Abstract
Although dysregulation of microRNAs (miRNAs/miRs) is a common feature of human malignancies, its involvement in gastrointestinal stromal tumors (GISTs) is not fully understood. The present study aimed to identify the miRNAs that perform a role in GIST metastasis. miRNA expression profiles from a series of 32 primary GISTs were analyzed using microarrays, and miR-186 was observed to be downregulated in tumors exhibiting metastatic recurrence. Reverse transcription-quantitative polymerase chain reaction analysis of an independent cohort of 100 primary GISTs revealed that low miR-186 expression is associated with metastatic recurrence and a poor prognosis. Inhibition of miR-186 in GIST-T1 cells promoted cell migration. Gene expression microarray analysis demonstrated that miR-186 inhibition upregulated a set of genes implicated in cancer metastasis, including insulin-like growth factor-binding protein 3, AKT serine/threonine kinase 2, hepatocyte growth factor receptor, CXC chemokine receptor 4 and epidermal growth factor-containing fibulin-like extracellular matrix protein 1. These results suggest that the downregulation of miR-186 is involved in the metastatic recurrence of GISTs, and that miR-186 levels could potentially be a predictive biomarker for clinical outcome.
Keywords: gastrointestinal stromal tumor, metastasis, recurrence, microRNA
Introduction
Gastrointestinal stromal tumors (GISTs) arise from the interstitial cells of Cajal and are the most frequently occurring mesenchymal tumors of the gastrointestinal tract (1). The majority of GISTs (~90%) exhibit constitutively activating KIT mutations, while ~5% show mutations in another tyrosine kinase receptor gene, platelet-derived growth factor receptor α. In addition to the abnormal activation of tyrosine kinase receptors observed in a majority of GISTs, a previous study revealed succinate dehydrogenase mutations to perform a critical role in the induction of aberrant DNA methylation in pediatric GISTs (2). These findings indicate that genetic and epigenetic dysregulation are causally associated with the pathogenesis of GISTs.
MicroRNAs (miRNAs/miRs) are small noncoding RNAs that perform critical roles in physiological and pathological settings (3). Dysregulation of miRNAs is commonly observed in malignancies, and numerous miRNAs are thought to act as oncogenes or tumor suppressors (4,5). We previously screened miRNAs to identify epigenetically silenced miRNA genes in GIST cells, and revealed that miR-34a and miR-335 are silenced in association with CpG island methylation in GISTs (6). In addition, functional analyses suggested that miR-34a and miR-335 act as tumor suppressors in GISTs and that the two genes are frequently methylated in primary GIST specimens. In addition, we observed that elevated expression of miR-196a is clearly associated with aggressiveness and poor prognosis of GISTs, which is indicative of its oncogenicity (7). These results highlight the critical impact of miRNA dysregulation in the pathogenesis of GISTs.
Distant metastasis is a critical event that prevents complete cure of malignant tumors. The process of tumor metastasis involves several steps: Detachment of cells from the primary tumor; tissue invasion by the tumor cells; intravasation, arrest and extravasation of the cells; and proliferation of the cells at metastatic sites (8). It is well known that a subset of miRNAs perform key roles in cancer metastasis. For instance, miR-10b is strongly expressed in metastatic breast cancer cells and promotes cell migration and invasion by suppressing HOXD10, which results in increased expression of a pro-metastatic gene, RHOC (9). miR-21 is also reportedly oncogenic and targets genes that suppress invasion and metastasis (10). On the other hand, members of the miR-200 family suppress metastasis by downregulating ZEB1, a negative modulator of E-cadherin (11). However, dysregulation of these miRNAs has not been reported in GISTs, and the mechanism underlying GIST metastasis is not fully understood. The present study therefore aimed to identify miRNAs involved in GIST metastasis. By analyzing miRNA expression profiles in a series of primary GISTs, it was determined that the downregulation of miR-186 is associated with metastatic recurrence and poor prognosis, which suggests that a loss of miR-186 could potentially act to drive GIST metastasis.
Materials and methods
Tissue samples
A total of 32 fresh frozen GIST specimens were obtained from Sapporo Medical University Hospital (Sapporo, Japan) and Osaka University Hospital (Osaka, Japan) as described previously (7). In addition, formalin-fixed paraffin-embedded tissue sections from 100 GIST specimens were obtained from Niigata University Hospital (Niigata, Japan), as described previously (7). Informed consent was obtained from all patients prior to the collection of the specimens, and the present study was approved by the institutional review board. Risk grade was assessed based on tumor size and mitotic activity, according to the risk classification system proposed by Fletcher et al (12). Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Cell line and transfection
GIST-T1 cells were cultured as described previously (13). Cells (3×106) were transfected with 100 pmol mirVana miRNA inhibitor (Thermo Fisher Scientific, Inc., Waltham, MA, USA) or 100 pmol mirVana miRNA inhibitor Negative Control (Thermo Fisher Scientific, Inc.) using a Cell Line Nucleofector kit V (Lonza Group, Ltd., Basel, Switzerland) and a Nucleofector I electroporation device (Lonza Group, Ltd.) according to the manufacturer's protocol.
