Abstract
The present study aimed to identify the regulatory mechanisms associated with the metastasis of prostate cancer (PC). The microRNA (miRNA/miR) microarray dataset GSE21036 and gene transcript dataset GSE21034 were downloaded from the Gene Expression Omnibus database. Following pre-processing, differentially expressed miRNAs (DEMs) and differentially expressed genes (DEGs) between samples from patients with primary prostate cancer (PPC) and metastatic prostate cancer (MPC) with |log2 fold change (FC)| >1 and a false discovery rate <0.05 were selected using the Linear Models for Microarray and RNA-seq Data 4 package of R. Next, a DEM-DEG regulatory network was constructed by downloading miRNA-DEG pairs from the miRNA.org database. Finally, functional annotation of each DEM-DEG module was performed using the Database for Annotation, Visualization and Integrated Discovery based on the Gene Ontology database. The upregulated miRNAs, including miR-144, miR-494 and miR-181a, exhibited a higher degree of connections compared with other nodes, including in the DEM-DEG regulatory network, and regulated a number of downregulated DEGs. According to the functional annotation of the DEM-DEG modules, miR-144 and its targeted DEGs enriched the highest number of biological process terms (36 terms), followed by miR-494 (24 terms), miR-30d (18 terms), miR-181a (15 terms), hsa-miR-196a (8 terms), miR-708 (7 terms) and miR-486-5p (2 terms). Therefore, these miRNAs may serve roles in the metastasis of PC cells via downregulation of their corresponding target DEGs.
Keywords: metastatic prostate cancer, microRNA, differentially expressed genes, DEM-DEG regulatory network
Introduction
Prostate cancer (PC) is the most common cancer in men and the second leading cause of cancer-associated mortality in USA (1). PC is challenging to treat due to its high metastasis rate (2); despite considerable advances in the treatment of PC, metastatic prostate cancer (MPC) remains incurable (3). The molecular mechanisms underlying the metastasis of PC cells remain largely unknown and require further study.
MicroRNAs (miRNAs/miRs) are a group of non-coding RNAs of 17–27 nucleotides in length that regulate gene expression by binding to the 3′untranslated regions of messenger RNAs (mRNAs) (4). miRNAs have been demonstrated to serve important roles in a number of cellular processes as post-transcriptional regulators, in addition to roles in cancer development and progression (5,6). Dysregulation of miRNAs has been demonstrated to contribute to tumorigenesis by stimulating proliferation, angiogenesis and invasion (7,8). Previous studies have investigated miRNA expression profiles in primary prostate cancer (PPC) or MPC and several miRNAs have been suggested as diagnostic markers for PC (9,10). However, the molecular mechanisms underlying the roles of miRNAs and their target differentially expressed genes (DEGs) in PC metastasis remain unclear.
Based on 218 prostate tumor samples (181 primaries and 37 metastases), Taylor et al (11) conducted an integrated analysis (including concordant assessment of DNA copy number, mRNA expression and focused exon resequencing), and revealed that nuclear receptor coactivator NCOA2 functions as an oncogene in ~11% of PC tumors, and FOXP, RYBP and SHQ1 serve as potential cooperative tumor suppressors in human PC. Using the same microarray datasets, the aim of the present study was to identify miRNAs and DEGs that are associated with the metastasis of PC cells by screening miRNAs and genes that are differentially expressed between MPC and PPC samples, with the objective to further understand the molecular mechanisms of MPC.
Materials and methods
Source of microarray data
The raw microarray datasets GSE21036 and GSE21034 were downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database. The miRNA expression dataset GSE21036 was collected from 141 patients with PC treated by radical prostatectomy, including 14 metastatic samples, 99 primary non-metastatic tumor samples and 28 normal adjacent benign prostate samples. The annotation platform was the Agilent-019118 Human miRNA Microarray 2.0 G4470B (miRNA ID version) (Agilent Technologies, Inc., Santa Clara, CA, USA). The GSE21034 transcript dataset was collected form 179 samples, including 29 normal samples, 131 primary samples and 19 metastatic samples. The annotation platform was the Affymetrix Human Exon 1.0 ST Array (Affymetrix, Inc., Santa Clara, CA, USA). As illustrated in Table I, 139 samples from datasets of GSE21036 and GSE21034 were overlapped and used for the following analyses.
