Abstract
Serous ovarian carcinoma (SOC) is one of the most life-threatening types of gynecological malignancy, but the pathogenesis of SOC remains unknown. Previous studies have indicated that differentially expressed genes and microRNAs (miRNAs) serve important functions in SOC. However, genes and miRNAs are identified in a disperse form, and limited information is known about the regulatory association between miRNAs and genes in SOC. In the present study, three regulatory networks were hierarchically constructed, including a differentially-expressed network, a related network and a global network to reveal associations between each factor. In each network, there were three types of factors, which were genes, miRNAs and transcription factors that interact with each other. Focus was placed on the differentially-expressed network, in which all genes and miRNAs were differentially expressed and therefore may have affected the development of SOC. Following the comparison and analysis between the three networks, a number of signaling pathways which demonstrated differentially expressed elements were highlighted. Subsequently, the upstream and downstream elements of differentially expressed miRNAs and genes were listed, and a number of key elements (differentially expressed miRNAs, genes and TFs predicted using the P-match method) were analyzed. The differentially expressed network partially illuminated the pathogenesis of SOC. It was hypothesized that if there was no differential expression of miRNAs and genes, SOC may be prevented and treatment may be identified. The present study provided a theoretical foundation for gene therapy for SOC.
Keywords: serous ovarian carcinoma, gene, microRNA, transcription factor, regulatory network
Introduction
Epithelial ovarian cancer is a heterogeneous group of neoplasms that is divided into histological subgroups, each with their own underlying molecular genetic events (1). The serous type of epithelial ovarian cancer accounts for between 75 and 80% of epithelial ovarian carcinomas; it is the most common type of ovarian cancer and is the most life-threatening type of gynecological malignancy (2). However, relatively limited information is known about the molecular genetics of the initiation and progression of serous ovarian cancer (SOC).
Experimentally validated data have demonstrated that differentially expressed genes and microRNAs (miRNAs/miRs) serve key functions in the pathogenesis of SOC (1,3,4). Delineation of the underlying molecular mechanisms involved in the initiation of SOC may increase understanding of the pathogenesis of SOC, and may serve as the theoretical basis of the development of novel diagnostic tests and therapeutic strategies (2).
Transcription factors (TFs) and miRNAs are principal regulators of gene expression (5). TFs are specific proteins that may activate gene transcription independently or indirectly (6). In addition, TFs are primary and important factors that may promote or suppress gene expression at the transcriptional level (7).
miRNAs are non-coding single-stranded RNAs of 22 nucleotides in length that constitute a novel class of gene regulators. miRNAs affect the expression of genes at a post-transcriptional level by binding to complementary sequences on target mRNAs (8). A previous study demonstrated that miRNAs control a variety of biological processes, including cell differentiation, cell proliferation, apoptosis, stress-resistance and fat metabolism (9). The study of the associations between miRNAs and cancer has become an active topic in recent studies.
Genes that are regulated by miRNA are known as target genes. miRNA affects the expression of proteins by regulating target genes; therefore, identifying and validating target genes of specific miRNAs may be a useful strategy for developing novel treatments (10). Host genes are a class of genes within which miRNAs are embedded (11). A previous study revealed that miRNAs are transcribed in parallel with host transcripts, and two transcriptional classes of miRNAs, exonic and intronic, were identified (12). Baskerville and Bartel (13) demonstrated that intronic miRNA and its host gene exhibit a close association. miRNAs and host genes may cooperate to regulate a number of biological functions; dysregulation of this system may affect the development of cancer (14).
A number of genes and miRNAs have been demonstrated to be expressed at different levels in SOC compared with healthy tissue, but were identified in a dispersed form and not as part of a regulatory network. Previous studies have focused on one or a number of genes or miRNAs (1,3,4) In the present study, all genes or miRNAs and the experimentally validated associations between these molecules, including miRNA gene targeting, TF regulation of miRNAs and miRNAs located in host genes, were analyzed. Three regulatory networks were constructed: A differentially expressed network, a related network and a global network. Except for host genes, all genes and miRNAs in the differentially expressed network were differentially expressed. The signaling pathways in the differentially expressed network were extracted and compared in three networks to reveal the pathogenesis of SOC. In addition, the upstream and the downstream elements of the differentially expressed genes and miRNAs in three networks (which include genes, miRNAs and targets) were analyzed, and focus was placed on the similarities and differences between these elements to identify key elements that may affect the development of SOC.
Materials and methods
Material collection and data processing
All SOC data selected were obtained from databases and relevant studies. The National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/gene) was used to ensure each miRNA and gene was referred to by its official name only. The experimentally validated dataset of human miRNAs and the corresponding target genes were selected from the Tarbase (diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index) (15), miRTarBase (mirtarbase.mbc.nctu.edu.tw) (16) and miRecords (c1.accurascience.com/miRecords) (17), and the dataset were termed set U1. Due to increasing interest on the interactions between miRNAs and human TFs, the dataset from TransmiR, a manually built database of TF-miRNA-regulating associations (http://www.cuilab.cn/transmir), was selected, and the dataset was termed set U2. The host genes of human miRNAs were selected from NCBI and miRbase (http://www.mirbase.org/) (18), and the dataset were termed set U3. Differentially expressed genes of SOC were selected primarily from relevant studies (cited below where appropriate) and a limited number were selected from the NCBI Single Nucleotide Polymorphism database, and the dataset of differentially expressed genes were termed U4. Differentially expressed miRNAs of SOC were primarily selected from mir2Disease (http://www.mir2disease.org/) (19) and relevant studies (20–75), and this dataset were termed set U5. Similarly, the SOC-related miRNAs were primarily selected from relevant studies (20–75) and this dataset were termed set U7.
The SOC-associated genes were selected through three different methods. A number of SOC-associated genes were selected from the GeneCards database (http://www.genecards.org) and others were identified in relevant studies (20–75). In addition, there are 31 TFs predicted using the P-match method and were considered to be SOC-associated genes. The UCSC database (genome.ucsc.edu) (76) was used to download the 1,000-nt promoter region sequences of the targets of differentially expressed miRNAs, which were used as the input to predict the TFs and miRNAs they regulate. The P-match method combines pattern matching and weight matrix approaches (77). P-match was used in the present study to identify transcription factor binding sites (TFBSs) in 1,000 nt promoter region sequences and map TFBSs onto promoter region of targets. The matrix library of P-match comes from the TRANScription FACtor database (78), which enables a large variety of different TF binding sites to be searched. The dataset of the SOC-associated genes was termed set U6.