miRNA expression analysis
miRNA expression profiles were analyzed using the miRNA microarray data obtained in our previous study (7). The Gene Expression Omnibus accession number for the microarray data is GSE31741. Expression of selected miRNAs was examined using TaqMan microRNA assays (Thermo Fisher Scientific, Inc.). DNase treatment was performed prior to the reverse transcription quantitative polymerase chain reaction (RT-qPCR), which was performed in a 7500HT Fast Real-Time PCR System (Thermo Fisher Scientific, Inc.) according to the manufacture's protocol, using primers supplied from the manufacturer (Thermo Fisher Scientific, Inc.). SDS v1.4 software (Thermo Fisher Scientific, Inc.) was used for comparative ∆Cq analysis (14). U6 snRNA (RNU6B; Thermo Fisher Scientific, Inc.) was used as an endogenous control. Experiments were performed in triplicate.
Cell viability assay
GIST-T1 cells were transfected with miRNA inhibitor or negative control as described above, and seeded into a 96-well plate to a density of 1×105 cells per well. Cell viabilities were examined every 24 h for 96 h using a Cell Counting kit-8 (Dojindo Molecular Technologies, Inc., Tokyo, Japan) according to the manufacturer's protocol.
Wound healing assays
GIST-T1 cells were transfected with the miRNA inhibitor or a negative control as described above. Subsequently, cells were suspended in 500 µl of Dulbecco's Modified Eagle's Medium (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with 10% fetal bovine serum, and 70 µl aliquots of cell suspension were added to the wells of Culture-Insert 2 plates (Ibidi, Munich, Germany). Subsequent to an incubation for 24 h at 37°C, the culture inserts were removed, and migration was assessed subsequent to incubation for an additional 16 h at 37°C. Cell images were captured using an IX81 light microscope system (Olympus, Tokyo, Japan) and wound area was measured using ImageJ software version 1.47 (National Institutes of Health, Bethesda, Maryland, USA).
Gene expression microarray analysis
GIST-T1 cells were transfected with the miRNA inhibitor or a negative control as described above, and total RNA was extracted 48 h after transfection. One-color microarray-based gene expression array analysis was then carried out according to the manufacturer's protocol (Agilent Technologies, Inc., Santa Clara, CA, USA). Briefly, 100 ng of total RNA was amplified and labeled using a Low-input Quick Amp Labeling kit One-color (Agilent Technologies, Inc.). The synthesized cRNA was subsequently hybridized to a SurePrint G3 Human GE microarray v2 (G4851; Agilent Technologies, Inc.). The microarray experiments were performed in duplicate, and data were analyzed using GeneSpring GX version 13 (Agilent Technologies, Inc.). The Gene Expression Omnibus accession number for the microarray data is GSE85613.
Statistical analysis
Quantitative variables were analyzed using a Student's t-test or one-way analysis of variance. Categorical values were compared using Fisher's exact test. Logistic regression analysis was used to assess the correlation between clinicopathological factors and gene expression. Survival was analyzed using a Cox's regression model; the log-rank test was used for two-group comparisons. All data were analyzed using EZR version 1.32 (15).
Results
Identification of miRNAs associated with GIST metastasis
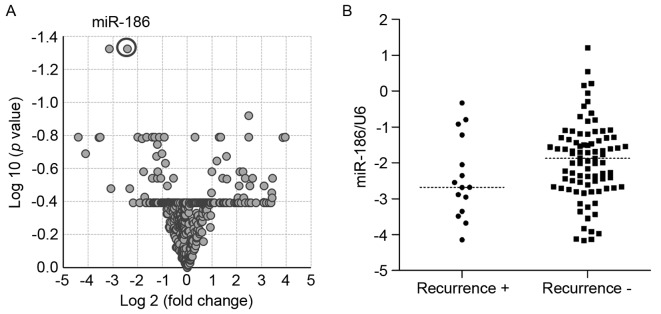
To identify miRNAs associated with GIST metastasis, the present study first analyzed miRNA microarray data previously obtained from 32 primary GIST specimens (7). Volcano plot analysis revealed that levels of miR-186 and miR-99a expression differed between tumors with or without metastatic recurrence (P<0.05, >2-fold) (Fig. 1A, Table I). The present study then validated the microarray results using RT-qPCR with an independent cohort of primary GISTs. Since the preliminary analysis suggested that expression of miR-99a was not associated with metastatic recurrence and poor prognosis in GISTs, miR-186 was selected for additional studies (data not shown). RT-qPCR analysis of the new cohort of GIST specimens (n=100) demonstrated that levels of miR-186 expression tended to be lower in GISTs with metastatic recurrence compared with those without recurrence, although the difference was not observed to be statistically significant (Fig. 1B).
Figure 1.
Low miR-186 expression is associated with metastatic recurrence of GISTs. (A) Volcano plot of miRNA microarray data used to identify differentially expressed miRNAs between GISTs with metastatic recurrence (n=5) and those without recurrence (n=27). miR-186 expression is highlighted by a circle. (B) Summary of reverse transcription-quantitative polymerase chain reaction analysis of miR-186 in primary GISTs with (n=15) and without metastatic recurrence (n=85). miR, microRNA; GIST, gastrointestinal stromal tumor.