Table I.
Microarray datasets used from the Gene Expression Omnibus database, and the proportions of metastatic, primary and normal samples in each dataset.
| Microarray dataset | Metastatic | Primary | Normal | Total |
|---|---|---|---|---|
| miRNA (GSE21036) | 14 | 99 | 28 | 141 |
| Transcript (GSE21034) | 19 | 131 | 29 | 179 |
| miRNA and transcript (common to GSE21036 and GSE21034) | 13 | 98 | 28 | 139 |
miRNA, microRNA.
Microarray data pre-processing
Using the AgiMicroRna Bioconductor library of R (http://bioconductor.org/help/search/index.html?q=AgiMicroRna), the GSE21036 raw data were subject to pre-processing as previously described (12), including removing the probes with a low detection rate (failure in >75% samples), background adjustment and quantile normalization by the Robust Multiarray Averaging (RMA) method (13). The GSE21034 gene transcript data had been pre-processed (by background adjustment and quantile normalization using the RMA method) prior to being downloaded, and were subsequently subjected to log2-transformation.
Differential expression analysis of miRNA and genes
The differential expression analyses of miRNAs and gene transcripts between patients with MPC and patients with PPC were performed using the Linear Models for Microarray and RNA-Seq Data 4 package of R (14). miRNAs or genes with |log2 fold change (FC)| >1 and a false discovery rate (FDR) value <0.05 were selected as differentially expressed miRNAs (DEMs) or DEGs. The FDR value was obtained by adjusting the raw P-values with the Benjamini and Hochberg method (15).
Construction of a DEM-DEG regulatory network
First, miRNA-gene pairs predicted by the miRanda method were downloaded from the miRNA.org database (16). Then, miRNA-gene interaction pairs of upregulated DEM to downregulated DEG, and downregulated DEM to upregulated DEG were screened to construct a DEM-DEG regulatory network. Cytoscape (version 3.2.0; http://www.cytoscape.org/release_notes_3_2_0.html) was used to visualize the resulting network (17).
Functional annotation of DEM-DEG modules
In the constructed DEM-DEG network, a miRNA and its target genes were defined as a module. For the DEGs in each module, functional annotation analysis was performed using the Database for Annotation, Visualization and Integrated Discovery online tool based on the Gene Ontology database (P<0.01) (18).
Results
Differentially expressed miRNA and genes between patients with MPC and PPC
Based on the analytical threshold, DEMs/DEGs between MPC and PPC were screened out. The numbers of DEMs/DEGs between patients with MPC and those with PPC are presented in Table II, in addition to the numbers of up- and downregulated DEMs/DEGs. The number of downregulated DEMs/DEGs was markedly greater compared with the number of upregulated DEM/DEGs identified between patients with MPC and PPC. DEGs with |log2 FC| >2 are presented in Table III.
Table II.
Overview of differentially expressed microRNAs and genes.
| Differential expression | Upregulated | Downregulated | Total |
|---|---|---|---|
| DEM | 25 | 48 | 73 |
| DEG | 22 | 191 | 213 |
DEM, differentially expressed miRNA; DEG, differentially expressed gene.
Table III.
