Abstract
As a main treatment of prostate cancer, castration therapy has been widely applied in the clinic. However, the therapeutic strategy for hormone-independent prostate cancer (HIPC) was not satisfied. Gemcitabine is an important chemotherapeutic agent that has been approved for the treatment of numerous human solid tumors, including HIPC, whereas the gemcitabine resistance has become a serious problem in clinical chemotherapy. In the present study, the mechanisms of resistance to gemcitabine were investigated in HIPC cell lines. The results demonstrated that the autophagy markers were induced significantly in HIPC cells subsequent to gemcitabine treatment. Meanwhile, administration of gemcitabine to HIPC cells increased the expression of high mobility group box1 (HMGB1). Furthermore, the gemcitabine-induced autophagy response was attenuated in stable HIPC cells harboring HMGB1 shRNA. Notably, the HIPC cells stably transfected with HMGB1 shRNA or treated with autophagy inhibitors were more sensitive to gemcitabine compared with the control group. These data suggested that inhibition of HMGB1 increased the sensitivity to gemcitabine by decreasing autophagy response in HIPC cells. Overall, the present findings demonstrate a new mechanism for the resistance to gemcitabine in HIPC cell lines.
Keywords: hormone-independent prostate cancer, autophagy, HMGB1, gemcitabine, resistance
Introduction
Prostate cancer is known as a main cancer type among men worldwide, and causes a high rate of death at the advanced stage of disease (1). Since understanding that abnormal activation of prostate-specific antigen/androgen receptor signaling promotes the aggressiveness of prostate cancer (2), androgen deprivation therapy has been applied as the main treatment for prostate cancer (3). However, patients with hormone-independent prostate cancer (HIPC) have a poor prognosis subsequent to androgen deprivation. Gemcitabine, a deoxycytidine analog, has been used as a first-line drug for HIPC (4). Gemcitabine exhibits anti-cancer activity in numerous cancer types by disturbing DNA biosynthesis (5). In addition, gemcitabine resistance has become a major obstacle for its application as a treatment for HIPC. However, the underlying mechanisms of resistance to gemcitabine remain unclear.
As a non-chromosomal DNA binding protein, high mobility group box1 (HMGB1) was described in numerous cellular biological progressions, including cell differentiation, cell autophagy and cell migration (6,7). Different functions of HMGB1 depend on its various target genes or pathways in different tissues. For example, HMGB1 could participate in inflammatory reactions by regulating nuclear factor-κB expression (8), while HMGB1 induced upregulation of c-myc contributes to tumor malignancies. In autophagy, HMGB1 is involved at several levels. HMGB1 regulates the expression of heat shock protein β-1 (HSPB1), which is important for dynamic intracellular trafficking during autophagy and mitophagy (9). Additionally, HMGB1 could disturb the Beclin-1-B cell lymphoma-2 (BCL-2) complex and then activate autophagy through interacting with Beclin-1 (10). A number of studies have demonstrated a crucial role for autophagy in cancer development and therapy (11,12). Notably, HMGB1 exhibits a significant function in facilitating autophagy in response to cytotoxic insults (7,13). Therefore, HMGB1-mediated autophagy is worthy of investigating drug resistance.
In the present study, gemcitabine was shown to induce the expression of HMGB1, which in turn decreased the sensitivity to this drug in HIPC cells. Further experiments indicated that HMGB1 upregulation enhances the expression of HSPB1, which may function as an effector targeted by HMGB1 in gemcitabine induced autophagy progression. Thus, the present findings provide potential targets for the treatment of HIPC.
Materials and methods
Cell culture and chemicals
The human prostate cancer cell line PC-3 was purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in RPMI-1640 (Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% FBS in a 37°C incubator in 5% CO2. Gemcitabine or 3-methyladenosine (3-MA) was purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany) and dissolved in dimethyl sulfoxide.