Table I.
miRNAs associated with metastatic recurrence in GIST.
| miRNA | P-value | Corrected P-value | Fold change |
|---|---|---|---|
| hsa-miR-186 | 1.01×10−4 | 0.047 | −5.33 |
| hsa-miR-99a | 6.67×10−5 | 0.047 | −8.89 |
| hsa-miR-299-3p | 3.85×10−5 | 0.120 | 5.61 |
| hsa-miR-320d | 9.17×10−4 | 0.162 | 2.44 |
| hsa-miR-23b | 2.76×10−3 | 0.162 | −3.12 |
| hsa-miR-145* | 2.30×10−3 | 0.162 | −2.48 |
| hsa-miR-34b | 2.37×10−3 | 0.162 | 5.58 |
| hsa-miR-144 | 1.81×10−3 | 0.162 | 15.53 |
| hsa-miR-30c | 1.54×10−3 | 0.162 | −1.91 |
| hsa-miR-647 | 2.40×10−3 | 0.162 | 1.25 |
| hsa-miR-30e | 1.15×10−3 | 0.162 | −2.19 |
| hsa-miR-27b | 2.55×10−3 | 0.162 | −2.64 |
| hsa-miR-204 | 1.62×10−3 | 0.162 | −21.18 |
| hsa-miR-30e* | 1.94×10−3 | 0.162 | −4.00 |
| hsa-miR-10a | 2.63×10−3 | 0.162 | −11.49 |
| hsa-miR-320c | 1.73×10−3 | 0.162 | 2.54 |
| hsa-miR-199b-5p | 3.00×10−3 | 0.163 | −11.79 |
| hsa-miR-431 | 3.12×10−3 | 0.163 | 14.61 |
| hsa-miR-148a | 3.36×10−3 | 0.166 | −3.54 |
| hsa-miR-34a | 3.84×10−3 | 0.180 | −2.30 |
miRNA/miR, microRNA; GIST, gastrointestinal stromal tumor; hsa, Homo sapiens; hsa-miR-30e*, hsa-miR-30e* gene.
The association between clinicopathological features and levels of miR-186 expression in the validation cohort is summarized in Table II. The mean levels of miR-186 expression were not associated with age, gender, tumor location or risk classifications of the primary GISTs (Table II). However, logistic regression analysis revealed that low levels of miR-186 expression were associated with an elevated risk of metastatic recurrence (Table III). In addition, Cox's hazard analysis revealed that the hazard ratio was highest for patients with downregulated miR-186 expression when a cutoff value of miR186/U6 ≤0.143 was employed (Table IV). Kaplan-Meyer analysis demonstrated that lower expression of miR-186 is associated with poorer overall survival among patients with GIST (Fig. 2).
Table II.
Correlation between miR-186 expression and the clinicopathological features of GISTs.
| Clinicopathological feature | n | miR-186/U6 (mean ± 95% CI) | P-value |
|---|---|---|---|
| Age, years | |||
| >65 | 44 | 0.30±0.21 | 0.34 |
| ≤65 | 56 | 0.36±0.38 | |
| Gender | |||
| Male | 44 | 0.39±0.32 | 0.07 |
| Female | 56 | 0.28±0.31 | |
| Tumor location | |||
| Stomach | 84 | 0.31±0.30 | 0.17 |
| Intestine | 8 | 0.28±0.37 | |
| Colon | 3 | 0.44±0.29 | |
| Esophagus | 5 | 0.62±0.50 | |
| Risk classificationa | |||
| Very low | 1 | 0.31 | 0.95 |
| Low | 45 | 0.33±0.23 | |
| Intermediate | 25 | 0.37±0.34 | |
| High | 26 | 0.32±0.43 | |
| Metastatic recurrence | |||
| + | 15 | 0.25±0.22 | 0.32 |
| − | 85 | 0.34±0.33 |
Risk classification of 3 patients were unavailable due to insufficient clinical information. miR, microRNA; GIST, gastrointestinal stromal tumor; CI, confidence interval.
Table III.
miR-186 expression is associated with metastatic recurrence risk in GIST patients.
| Features | OR (95% CI) | P-value | Adjusted OR (95% CI) | P-value |
|---|---|---|---|---|
| Tumor location | ||||
| Stomach | 1 | |||
| Other (intestine, esophagus, colon) | 0.78 (0.16–3.85) | 0.76 | 3.12 (0.47–20.9)a | 0.24 |
| Risk classification | ||||
| Low or intermediate risk | 1 | |||
| High risk | 12.8 (3.59–45.8) | <0.001 | 13.4 (3.47–51.7)b | <0.001 |
| miR-186/U6 | ||||
| >0.172 | 1 | |||
| ≤0.172 | 4.04 (1.30–12.6) | <0.05 | 3.84 (1.02–14.4)c | <0.05 |
Risk classification and miR-186/U6 adjusted OR.
Tumor location and miR-186/U6 adjusted OR.