Differentially expressed genes with |log2 FC| >2 between metastatic and primary prostate cancer samples.
| Gene transcript | log2 FC value | Adjusted P-value | Gene transcript | log2 FC value | Adjusted P-value |
|---|---|---|---|---|---|
| TAGLN | −2.4964 | 1.1987×10−31 | SORBS1 | −2.24347 | 1.4898×10−23 |
| SLC22A3 | −2.0225 | 5.9268×10−10 | SLC14A1 | −2.0417 | 4.5910×10−12 |
| SERPINA3 | −2.1640 | 1.0787×10−08 | PI15 | −2.5879 | 1.0048×10−13 |
| PGM5 | −2.1181 | 5.0371×10−23 | PDE5A | −2.1878 | 5.7525×10−19 |
| PCP4 | −2.3182 | 1.0747×10−24 | MYLK | −2.5515 | 1.9944×10−29 |
| MYH11 | −3.7948 | 1.5295×10−42 | MYBPC1 | −2.3552 | 1.8570×10−12 |
| MSMB | −3.1696 | 1.6542×10−15 | LTF | −2.5535 | 4.2645×10−07 |
| IGJ | −2.0615 | 2.2689×10−07 | PAM3B | −2.1011 | 5.5298×10−11 |
| DDP4 | −2.4582 | 1.0125×10−12 | CSRP1 | −2.0686 | 7.1619×10−22 |
| CNN1 | −2.7962 | 1.4001×10−35 | CHRDL1 | −2.2994 | 1.2660×10−22 |
| AZGP1 | −2.1882 | 2.8966×10−18 | ACTG2 | −3.2887 | 3.1041×10−33 |
DEM-DEG regulatory network
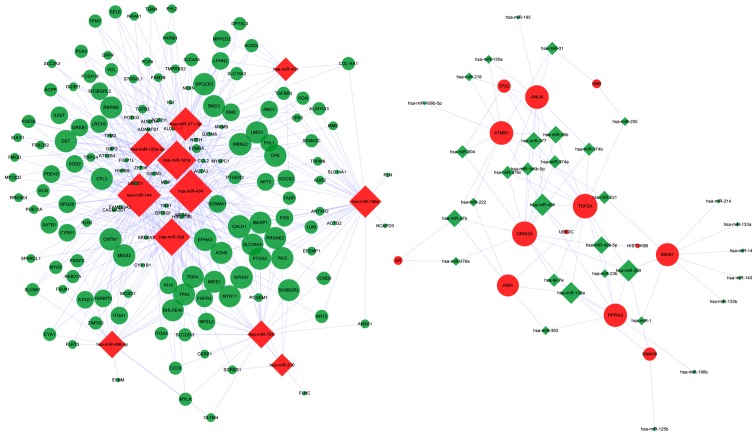
The resulting DEM-DEG regulatory network is illustrated in Fig. 1. The average connection degree of DEM was 12.4 (523/43), and that of DEG was 3.2 (523/166) (Table IV). Compared with that of the downregulated miRNAs, upregulated miRNAs had a higher degree of connection, that is, the latter regulate more downregulated DEGs, including miR-144, miR-494 and miR-181a. Certain DEGs were regulated by several miRNAs, including glutamate ionotropic receptor NMDA type subunit 3A (GRIN3A), topoisomerase II alpha (TOP2A) and caldesmon 1 (CALD1). GRIN3A and TOP2A were upregulated by 14 and 13 downregulated miRNAs, respectively, while CALD1 was downregulated by 8 upregulated DEMs.
Figure 1.
The DEM-DEG regulatory network. The rhombi represent upregulated DEMs; the circles represent DEGs. The green color indicates downregulation of a miRNA or gene; the red color indicates upregulation of a miRNA or gene. DEM, differentially expressed miRNA; DEG, differentially expressed gene; hsa, Homo sapiens; miR/miRNA, microRNA.
Table IV.
Nodes and regulation pair statistics of the DEM-DEG regulatory network.
| Nodes | ||||
|---|---|---|---|---|
| Regulation | DEM | DEG | Edges | Total |
| Up | 11 | 13 | DEM.up-DEG.down | 442 |
| Down | 32 | 153 | DEM.down-DEG.up | 81 |
DEM, differentially expressed miRNA; DEG, differentially expressed gene; up, upregulated; down, downregulated.