Antibodies and shRNA
Antibodies against light chain 3 (LC3)-I/II (catalog no. sc-16756), sequestosome 1 (SQSTM1) (catalog no. sc-25575), Beclin-1 (catalog no. sc-11427), HSPB1 (catalog no. sc-1048), HMGB1 (catalog no. sc-56698) and GAPDH (catalog no. sc-25778) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). IRDye® 800CW- or IRDye® 680RD-conjugated secondary antibodies (926–32210; 926-68071; 926-32211; 926-68070) were purchased from LI-COR Biosciences (Lincoln, NE, USA). HMGB1-shRNA (Sigma-Aldrich; Merck KGaA) and pcDNA3.1-HMGB1 (self-constructed; HMGB1 CDS; CCDS9335.1; GenBank; National Institute of Health, Bethesda, MD, USA) were stably transfected with the Lipofectamine 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according the manufacturer's instructions.
Western blot analysis
The whole cell lysates from PC-3 or shHMGB1 PC-3 cells treated with 10 µg/ml gemcitabine were prepared with cold cell radioimmunoprecipitation assay lysis buffer (20–188; EMD Millipore, Billerica, MA, USA) and the total protein concentration was measured using a Bradford assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Acquired proteins (35 µg/lane) were separated by 10% SDS-PAGE gel and then transferred to a nitrocellulose membrane. The membrane was blocked with 5% milk for 2 h at room temperature and incubated with primary antibodies against LC3-I/II (dilution, 1:1,000), SQSTM1 (dilution, 1:1,000), HMGB1 (dilution, 1:1,000), Beclin-1 (dilution, 1:500), HSPB1 (dilution, 1:500) and GAPDH antibody (dilution, 1:2,000) at 4°C overnight. Subsequently, the membranes were incubated with appropriate secondary antibodies at 1:10,000 dilution at room temperature for 1 h. IRDye® 800CW- or IRDye® 680-conjugated secondary antibodies (IRDye® 800CW goat anti-Mouse IgG (H + L), cat. no. 926-32210; IRDye® 680RD goat anti-Rabbit IgG (H + L), cat. no. 926-68071; IRDye® 800CW goat anti-Rabbit IgG (H + L), cat. no. 926-32211; IRDye® 680RD goat anti-Mouse IgG (H + L), cat. no. 926-68070) (LI-COR Biosciences, Lincoln, NE, USA) were used for staining, and then the secondary antibodies were detected by an Odyssey® infrared imaging system (LI-COR Biosciences).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from the PC-3 cells with or without gemcitabine treatment was obtained with the RNAiso™ Plus reagent (Takara Bio, Inc., Otsu, Japan) and reverse-transcribed by a PrimeScript™ RT reagent kit (Takara Bio, Inc.). qPCR was performed using SYBR Green mix (Roche Diagnostics, Basel, Switzerland), according to the manufacturer's instructions. β-actin was used as a loading control. The thermocycling conditions for RT-qPCR reactions were as follows: 95°C 5 min, followed by 40 cycles of 95°C 45 sec, annealing at 55°C 45 sec and extension 72°C 1 min. The following primers were used: HSPB1 forward, 5′-GCGTGTCCCTGGATGTCAAC-3′; HSPB1 reverse, 5′-TGTATTTCCGCGTGAAGCAC-3′; Beclin1 forward, 5′-TGTGAGGAATGCACAGATAC-3′; Beclin1 reverse, 5′-TGTCCACTGTGCCAGATGT-3′; HMGB1 forward, 5′-TGTCCATTGGTGATGTTGCG-3′; HMGB1 reverse, 5′-GGACAGGGCTATCTAAAGACACA-3′; β-actin forward, 5′-CTCCATCCTGGCCTCGCTGT-3′; β-actin reverse, 5′-GCTGTCACCTTCACCGTTCC-3′. The relative mRNA level was calculated using the 2−ΔΔCq method (14).
Cell viability assay
Cell viability of PC-3 treated with 2, 4, 8, 16, 32 or 64 µg/ml of gemcitabine for 48 h was detected using the Cell Counting Kit-8 (CCK8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer's protocol. Absorbance was then measured by a microplate reader at 450 nm. Data were obtained from at least three separate experiments performedin triplicate.
