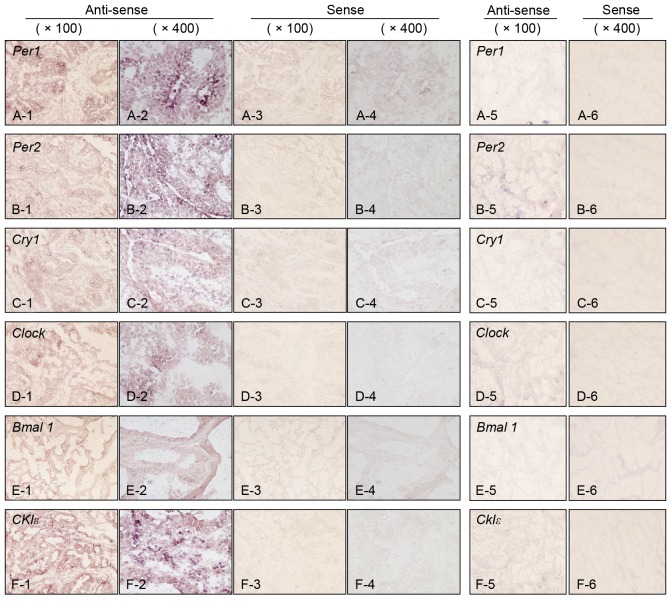
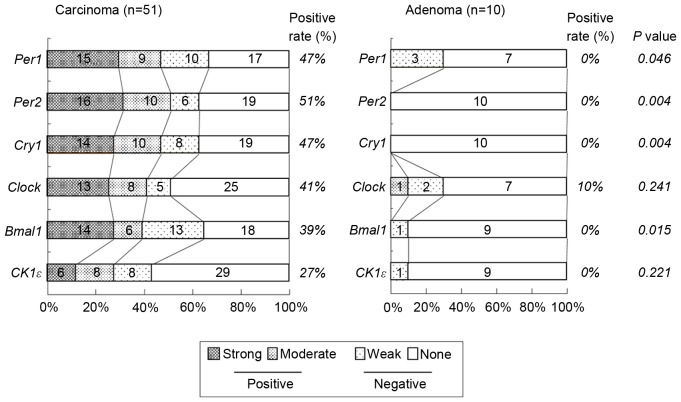
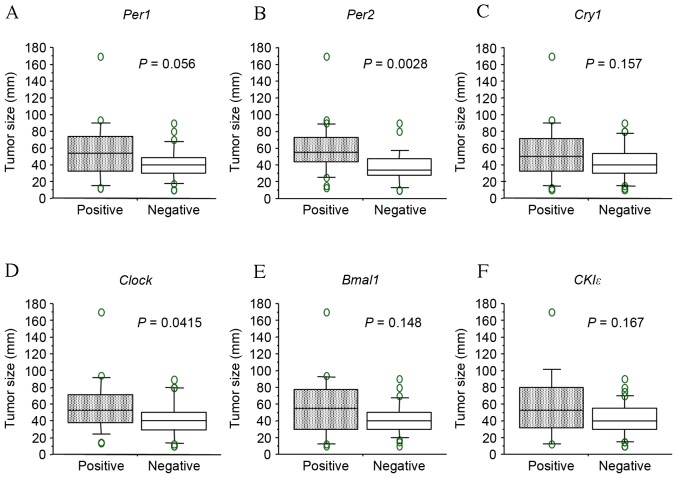
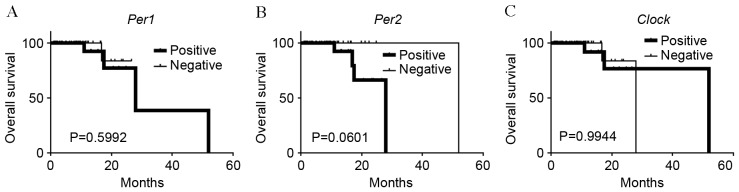
mRNA expression of the clock genes, including Per1 and 2, cryptochrome 1 (Cry1), circadian locomoter output cycles protein kaput (Clock), brain and muscle ARNT-like protein 1 (Bmal1) and casein kinase 1ε (CK1ε), was examined in colorectal cancer tissues by in situ hybridization. As presented in Fig. 1, sense probes as negative controls exhibited no staining (Fig. 1 panels 3–4), whereas various levels of staining were observed in tumor cells detected by anti-sense probes (Fig. 1, panels 1–2). Normal epithelial areas were evaluated in a number of specimens where the surrounding normal mucosa was available, however, no clear staining was detected by any of the probes (Fig. 1. panels 5–6). The levels of expression of clock genes in tumor cells were classified into four groups (none, weak, moderate or strong), based on the intensity of staining (Fig. 2). Tumors with no or weak staining were further defined as a negative group, while tumors with moderate and strong staining were a positive group. Of the 51 colorectal carcinomas evaluated, positive staining for Per1, Per2, Cry1, Clock, Bmal1 and CK1ε was observed in 24 (47%), 26 (51%), 24 (47%), 21 (41%), 20 (39%) and 14 (27%) tumors, respectively (Table I and Fig. 3). However, no significant associations were observed between levels of clock gene expression and histopathological type, depth of invasion, lymph node metastasis or disease stage (Table I). Although positive Per2 and positive Clock