Functional annotation of DEM-DEG modules
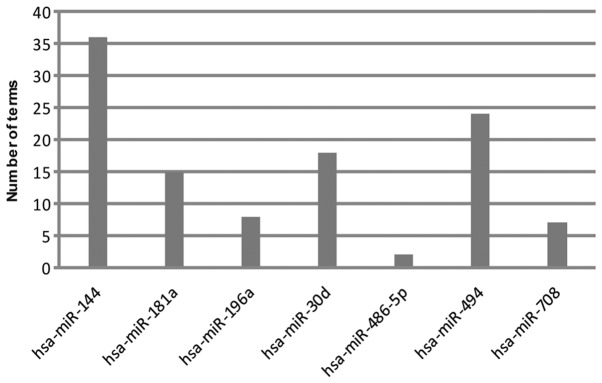
Seven modules were obtained (P<0.01), and their enriched biological processes (BPs) are presented in Fig. 2. DEGs regulated by miR-144 enriched the most BP terms (36), followed by miR-494 (24), miR-30d (18), miR-181a (15), miR-196a (8), miR-708 (7) and miR-486-5p (2). All these miRNAs were upregulated.
Figure 2.
The number of enriched biological process terms of each differentially expressed miRNA-differentially expressed gene module, as analyzed using the Database for Annotation, Visualization and Integrated Discovery online tool. hsa, Homo sapiens; miR, microRNA.
Discussion
In the present study, DEMs and DEGs between patients with PPC and MPC may function as biomarkers for the occurrence of MPC. Furthermore, miRNAs and genes (particularly those with large differential expression values) involved in the seven resulting DEM-DEG regulatory modules may serve significant roles in the metastasis of PC cells.
The target DEGs of miR-144 were enriched in the most BP terms, implying that this integrated analysis of multidimensional data miRNA and its targeted DEGs may perform important roles in the occurrence of MPC. miR-144 has been reported to improve the growth of HeLa cells (19), suggesting its role in tumor cell proliferation. Zhang et al (20) have proposed that miR-144 promotes the malignant progression of nasopharyngeal carcinoma cells by targeting the tumor-suppressor gene phosphatase and tensin homolog; however, its role in PC has not been reported thus far, to the best of our knowledge. In the present study, numerous DEGs were observed to be downregulated by miR-144. Among them, myosin heavy chain 11 (MYH11), solute carrier family 22 member 3 (SLC22A3) and dipeptidyl peptidase 4 (DPP4) exhibited large |log2 FC| values. MYH11 (also known as SMMHC) encodes the smooth muscle myosin heavy chain, which serves a key role in smooth muscle contraction (21). Its downregulation has been reported by Lin et al (22) in proliferating smooth muscle cells of human prostate tissue. SLC22A3 is one of three similar cation transporter genes located in a cluster on chromosome 6 and has previously been suggested to be associated with PC pathogenesis (23). DPP4 encodes a serine exopeptidase that has been implicated in cell-extracellular matrix interactions and bioactive peptide/cytokine/growth factor metabolism (24,25). DPP4 activity is elevated in PC and adjacent benign hyperplastic glands (26). Taken together, miR-144 may serve roles in the metastasis of PC cells by downregulating a number of target genes, including MYH11, SLC22A3 and DPP4.
The regulator miR-494 may also serve an important role in the metastasis of PC. MYH11 was the only target gene with |log2 FC| >2 among the downregulated DEGs of miR-494 in the present study. In a previous study, using multiple experimental methods, Shen et al (27) demonstrated that miRNA-494-3p could suppress the proliferation, invasion and migration of PC by downregulating C-X-C motif chemokine receptor 4 (CXCR4), which was overexpressed in PC. This is consistent with the upregulation of this miRNA observed in the present study, although differential expression of CXCR4 was not observed.