Network construction
The following three regulatory networks of SOC were constructed: The differentially expressed network, the related network and the global network. The differentially expressed network contained the key elements and pathways and was considered the core network. All associations between TFs, miRNAs, target genes and host genes were combined to construct the global network. All associations between differentially expressed miRNAs and the corresponding host genes in the set U3 were included in the differentially expressed network. All differentially expressed genes and differentially expressed miRNAs were mapped to the global network and their associations were combined. These associations also belong to differentially expressed network. The two parts of these associations were combined to obtain the differentially expressed network. Using the same method as that used for the related elements, the related network was constructed. Cytoscape software (version 3.0.0; Institute for Systems Biology, Seattle, WA, USA) was used to present the network graphically and in order to analyze the regulatory pathways more easily.
Results
Differentially expressed network of SOC
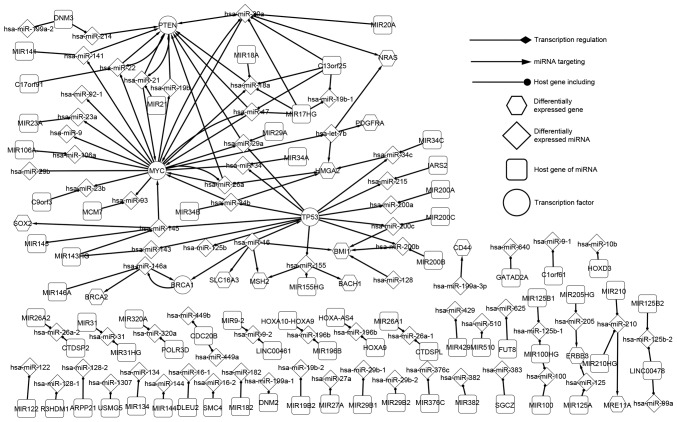
The differentially expressed network is the core SOC network because, except for host genes, all elements were differentially expressed. As such, the differentially expressed network is the core network, which may reveal the pathogenic mechanism of SOC. This network consisted of four TFs [BRCA1, DNA repair associated (BRCA1), MYC proto-oncogene, bHLH transcription factor (MYC), phosphatase and tensin homolog (PTEN) and tumor protein p53 (TP53)], 12 differentially expressed genes (which are targets of miRNA), 65 miRNAs and 72 host genes. As presented in Fig. 1, the essential regulatory associations between significant factors were observed. The network was composed of three associations, including TFs regulating miRNAs, miRNAs targeting target genes and miRNAs located in host genes. In the differentially expressed network, a number of data linkages present special characteristics and, when SOC emerges, the network balance may be broken. The linkages between elements may provide information of the regulatory associations of SOC.
Figure 1.
Differentially expressed network of genes and miRNAs in SOC. All elements are involved in various progressions of SOC. The associations between the differentially expressed genes and miRNAs are presented in this network, which partially revealed the underlying mechanism of SOC. SOC, serous ovarian carcinoma.
The four TF-associated pathways are marked in Fig. 1. TP53 is an extensively studied tumor-suppressor gene and it has been demonstrated that TP53 is commonly mutated in human cancers (18,79). TP53 serves a key function in the development of SOC and it has been identified that high-grade SOC is characterized by TP53 mutations in almost all tumors (96%) (80). In the differentially expressed network, human (hsa-)miR-16 targets TP53 which regulates 12 miRNAs (hsa-miR-125b, −143, −145, −155, −200a, −200b, −200c, −215, −29a, −34, −34b and −34c). MYC contributes to the genesis of a number of types of human cancer and a previous insight into the expression and function of MYC led to therapeutic opportunities (20). In the differentially expressed network, 6 miRNAs (hsa-miR-145, hsa-miR-17, hsa-miR-20a, hsa-miR-21, hsa-miR-26a and hsa-miR-34b) target MYC, which regulates 17 miRNAs (hsa-let-7b, −106a, −141, −17, −18a, −19b, −20a, −22, −23a, −23b, −26a, −29a, −29b, −34, −9, −92-1 and −93). TFs were identified to be core elements in the regulatory network.
As presented in Fig. 1, PTEN regulates hsa-miR-21 and hsa-miR-21 targets PTEN in return, which is an association known as a feedback-loop (FBL). Other FBLs were identified in the differentially expressed network including PTEN and hsa-miR-22, MYC and hsa-miR-20a (hsa-miR-17), and BRCA1 and hsa-miR-146a. It was identified that hsa-miR-200b is regulated by TP53 and targets BMI1 proto-oncogene, polycomb ring finger (BMI1). Therefore, the mutation of TP53 may influence the expression of BMI1 indirectly. The results of the present study indicate that precursors influence their successors in an orderly manner (e.g., hsa-miR-200b is regulated by TP53, and hsa-miR-200b also targets BMI1. When TP53 is differentially expressed, hsa-miR-200b will be differentially expressed which may affect the expression of BMI1). Furthermore, targets of miRNAs, including BTB domain and CNC homolog 1 and BMI1, do not regulate any miRNA, and some miRNAs such as hsa-miR-125b, hsa-miR-215 which are regulated by TFs do not target any gene in the networks outlined in the present study. These factors may be the last actors in the network (elements which are regulated by other genes but not regulate any other elements in the SOC network) and may affect the tumorigenesis of SOC. The core transcriptional network partially revealed the mechanism of SOC and the present study may contribute to the development of cancer prevention and gene therapy.
SOC-related network
The SOC-related network included differentially expressed genes and miRNAs and associated genes and miRNAs. As can be observed in Fig. 1, the differentially expressed network is part of the related network because the SOC-related network contains an increased number of elements and pathways, which may influence the development of SOC, and it is complicated compared with the differentially expressed network. In the related network, there were 38 TFs including 4 differentially expressed TFs, 110 miRNAs and a number of targets of miRNAs. Due to the complexity of the related network, primary focus was placed on the regulatory associations between differentially expressed elements and associated elements.
The present study found that MYC, a differentially expressed gene, regulates 2 SOC-associated miRNAs (hsa-let-7a and hsa-miR-146a) in the related network. In the differentially expressed network, hsa-miR-34c targeted HMGA2. hsa-miR-34c targets 3 genes in the related network, enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), MYB proto-oncogene like 2 (MYBL2) and B cell lymphoma 2. It has been suggested that miR-34c may suppress the expression of EZH2 and MYBL2 in SOC (79). Hsa-miR-31 is a differentially expressed miRNA in SOC (81). In the differentially expressed network, hsa-miR-31 is only located in 2 host genes. No genes regulate hsa-miR-31 and it target no genes; however, in the related network, it targets several genes. Creighton et al (81) identified that functional overexpression of miR-31, the most under-expressed miRNA in serous ovarian cancer, repressed predicted miR-31 gene targets including the cell cycle regulator E2F2, miR31 and cyclin dependent kinase inhibitor 2A. Furthermore, miR-31 overexpression may affect many genes underlying SOC disease progression (81). Without differentially expressed factors, the regulatory associations between elements may influence the tumorigenesis of SOC. Yang et al (82) suggested that miR-506 expression was associated with decreased Snail family transcriptional repressor 2 and vimentin, elevated epithelial-cadherin, and improved prognosis for patients with SOC. The related network enables the underlying molecular mechanism of SOC to be explored.