groups tended to be associated with a deeper depth of invasion and advanced stage, respectively, these associations were not significance (P=0.052 and P=0.064, respectively). By contrast, positive-Per1, Per2 and Clock groups were each associated with larger tumor size (>50 mm; P=0.012, P=0.011 and P=0.009, respectively; Table I). Similar results were obtained when tumor size was treated as a continuous variable (Fig. 4), therefore, the potential prognostic significance of Per1, Per2 and Clock was investigated. No association was observed between Per1 or Clock positive expression and overall survival rates (P=0.0599 and P=0.994, respectively; Fig. 5A and B). On the other hand, patients with carcinomas exhibiting positive-Per2 expression tended to have lower rates of survival than patients with negative-Per2 carcinomas, although this association was not significant (P=0.060; Fig. 5C).
Figure 1.
Expression of clock genes detected by in situ hybridization in colorectal carcinoma and surrounding normal mucosa. (A) Per1, (B) Per2, (C) Cry1, (D) Clock, (E) Bmal1 and (F) CK1ε. Panels 1 and 2 represent carcinoma tissue with anti-sense probe (magnification, ×100 and ×400, respectively). Panels 3 and 4 represent carcinoma tissues with sense probe (magnification, ×100 and ×400, respectively). Panels 5 and 6 represent normal mucosa tissues with anti-sense and sense probes, respectively (magnification, ×100 for both). Per, Period circadian protein homolog; Cry1, Cryptochrome 1; Clock, Circadian locomoter output cycles protein kaput; Bmal1, Brain and Muscle ARNT-like Protein 1; CK1ε, Casein Kinase 1ε.
Figure 2.