Flow cytometry
PC-3 cells were seeded into 6-well plates (5×105 cells/well) and pre-treated with 2 mM 3-MA or DMSO for 24 h. The PC-3 cells treated with 3-MA or stably transfected with shHMGB1 were stimulated with 10 µg/ml gemcitabine for 48 h. The cells were then collected and stained using the Annexin-V-FLUOS staining kit (Roche Diagnostics), according to the manufacturer's protocol. The stained cells were analyzed immediately on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA) using the CellQuest 3.0 software system (BD Biosciences).
Statistical analysis
SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA) was used in statistical analyses. Group distributions were performed with the Student's t-test or one-way and two-way analysis of variance (Post-hoc test one-way, Tukey's test; post-hoc test two-way, Sidak's test). P<0.05 was considered to indicate a statistically significant difference.
Results
Autophagy was induced by gemcitabine treatment in HIPC cells
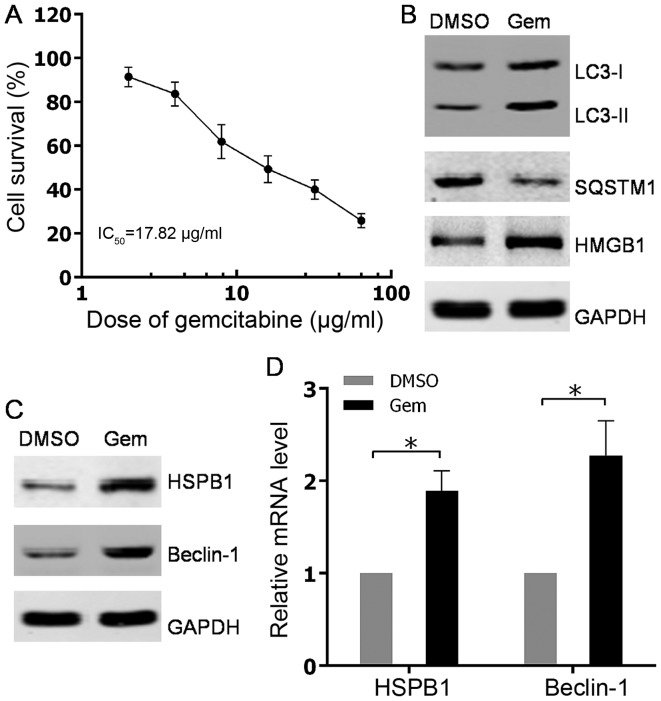
The human hormone-independent prostate cancer PC3 cell line was used to determine whether autophagy could be induced by gemcitabine. Firstly, the sensitivity to different concentrations of gemcitabine was examined using the CCK8 cell viability assay in PC3 cells. The results indicated that gemcitabine suppressed PC3 cell viability in a concentration-dependent manner, with a 50% inhibitory concentration (IC50) of 17.82 µg/ml (Fig. 1A). Based on the cell viability curve of PC3 to gemcitabine, a nonlethal concentration of 10 µg/ml was adopted as the treatment concentration of gemcitabine to induce autophagy in subsequent experiments. Western blot analysis subsequently showed that the conversion of LC3-I to LC3-II was increased and the SQSTM1 level was decreased, which suggested the role of gemcitabine in promoting autophagy in PC3 cells (Fig. 1B). Considering the upregulation of HMGB1 subsequent to gemcitabine treatment (Fig. 1B), HMGB1-dependent autophagy was detected by testing the expression levels of its effectors, HSPB1 and Beclin-1, in PC3 cells. The present findings demonstrated that upregulation of HSPB1 and Beclin-1 occurred at both mRNA and protein levels in PC3 cells with gemcitabine stimulation (Fig. 1C and D). Overall, the results showed that the chemotherapeutic agent gemcitabine could activate autophagy in HIPC cells.
Figure 1.
Autophagy was induced by gemcitabine treatment in HIPC cells. (A) PC-3 cells were treated with 2, 4, 8, 16, 32 or 64 µg/ml gemcitabine for 48 h. Cell survival was determined using Cell Counting Kit-8 assays. All experiments were independently repeated at least three times. *P<0.05. (B and C) PC3 cells were treated with 10 µg/ml gemcitabine or DMSO for 48 h. The expression levels of indicated proteins were analyzed via western blotting. (D) PC3 cells were treated with 10 µg/ml gemcitabine or DMSO for 48 h. HSPB1 and Beclin-1 mRNA levels were measured with quantitative PCR. All experiments were independently repeated at least three times. *P<0.05. DMSO, dimethyl sulfoxide; Gem, gemcitabine; LC3, light chain 3; SQSTM1, sequestosome 1; HMGB1, high mobility group box 1; HSPB1, heat shock protein β-1.