Global network of SOC
A global network is an experimentally validated biological network in the human body, which contains all the experimentally validated interactions between miRNAs and genes. Therefore, the differentially expressed and the related networks were included in the global network. The global network was used as the reference when the two other networks were constructed and studied.
Host genes and miRNAs in SOC
If miRNAs were differentially expressed, their corresponding host genes were considered to be differentially expressed genes, as mutation of the host gene may make the miRNAs located within it similarly differentially expressed. The host genes and corresponding miRNAs demonstrate important characteristics. A host gene includes a number of miRNAs that target genes, either alone or together. MIR17HG is a cluster host gene that includes four miRNAs (hsa-miR-17, hsa-miR-18a, hsa-miR-19b-1 and hsa-miR-20a), of which three (hsa-miR-17, hsa-miR-18a, hsa-miR-20a) together target PTEN (Fig. 1). Furthermore, the three miRNAs are regulated by MYC. Therefore, miRNA may locate in several host genes. In addition, hsa-miR-20a and hsa-miR-18a locate in MIR20A and MIR18, respectively. Hsa-miR-31 locates in MIR31 and MIR31HG (Fig. 1). It has been hypothesized that overexpression of miR-31 results in decreased cell proliferation, clonogenic potential, cell migration and invasion in SOC (83). Hsa-miR-21 and hsa-miR-22, which form a self-adaption association with PTEN locate in MIR21 and C17orf91, respectively (Fig. 1).
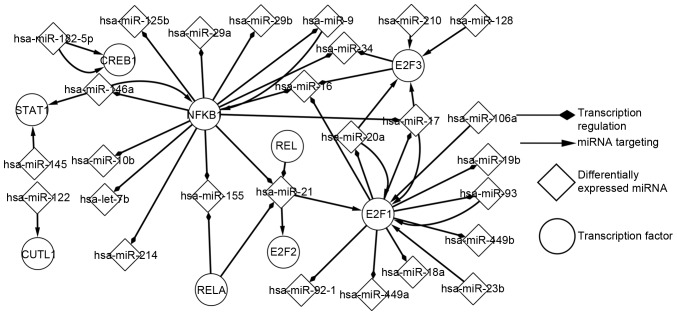
Network of predicted TFs and differentially expressed miRNAs in SOC
Using the P-match algorithm enabled the predicted TFs to be determined. Regulatory associations between predicted TFs and differentially expressed miRNAs were analysed (Fig. 2). As presented in Fig. 2, E2F1 and nuclear factor-κB subunit 1 (NFKB1) were identified to be more marked, compared with other elements, in the development of SOC. E2F1 regulates 10 miRNAs and it is regulated by 6 miRNAs. NFKB1 regulates 13 miRNAs and it is regulated by 2 miRNAs. The differential expression of a miRNA may indirectly influence a different miRNA, by targeting TFs; for example, hsa-miR-128 targets E2F3, which regulates hsa-miR-34. Similarly, the differential expression of a predicted TF may indirectly influence a predicted TF, by regulating a differentially expressed miRNA. Additionally, this network consisted of a number of FBLs; for instance, E2F1 separately forms FBLs with hsa-miR-106a, hsa-miR-20a, hsa-miR-17 and hsa-miR-93. NFKB1 separately form FBLs with hsa-miR-9 and hsa-miR-146a. In addition, CAMP responsive element binding protein 1 and hsa-miR-182-5p form FBLs. The transcription network of predicted TFs and miRNAs is important for the analysis of the pathogenesis of SOC.
Figure 2.
Network of predicted TFs and differentially expressed miRNAs in SOC. The associations between popular TFs and differentially expressed miRNAs are presented in this network, these TFs are typically involved in the transcriptional progression of cancer. SOC, serous ovarian carcinoma; TF, transcription factor.
Regulatory pathways of differentially expressed genes
In order to analyze the network of SOC, the focus was placed on the core elements and adjacent nodes. The upstream and downstream elements of differentially expressed genes, miRNAs and predicted TFs in three networks (differentially expressed network, related network and global network), were extracted and listed separately to analyze the characteristics.
The precursors and successors of differentially expressed genes are listed in three levels. The precursors are miRNAs that target genes and the successors are miRNAs that are regulated by genes. PTEN and MYC are the much more marked compared with other genes. As presented in Table I, the upstream and downstream elements of MYC and the regulatory associations were observed (some miRNAs in the global network are omitted as they are not associated with SOC). MYC has 6 types of adjacent nodes (three successors and three predecessors). In the differentially expressed network, 6 miRNAs target MYC, MYC regulates 17 miRNAs, and hsa-miR-17, hsa-miR-20a and hsa-miR-26a separately form FBLs with MYC. In the related network, seven miRNAs target MYC and MYC regulates 18 miRNAs. In the global network, 19 miRNAs target MYC which regulates 57 miRNAs. The precursors may regulate successors indirectly by influencing MYC. For example, the mutation of hsa-miR-21 may influence the expression of hsa-miR-18a by up- or downregulating the expression of MYC. As presented in Fig. 1, MYC may influence the expression of a number of other genes by regulating its successors. MYC regulates hsa-miR-20a and hsa-miR-20a targets neuroblastoma RAS viral oncogene homolog (NRAS); therefore, MYC may affect NRAS. BMI1 only has 3 types of predecessors and it does not regulate any miRNA; therefore, BMI1 may be the last actor in the network of SOC.
Table I.