miR-30d downregulated a number of genes in the present study. Kobayashi et al (28) observed significantly higher expression levels of miR-30d in three PC cell lines compared with those in two normal prostate cell lines using miRNA microarray and quantitative polymerase chain reaction analysis (28). Furthermore, the authors suggested that miR-30d mediated its effects in PC by downregulating suppressor of cytokine signaling 1. In the present study, among the target DEGs of miR-30d, sorbin and SH3 domain containing 1 (SORBS1), phosphodiesterase-5 (PDE5) and myosin light chain kinase (MYLK) exhibited large |log2 FC| values. SORBS1 encodes a Cbl-associated protein involved in the formation of actin stress fibers and focal adhesions (29). The observed downregulation of SORBS1 in the present study was consistent with that reported by Vanaja et al (30) in PC tissues. PDE5 encodes an enzyme that hydrolyzes the 3′,5′-phosphodiester bond in the second messenger molecule cyclic guanosine monophosphate (GMP) to form 5′-GMP (31). Its expression has been observed in the smooth muscles of the prostate, and PDE5 inhibitors (such as sildenafil) are able to relax the prostate (32). Therefore, the upregulation of PDE5 is suggested to be disadvantageous for patients with PC. MYLK has been demonstrated to be necessary for the invasiveness of MPC cells (33). However, PDE5 and MYLK were observed to be downregulated in the present study; therefore, further studies are required to elucidate the roles of PDE5 and MYLK in PC metastasis.
In addition to their observed downregulation by miR-144 MYH11 and SLC22A3 were downregulated by miR-181a, which was upregulated in the present study. miR-181a has been reported in previous studies to induce apoptosis in a number of cancer types by downregulating the apoptosis regulator B-cell lymphoma 2 (11,34), and it has been observed to mediate bufalin-induced apoptosis in PC-3 PC cells by Zhai et al (35). However, the implications of upregulation of miR-181a in MPC samples remains unclear. Additionally, Su et al (36) reported that the downregulation of miR-30d and miR-181a in prostate tumors cooperatively suppresses the expression of glucose-regulated protein, 78 kDa (GRP78), a major endoplasmic reticulum chaperone and signaling regulator that is typically overexpressed in cancer (37). Differential expression of GRP78 was not observed in the present study; however, MYH11 was revealed to be another common target of miR-30d and miR-181a, indicating that miR-30d and miR-181a may cooperate to regulate the metastasis of PC cells.
A previous study suggested that miR-196a regulates homeobox (Hox) gene expression during vertebrate embryogenesis (38). In addition, a correlation between aberrant HoxC8 expression and a malignant phenotype in human PC has been reported (39,40). Therefore, it can be inferred that miR-196a may serve a role in the occurrence of MPC. In the present study, one of the target genes of miR-196a, actin, gamma 2 (ACTG2), exhibited |log2 FC| >2, in agreement with the significant downregulation in metastatic and primary tumor samples observed by Chandran et al (41). The findings from the present study further suggest that miR-196a may serve a role in the metastasis of PC cells by downregulating ACTG2.
Regarding miR-708 and miR-486-5p, no DEGs with a |log2 FC| >2 were observed among their target genes in the present study, implying that these two miRNAs do not perform crucial roles in the occurrence of MPC. According to Watahiki et al (10), miR-708 exhibited a >5-fold decrease in an MPC cell line following comparative analysis of miRNAs libraries between MPC and PPC cell lines. This was not consistent with the upregulation of miR-708 observed in the present study. Therefore, the expression changes and role of miR-708 in MPC require further investigation. Additionally, Watahiki et al (10) also reported that elevated miR-486 level enhanced the invasiveness of MPC cells, which is consistent with its upregulation in the present study.
In conclusion, the significantly upregulated miR-144, miR-494, miR-30d, miR-181a, miR-196a, miR-708 and miR-486-5p screened in the present study may participate in the metastasis of PC cells via the downregulation of their corresponding target DEGs, particularly those with large |log2 FC| values, including MYH11, SLC22A3, DPP4, SORBS1, PDE5, MYLK and ACTG2. The effects on these target DEGs require further experimental verification. A number of these DEMs or DEGs have been associated with the occurrence of PC; however, the molecular mechanisms underlying their roles in the occurrence of MPC remain unclear and require further investigation.