Gemcitabine induced upregulation of HMGB1 contributes to autophagy in HIPC cells
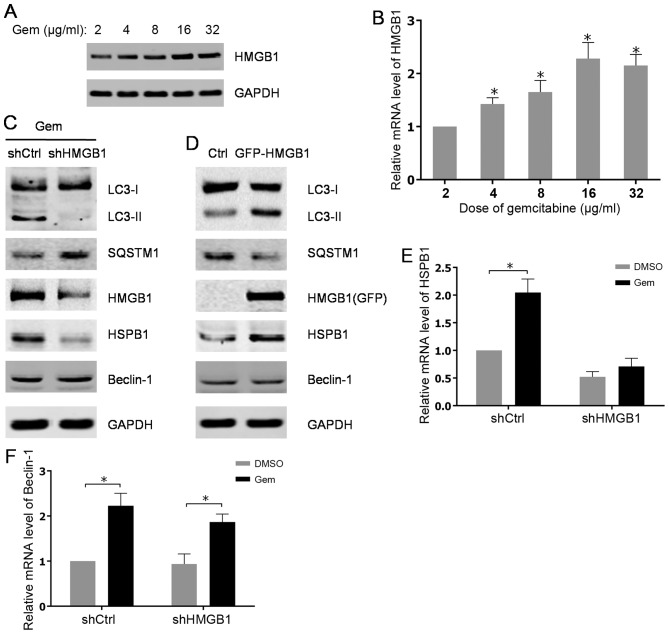
To understand the underlying mechanisms for gemcitabine-induced autophagy in HIPC cells, the roles of the important regulator of autophagy HMGB1 and its associated factors in gemcitabine-induced autophagy progression were investigated. The present data showed that the protein level of HMGB1was significantly increased in a dose-dependent manner in PC3 cells after treated with gemcitabine (Fig. 2A). Consistently, the elevated expression of HMGB1 was also verified at mRNA level after gemcitabine addition (Fig. 2B). Additionally, through examining autophagy-associated markers, it was found that gemcitabine-induced autophagy in PC3 cells was attenuated by stable knockdown of HMGB1 using shRNA (Fig. 2C). However, stably ectopic expression of HMGB1 in PC3 cells evidently promoted autophagy progression compared with parental PC3 cells (Fig. 2D). Notably, data indicated that abrogation of HMGB1 impaired the induction of HSPB1, but not Beclin-1 at both mRNA and protein levels subsequent to gemcitabine treatment (Fig. 2C, E and F). As expected, overexpression of HMGB1 raised the protein level of HSPB1, while no significant alterations of Beclin-1 expression were found in PC3 cells with stably expressing HMGB1 (Fig. 2D). Thus, the aforementioned findings revealed that HMGB1 may mediate gemcitabine-induced autophagy in PC3 cells via its target gene, HSPB1.
Figure 2.
Gemcitabine-induced upregulation of HMGB1 contributes to autophagy in HIPC cells. (A) PC-3 cells were treated with 2, 4, 8, 16 or 32 µg/ml of gemcitabine for 48 h. The expression levels of HMGB1 were analyzed by western blot analysis. (B) HMGB1 mRNA levels were measured using qPCR. All experiments were independently repeated at least three times. *P<0.05. (C and D) PC3 cells were transfected with shRNA or pcDNA3.1-HMGB1 and screened with G418. The PC3 cells transfected with shRNA were treated with 10 µg/ml gemcitabine for 48 h. The expression levels of indicated proteins were analyzed by western blot analysis. (E and F) The transfected PC3 cells were treated with 10 µg/ml gemcitabine or DMSO for 48 h. HSPB1 and Beclin-1mRNA levels were measured with qPCR. All experiments were independently repeated at least three times. *P<0.05. qPCR, quantitative polymerase chain reaction; Gem, gemcitabine; Ctrl, control; LC3, light chain 3; SQSTM1, sequestosome 1; HMGB1, high mobility group box1; HSPB1, heat shock protein β-1; shCtrl, control short hairpin RNA; shHMGB1, short hairpin RNA against HMGB1.