Upstream and downstream of MYC in three networks of serous ovarian cancer.
| Upstream | Downstream | |||||
|---|---|---|---|---|---|---|
| Differentially expressed network | Related network | Global network | Gene | Differentially expressed network | Related network | Global network |
| miR-145 | miR-145 | let-7a | MYC | let-7b | let-7b | let-7 |
| miR-17 | miR-17 | let-7c | miR-106a | miR-106a | let-7a | |
| miR-20a | miR-20a | let-7g | miR-141 | miR-141 | let-7a-1 | |
| miR-21 | miR-21 | miR-145 | miR-17 | miR-17 | let-7a-2 | |
| miR-26a | miR-26a | miR-17 | miR-18a | miR-18a | let-7a-3 | |
| miR-34b | miR-34b | miR-20a | miR-19b | miR-19b | let-7b | |
| miR-34a | miR-21 | miR-20a | miR-20a | let-7c | ||
| miR-24 | miR-22 | miR-22 | let-7d | |||
| miR-26a | miR-23a | miR-23a | let-7e | |||
| miR-34a | miR-23b | miR-23b | let-7f | |||
| miR-34a-5p | miR-26a | miR-26a | let-7f-1 | |||
| miR-34b | miR-29a | miR-29a | let-7f-2 | |||
| miR-34b* | miR-29b | miR-29b | let-7g | |||
| miR-34c-5p | miR-34 | miR-34 | let-7i | |||
| miR-371a-3p | miR-9 | miR-9 | miR-106 | |||
| miR-373-3p | miR-92–1 | miR-92–1 | miR-106a | |||
| miR-378 | miR-93 | miR-93 | miR-106b | |||
| miR-451a | miR-34a | miR-141 | ||||
| miR-98 | miR-15a | |||||
| miR-16–1 | ||||||
| miR-17 | ||||||
| miR-18a | ||||||
| miR-195 | ||||||
| miR-19a | ||||||
| miR-221 | ||||||
| miR-29b-2 | ||||||
miR, microRNA.
Regulatory pathways of differentially expressed miRNAs
Similarly, the pathways of each differentially expressed miRNA were extracted, compared and analyzed in the same way to the method by which the regulatory pathways of differentially expressed genes were analyzed. The precursors and successors of differentially expressed miRNAs were listed in three levels. To describe the results, hsa-miR-17 is used as an example. Table II presents the upstream and downstream elements of hsa-miR-17 and the regulatory associations. In the differentially expressed network, it was indicated that MYC regulates hsa-miR-17 and hsa-miR-17 targets PTEN and MYC. In addition, hsa-miR-17 and MYC form FBLs. In the related network, six more genes regulate hsa-miR-17, which itself targets 14 more genes. Cyclin D1 (CCND1) and E2F1 separately form FBLs with hsa-miR-17 in the related network, and CCND1 and E2F1 are TFs in the related network. Therefore, it is indicated that the mutation of hsa-miR-17 may influence a number of other elements. In the global network, there are 12 genes regulating hsa-miR-17 which, itself, targets 47 genes. The results of the present study suggested that hsa-miR-17 is a crucial miRNA in the progression of SOC. hsa-miR-128 deserves more attention because it is not regulated by any gene. That is to say hsa-miR-128 is the ‘starter’ in the network of SOC (it regulates elements but is not regulated by elements in the SOC network) and it is the switch of SOC. The regulation of those switches would be a notable issue.
Table II.
Upsteam and downstream of hsa-miR-17 in three networks of serous ovarian cancer.
| Upstream | Downstream | |||||
|---|---|---|---|---|---|---|
| Differentially expressed network | Related network | Global network | miRNA | Differentially expressed network | Related network | Global network |
| MYC | CCND1 | CCND1 | miR-17 | PTEN | CCND1 | APP |
| E2F1 | E2F1 | MYC | BCL2 | BCL2 | ||
| ESR1 | ESR1 | RUNX1 | BCL2L11 | |||
| MYC | MYC | CCND2 | BIM | |||
| MYCN | MYCN | CDKN1A | BMPR2 | |||
| NFKB1 | NFKB1 | E2F1 | CCL1 | |||
| TNF | NKX2-5 | CXCL8 | CCND1 | |||
| SPI1 | SMAD4 | CCND2 | ||||
| STAT5B | MYC | CDKN1A | ||||
| TLX1 | PTEN | DNAJC27 | ||||
| TLX3 | RB1 | E2F1 | ||||
| TNF | TGFBR2 | E2F3 | ||||
| THBS1 | S1PR1 | |||||
| VEGFA | FBXO31 | |||||
| VIM | GAB1 | |||||
| E2F3 | GPR137B | |||||
| ICAM1 | ||||||
| CXCL8 | ||||||
| JAK1 | ||||||
| MAP3K12 | ||||||
| MAPK9 | ||||||
| MEF2D | ||||||
| MUC17 | ||||||
| MYC | ||||||
| NCOA3 | ||||||
| NPAT | ||||||
| NABP1 | ||||||
| PKD2 | ||||||
| PRKD1 | ||||||
| PRKD2 | ||||||
| PTEN | ||||||
| PTPRO | ||||||
| RB1 | ||||||
| RBL1 | ||||||
| YES1 | ||||||
| ZNFX1 | ||||||
hsa, human; miRNA, microRNA; APP, amyloid beta precursor protein; BCL2, apoptosis regulator; BCL2L11, BCL2 like 11; BMPR2, bone morphogenetic protein receptor type 2; CCL1, C-C motif chemokine ligand 1; CCND1, cyclin D1; CCND2, cyclin D2; CDKN1A, cyclin dependent kinase inhibitor 1A; DNAJC27, DnaJ heat shock protein family (Hsp40) member C27; E2F1, E2F transcription factor 1; E2F3, E2F transcription factor 3; S1PR1, sphingosine-1-phosphate receptor 1; ESR1, estrogen receptor 1; FBXO31, F-box protein 31; GAB1, GRB2 associated binding protein 1; GPR137B, G protein-coupled receptor 137B; ICAM1, intercellular adhesion molecule 1; CXCL8, C-X-C motif chemokine ligand 8; JAK1, Janus kinase 1; MAP3K12, mitogen-activated protein kinase kinase kinase 12; MAPK9, mitogen-activated protein kinase 9; MEF2D, myocyte enhancer factor 2D; MUC17, mucin 17, cell surface associated; MYC, MYC proto-oncogene, bHLH transcription factor; MYCN, MYCN proto-oncogene, bHLH transcription factor; NCOA3, nuclear receptor coactivator 3; NFKB1, nuclear factor-κB subunit 1; NKX2-5, NK2 homeobox 5; NPAT, nuclear protein, coactivator of histone transcription; NABP1, nucleic acid binding protein 1; PKD2, polycystin 2, transient receptor potential cation channel; PRKD1, protein kinase D1; PRKD2, protein kinase D2; PTEN, phosphatase and tensin homolog; PTPRO, protein tyrosine phosphatase, receptor type O; RB1, RB transcriptional corepressor 1; RBL1, RB transcriptional corepressor like 1; RUNX1, runt related transcription factor 1; SMAD4, SMAD family member 4; SPI1, Spi-1 proto-oncogene; STAT5B, signal transducer and activator of transcription 5B; TGFBR2, transforming growth factor beta receptor 2; THBS1, thrombospondin 1; TLX1, T-cell leukemia homeobox 1; TLX3, T-cell leukemia homeobox 3; TNF, tumor necrosis factor; VEGFA, vascular endothelial growth factor A; VIM, vimentin; YES1, YES proto-oncogene 1, Src family tyrosine kinase; ZNFX1, zinc finger NFX1-type containing 1.