Tumor location and risk classification adjusted OR. miR, microRNA; GIST, gastrointestinal stromal tumor; OR, odds ratio; CI, confidence interval.
Table IV.
miR-186 expression is associated with poor clinical outcome in GIST patients.
| Outcome | |||||||
|---|---|---|---|---|---|---|---|
| miR-186/U6 | Mortality | Survival | Total | HR (95% CI) | P-value | Adjusted HRa (95% CI) | P-value |
| >0.143 | 11 | 69 | 80 | ||||
| ≤0.143 | 7 | 13 | 20 | 2.89 (1.12–7.49) | <0.05 | 2.73 (1.04–7.16) | <0.05 |
Age and gender adjusted HR. miR, microRNA; GIST, gastrointestinal stromal tumor; HR, hazard ratio; CI, confidence interval.
Figure 2.
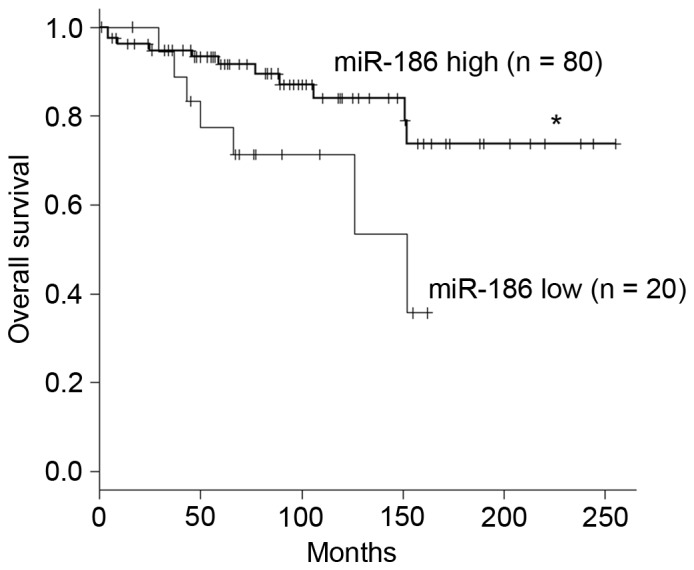
Low expression of miR-186 is associated with poor prognosis of patients with GIST. Kaplan-Meier curves demonstrate the effects of miR-186 expression (high, miR-186/U6 ≥0.143; low, miR-186/U6 <0.143) on survival among patients with GIST. *P<0.05. miR, microRNA; GIST, gastrointestinal stromal tumor.
Functional analysis of miR-186 in GIST-T1 cells
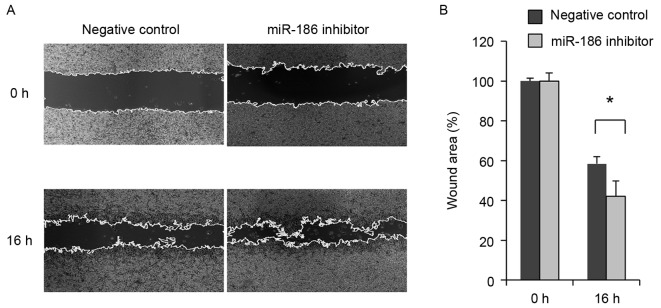
The association between low miR-186 expression and malignant characteristics of GISTs suggests that miR-186 may act as a tumor suppressor in these tumors. The present study therefore investigated the function of miR-186 in GIST cells. RT-qPCR revealed that GIST-T1 cells strongly express miR-186 (miR-186/U6=7.34). The present study therefore transiently transfected GIST-T1 cells with an inhibitor of miR-186 or a negative control and assessed their effects on cell viability and migration. It was revealed that inhibiting miR-186 in GIST-T1 cells did not affect cellular proliferation (Fig. 3). By contrast, wound healing assays revealed that inhibiting miR-186 promoted GIST-T1 cell migration (Fig. 4).
Figure 3.
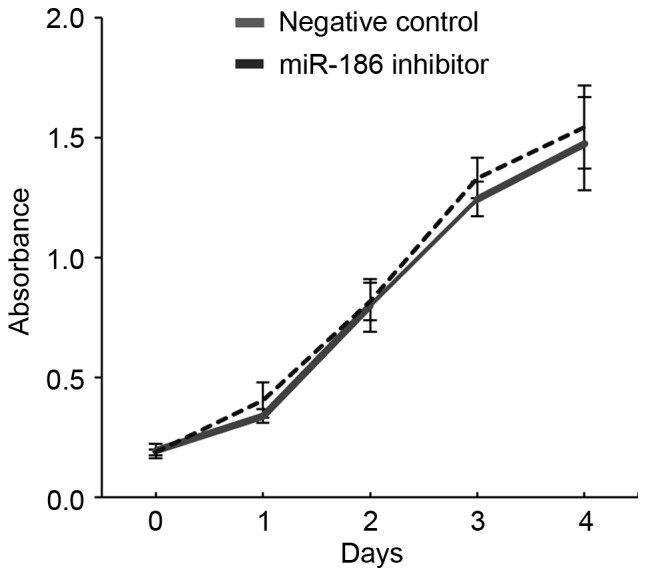
Inhibition of miR-186 did not affect GIST-T1 cellular proliferation. Growth curves are presented for GIST-T1 cells transfected with a miR-186 inhibitor or a negative control generated using cell viability assays. miR, microRNA; GIST, gastrointestinal stromal tumor.