Acknowledgements
The present study was supported by a grant from the Jilin Provincial Health Department (grant no. 3D5157343428).
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Bonci D, Coppola V, Patrizii M, Addario A, Cannistraci A, Francescangeli F, Pecci R, Muto G, Collura D, Bedini R, et al. A microRNA code for prostate cancer metastasis. Oncogene. 2016;35:1180–1192. doi: 10.1038/onc.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guinney J, Wang T, Laajala TD, Winner KK, Bare JC, Neto EC, Khan SA, Peddinti G, Airola A, Pahikkala T, et al. Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: Development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet Oncol. 2017;18:132–142. doi: 10.1016/S1470-2045(16)30560-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 5.Fabris L, Ceder Y, Chinnaiyan AM, Jenster GW, Sorensen KD, Tomlins S, Visakorpi T, Calin GA. The potential of MicroRNAs as prostate cancer biomarkers. Eur Urol. 2016;70:312–322. doi: 10.1016/j.eururo.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endzeliņš E, Melne V, Kalniņa Z, Lietuvietis V, Riekstiņa U, Llorente A, Linē A. Diagnostic, prognostic and predictive value of cell-free miRNAs in prostate cancer: A systematic review. Mol Cancer. 2016;15:41. doi: 10.1186/s12943-016-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watahiki A, Wang Y, Morris J, Dennis K, O'Dwyer HM, Gleave M, Gout PW, Wang Y. MicroRNAs associated with metastatic prostate cancer. PLoS One. 2011;6:e24950. doi: 10.1371/journal.pone.0024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.López-Romero P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC Genomics. 2011;12:64. doi: 10.1186/1471-2164-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 14.Smyth GK. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; 2005. Limma: Linear models for microarray data; pp. 397–420. [DOI] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 1995;57:289–300. [Google Scholar]
- 16.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang LY, Lee Ho-Fun V, Wong AM, Kwong DL, Zhu YH, Dong SS, Kong KL, Chen J, Tsao SW, Guan XY, Fu L. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis. 2013;34:454–463. doi: 10.1093/carcin/bgs346. [DOI] [PubMed] [Google Scholar]
- 21.Lopezcamacho C, van Wijnen AJ, Lian JB, Stein JL, Stein GS. (CBFβ) and the leukemogenic fusion protein CBFβ-SMMHC associate with mitotic chromosomes to epigenetically regulate ribosomal gene. J Cell Biochem. 2014;115:2155–2164. doi: 10.1002/jcb.24892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin VK, Wang D, Lee I, Vasquez D, Fagelson JE, McConnell JD. Myosin heavy chain gene expression in normal and hyperplastic human prostate tissue. Prostate. 2000;44:193–203. doi: 10.1002/1097-0045(20000801)44:3<193::AID-PROS3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 23.Grisanzio C, Werner L, Takeda D, Awoyemi BC, Pomerantz MM, Yamada H, Sooriakumaran P, Robinson BD, Leung R, Schinzel AC, et al. Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proc Natl Acad Sci USA. 2012;109:11252–11257. doi: 10.1073/pnas.1200853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zilleßen P, Celner J, Kretschmann A, Pfeifer A, Racké K, Mayer P. Metabolic role of dipeptidyl peptidase 4 (DPP4) in primary human (pre)adipocytes. Sci Rep. 2016;6:23074. doi: 10.1038/srep23074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong J, Kankanala S, Rajagopalan S. Dipeptidyl peptidase-4 inhibition: Insights from the bench and recent clinical studies. Curr Opin Lipidol. 2016;27:484–492. doi: 10.1097/MOL.0000000000000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson MJ, Ruhland AR, Quast BJ, Reddy PK, Ewing SL, Sinha AA. Dipeptidylpeptidase IV activities are elevated in prostate cancers and adjacent benign hyperplastic glands. J Androl. 2000;21:220–226. [PubMed] [Google Scholar]