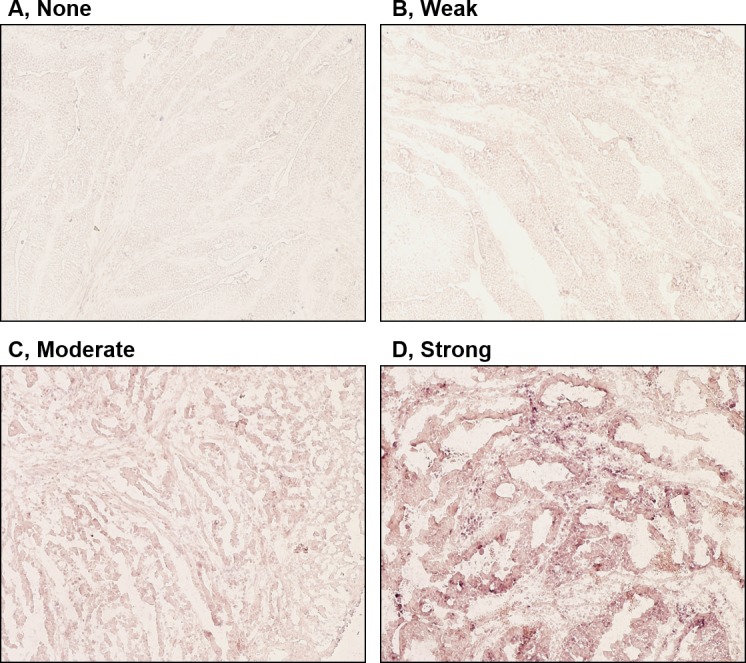
Classification of the expression of clock genes by in situ hybridization. The intensity of staining in tumor cells was classified into four groups as: (A) None, (B) weak, (C) moderate and (D) strong. Magnification, ×100.
Figure 3.
Proportion of expression levels of clock genes in colorectal carcinoma (left panel) and adenoma (right panel). Differences between positive and negative were analyzed by Fisher's exact test. Per, period circadian protein homolog; Cry1, cryptochrome 1; Clock, circadian locomoter output cycles protein kaput; Bmal1, brain and muscle ARNT-like protein 1; CK1ε, casein kinase 1ε.
Figure 4.
Comparison of tumor size between positive and negative staining of each gene. (A) Per1, (B) Per2, (C) Cry1, (D) Clock, (E) Bmal1 and (F) CKIε. Boxes correspond to the inter-quartile ranges, with the lower boundary of the box representing the 25th percentile and the upper boundary representing the 75th percentile. Differences between groups were analyzed by Student's t-test. Per, period circadian protein homolog; Cry1, cryptochrome 1; Clock, circadian locomoter output cycles protein kaput; Bmal1, brain and muscle ARNT-like protein 1; CK1ε, casein kinase 1ε.
Figure 5.
Kaplan-Meier survival curves for Per1 (A), Per2 (B) and Clock (C). Survival differences were assessed using the log-rank test. Per, period circadian protein homolog; Clock, circadian locomoter output cycles protein kaput.
The expression of clock genes in colorectal adenoma
To investigate the expression of clock genes in precancerous lesions compared with cancer tissues, 10 colorectal adenomas were examined by in situ hybridization. In contrast to cancer tissues, the expression of each clock gene was undetectable in the majority of adenoma tissue (Fig. 3). No adenomas (0%) exhibited positive staining for Per1, Per2, Cry1, Bmal1 or CK1ε and only 10% of adenomas exhibited positive expression of Clock. Hence, the proportion of tissues indicating positive Per1, Per2, Cry1 and Cry2 expression in colorectal carcinoma was significantly higher than in colorectal adenoma (P=0.046, P=0.004, P=0.004 and P=0.015, respectively; Fig. 3).
Discussion
Epidemiological studies have suggested that disruption of the circadian rhythm is associated with increased cancer incidence and poorer disease outcome (3–5,8). Previous studies have indicated that in Per2 mutant mice, Bmal1 expression decreased causing an increase in c-Myc transcription, thus disrupting the circadian rhythm and increasing cancer risk (10). In colorectal cancer, animal studies using chemically induced models as well as APCMin/+ mice, have suggested a link between alterations of circadian genes and colorectal tumor development and progression (16–18). Other studies have used RT-qPCR to demonstrate that clock gene expression is dysregulated in human colorectal cancer (9,19,20).