Figure 4.
miR-186 inhibition promoted migration of GIST-T1 cells. (A) Representative results from wound healing assays using GIST-T1 cells transfected with a miR-186 inhibitor or a negative control. The wound was made 24 h after transfection, and photographs were captured at the indicated time points. (B) Summary of the wound healing assay results obtained from three independent experiments. Error bar represent standard deviations. *P<0.05. miR, microRNA; GIST, gastrointestinal stromal tumor.
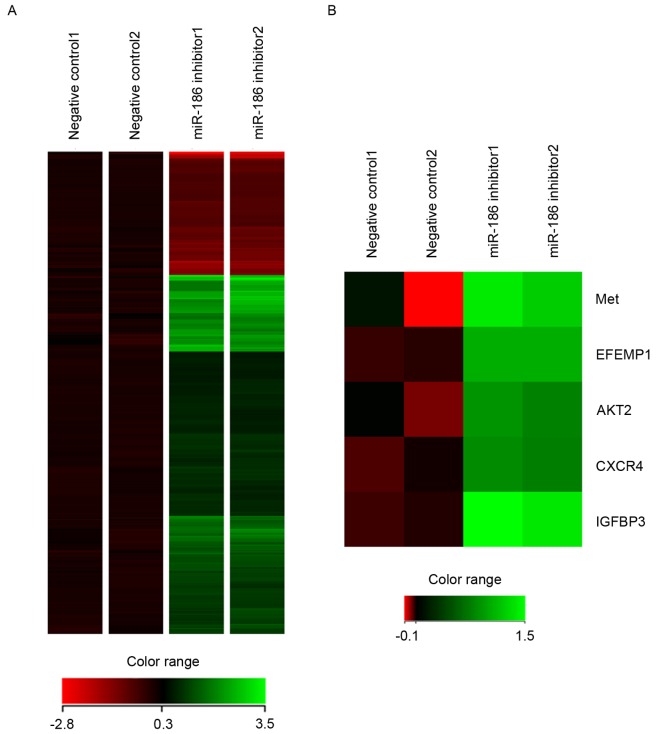
To identify downstream targets of miR-186 in GIST cells, gene expression microarray analysis was conducted in GIST-T1 cells transfected with the miR-186 inhibitor or negative control. It was revealed that 253 probe sets (corresponding to 229 unique genes) were upregulated and 87 probe sets (corresponding to 66 unique genes) were downregulated by the miR-186 inhibitor in GIST-T1 cells (Fig. 5A). Among them, it was noted that several genes were implicated in cancer metastasis, including MET (hepatocyte growth factor receptor), EFEMP1 (epidermal growth factor-containing fibulin-like extracellular matrix protein 1), AKT serine/threonine kinase 2, CXCR4 (CXC chemokine receptor 4) and IGFBP3 (insulin-like growth factor-binding protein 3), were all upregulated by inhibition of miR-186 (Fig. 5B).
Figure 5.
Inhibition of miR-186 affects gene expression profiles in GIST-T1 cells. (A) Heat map showing the changes in gene expression subsequent to miR-186 inhibition in GIST-T1 cells. Genes upregulated or downregulated by the miR-186 inhibitor (>1.5-fold) were selected, and hierarchical clustering was subsequently performed. (B) Heat map of metastasis-associated genes upregulated by miR-186 inhibition in GIST-T1 cells. miR, microRNA; GIST, gastrointestinal stromal tumor; Met, hepatocyte growth factor receptor; EFEMP1, epidermal growth factor-containing fibulin-like extracellular matrix protein 1; AKT2, AKT serine/threonine kinase 2; CXCR4, CXC chemokine receptor 4; IGFBP3, insulin-like growth factor-binding protein 3.
Discussion
Numerous miRNAs are reportedly implicated in tumorigenesis, acting as oncogenes or tumor suppressors (4,5). The present study revealed that low levels of miR-186 expression were associated with metastatic recurrence and a worse outcome in patients with GISTs. Functional analysis revealed that inhibiting miR-186 in GIST cells promotes cell migration and upregulates a set of genes involved in cancer metastasis, which supports our hypothesis that the downregulation of miR-186 is, at least in part, associated with metastatic recurrence of GISTs.
