Abstract
Previous studies have suggested that semaphorin 3C (SEMA3C) is involved in the tumorigenesis and metastasis of a number of types of cancer. The aim of the present study was to investigate the role of SEMA3C in the proliferation and migration of MCF-7 breast cancer cells. Small interfering (si)RNA sequences targeting SEMA3C were constructed and transfected into MCF-7 cells in order to silence the expression of SEMA3C. Cell proliferation and migration were measured using CCK-8 and Transwell assays, respectively. Transfection with SEMA3C siRNA significantly downregulated the expression of SEMA3C in MCF-7 cells, and significantly suppressed cell proliferation and migration. Therefore, SEMA3C-targeted siRNA may be of potential use for the early diagnosis and treatment of breast cancer.
Keywords: breast cancer, proliferation, migration, MCF-7 cells, semaphorin 3C
Introduction
Breast cancer is the most common type of malignancy in women and its incidence rates are increasing (1). It is estimated that ~1100,000 new cases of female breast cancer are diagnosed worldwide each year, and 37% of patients (410,000 cases) succumb to the disease each year (2–4). Targeted therapy, including RNA interference (RNAi) technology, has gained interest in recent years as a potential treatment due to its low toxicity, specificity and efficiency (5). The use of small interfering (si)RNA has several advantages, including simple sequence design and fewer adverse effects on cells or tissues. Therefore siRNA could be a more promising candidate for the diagnosis and treatment of diseases compared with shRNA (6). A number of cancer-associated genes, including B-cell lymphoma 2, tumor protein p53, hypoxia-inducible factor and vascular endothelial growth factor have previously been identified as potential targets for RNAi (7–9). Semaphorin 3C (SEMA3C) is a member of the semaphorin family that serves important roles in a number of physiological processes, including axonal growth, immune response, cell adhesion, migration and bone remodeling (10). Numerous studies have demonstrated that semaphorins are overexpressed in a variety of malignant tumors, including glioma, gastric cancer and lung cancer (11). In addition, upregulation of semaphorins is associated with cancer metastasis and angiogenesis, and affects the prognosis and life quality of patients (12,13). In the present study, siRNA was used to silence SEMA3C, which resulted in significantly suppressed cell proliferation and migration in MCF-7 cells. These results suggest that SEMA3C may be a potential target for breast cancer therapy.
Materials and methods
Cells and reagents
The human breast cancer cell line MCF-7 was obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Fetal bovine serum (FBS) and Dulbecco's modified Eagle's medium (DMEM) were obtained from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA). RNAiso Plus, PrimeScript RT Reagent kit, and SYBR Premix Ex Taq II were from Takara Biotechnology, Co., Ltd. (Dalian, China). A SEMA3C rabbit polyclonal antibody (catalog number: ARP38906) was purchased from BD Biosciences (San Jose, CA, USA). GAPDH and α-tubulin mouse monoclonal antibodies (catalog numbers: ABIN268426 and AB9354) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). The horseradish peroxidase (HRP)-conjugated secondary antibodies, RIPA buffer, SDS-PAGE Gel Preparation kit, BCA Protein Assay kit, crystal violet, and Cell Counting Kit-8 were obtained from Beyotime Institute of Biotechnology (Haimen, China). Polyvinylidene difluoride (PVDF) membranes and Transwell plates were purchased from EMD Millipore (Billerica, MA, USA). Lipofectamine® 2000 was obtained from Invitrogen (Thermo Fisher Scientific, Inc.).
siRNA sequences
Three siRNA sequences targeting the SEMA3C gene were designed using the SEMA3C full-length complementary (c)DNA sequence (XM_009456869.1) as a template. The SEMA3C siRNA (siRNA-1, siRNA-2 and siRNA-3), fluorescein amidite (FAM)-labeled negative control siRNA (siRNA-FAM), GAPDH siRNA (siRNA-GAPDH), and negative control siRNA (siRNA-NC) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China) and the sequences are listed in Table I.
Table I.