- 27.Shen PF, Chen XQ, Liao YC, Chen N, Zhou Q, Wei Q, Li X, Wang J, Zeng H. MicroRNA-494-3p targets CXCR4 to suppress the proliferation, invasion, and migration of prostate cancer. Prostate. 2014;74:756–767. doi: 10.1002/pros.22795. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi N, Uemura H, Nagahama K, Okudela K, Furuya M, Ino Y, Ito Y, Hirano H, Inayama Y, Aoki I, et al. Identification of miR-30d as a novel prognostic maker of prostate cancer. Oncotarget. 2012;3:1455–1471. doi: 10.18632/oncotarget.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribon V, Herrera R, Kay BK, Saltiel AR. A role for CAP, a novel, multifunctional Src homology 3 domain-containing protein in formation of actin stress fibers and focal adhesions. J Biol Chem. 1998;273:4073–4080. doi: 10.1074/jbc.273.7.4073. [DOI] [PubMed] [Google Scholar]
- 30.Vanaja DK, Ballman KV, Morlan BW, Cheville JC, Neumann RM, Lieber MM, Tindall DJ, Young CY. PDLIM4 repression by hypermethylation as a potential biomarker for prostate cancer. Clin Cancer Res. 2006;12:1128–1136. doi: 10.1158/1078-0432.CCR-05-2072. [DOI] [PubMed] [Google Scholar]
- 31.Stoclet JC, Keravis T, Komas N, Lugnier C. Section Review: Cardiovascular & Renal: Cyclic nucleotide phosphodiesterases as therapeutic targets in cardiovascular diseases. Expert Opinion Investigational Drugs. 2008;4:1995. [Google Scholar]
- 32.Uckert S, Küthe A, Jonas U, Stief CG. Characterization and functional relevance of cyclic nucleotide phosphodiesterase isoenzymes of the human prostate. J Urol. 2001;166:2484–2490. doi: 10.1016/S0022-5347(05)65621-2. [DOI] [PubMed] [Google Scholar]
- 33.Tohtong R, Phattarasakul K, Jiraviriyakul A, Sutthiphongchai T. Dependence of metastatic cancer cell invasion on MLCK-catalyzed phosphorylation of myosin regulatory light chain. Prostate Cancer Prostatic Dis. 2003;6:212–216. doi: 10.1038/sj.pcan.4500663. [DOI] [PubMed] [Google Scholar]
- 34.Zhu DX, Zhu W, Fang C, Fan L, Zou ZJ, Wang YH, Liu P, Hong M, Miao KR, Liu P, et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis. 2012;33:1294–1301. doi: 10.1093/carcin/bgs179. [DOI] [PubMed] [Google Scholar]
- 35.Zhai XF, Fang FF, Liu Q, Meng YB, Guo YY, Chen Z. MiR-181a contributes to bufalin-induced apoptosis in PC-3 prostate cancer cells. BMC Complement Altern Med. 2013;13:325. doi: 10.1186/1472-6882-13-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su SF, Chang YW, Andreu-Vieyra C, Fang JY, Yang Z, Han B, Lee AS, Liang G. miR-30d, miR-181a and miR-199a-5p cooperatively suppress the endoplasmic reticulum chaperone and signaling regulator GRP78 in cancer. Oncogene. 2013;32:4694–4701. doi: 10.1038/onc.2012.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen WT, Zhu G, Pfaffenbach K, Kanel G, Stiles B, Lee AS. GRP78 as a regulator of liver steatosis and cancer progression mediated by loss of the tumor suppressor PTEN. Oncogene. 2014;33:4997–5005. doi: 10.1038/onc.2013.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 39.Waltregny D, Alami Y, Clausse N, de Leval J, Castronovo V. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate. 2002;50:162–169. doi: 10.1002/pros.10045. [DOI] [PubMed] [Google Scholar]
- 40.Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, Schirripa O, Lucia MS, Nordeen SK. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res. 2003;63:5879–5888. [PubMed] [Google Scholar]
- 41.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]