Studies have revealed suggested that miR-186 acts as a tumor suppressor in lung adenocarcinoma (16), esophageal squamous cell carcinoma (17), hepatocellular carcinoma (HCC) (18) and bladder cancer (19). In lung adenocarcinoma, downregulated expression of miR-186 is associated with poor survival, whilst overexpression of miR-186 inhibits cellular proliferation by inducing G1-S cell cycle arrest (16). miR-186 also reportedly inhibits the migration and invasion of non-small cell lung cancer (NSCLC) cells by targeting the pituitary tumor transforming gene (20). Another study has reported that miR-186 suppresses the growth and metastasis of NSCLC cells by targeting Rho-associated protein kinase 1 (21). In bladder cancer, miR-186 suppresses cellular proliferation and invasion by targeting nucleosomal binding protein 1 (also termed high mobility group nucleosome binding domain 5) (19). miR-186 is also downregulated in several HCC cell lines, where it suppresses cell migration, invasion and proliferation by targeting Yes-associated protein 1 (18). The introduction of miR-186 into ovarian cancer cells downregulates Twist1, which leads to induction of mesenchymal-to-epithelial transition, G1 cell cycle arrest and apoptosis (22). By contrast, miR-186 is reportedly overexpressed in pancreatic ductal adenocarcinoma and is associated with a poor prognosis (23). These results clearly suggest that altered expression of miR-186 affects tumor growth, invasion and metastasis in various cancers, although its function may differ among malignancies of different origins.
Microarray analysis in the present study revealed that inhibiting miR-186 in GIST cells upregulated a set of genes implicated in cancer metastasis. IGFBP3 interacts with insulin-like growth factor (IGF), which antagonizes IGF1 activities or, conversely, stabilizes tumorigenic potential (24). An association between high IGFBP3 expression and distant metastasis has been reported in oral squamous cell carcinoma (25), colorectal cancer (26) and pancreatic endocrine neoplasms (27). IGF signaling has also been implicated in GIST. IGF1 receptor is reportedly overexpressed in all GIST cases, and higher levels of IGF1 and IGF2 expression are positively associated with malignant clinicopathological characteristics, including larger tumor size, higher mitotic count, higher risk and metastasis (28). However, functional analysis of IGFBP3 in GIST cells yielded complicated results. Loss of IGFBP3 from GIST882 cells, where IGFBP3 is abundantly expressed, suppressed cell viability, whereas overexpression of IGFBP3 in GIST-T1 cells, where IGFBP3 is not endogenously expressed, had a cytotoxic effect (29). Although these results suggest that IGFBP3 has dual opposing effects on GIST cell viability, its effects on GIST recurrence and metastasis remains to be clarified.
It is known that hepatocyte growth factor receptor (c-Met)/phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) signaling performs a pivotal role in promoting epithelial-mesenchymal transition (EMT), and miRNAs reportedly affect the process. For instance, miR-206 inhibits hepatocyte growth factor-induced EMT by directly targeting c-Met and suppressing its downstream PI3K/Akt/mechanistic target of rapamycin signaling pathway (30). In gastric cancer, miR-338-3p directly targets ZEB2 and metastasis-associated in colon cancer-1 (MACC1), which results in repression of MACC1/Met/Akt signaling and EMT (31). By contrast, miR-93 activates Met/PI3K/Akt activity in HCC by directly inhibiting phosphatase and tensin homolog and cyclin dependent kinase inhibitor 1A (32). Taken together with these observations, our results suggest that miR-186 may regulate GIST metastasis by modulating Met/Akt signaling. CXCR4 is a receptor for chemokine CXC ligand 12 (also termed stromal cell-derived factor 1), and their binding leads to the activation of several key signal pathways that promote cancer growth and metastasis (33). Elevated expression of CXCR4, for example, has been observed in breast (34) and colorectal cancer (35), and head and neck squamous cell carcinoma (HNSCC) (36). Activation of CXCR4 in estrogen receptor-positive breast cancer cells drives a metastatic and endocrine therapy-resistant phenotype by increasing MAPK signaling (34). Overexpression of CXCR4 is associated with lymph node metastasis and poor disease-free survival in colorectal cancer (35). In addition, high CXCR4 expression is associated with distant metastasis in HNSCC (36). Notably, one recent study demonstrated that deletion of exon 11 codons 557–558 from KIT is associated with GIST liver metastasis (37). Mechanistically, KIT exon 11 557–558 deletion upregulates CXCR4 through increased binding of ETS variant 1 to the CXCR4 promoter. This in turn leads to increased GIST cell motility and liver metastasis. EFEMP1 is member of the fibulin family of extracellular matrix proteins, and its overexpression correlates with lymph node metastasis in ovarian cancer (38). EFEMP1 has also been demonstrated to promote migration, invasion and metastasis in osteosarcoma, and upregulation of EFEMP1 is an indicator of poor prognosis (39). Upregulation of CXCR4 and EFEMP1 through miR-186 inhibition in GIST-T1 cells suggests that downregulation of miR-186 is associated with an increased risk of metastatic recurrence in GIST.
In summary, the present study revealed that low miR-186 expression is associated with metastatic recurrence and a poor prognosis in patients with GIST. Downregulation of miR-186 may promote GIST metastasis by inducing expression of metastasis-associated genes. miR-186 may thus be a useful biomarker for predicting recurrence and assessing prognosis in patients with GIST, and could potentially be an effective therapeutic agent with which to treat malignant GISTs.