In the present study, unlike previous studies, the in situ hybridization technique was utilized to detect clock gene mRNA expression in colorectal tumor tissues, including precancerous and cancerous lesions. The proportion of colorectal carcinomas with positive Per1, Per2, Cry1 and Cry2 expression was observed to be significantly higher than the proportion of adenomas, suggesting that dysregulated clock gene expression may be involved in colorectal tumorigenesis. Colorectal carcinoma tumors exhibiting positive staining of Per1, Per2 and Clock were significantly larger than those exhibiting negative staining. Correspondingly, tumors with positive staining of Per2 and Clock tended to be associated with deeper depth of invasion and a more advanced stage of cancer. Furthermore, an association was observed between positive-Per2 expression and poorer overall survival outcome, though this was not technically significant. However, due to the relatively small sample size and short follow-up time of the present study, the prognostic impact of positive-Per2 expression remains to be fully determined.
Clock genes are involved in cell cycle regulation (21). A positive factor, the Clock-Bmal1 dimer, is required for transcription initiation of the Per and Cry genes oscillating mechanism in the feedback mechanisms of the clock genes (21). By contrast, Per and Cry proteins are supposed to act as negative factors and promote oscillation (22). The Clock-Bmal1 dimer promotes transcription through the E-box of Wee1, suppressing the cell cycle at M-phase (22). The results of previous studies have demonstrated a connection between the alterations of clock genes, and cell cycle progression and proliferation through c-Myc/p21 signaling and the Wnt/β-catenin pathway, which are implicated in the molecular pathogenesis of colorectal cancer (11,17,18). Taken together with the results of the present study, this indicates that the imbalance of clock gene expression levels, which results in the dysregulation of cell cycle, may stimulate the adenoma-carcinoma transition and tumor progression during colorectal carcinogenesis. However, the current study did not address whether clock gene expression directly contributes to cell cycle dysregulation and tumorigenesis. The biological significance of in situ clock gene expression remains to be elucidated. Clock gene expression in situ may at least in part represent the dysregulated rhythms in carcinomas, therefore, future studies are required to address the molecular mechanisms by which the imbalance of clock genes expression contributes to dysregulated circadian rhythms and consequently, to tumorigenesis.
In conclusion, mRNA expression of key clock genes, including Per1, Per2, Cry1 and Cry2 was frequently found in carcinomas, but not in adenomas, using in situ hybridization. Also, the expression of some clock genes were associated with tumor size, and tended to be associated with depth of invasion and survival outcome. Therefore, the present study suggests that dysregulated clock gene expression may serve an important role in human colorectal tumorigenesis.
Glossary
Abbreviations
- SCN
suprachiasmatic nuclei
- Per
period circadian protein homolog
- Bmal1
brain and Muscle ARNT-Like Protein 1
- Clock
circadian locomoter output cycles protein kaput
- Cry
cryptochrome
- CK1ε
casein Kinase 1ε
References
- 1.Fu L, Lee CC. The circadian clock: Pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 2.Hunt T, Sassone-Corsi P. Riding tandem: Circadian clocks and the cell cycle. Cell. 2007;129:461–464. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Devilee P, Schuuring E, van de Vijver MJ, Cornelisse CJ. Recent developments in the molecular genetic understanding of breast cancer. Crit Rev Oncog. 1994;5:247–270. doi: 10.1615/CritRevOncog.v5.i2-3.70. [DOI] [PubMed] [Google Scholar]