Oligonucleotide sequences of the siRNAs used in the study.
| Name | Sequence (5′-3′) |
|---|---|
| siRNA-1 | Sense: 5′-GCCCAGCUUAAUCAAGAAATT-3′ |
| Antisense: 5′-UUUGUUGAUUAACCUGGGCTT-3′ | |
| siRNA-2 | Sense: 5′-GCGCUACUAAUUGGGAAGATT-3′ |
| Antisense: 5′-UCUUCGCAAUUAGUUAGGGCTT-3′ | |
| siRNA-3 | Sense: 5′-GGGCUGAGGACCUUGCAGAAGATT-3′ |
| Antisense: 5′-UCUUCCGCAAGGUCCUCAGGCCTT-3′ | |
| siRNA-FAM | Sense: 5′-UUCUGCGAACGUGUCACGUTT-3′ |
| Antisense: 5′-ACGUCACACGUUCGGAGAATT-3′ | |
| siRNA-NC | Sense: 5′-UUCUCCGAACGUGUCACGUTT-3′ |
| Antisense: 5′-ACGUGACACGUUCGGAGAATT-3′ | |
| siRNA-GADPH | Sense: 5′-GUAUCACAACAGCCUCAAGTT-3′ |
| Antisense: 5′-CUUGAGGCUGUUGUCAUACTT-3′ |
siRNA, small interfering RNA; NC, negative control.
Cell culture and siRNA transfection
Human MCF-7 breast cancer cells were cultured in DMEM containing 10% FBS, 100 µg/ml streptomycin, and 100 U/ml penicillin, in a humidified 37°C incubator with 5% CO2. MCF-7 cells (5×104) in the logarithmic growth phase were seeded into 24-well plates 24 h prior to transfection. Cells were transfected with siRNA (siRNA-1, siRNA-2, siRNA-3, siRNA-FAM or siRNA-NC) using Lipofectamine® 2000 reagent according to the manufacturer's protocol. RNA and protein were isolated at 48 and 72 h following transfection, respectively.
Determination of the optimal siRNA transfection concentration
MCF-7 cells (5×104) in the logarithmic growth phase were seeded in 24-well plates. Following 24 h incubation at 37°C, cells were transfected with increasing concentrations of siRNA-FAM (0, 10, 25, 50, 75, 100, 150, 200 nmol/l). The fluorescent signal in the cells was detected under fluorescence microscopy 24 h post-transfection. For the positive control, cells were transfected with 0, 25, 50, 100, 200 nmol/l siRNA-GAPDH. At 72 h following transfection, cells were harvested and protein was collected. The protein level of GAPDH was detected by western blotting with α-tubulin as an internal control. The band density was quantified using ImageJ software (version 1.41; National Institutes of Health, Bethesda, MD, USA). Based on the fluorescence signal and GAPDH protein level, the optimal siRNA transfection concentration was defined.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The knockdown efficiencies of the SEMA3C-targeting siRNAs were evaluated by RT-qPCR. Total RNA was isolated from MCF-7 cells using RNAiso Plus reagent 48 h after transfection, and reverse transcribed to cDNA using the PrimeScript RT Reagent kit. The total RNA of the cultured cells was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. A total of 1 µg total RNA was reverse-transcribed to cDNA using AMV reverse transcriptase (Takara Biotechnology, Co., Ltd.) and a stem-loop RT primer (Applied Biosystems; Thermo Fisher Scientific, Inc.), which was performed with the following conditions: 16°C for 15 min, 42°C for 60 min and 85°C for 5 min. Each cDNA sample was analyzed in triplicate using SYBR Premix Ex Taq II according to the manufacturer's protocol. Real-time PCR was performed using a TaqMan PCR kit on an Applied Biosystems 7500 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reactions were incubated at 95°C for 5 min followed by 35 cycles of 94°C for 15 sec and 72°C for 1 min. GAPDH was used as an internal control. The primer sequences were as follows: GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′, and reverse, 5′-GAAGATGGTGATGGGATTTC-3′; SEMA3C forward, 5′-GCGAAGCAGCATGAGGTGTATTGGA-3′, and reverse, 5′-CGATGTAGTTGTGGCACTCTGTCTG-3′. Relative mRNA quantification was assessed using the 2−∆∆Cq method (14).