Autophagy induced by gemcitabine protects HIPC cells from the cytotoxicity of gemcitabine
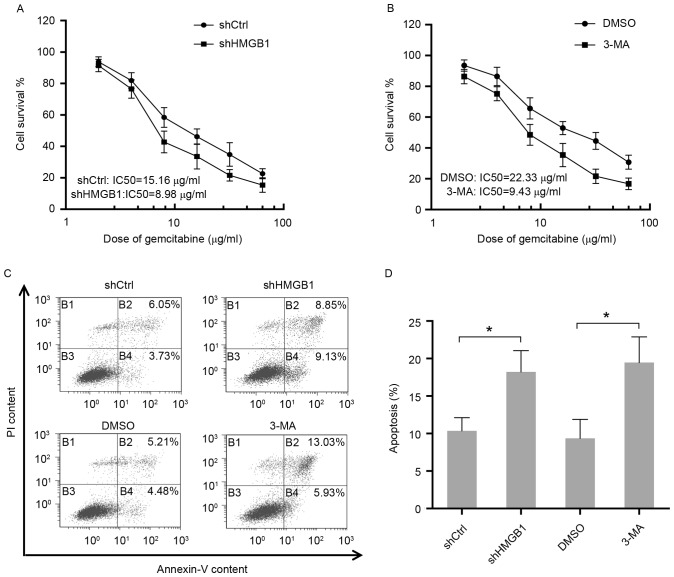
To determine the function of autophagy in response to gemcitabine, the sensitivity to gemcitabine in HIPC cells with different treatments was measured by cell viability assay. The results suggested that elimination of HMGB1 by shRNA in PC3 cells resulted in an apparent reduction of IC50 for gemcitabine treatment (Fig. 3A). Additionally, PC3 cells were either treated with gemcitabine alone or co-treated with gemcitabine and the inhibitor of autophagy 3-MA. The present findings showed that the group treated with only gemcitabine was more resistant to gemcitabine compared to the group that received co-treatment, indicating the protective role of autophagy in HIPC cells exposed to gemcitabine (Fig. 3B). Additionally, consistent with the aforementioned results, inhibition of autophagy in PC3 cells by HMGB1 shRNA or 3-MA-enhanced cell apoptosis induced by gemcitabine (Fig. 3C and D). Therefore, these data elucidated that HMGB1-mediated autophagy reduced the sensitivity of HIPC cells to the drug gemcitabine.
Figure 3.
Autophagy induced by gemcitabine protects HIPC cells from the cytotoxicity of gemcitabine. (A) PC-3 cells stably transfected with HMGB1-shRNA or Ctrl-shRNA were treated with 2, 4, 8, 16, 32 or 64 µg/ml of gemcitabine for 48 h. Cell survival was determined using CCK-8 assays. All experiments were independently repeated at least three times. (B) PC-3 cells pre-treated with 2 mM 3-MA or DMSO for 24 h were then treated with 2, 4, 8, 16, 32 or 64 µg/ml gemcitabine for 48 h. Cell survival was determined using CCK-8 assays. All experiments were independently repeated at least three times. (C) Apoptosis in transfected or treated PC3 cells were analyzed by flow cytometry. (D) Quantitation of apoptosis data. Gem, gemcitabine; Ctrl, control; LC3, light chain 3; SQSTM1, sequestosome 1; HMGB1, high mobility group box1; HSPB1, heat shock protein β-1; shCtrl, control short hairpin RNA; shHMGB1, short hairpin RNA against HMGB1; 3-MA, 3-methyladenosine; PI, propidium iodide; IC50, 50% inhibitory concentration.