- 4.Ronco A, De Stefani E, Mendilaharsu M, Deneo-Pellegrini H. Meat, fat and risk of breast cancer: A case-control study from Uruguay. Int J Cancer. 1996;65:328–331. doi: 10.1002/(SICI)1097-0215(19960126)65:3<328::AID-IJC9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Stevens RG, Brainard GC, Blask DE, Lockley SW, Motta ME. Breast cancer and circadian disruption from electric lighting in the modern world. CA Cancer J Clin. 2014;64:207–218. doi: 10.3322/caac.21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambrosone CB, Freudenheim JL, Graham S, Marshall JR, Vena JE, Brasure JR, Michalek AM, Laughlin R, Nemoto T, Gillenwater KA, Shields PG. Cigarette smoking, N-acetyltransferase 2 genetic polymorphisms, and breast cancer risk. JAMA. 1996;276:1494–1501. doi: 10.1001/jama.276.18.1494. [DOI] [PubMed] [Google Scholar]
- 7.Magnusson C, Baron J, Persson I, Wolk A, Bergström R, Trichopoulos D, Adami HO. Body size in different periods of life and breast cancer risk in post-menopausal women. Int J Cancer. 1998;76:29–34. doi: 10.1002/(SICI)1097-0215(19980330)76:1<29::AID-IJC6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 8.Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, Fuchs CS, Colditz GA. Night-shift work and risk of colorectal cancer in the nurses' health study. J Natl Cancer Inst. 2003;95:825–828. doi: 10.1093/jnci/95.11.825. [DOI] [PubMed] [Google Scholar]
- 9.Karantanos T, Theodoropoulos G, Pektasides D, Gazouli M. Clock genes: Their role in colorectal cancer. World J Gastroenterol. 2014;20:1986–1992. doi: 10.3748/wjg.v20.i8.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/S0092-8674(02)01223-0. [DOI] [PubMed] [Google Scholar]
- 11.Huisman SA, Oklejewicz M, Ahmadi AR, Tamanini F, Ijzermans JN, van der Horst GT, de Bruin RW. Colorectal liver metastases with a disrupted circadian rhythm phase shift the peripheral clock in liver and kidney. Int J Cancer. 2014;136:1024–1032. doi: 10.1002/ijc.29089. [DOI] [PubMed] [Google Scholar]
- 12.Yeh KT, Yang MY, Liu TC, Chen JC, Chan WL, Lin SF, Chang JG. Abnormal expression of period 1 (PER1) in endometrial carcinoma. J Patol. 2005;206:111–120. doi: 10.1002/path.1756. [DOI] [PubMed] [Google Scholar]
- 13.Shih HC, Choo KB, Chang TJ, Yang MY, Shih MC, Yeh KT, Liu TC, Lin SF, Chang JG. Disturbance of circadian gene expression in endometrial cancer: Detection by real-time quantitative RT-PCR. Oncol Rep. 2005;14:1533–1538. [PubMed] [Google Scholar]
- 14.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 15.Sobin LH. World Health Organization international histological classification of tumors: Histological typing of intestinal tumors. 2nd. New York: Springer-Verlag; 1989. [Google Scholar]
- 16.Wood PA, Yang X, Taber A, Oh EY, Ansell C, Ayers SE, Al-Assaad Z, Carnevale K, Berger FG, Peña MM, Hrushesky WJ. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res. 2008;6:1786–1793. doi: 10.1158/1541-7786.MCR-08-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Wood PA, Ansell CM, Ohmori M, Oh EY, Xiong Y, Berger FG, Peña MM, Hrushesky WJ. Beta-catenin induces beta-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem. 2009;145:289–297. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- 18.Soták M, Polidarová L, Ergang P, Sumová A, Pácha J. An association between clock genes and clock-controlled cell cycle genes in murine colorectal tumors. Int J Cancer. 2013;132:1032–1041. doi: 10.1002/ijc.27760. [DOI] [PubMed] [Google Scholar]
- 19.Oshima T, Takenoshita S, Akaike M, Kunisaki C, Fujii S, Nozaki A, Numata K, Shiozawa M, Rino Y, Tanaka K, et al. Expression of circadian genes correlates with liver metastasis and outcomes in colorectal cancer. Oncol Rep. 2011;25:1439–1446. doi: 10.3892/or.2011.1207. [DOI] [PubMed] [Google Scholar]
- 20.Karantanos T, Theodoropoulos G, Gazouli M, Vaiopoulou A, Karantanou C, Lymberi M, Pektasides D. Expression of clock genes in patients with colorectal cancer. Int J Biol Markers. 2013;28:280–285. doi: 10.5301/jbm.5000033. [DOI] [PubMed] [Google Scholar]
- 21.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 22.Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]