Western blot analysis
The protein level of SEMA3C was detected by western blot analysis. MCF-7 cells were harvested and lysed in RIPA buffer at 72 h post-transfection. The protein concentration was determined using the BCA Protein Assay kit. Protein (50 µg) was separated using 8% SDS-PAGE and transferred to PVDF membranes. Following blocking with 5% bovine serum albumin (Takara Biotechnology, Co., Ltd.) for 1 h at room temperature, the membranes were incubated with primary antibodies (dilutions: SEMA3C, 1:800; GAPDH, 1:1,000) overnight at 4°C. Following three washes with TBS-Tween-20 (TBST), the membranes were incubated with HRP-conjugated secondary antibodies for 2 h at room temperature. Following a second round of washing with TBST, protein bands were detected with an enhanced chemiluminescence assay kit (GE Healthcare, Chicago, IL, ISA). ImageJ software was used for densitometric analysis with GAPDH as an internal control.
CCK-8 assay
The effect of SEMA3C knockdown on MCF-7 cell proliferation was accessed using a CCK-8 assay. At 24 h prior to siRNA transfection, MCF-7 cells in the logarithmic growth phase were seeded at 2,000 cells/well in 96-well plates containing 100 µl medium. At the indicated time points (24, 48 and 72 h following transfection), 10 µl CCK-8 was added into each well and the plates were incubated at 37°C for another 3 h. The absorbance (optical density; OD) was measured at a wavelength of 450 nm. The experiment was performed in triplicate. The relative growth rate was calculated as follows: Relative growth rate (%)=(OD of treated cells/OD of control cells)x100.
Migration assay
The effect of SEMA3C knockdown on MCF-7 cell migration was determined using a Transwell migration assay. MCF-7 cells were plated at 2.5×105/well in 6-well plates. Following transfection with siRNA for 72 h, cells (5×104) in 800 µl serum-free medium were seeded in the upper chamber of 12-well plates and 1 ml DMEM containing 10% FBS was placed in the lower chamber. Following incubation for 24 h, the upper chamber was washed twice with PBS and fixed with 4% paraformaldehyde for 15 min. Cells on the top of the membrane were wiped off with cotton swabs. Cells that migrated to the bottom were stained with 0.1% crystal violet for 15 min. Four randomly selected fields of the membrane were counted under an inverted microscope (magnification, ×100).
Statistical analysis
All experiments were performed in triplicate. Statistical analyses were carried out using SPSS software (version 19.0; IBM Corp., Armonk, NY, USA). All data are presented as the mean ± standard deviation. Experiments were performed in triplicate. Differences between groups were determined by a one-way analysis of variance. ANOVA was used for comparison between >2 groups and the t-test was used for comparison between 2 groups. Differences between two groups were accessed by least significant difference t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Determination of the optimal effective siRNA concentration
The siRNA transfection efficiency can vary for different cell types. To achieve the optimal effective siRNA concentration, the negative fluorescence control siRNA (siRNA-FAM) and the positive control siRNA (siRNA-GAPDH) were transfected into MCF-7 cells. Following transfection with siRNA-FAM for 24 h, a fluorescent signal was observed, which increased in a dose-dependent manner. Following transfection with 25 nmol/l siRNA, 30% of cells exhibited fluorescence, and ~90% of cells exhibited fluorescence following transfection with 50 nmol/l siRNA. The knockdown efficiencies of siRNA-GAPDH were 35.71±5.42% for 25 nmol/l siRNA and 51.6±7.59% for 100 nmol/l siRNA (data not shown). However, more apoptotic cells were detected with the increased amount of siRNA. Based on these results, 100 nmol/l siRNA was selected as the treatment dose for further experiments.
siRNA mediates SEMA3C silencing in breast cancer cells
To assess the knockdown efficiency of SEMA3C, SEMA3C mRNA levels were determined by RT-qPCR 48 h after transfection. The mRNA level of SEMA3C was significantly decreased in MCF-7 cells following transfection with SEMA3C siRNAs compared with non-transfected cells (P<0.05; Fig. 1). The mRNA levels of SEMA3C were 71.13±3.15% (siRNA-1), 58.26±2.04% (siRNA-2) and 37.11±2.53% (siRNA-3) of the level in the control group. In agreement this result, western blot analysis demonstrated that the protein levels of SEMA3C were significantly decreased following siRNA knockdown (Fig. 2). The knockdown efficiencies were 47.37±6.02, 50.87±4.61 and 65.27±3.15% for siRNA-1, siRNA-2, and siRNA-3, respectively, relative to the control (data not shown). Together, these results suggest that all three siRNAs effectively suppress SEMA3C expression. siRNA-3, which exhibited the highest knockdown efficiency, was selected for further experiments.
Figure 1.