Acknowledgements
The authors would like to thank Ms. Mutsumi Toyota and Ms. Tomo Hatahira for technical assistance and Dr William F. Goldman for editing the manuscript. The present study was supported in part by ‘Grants-in-Aid for Young Investigators (B)’ from Japan Society for the Promotion of Science (grant no. JSPS KAKENHI 16K19352) and Grant-in-Aid for Scientific Research (B) from Japan Society for the Promotion of Science (grant no. JSPS KAKENHI 15H04299).
References
- 1.Joensuu H, Hohenberger P, Corless CL. Gastrointestinal stromal tumour. Lancet. 2013;382:973–983. doi: 10.1016/S0140-6736(13)62398-3. [DOI] [PubMed] [Google Scholar]
- 2.Killian JK, Kim SY, Miettinen M, Smith C, Merino M, Tsokos M, Quezado M, Smith WI, Jr, Jahromi MS, Xekouki P, et al. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013;3:648–657. doi: 10.1158/2159-8290.CD-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Isosaka M, Niinuma T, Nojima M, Kai M, Yamamoto E, Maruyama R, Nobuoka T, Nishida T, Kanda T, Taguchi T, et al. A Screen for epigenetically silenced microRNA genes in gastrointestinal stromal tumors. PLoS One. 2015;10:e0133754. doi: 10.1371/journal.pone.0133754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 8.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 10.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 11.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, Miettinen M, O'Leary TJ, Remotti H, Rubin BP, et al. Diagnosis of gastrointestinal stromal tumors: A consensus approach. Hum Pathol. 2002;33:459–465. doi: 10.1053/hupa.2002.123545. [DOI] [PubMed] [Google Scholar]
- 13.Taguchi T, Sonobe H, Toyonaga S, Yamasaki I, Shuin T, Takano A, Araki K, Akimaru K, Yuri K. Conventional and molecular cytogenetic characterization of a new human cell line, GIST-T1, established from gastrointestinal stromal tumor. Lab Invest. 2002;82:663–665. doi: 10.1038/labinvest.3780461. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.http://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmedEN.html [Google Scholar]
- 16.Cai J, Wu J, Zhang H, Fang L, Huang Y, Yang Y, Zhu X, Li R, Li M. miR-186 downregulation correlates with poor survival in lung adenocarcinoma, where it interferes with cell-cycle regulation. Cancer Res. 2013;73:756–766. doi: 10.1158/0008-5472.CAN-12-2651. [DOI] [PubMed] [Google Scholar]
- 17.He W, Feng J, Zhang Y, Wang Y, Zang W, Zhao G. microRNA-186 inhibits cell proliferation and induces apoptosis in human esophageal squamous cell carcinoma by targeting SKP2. Lab Invest. 2016;96:317–324. doi: 10.1038/labinvest.2015.134. [DOI] [PubMed] [Google Scholar]
- 18.Ruan T, He X, Yu J, Hang Z. MicroRNA-186 targets Yes-associated protein 1 to inhibit Hippo signaling and tumorigenesis in hepatocellular carcinoma. Oncol Lett. 2016;11:2941–2945. doi: 10.3892/ol.2016.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao K, He L, Gan Y, Zeng Q, Dai Y, Tan J. MiR-186 suppresses the growth and metastasis of bladder cancer by targeting NSBP1. Diagn Pathol. 2015;10:146. doi: 10.1186/s13000-015-0372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Yin C, Zhang B, Sun Y, Shi L, Liu N, Liang S, Lu S, Liu Y, Zhang J, et al. PTTG1 promotes migration and invasion of human non-small cell lung cancer cells and is modulated by miR-186. Carcinogenesis. 2013;34:2145–2155. doi: 10.1093/carcin/bgt158. [DOI] [PubMed] [Google Scholar]
- 21.Cui G, Cui M, Li Y, Liang Y, Li W, Guo H, Zhao S. MiR-186 targets ROCK1 to suppress the growth and metastasis of NSCLC cells. Tumour Biol. 2014;35:8933–8937. doi: 10.1007/s13277-014-2168-6. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, Shen H, Yin X, Long L, Xie C, Liu Y, Hui L, Lin X, Fang Y, Cao Y, et al. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene. 2016;35:323–332. doi: 10.1038/onc.2015.84. [DOI] [PubMed] [Google Scholar]
- 23.Zhang ZL, Bai ZH, Wang XB, Bai L, Miao F, Pei HH. miR-186 and 326 predict the prognosis of pancreatic ductal adenocarcinoma and affect the proliferation and migration of cancer cells. PLoS One. 2015;10:e0118814. doi: 10.1371/journal.pone.0118814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baxter RC. IGF binding proteins in cancer: Mechanistic and clinical insights. Nat Rev Cancer. 2014;14:329–341. doi: 10.1038/nrc3720. [DOI] [PubMed] [Google Scholar]