Discussion
Autophagy is a biological process through which cells use the lysosome to degrade damaged organelles and macromolecules. Therefore, autophagy may prevent the accumulation of damaged or useless components in cells, which confers cell survival under unfavorable conditions. The dysregulation of autophagy has been associated with numerous cancer types, and enhanced autophagy following chemotherapy and radiotherapy was observed in various cancer cells (15–17). Previous studies have indicated that acquired autophagy is closely associated with multidrug resistance by decreasing the permeability of cell membranes or slowing down cell metabolism (18). In the present study, using LC3-II and SQSTM1 (p62) as markers of autophagy, it was found that the first-line chemotherapeutic gemcitabine induced autophagy in HIPC cells. Additionally, disruption of autophagy by the autophagy inhibitor 3-MA increased the sensitivity to gemcitabine in HIPC cells. Thus, the induction of autophagy has an important role in gemcitabine resistance of HIPC cells. Consistently, it has been reported that induced autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin (19) and mediates multi-drug resistance in osteosarcoma (20).
Autophagy is a complex progression associated with various factors and signaling pathways. Among these factors, HMGB1 is a critical regulator of autophagy, which was the focus of the present study. The results showed that the mRNA and protein levels of HMGB1 were increased in a dose-dependent manner following gemcitabine treatment. In addition, stable knockdown of HMGB1 by shRNA reversed the autophagy induced by gemcitabine in HIPC cells. Stable overexpression of HMGB1 in HIPC cells without treatment also enhanced the autophagy progress, which indicated an increased level of LC3-II and a reduction of SQSTM1. In addition, attenuation of HMGB1 weakened the resistance to gemcitabine in HIPC cells. Therefore, it was hypothesized that gemcitabine resistance in HIPC cells results from HMGB1-associated autophagy.
The roles of HMGB1 in autophagy are performed via two well-known factors, HSPB1 and Beclin-1. Nuclear localization of HMGB1 activates HSPB1 expression (21), which affects dynamic intracellular trafficking during autophagy (22). Additionally, cytosolic localization of HMGB1 interacts with Beclin-1 and dissolved it from BCL-2 (10). Released Beclin-1 regains its ability to promote autophagy (23). Notably, exposure to gemcitabine stimulates the expressions of HSPB1 and Beclin-1 in PC3 cells. However, alterations of HMGB1 level by knockdown or overexpression had an effect on the level of HSPB1, but not Beclin-1, in PC3 cells. This is coincident with previous studies that HSPB1 is tightly regulated by HMGB1 (24). Therefore, upregulation of Beclin-1 in response to gemcitabine may be activated by other autophagy-associated factors or pathways, and the functions of HSPB1 and Beclin-1 in gemcitabine-induced autophagy of HIPC cells require additional investigation.
In summary, it was demonstrated that HMGB1 expression is increased in the HIPC PC3 cell line after gemcitabine treatment. Subsequently, the increasing HMGB1 expression causes autophagy and decreases the sensitivity to gemcitabine in HIPC cells. Therefore, the present findings provided a novel mechanism for gemcitabine resistance in HIPC cells, which could be used as a target for the diagnosis and treatment of patients with HIPC.
Acknowledgements
The present study was supported by the Shenzhen Science and innovation Commission Research Foundation (grant no. JCYJ20150403101028172), Guangdong Provincial Medical Scientific Research Foundation (grant no. A2014634) and Shenzhen Health and Family Planning Commission Research Foundation (grant no. 201401002).