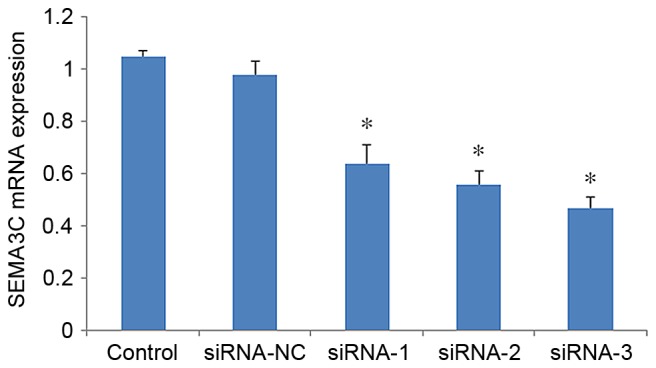
SEMA3C is knocked down at the mRNA level following siRNA transfection. Reverse transcription-quantitative polymerase chain reaction analysis of mRNA levels of SEMA3C in MCF-7 cells 48 h after SEMA3C knockdown. Data are presented as the mean ± standard deviation. Data are presented as the mean ± standard deviation of three independent experiments. siRNA-NC group represents a value of 1. *P<0.05 vs. control. SEMA3C, semaphorin 3C; siRNA, small interfering RNA; control, mock control; NC, negative control.
Figure 2.
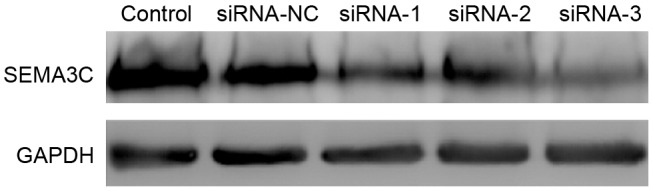
SEMA3C is knocked down at the protein level following siRNA transfection. Western blot analysis of protein levels of SEMA3C in MCF-7 cells 48 h following siRNA transfection. SEMA3C, semaphorin 3C; siRNA, small interfering RNA; control, mock control; NC, negative control.
SEMA3C knockdown inhibits MCF-7 cell proliferation
To understand the function of SEMA3C in breast cancer, the effect of SEMA3C knockdown on breast cancer cell proliferation was examined. A CCK-8 assay demonstrated that the relative growth rate [(OD of treated cells/OD of control cells)x100] was significantly decreased in SEMA3C siRNA-transfected cells compared with that of control cells (control or siRNA-NC; P<0.05; Fig. 3). The numbers of cells in the SEMA3C-siRNA group were 81.06±9.43, 63.47±7.81 and 55.12±5.03% relative to the siRNA-NC group at 24, 48 and 72 h post-transfection (data not shown). No significant differences were observed between the growth rates of the control cells and cells transfected with siRNA-NC (P>0.05; Fig. 3). These results suggest that SEMA3C knockdown inhibits MCF-7 cell proliferation.
Figure 3.
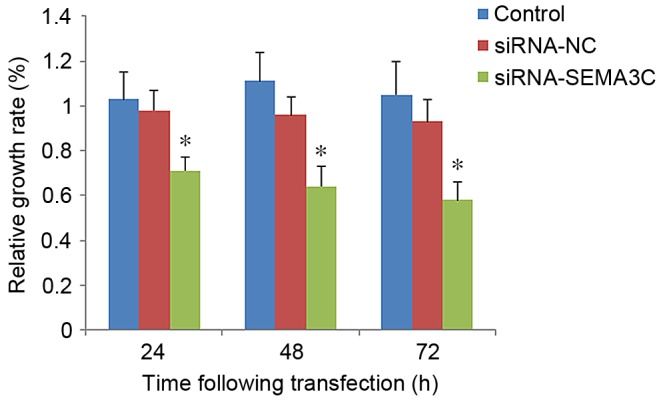
SEMA3C knockdown inhibits MCF-7 cell proliferation. Results of the Cell Counting Kit-8 assay on MCF-7 cells 24, 48 and 72 h following siRNA treatment. Data are presented as the mean ± standard deviation. *P<0.05 vs. control at same time-point. SEMA3C, semaphorin 3C; siRNA, small interfering RNA; control, mock control; NC, negative control.