- 25.Yen YC, Hsiao JR, Jiang SS, Chang JS, Wang SH, Shen YY, Chen CH, Chang IS, Chang JY, Chen YW. Insulin-like growth factor-independent insulin-like growth factor binding protein 3 promotes cell migration and lymph node metastasis of oral squamous cell carcinoma cells by requirement of integrin b1. Oncotarget. 2015;6:41837–41855. doi: 10.18632/oncotarget.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georges RB, Adwan H, Hamdi H, Hielscher T, Linnemann U, Berger MR. The insulin-like growth factor binding proteins 3 and 7 are associated with colorectal cancer and liver metastasis. Cancer Biol Ther. 2011;12:69–79. doi: 10.4161/cbt.12.1.15719. [DOI] [PubMed] [Google Scholar]
- 27.Hansel DE, Rahman A, House M, Ashfaq R, Berg K, Yeo CJ, Maitra A. Met proto-oncogene and insulin-like growth factor binding protein 3 overexpression correlates with metastatic ability in well-differentiated pancreatic endocrine neoplasms. Clin Cancer Res. 2004;10:6152–6158. doi: 10.1158/1078-0432.CCR-04-0285. [DOI] [PubMed] [Google Scholar]
- 28.Braconi C, Bracci R, Bearzi I, Bianchi F, Sabato S, Mandolesi A, Belvederesi L, Cascinu S, Valeri N, Cellerino R. Insulin-like growth factor (IGF) 1 and 2 help to predict disease outcome in GIST patients. Ann Oncol. 2008;19:1293–1298. doi: 10.1093/annonc/mdn040. [DOI] [PubMed] [Google Scholar]
- 29.Dupart JJ, Trent JC, Lee HY, Hess KR, Godwin AK, Taguchi T, Zhang W. Insulin-like growth factor binding protein-3 has dual effects on gastrointestinal stromal tumor cell viability and sensitivity to the anti-tumor effects of imatinib mesylate in vitro. Mol Cancer. 2009;8:99. doi: 10.1186/1476-4598-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen QY, Jiao DM, Wu YQ, Chen J, Wang J, Tang XL, Mou H, Hu HZ, Song J, Yan J, et al. MiR-206 inhibits HGF-induced epithelial-mesenchymal transition and angiogenesis in non-small cell lung cancer via c-Met/PI3k/Akt/mTOR pathway. Oncotarget. 2016;7:18247–18261. doi: 10.18632/oncotarget.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang N, Wu Z, Lin L, Zhou M, Wang L, Ma H, Xia J, Bin J, Liao Y, Liao W. MiR-338-3p inhibits epithelial-mesenchymal transition in gastric cancer cells by targeting ZEB2 and MACC1/Met/Akt signaling. Oncotarget. 2015;6:15222–15234. doi: 10.18632/oncotarget.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohta K, Hoshino H, Wang J, Ono S, Iida Y, Hata K, Huang SK, Colquhoun S, Hoon DS. MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget. 2015;6:3211–3224. doi: 10.18632/oncotarget.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo F, Wang Y, Liu J, Mok SC, Xue F, Zhang W. CXCL12/CXCR4: A symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene. 2016;35:816–826. doi: 10.1038/onc.2015.139. [DOI] [PubMed] [Google Scholar]
- 34.Rhodes LV, Short SP, Neel NF, Salvo VA, Zhu Y, Elliott S, Wei Y, Yu D, Sun M, Muir SE, et al. Cytokine receptor CXCR4 mediates estrogen-independent tumorigenesis, metastasis, and resistance to endocrine therapy in human breast cancer. Cancer Res. 2011;71:603–613. doi: 10.1158/0008-5472.CAN-10-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ottaiano A, Franco R, Talamanca Aiello A, Liguori G, Tatangelo F, Delrio P, Nasti G, Barletta E, Facchini G, Daniele B, et al. Overexpression of both CXC chemokine receptor 4 and vascular endothelial growth factor proteins predicts early distant relapse in stage II–III colorectal cancer patients. Clin Cancer Res. 2006;12:2795–2803. doi: 10.1158/1078-0432.CCR-05-2142. [DOI] [PubMed] [Google Scholar]
- 36.Katayama A, Ogino T, Bandoh N, Nonaka S, Harabuchi Y. Expression of CXCR4 and its down-regulation by IFN-gamma in head and neck squamous cell carcinoma. Clin Cancer Res. 2005;11:2937–2946. doi: 10.1158/1078-0432.CCR-04-1470. [DOI] [PubMed] [Google Scholar]
- 37.Wang HC, Li TY, Chao YJ, Hou YC, Hsueh YS, Hsu KH, Shan YS. KIT exon 11 codons 557–558 deletion mutation promotes liver metastasis through the CXCL12/CXCR4 axis in gastrointestinal stromal tumors. Clin Cancer Res. 2016;22:3477–3487. doi: 10.1158/1078-0432.CCR-15-2748. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Wei D, Zhao Y, Liu X, Zhang J. Overexpression of EFEMP1 correlates with tumor progression and poor prognosis in human ovarian carcinoma. PLoS One. 2013;8:e78783. doi: 10.1371/journal.pone.0078783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Cao CJ, Huang LL, Ke ZF, Luo CJ, Lin ZW, Wang F, Zhang YQ, Wang LT. EFEMP1 promotes the migration and invasion of osteosarcoma via MMP-2 with induction by AEG-1 via NF-κB signaling pathway. Oncotarget. 2015;6:14191–14208. doi: 10.18632/oncotarget.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]