References
- 1.Trewartha D, Carter K. Advances in prostate cancer treatment. Nat Rev Drug Discov. 2013;12:823–824. doi: 10.1038/nrd4068. [DOI] [PubMed] [Google Scholar]
- 2.Lonergan PE, Tindall DJ. Androgen receptor signaling in prostate cancer development and progression. J Carcinog. 2011;10:20. doi: 10.4103/1477-3163.83937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madan RA, Arlen PM. Recent advances revolutionize treatment of metastatic prostate cancer. Future Oncol. 2013;9:1133–1144. doi: 10.2217/fon.13.65. [DOI] [PubMed] [Google Scholar]
- 4.el-Rayes BF, Shields AF, Vaitkevicius V, Philip PA. Developments in the systemic therapy of pancreatic cancer. Cancer Invest. 2003;21:73–86. doi: 10.1081/CNV-120016406. [DOI] [PubMed] [Google Scholar]
- 5.Gray SG, Baird AM, O'Kelly F, Nikolaidis G, Almgren M, Meunier A, Dockry E, Hollywood D, Ekström TJ, Perry AS, O'Byrne KJ. Gemcitabine reactivates epigenetically silenced genes and functions as a DNA methyltransferase inhibitor. Int J Mol Med. 2012;30:1505–1511. doi: 10.3892/ijmm.2012.1138. [DOI] [PubMed] [Google Scholar]
- 6.Srinivasan M, Banerjee S, Palmer A, Zheng G, Chen A, Bosland MC, Kajdacsy-Balla A, Kalyanasundaram R, Munirathinam G. HMGB1 in hormone-related cancer: A potential therapeutic target. Horm Cancer. 2014;5:127–139. doi: 10.1007/s12672-014-0175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, Benschop R, Sparvero LJ, Amoscato AA, Tracey KJ, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299–5310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bierhaus A, Schiekofer S, Schwaninger M, Andrassy M, Humpert PM, Chen J, Hong M, Luther T, Henle T, Klöting I, et al. Diabetes-associated sustained activation of the transcription factor nuclear factor-kappaB. Diabetes. 2001;50:2792–2808. doi: 10.2337/diabetes.50.12.2792. [DOI] [PubMed] [Google Scholar]
- 9.Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B, Zeh HJ, III, Lotze MT. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab. 2011;13:701–711. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, III, Lotze MT. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livesey KM, Tang D, Zeh HJ, Lotze MT. Autophagy inhibition in combination cancer treatment. Curr Opin Investig Drugs. 2009;10:1269–1279. [PubMed] [Google Scholar]
- 12.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, Lotze MT, et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia. 2011;25:23–31. doi: 10.1038/leu.2010.225. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Qian HR, Yang Y. Functional role of autophagy in gastric cancer. Oncotarget. 2016;7:17641–17651. doi: 10.18632/oncotarget.7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mancias JD, Kimmelman AC. Mechanisms of selective autophagy in normal physiology and cancer. J Mol Biol. 2016;428:1659–1680. doi: 10.1016/j.jmb.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen K, Shi W. Autophagy regulates resistance of non-small cell lung cancer cells to paclitaxel. Tumour Biol. 2016;37:10539–10544. doi: 10.1007/s13277-016-4929-x. [DOI] [PubMed] [Google Scholar]
- 18.Rocchi A, He C. Emerging roles of autophagy in metabolism and metabolic disorders. Front Biol (Beijing) 2015;10:154–164. doi: 10.1007/s11515-015-1354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, Zhang Z, Zhang Y, Chen X, Guo S, Lei Y, Xu Y, Ji C, Bi Z, Wang K. HMGB1-mediated autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin via MEK/ERK signaling pathway. Cancer Biol Ther. 2015;16:511–517. doi: 10.1080/15384047.2015.1017691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R, Vernon P, Cao L, Tang D. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72:230–238. doi: 10.1158/0008-5472.CAN-11-2001. [DOI] [PubMed] [Google Scholar]
- 21.Narumi T, Shishido T, Otaki Y, Kadowaki S, Honda Y, Funayama A, Honda S, Hasegawa H, Kinoshita D, Yokoyama M, et al. High-mobility group box 1-mediated heat shock protein beta 1 expression attenuates mitochondrial dysfunction and apoptosis. J Mol Cell Cardiol. 2015;82:1–12. doi: 10.1016/j.yjmcc.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 22.Acunzo J, Katsogiannou M, Rocchi P. Small heat shock proteins HSP27 (HspB1), αB-crystallin (HspB5) and HSP22 (HspB8) as regulators of cell death. Int J Biochem Cell Biol. 2012;44:1622–1631. doi: 10.1016/j.biocel.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Wirth M, Joachim J, Tooze SA. Autophagosome formation-the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23:301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Kang R, Livesey KM, Zeh HJ, III, Lotze MT, Tang D. Metabolic regulation by HMGB1-mediated autophagy and mitophagy. Autophagy. 2011;7:1256–1258. doi: 10.4161/auto.7.10.16753. [DOI] [PubMed] [Google Scholar]