Regulatory pathways of predicted TFs in SOC
The same method as that used to analyze the regulatory pathways of differentially expressed genes was used to extract, compare and analyze the pathways of TFs predicted using the P-match method in SOC. The miRNAs associated with these TFs were listed in three networks and not all TFs have precursors and successors. Here, the focus is on NFKB1 as a representative example because its precursors and successors are uniformly distributed in three networks. Table III presents the upstream and downstream elements of hsa-miR-17 and the regulatory associations. In the differentially expressed network, 2 miRNAs target NFKB1, NFKB1 regulates 13 miRNAs, and hsa-miR-146a and hsa-miR-9 separately form FBLs with NFKB1. In the related network, NFKB1 only regulates one additional miRNA compared with the differentially expressed network. In the global network, NFKB1 was the target of 8 miRNAs and it regulates 34 miRNAs. The results of the present study indicated that NFKB1 exhibits an increased likelihood to influence the development of SOC. A number of TFs, including E2F4, do not have precursors or successors and these TFs exhibit small effects on the development of SOC.
Table III.
Upstream and downstream of NFKB1 in three networks of serous ovarian carcinoma.
| Upstream | Downstream | |||||
|---|---|---|---|---|---|---|
| Differentially expressed network | Related network | Global network | NFKB1 | Differentially expressed network | Related network | Global network |
| miR-146a | miR-146a | miR-let-7a | NFKB1 | miR-let-7b | miR-let-7b | miR-let-7a-3 |
| miR-9 | miR-9 | miR-146a | NFKB1 | miR-10b | miR-10b | miR-let-7b |
| miR-146b-5p | NFKB1 | miR-125b | miR-125b | miR-10b | ||
| miR-15a | NFKB1 | miR-146a | miR-146a | miR-125b | ||
| miR-16-5p | NFKB1 | miR-155 | miR-155 | miR-125b-1 | ||
| miR-21-5p | NFKB1 | miR-16 | miR-16 | miR-125b-2 | ||
| miR-9 | NFKB1 | miR-17 | miR-17 | miR-146a | ||
| miR-9-5p | NFKB1 | miR-21 | miR-21 | miR-155 | ||
| NFKB1 | miR-214 | miR-214 | miR-16 | |||
| NFKB1 | miR-29a | miR-29a | miR-16–1 | |||
| NFKB1 | miR-29b | miR-29b | miR-16–2 | |||
| NFKB1 | miR-34 | miR-34 | miR-17 | |||
| NFKB1 | miR-9 | miR-34a | miR-199a-2 | |||
| NFKB1 | miR-9 | miR-21 | ||||
| NFKB1 | miR-214 | |||||
| NFKB1 | miR-224 | |||||
| NFKB1 | miR-29a | |||||
| NFKB1 | miR-29b | |||||
| NFKB1 | miR-29b-1 | |||||
| NFKB1 | miR-29b-2 | |||||
| NFKB1 | miR-29c | |||||
| NFKB1 | miR-34 | |||||
| NFKB1 | miR-34a | |||||
| NFKB1 | miR-365 | |||||
| NFKB1 | miR-365–1 | |||||
| NFKB1 | miR-365–2 | |||||
| NFKB1 | miR-365a | |||||
| NFKB1 | miR-365b | |||||
| NFKB1 | miR-448 | |||||
| NFKB1 | miR-9 | |||||
| NFKB1 | miR-91 | |||||
| NFKB1 | miR-9–1 | |||||
| NFKB1 | miR-9–2 | |||||
| NFKB1 | miR-9–3 | |||||
NFKB1, nuclear factor-κB subunit 1; miR, microRNA.
Discussion
In the present study, differentially expressed genes, differentially expressed miRNAs, TFs predicted using the P-match method and the interactions between them were focused on. From the aforementioned networks, a number of signaling pathways include ≥3 elements. For example, hsa-miR-16 targets TP53 and TP53 regulates hsa-miR-29a. All these pathways have important functions in SOC as elements within them are differentially expressed in SOC, but some pathways are only proposed notionally and their functions remain unclear in SOC. In other types of carcinoma, miRNAs serve a key role; for example, in human breast cancer, miR-145 exhibited a pro-apoptotic effect, dependent on TP53 activation, and that TP53 activation may, in turn, stimulate miR-145 expression (84). Data from the present study suggests that TFs that are predicted using the P-match method exhibit potential associations with differentially expressed miRNAs. However, whether they are closely associated with SOC remains unknown.
In conclusion, the present study constructed 3 regulatory networks, the differentially expressed network, the related network and the global network, to analyze the associations between genes and miRNAs in SOC. Certain signaling pathways and elements were highlighted to analyze the potential regulation mechanism of SOC. The results of the present study identified pathways associated with SOC and may be used to assist gene therapy of SOC. The function of a number of elements and signaling pathways remain unknown.
Acknowledgements
The present study was supported by the Science and Technology Development Project of Jilin Province (grant no. 20160414009GH), the National Natural Science Foundation of China (grant nos. 61300147 and 61472159), the China Postdoctoral Science Foundation (grant no. 2014M551185), the Electronic Commerce Engineering Laboratory Project of Jilin Province (grant no. 2014N143), the Science and Technology Program of Changchun (grant no. 14GH014), the Premier-Discipline Enhancement Scheme supported by Zhuhai Government and the Premier Key-Discipline Enhancement Scheme supported Guangdong Government Funds.