SEMA3C knockdown suppresses MCF-7 cell migration
A Transwell migration assay demonstrated that the number of migrated cells was significantly decreased following SEMA3C knockdown (104.71±3.01) compared with the number of migrated cells in the control (198.16±9.07) and siRNA-NC groups (179.34±6.48) (P<0.05; Fig. 4). No significant differences in the number of migrated cells were observed between the control and siRNA-NC groups (P>0.05). These results suggest that SEMA3C knockdown inhibits MCF-7 breast cancer cell migration.
Figure 4.
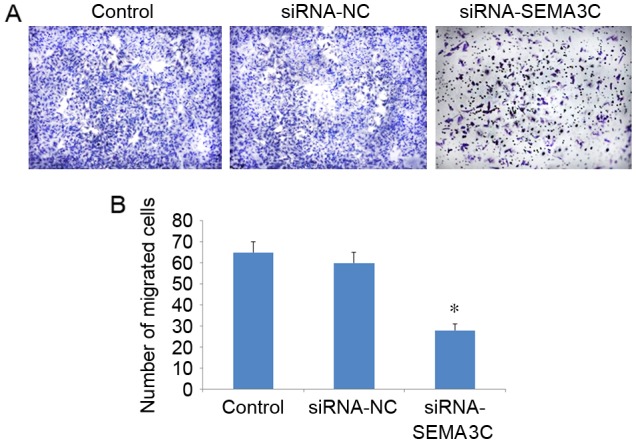
SEMA3C knockdown inhibits MCF-7 cell migration. (A) Representative images of migrated control MCF-7 cells, or MCF-7 cells transfected with NC or SEMA3C-specific siRNAs (magnification, ×100). (B) Quantification of migrated MCF-7 cells. Data are presented as the mean ± standard deviation. All the experiments were performed in triplicate. *P<0.05 vs. control. SEMA3C, semaphorin 3C; siRNA, small interfering RNA; control, mock control; NC, negative control.
Discussion
Semaphorins are a family of secreted proteins that have been identified as novel tumor-associated factors (15). Based on their structural similarity, semaphorins are divided into eight classes that contain ~25 proteins. Semaphorins serve crucial roles in the nervous system, immune system, bone remodeling and cancer (8,16). Previous studies have demonstrated that in the tumor microenvironment, SEMA3C promotes endotheliocyte migration, cancer metastasis and angiogenesis (17–20). By contrast, deletion of SEMA3C suppresses tumorigenesis and angiogenesis (21). However, the functions of SEMA3C in breast cancer cell growth and migration remain unknown. In the present study, the expression of SEMA3C in MCF-7 breast cancer cells was suppressed using RNAi. Treating MCF-7 cells with SEMA3C siRNA significantly inhibited cell proliferation and migration. These results, together with previous studies, suggest that SEMA3C serves an important role in breast tumorigenesis and metastasis, indicating that SEMA3C may be a potential target for breast cancer therapy.
The combination of specific molecular targeted therapy and conventional chemotherapy may allow a reduction of the chemotherapy dose, increase the sensitivity of tumors to therapy, and reduce adverse reactions (22,23). Data from the present study demonstrated that an siRNA sequence effectively suppresses SEMA3C expression in vitro. Knockdown of SEMA3C significantly inhibited breast cancer cell growth and migration. These findings suggest that SEMA3C may be a promising target for breast cancer therapy and SEMA3C siRNA may be a novel therapy for the treatment of breast cancer. However, further studies are required to investigate the molecular mechanisms underlying the effect of SEMA3C on breast cancer cell growth and migration.
Acknowledgements
The current study was supported by the Science & Technology Action Plan for Prevention and Control of Major Diseases: Special Fund for Wound Repair (The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China; grant no. ZX-01-C2015051).