Glossary
Abbreviations
- miRNA
microRNA
- TFs
transcription factors
- targets
target genes
- SOC
serous ovarian carcinoma
- NCBI
national center for biotechnology information
- TFBSs
transcription factor binding sites
- FBLs
feedback-loops
References
- 1.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. microRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 2.Singer G, Kurman RJ, Chang HW, Cho SK, Shih IeM. Diverse tumorigenic pathways in ovarian serous carcinoma. Am J Pathol. 2002;160:1223–1228. doi: 10.1016/S0002-9440(10)62549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soussi T, Béroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1:233–240. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 4.Yu Z, Kim J, He L, Creighton CJ, Gunaratne PH, Hawkins SM, Matzuk MM. Functional analysis of miR-34c as a putative tumor suppressor in high-grade serous ovarian cancer. Biol Reprod. 2014;91:113. doi: 10.1095/biolreprod.114.121988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 6.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;4:1837–1851. [PubMed] [Google Scholar]
- 7.Tran DH, Satou K, Ho TB, Pham TH. Computational discovery of miR-TF regulatory modules in human genome. Bioinformation. 2010;4:371–377. doi: 10.6026/97320630004371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. microRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. microRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/S0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Li J, Ding X, He M, Cheng SY. microRNA and cancer. AAPS J. 2010;2:309–317. doi: 10.1208/s12248-010-9194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao X, Qiao Y, Han D, Zhang Y, Ma N. Enemy or partner: Relationship between intronic micrornas and their host genes. IUBMB Life. 2012;64:835–840. doi: 10.1002/iub.1079. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia W, Li J, Li S, Chen L, Ding H, et al. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res Commun. 2010;396:978–982. doi: 10.1016/j.bbrc.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 15.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: A functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al. miRTarBase update 2014: An information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–D85. doi: 10.1093/nar/gkt1266. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivan C, Hu W, Bottsford-Miller J, Zand B, Dalton HJ, Liu T, Huang J, Nick AM, Lopez-Berestein G, Coleman RL, et al. Epigenetic analysis of the Notch superfamily in high-grade serous ovarian cancer. Gynecol Oncol. 2013;128:506–511. doi: 10.1016/j.ygyno.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singer G, Kurman RJ, Chang HW, Cho SK, Shih IeM. Diverse tumorigenic pathways in ovarian serous carcinoma. Am J Pathol. 2002;160:1223–1228. doi: 10.1016/S0002-9440(10)62549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmid S, Bieber M, Zhang F, Zhang M, He B, Jablons D, Teng NN. Wnt and hedgehog gene pathway expression in serous ovarian cancer. Int J Gynecol Cancer. 2011;21:975–980. doi: 10.1097/IGC.0b013e31821caa6f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh S, Albitar L, LeBaron R, Welch WR, Samimi G, Birrer MJ, Berkowitz RS, Mok SC. Up-regulation of stromal versican expression in advanced stage serous ovarian cancer. Gynecol Oncol. 2010;119:114–120. doi: 10.1016/j.ygyno.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouellet V, Guyot MC, Le Page C, Filali-Mouhim A, Lussier C, Tonin PN, Provencher DM, Mes-Masson AM. Tissue array analysis of expression microarray candidates identifies markers associated with tumor grade and outcome in serous epithelial ovarian cancer. Int J Cancer. 2006;119:599–607. doi: 10.1002/ijc.21902. [DOI] [PubMed] [Google Scholar]
- 26.Thériault BL, Cybulska P, Shaw PA, Gallie BL, Bernardini MQ. The role of KIF14 in patient-derived primary cultures of high-grade serous ovarian cancer cells. J Ovarian Res. 2014;7:123. doi: 10.1186/s13048-014-0123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo LY, Kim E, Cheung HW, Weir BA, Dunn GP, Shen RR, Hahn WC. The tyrosine kinase adaptor protein FRS2 is oncogenic and amplified in high-grade serous ovarian cancer. Mol Cancer Res. 2015;13:502–509. doi: 10.1158/1541-7786.MCR-14-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao M, Sun J, Zhao Z. Synergetic regulatory networks mediated by oncogene-driven microRNAs and transcription factors in serous ovarian cancer. Mol Biosyst. 2013;9:3187–3198. doi: 10.1039/c3mb70172g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiangjun He, Jing Yang, Qi Zhang, Heng Cui, Yujun Zhang. Shortening of the 3′untranslated region: An important mechanism leading to overexpression of HMGA2 in serous ovarian cancer. Chin Med J. 2014;127:494–499. [PubMed] [Google Scholar]
- 30.Ouellet V, Le Page C, Guyot MC, Lussier C, Tonin PN, Provencher DM, Mes-Masson AM. SET complex in serous epithelial ovarian cancer. Int J Cancer. 2006;119:2119–2126. doi: 10.1002/ijc.22054. [DOI] [PubMed] [Google Scholar]
- 31.Kwon JY, Seo YR, Ahn WS. Recognition of potential predictive markers for diagnosis in Korean serous ovarian cancer patients at stage IIIc using array comparative genomic hybridization with high resolution. Mol Cell Toxicol. 2011;7:77. doi: 10.1007/s13273-011-0011-3. [DOI] [Google Scholar]
- 32.Bi FF, Li D, Yang Q. Promoter hypomethylation, especially around the E26 transformation-specific motif and increased expression of poly (ADP-ribose) polymerase 1 in BRCA-mutated serous ovarian cancer. BMC Cancer. 2013;13:90. doi: 10.1186/1471-2407-13-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai W, Zeller C, Masrour N, Siddiqui N, Paul J, Brown R. Promoter CpG island methylation of genes in key cancer pathways associates with clinical outcome in high-grade serous ovarian cancer. Clin Cancer Res. 2013;19:5788–5797. doi: 10.1158/1078-0432.CCR-13-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montavon C, Gloss BS, Warton K, Barton CA, Statham AL, Scurry JP, Tabor B, Nguyen TV, Qu W, Samimi G, et al. Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecol Oncol. 2012;124:582–588. doi: 10.1016/j.ygyno.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 35.Amankwah EK, Wang Q, Schildkraut JM, Tsai YY, Ramus SJ, Fridley BL, Beesley J, Johnatty SE, Webb PM, Chenevix-Trench G, et al. Polymorphisms in stromal genes and susceptibility to serous epithelial ovarian cancer: A report from the ovarian cancer association consortium. PLoS One. 2011;6:e19642. doi: 10.1371/journal.pone.0019642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashuba V, Dmitriev AA, Krasnov GS, Pavlova T, Ignatjev I, Gordiyuk VV, Gerashchenko AV, Braga EA, Yenamandra SP, Lerman M, et al. NotI microarrays: Novel epigenetic markers for early detection and prognosis of high grade serous ovarian cancer. Int J Mol Sci. 2012;13:13352–13377. doi: 10.3390/ijms131013352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurita T, Izumi H, Kagami S, Kawagoe T, Toki N, Matsuura Y, Hachisuga T, Kohno K. Mitochondrial transcription factor A regulates BCL2L1 gene expression and is a prognostic factor in serous ovarian cancer. Cancer Sci. 2012;103:239–244. doi: 10.1111/j.1349-7006.2011.02156.x. [DOI] [PubMed] [Google Scholar]
- 38.Berchuck A, Iversen ES, Luo J, Clarke JP, Horne H, Levine DA, Boyd J, Alonso MA, Secord AA, Bernardini MQ, et al. Microarray analysis of early stage serous ovarian cancers demonstrates profiles predictive of favorable outcome. Clin Cancer Res. 2009;15:2448–2455. doi: 10.1158/1078-0432.CCR-08-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hjerpe E, Brage SE, Stolt Frostvik M, Johansson H, Shoshan M, Avall-Lundqvist E. Metabolic markers and HSP60 in chemonaive serous solid ovarian cancer versus ascites. Int J Gynecol Cancer. 2014;24:1389–1394. doi: 10.1097/IGC.0000000000000246. [DOI] [PubMed] [Google Scholar]
- 40.Cowin PA, George J, Fereday S, Loehrer E, Van Loo P, Cullinane C, Etemadmoghadam D, Ftouni S, Galletta L, Anglesio MS, et al. LRP1B deletion in high-grade serous ovarian cancers is associated with acquired chemotherapy resistance to liposomal doxorubicin. Cancer Res. 2012;72:4060–4073. doi: 10.1158/0008-5472.CAN-12-0203. [DOI] [PubMed] [Google Scholar]
- 41.Tanwar PS, Mohapatra G, Chiang S, Engler DA, Zhang L, Kaneko-Tarui T, Ohguchi Y, Birrer MJ, Teixeira JM. Loss of LKB1 and PTEN tumor suppressor genes in the ovarian surface epithelium induces papillary serous ovarian cancer. Carcinogenesis. 2014;35:546–553. doi: 10.1093/carcin/bgt357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamir A, Jag U, Sarojini S, Schindewolf C, Tanaka T, Gharbaran R, Patel H, Sood A, Hu W, Patwa R, et al. Kallikrein family proteases KLK6 and KLK7 are potential early detection and diagnostic biomarkers for serous and papillary serous ovarian cancer subtypes. J Ovarian Res. 2014;7:109. doi: 10.1186/s13048-014-0109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He QZ, Luo XZ, Wang K, Zhou Q, Ao H, Yang Y, Li SX, Li Y, Zhu HT, Duan T. Isolation and characterization of cancer stem cells from high-grade serous ovarian carcinomas. Cell Physiol Biochem. 2014;33:173–184. doi: 10.1159/000356660. [DOI] [PubMed] [Google Scholar]
- 44.Callahan MJ, Nagymanyoki Z, Bonome T, Johnson ME, Litkouhi B, Sullivan EH, Hirsch MS, Matulonis UA, Liu J, Birrer MJ, et al. Increased HLA-DMB expression in the tumor epithelium is associated with increased CTL infiltration and improved prognosis in advanced-stage serous ovarian cancer. Clin Cancer Res. 2008;23:7667–7673. doi: 10.1158/1078-0432.CCR-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li YL, Ye F, Hu Y, Lu WG, Xie X. Identification of suitable reference genes for gene expression studies of human serous ovarian cancer by real-time polymerase chain reaction. Anal Biochem. 2009;394:110–116. doi: 10.1016/j.ab.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 46.Li YL, Ye F, Cheng XD, Hu Y, Zhou CY, Lü WG, Xie X. Identification of glia maturation factor beta as an independent prognostic predictor for serous ovarian cancer. Eur J Cancer. 2010;46:2104–2118. doi: 10.1016/j.ejca.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 47.Borley J, Ghaem-Maghami S, Honeyfield L, Williamson R, Brown R. Hypomethylation of MSX1 is associated with decreased gene expression, poor progression free survival and chemotherapy resistance in serous ovarian cancer. An Int J Obstetr Gynaecol. 2013;120:249. [Google Scholar]
- 48.Singh H, Li Y, Fuller PJ, Harrison C, Rao J, Stephens AN, Nie G. HtrA3 is downregulated in cancer cell lines and significantly reduced in primary serous and granulosa cell ovarian tumors. J Cancer. 2013;4:152–164. doi: 10.7150/jca.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumtepe Y, Halici Z, Sengul O, Kunak CS, Bayir Y, Kilic N, Cadirci E, Pulur A, Bayraktutan Z. High serum HTATIP2/TIP30 level in serous ovarian cancer as prognostic or diagnostic marker. Eur J Med Res. 2013;18:18. doi: 10.1186/2047-783X-18-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sayer RA, Lancaster JM, Pittman J, Gray J, Whitaker R, Marks JR, Berchuck A. High insulin-like growth factor-2 (IGF-2) gene expression is an independent predictor of poor survival for patients with advanced stage serous epithelial ovarian cancer. Gynecol Oncol. 2005;96:355–361. doi: 10.1016/j.ygyno.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 51.Emmanuel C, Chiew YE, George J, Etemadmoghadam D, Anglesio MS, Sharma R, Russell P, Kennedy C, Fereday S, Hung J, et al. Genomic classification of serous ovarian cancer with adjacent borderline differentiates RAS pathway and TP53-mutant tumors and identifies NRAS as an oncogenic driver. Clin Cancer Res. 2014;20:6618–6630. doi: 10.1158/1078-0432.CCR-14-1292. [DOI] [PubMed] [Google Scholar]
- 52.Yoshihara K, Tajima A, Komata D, Yamamoto T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, et al. Gene expression profiling of advanced-stage serous ovarian cancers distinguishes novel subclasses and implicates ZEB2 in tumor progression and prognosis. Cancer Sci. 2009;100:1421–1428. doi: 10.1111/j.1349-7006.2009.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Z, Kim J, He L, Creighton CJ, Gunaratne PH, Hawkins SM, Matzuk MM. Functional analysis of miR-34c as a putative tumor suppressor in high-grade serous ovarian cancer. Biol Reprod. 2014;91:113. doi: 10.1095/biolreprod.114.121988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merritt MA, Parsons PG, Newton TR, Martyn AC, Webb PM, Green AC, Papadimos DJ, Boyle GM. Expression profiling identifies genes involved in neoplastic transformation of serous ovarian cancer. BMC Cancer. 2009;9:378. doi: 10.1186/1471-2407-9-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newton TR, Parsons PG, Lincoln DJ, Cummings MC, Wyld DK, Webb PM, Green AC, Boyle GM. Expression profiling correlates with treatment response in women with advanced serous epithelial ovarian cancer. Int J Cancer. 2006;119:875–883. doi: 10.1002/ijc.21823. [DOI] [PubMed] [Google Scholar]