References
- 1.Eccles SA, Aboagye EO, Ali S, Anderson AS, Armes J, Berditchevski F, Blaydes JP, Brennan K, Brown NJ, Bryant HE, et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013;15:R92. doi: 10.1186/bcr3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold M, Karim-Kos HE, Coebergh JW, Byrnes G, Antilla A, Ferlay J, Renehan AG, Forman D, Soerjomataram I. Recent trends in incidence of five common cancers in 26 European countries since 1988: Analysis of the European Cancer Observatory. Eur J Cancer. 2015;51:1164–1187. doi: 10.1016/j.ejca.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 4.Stewart BW, Kleihues Paul P. World Cancer Report. Lyon, France: International Agency Research on Cancer; 2003. [Google Scholar]
- 5.Shao S, Zhao X, Zhang X, Luo M, Zuo X, Huang S, Wang Y, Gu S, Zhao X. Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol Cancer. 2015;14:28. doi: 10.1186/s12943-015-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrio-Real L, Benedetti LG, Engel N, Tu Y, Cho S, Sukumar S, Kazanietz MG. Subtype-specific overexpression of the Rac-GEF P-REX1 in breast cancer is associated with promoter hypomethylation. Breast Cancer Res. 2014;16:441. doi: 10.1186/s13058-014-0441-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J, Cai Q, Lu J, Jha HC, Robertson ES. Upregulation of cellular Bcl-2 by the KSHV encoded rta promotes virion production. PLoS One. 2011;6:e23892. doi: 10.1371/journal.pone.0023892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie L, Gazin C, Park SM, Zhu LJ, Debily MA, Kittler EL, Zapp ML, Lapointe D, Gobeil S, Virbasius CM, Green MR. A synthetic interaction screen identifies factors selectively required for proliferation and TERT transcription in p53-deficient human cancer cells. PLoS Genet. 2012;8:e1003151. doi: 10.1371/journal.pgen.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harper SJ, Bates DO. VEGF-A splicing: The key to anti-angiogenic therapeutics? Nat Rev Cancer. 2008;8:880–887. doi: 10.1038/nrc2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzolio S, Tamagnone L. Semaphorin signals on the road to cancer invasion and metastasis. Cell Adh Migr. 2007;1:62–68. doi: 10.4161/cam.1.2.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyato H, Tsuno NH, Kitayama J. Semaphorin 3C is involved in the progression of gastric cancer. Cancer Sci. 2012;103:1961–1966. doi: 10.1111/cas.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Satué M, Blanco J. Identification of semaphorin E gene expression in metastatic human lung adenocarcinoma cells by mRNA differential display. J Surg Oncol. 1999;72:18–23. doi: 10.1002/(SICI)1096-9098(199909)72:1<18::AID-JSO5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 13.Rieger J, Wick W, Weller M. Human malignant glioma cells express semaphorins and their receptors, neuropilins and plexins. Glia. 2003;42:379–389. doi: 10.1002/glia.10210. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Neufeld G, Mumblat Y, Smolkin T, Toledano S, Nir-Zvi I, Ziv K, Kessler O. The semaphorins and their receptors as modulators of tumor progression. Drug Resist Updat. 2016;29:1–12. doi: 10.1016/j.drup.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Feiner L, Webber AL, Brown CB, Lu MM, Jia L, Feinstein P, Mombaerts P, Epstein JA, Raper JA. Targeted disruption of semaphorin 3C leads to persistent truncus arteriosus and aortic arch interruption. Development. 2001;128:3061–3070. doi: 10.1242/dev.128.16.3061. [DOI] [PubMed] [Google Scholar]
- 17.Brown CB, Feiner L, Lu MM, Li J, Ma X, Webber AL, Jia L, Raper JA, Epstein JA. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development. 2001;128:3071–3080. doi: 10.1242/dev.128.16.3071. [DOI] [PubMed] [Google Scholar]
- 18.Herman JG, Meadows GG. Increased class 3 semaphorin expression modulates the invasive and adhesive properties of prostate cancer cells. Int J Oncol. 2007;30:1231–1238. [PubMed] [Google Scholar]
- 19.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 20.Esselens C, Malapeira J, Colomé N, Casal C, Rodríguez-Manzaneque JC, Canals F, Arribas J. The cleavage of semaphorin 3C induced by ADAMTS1 promotes cell migration. J Biol Chem. 2010;285:2463–2473. doi: 10.1074/jbc.M109.055129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra R, Kumar D, Tomar D, Chakraborty G, Kumar S, Kundu GC. The potential of class 3 semaphorins as both targets and therapeutics in cancer. Expert Opin Ther Targets. 2015;19:427–442. doi: 10.1517/14728222.2014.986095. [DOI] [PubMed] [Google Scholar]
- 22.Hong CA, Nam YS. Functional nanostructures for effective delivery of small interfering RNA therapeutics. Theranostics. 2014;4:1211–1232. doi: 10.7150/thno.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J. 2011;6:1130–1146. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]