- 56.Bateman NW, Jaworski E, Ao W, Wang G, Litzi T, Dubil E, Marcus C, Conrads KA, Teng PN, Hood BL, et al. Elevated AKAP12 in paclitaxel-resistant serous ovarian cancer cells is prognostic and predictive of poor survival in patients. J Proteome Res. 2015;14:1900–1910. doi: 10.1021/pr5012894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bashashati A, Ha G, Tone A, Ding J, Prentice LM, Roth A, Rosner J, Shumansky K, Kalloger S, Senz J, et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol. 2013;231:21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jannesari-Ladani F, Hossein G, Izadi-Mood N. Differential Wnt11 expression related to Wnt5a in high- and low-grade serous ovarian cancer: Implications for migration, adhesion and survival. Asian Pac J Cancer Prev. 2014;15:1489–1495. doi: 10.7314/APJCP.2014.15.3.1489. [DOI] [PubMed] [Google Scholar]
- 59.Ye Q, Chen L, Yin X, Liu YJ, Ji Q, Zhao E. Development of serous ovarian cancer is associated with the expression of homologous recombination pathway proteins. Pathol Oncol Res. 2014;20:931–938. doi: 10.1007/s12253-014-9776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karst AM, Jones PM, Vena N, Ligon AH, Liu JF, Hirsch MS, Etemadmoghadam D, Bowtell DD, Drapkin R. Cyclin E1 deregulation occurs early in secretory cell transformation to promote formation of fallopian tube-derived high-grade serous ovarian cancers. Cancer Res. 2014;74:1141–1152. doi: 10.1158/0008-5472.CAN-13-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martins FC, Santiago Id, Trinh A, Xian J, Guo A, Sayal K, Jimenez-Linan M, Deen S, Driver K, Mack M, et al. Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome Biol. 2014;15:526. doi: 10.1186/s13059-014-0526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tashiro H, Miyazaki K, Okamura H, Iwai A, Fukumoto M. c-myc over-expression in human primary ovarian tumours: Its relevance to tumour progression. Int J Cancer. 1992;50:828–833. doi: 10.1002/ijc.2910500528. [DOI] [PubMed] [Google Scholar]
- 63.Kannan K, Coarfa C, Rajapakshe K, Hawkins SM, Matzuk MM, Milosavljevic A, Yen L. CDKN2D-WDFY2 is a cancer-specific fusion gene recurrent in high-grade serous ovarian carcinoma. PLoS Genet. 2014;10:e1004216. doi: 10.1371/journal.pgen.1004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schildkraut JM, Iversen ES, Wilsonv MA, Clyde MA, Moorman PG, Palmieri RT, Whitaker R, Bentley RC, Marks JR, Berchuck A. Association between DNA damage response and repair genes and risk of invasive serous ovarian cancer. PLoS One. 2010;5:e10061. doi: 10.1371/journal.pone.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shah NR, Tancioni I, Ward KK, Lawson C, Chen XL, Jean C, Sulzmaier FJ, Uryu S, Miller NL, Connolly DC, Schlaepfer DD. Analyses of merlin/NF2 connection to FAK inhibitor responsiveness in serous ovarian cancer. Gynecol Oncol. 2014;134:104–111. doi: 10.1016/j.ygyno.2014.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koti M, Siu A, Clément I, Bidarimath M, Turashvili G, Edwards A, Rahimi K, Mes-Masson AM, Squire JA. A distinct pre-existing inflammatory tumour microenvironment is associated with chemotherapy resistance in high-grade serous epithelial ovarian cancer. Br J Cancer. 2015;112:1215–1222. doi: 10.1038/bjc.2015.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheon DJ, Tong Y, Sim MS, Dering J, Berel D, Cui X, Lester J, Beach JA, Tighiouart M, Walts AE, et al. A collagen-remodeling gene signature regulated by TGF-β signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin Cancer Res. 2014;20:711–723. doi: 10.1158/1078-0432.CCR-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen Y, Wang J, Liu Y, Chen P, Wu X, Wen J. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. 2015;33:2915–2923. doi: 10.3892/or.2015.3937. [DOI] [PubMed] [Google Scholar]
- 69.Li J, Li L, Li Z, Gong G, Chen P, Liu H, Wang J, Liu Y, Wu X. The role of miR-205 in the VEGF-mediated promotion of human ovarian cancer cell invasion. Gynecol Oncol. 2015;137:125–133. doi: 10.1016/j.ygyno.2015.01.531. [DOI] [PubMed] [Google Scholar]
- 70.Swiercz A, Dansonka-Mieszkowska A, Goryca K, Kulinczak M, Zajdel M, Chechlinska M, Rembiszewska A, Kupryjańczyk J, Siwicki KJ. 273 MiR-7 Expression depends on TP53 mutational status in primary serous ovarian cancer. Eur J Cancer. 2012;48(Suppl 5):S66–S67. doi: 10.1016/S0959-8049(12)70968-1. [DOI] [Google Scholar]
- 71.Furlong F, Fitzpatrick P, O'Toole S, Phelan S, McGrogan B, Maguire A, O'Grady A, Gallagher M, Prencipe M, McGoldrick A, et al. Low MAD2 expression levels associate with reduced progression-free survival in patients with high-grade serous epithelial ovarian cancer. J Pathol. 2012;226:746–755. doi: 10.1002/path.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jang SG, Yoo CW, Park SY, Kang S, Kim HK. Low expression of miR-449 in gynecologic clear cell carcinoma. Int J Gynecol Cancer. 2014;24:1558–1563. doi: 10.1097/IGC.0000000000000267. [DOI] [PubMed] [Google Scholar]
- 73.Kan CW, Hahn MA, Gard GB, Maidens J, Huh JY, Marsh DJ, Howell VM. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer. 2012;12:627. doi: 10.1186/1471-2407-12-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chong GO, Jeon HS, Han HS, Son JW, Lee YH, Hong DG, Lee YS, Cho YL. Differential microRNA expression profiles in primary and recurrent epithelial ovarian cancer. Anticancer Res. 2015;35:2611–2617. [PubMed] [Google Scholar]
- 75.Zhang P, Wang M, Jie ZH, Shuang T, Yan XY, Zhou YY, Wu JL. Detection and significance of miR-210 in chemotherapy resistant and chemotherapy sensitive ovarian serous carcinoma. J China Med Univ. 2014;43:487–492. [Google Scholar]
- 76.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC genome browser database: Update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chekmenev DS, Haid C, Kel AE. P-Match: Transcription factor binding site search by combining patterns and weight matrices. Nucleic Acids Res. 2005;33:W432–W437. doi: 10.1093/nar/gki441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wingender E, Dietze P, Karas H, Knüppel R. TRANSFAC: A database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 80.Cancer Genome Atlas Research Network: Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Creighton CJ, Fountain MD, Yu Z, Nagaraja AK, Zhu H, Khan M, Olokpa E, Zariff A, Gunaratne PH, Matzuk MM, Anderson ML. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 2013;23:186–199. doi: 10.1016/j.ccr.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ibrahim FF, Jamal R, Syafruddin SE, Mutalib Ab NS, Saidin S, MdZin RR, Mollah Hossain MM, Mokhtar NM. microRNA-200c and microRNA-31 regulate proliferation, colony formation, migration and invasion in serous ovarian cancer. J Ovarian Res. 2015;8:56. doi: 10.1186/s13048-015-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, Zagatti B, Fabbri M, Veronese A, Liu X, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 2010;17:246–254. doi: 10.1038/cdd.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]