Abstract
Liver disease represents a growing global health burden. The development of in vitro liver models which allow the study of disease and the prediction of metabolism and drug-induced liver injury in humans remains a challenge. The maintenance of functional primary hepatocytes cultures, the parenchymal cell of the liver, has historically been difficult with dedifferentiation and the consequent loss of hepatic function limiting utility. The desire for longer term functional liver cultures sparked the development of numerous systems, including collagen sandwiches, spheroids, micropatterned co-cultures and liver microphysiological systems. This review will focus on liver microphysiological systems, often referred to as liver-on-a-chip, and broaden to include platforms with interconnected microphysiological systems or multi-organ-chips. The interconnection of microphysiological systems presents the opportunity to explore system level effects, investigate organ cross talk, and address questions which were previously the preserve of animal experimentation. As a field, microphysiological systems have reached a level of maturity suitable for commercialization and consequent evaluation by a wider community of users, in academia and the pharmaceutical industry. Here scientific, operational, and organizational considerations relevant to the wider adoption of microphysiological systems will be discussed. Applications in which microphysiological systems might offer unique scientific insights or enable studies currently feasible only with animal models are described, and challenges which might be addressed to enable wider adoption of the technologies are highlighted. A path forward which envisions the development of microphysiological systems in partnerships between academia, vendors and industry, is proposed.
Impact statement
Microphysiological systems are in vitro models of human tissues and organs. These systems have advanced rapidly in recent years and are now being commercialized. To achieve wide adoption in the biological and pharmaceutical research communities, microphysiological systems must provide unique insights which translate to humans. This will be achieved by identifying key applications and making microphysiological systems intuitive to use.
Keywords: Microphysiological systems, in vitro models, liver, microfluidic, interconnected, tissue culture
Introduction
The continuing high attrition rate in the pharmaceutical industry has led to an on-going search for new technologies, which can be incorporated into pre-clinical drug development to de-risk compounds before they enter the clinic. In the broadest terms, the goal is to gain the most translationally relevant information, enabling an accurate prediction at the earliest possible stage, of the behavior of new medicines in humans. These new technologies will either replace, or more likely augment, the current range of in vitro cell culture and animal models available to assess drug safety and efficacy. One such technology is microphysiological systems (MPS) or in common parlance organ-on-a-chip. These are small-scale, in vitro cell cultures, which model facets of tissue or organ level function. The term organ-on-a-chip is somewhat misleading as the modeling of whole organ function is not possible with current systems, which are typically capable of recapitulating tissue level effects, hence the term microphysiological systems is preferred. MPS typically contain multiple cell types, co-cultured to recapitulate in some manner the histoarchitecture of the tissue, either through controlled positioning of cells in the device, and/or the use of three-dimensional scaffolds. To obtain a highly functional model and improve translational relevance, primary human cells are frequently used. In MPS a flow of culture medium seeks to ensure that cells are adequately supplied with nutrients and subjects cells to shear forces such as those experienced in vivo. This has led to MPS often being termed microfluidic cell culture systems, and indeed microfluidics is often a key component of MPS. Additional mechanical stimulation may also be imposed on the cells, to simulate normal body movements (e.g. breathing, gut peristalsis, blood pressure shear). MPS may be utilized as fluidically isolated single systems, or connected through fluidic circuits to model the function of interconnecting tissues.
Demonstrating translational relevance will be key to driving wider adoption of MPS. Do MPS more faithfully mimic human physiology? The design of experiments to answer this critical question is challenging, when the human biology is itself not fully understood. It is unrealistic to expect a MPS to fully recapitulate all functional aspects of a given organ or tissue, since all models are inherently a simplification of the system or process they seek to mimic. The aim is to capture the critical features of the system while in the interests of tractability neglecting less important components. The demonstration of translation relevance is best achieved by focusing on specific applications and showing the utility of MPS in the chosen application. Examples for the liver include: expression and maintenance of a specific disease-related target; prediction of drug-induced liver injury; maintenance of metabolic competence to enable drug metabolism studies; response of a diseased model to currently available therapies. In almost all applications, there will be an incumbent technology, or assay, already in use within the pharmaceutical industry which addresses the need to some degree. In drug metabolism, suspension and plated primary hepatocytes are routinely used; in safety testing, animal studies are mandated by the regulator; in target discovery and validation, genetically modified small animal models are widely employed. Against this backdrop of well-established technologies, albeit with certain limitations, MPS must demonstrate unique value in specific applications before end users will adopt this new technology.
The end users for MPS will be biological and pharmaceutical scientists working in academia and industry, whilst those developing the systems, usually in academia, often come from an engineering, physics or materials background. This meeting of disciplines creates the opportunity for innovation and radical steps forward, but also carries the risk that systems developed primarily by engineers will be alien to biologists, limiting utility and uptake. It is here that companies commercializing MPS technologies can bridge the gap and develop or productize academic prototypes, into systems which are user-friendly, affordable and compatible with current work flows.
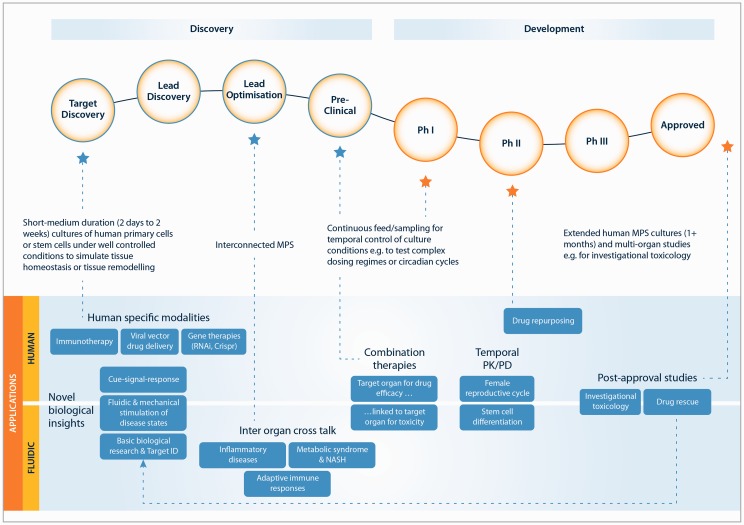
This review will consider the opportunities to increase the adoption of MPS in industry and academia, with a focus upon liver MPS and interconnected or multi-MPS. MPS applications in the drug discovery and development process, broadly stem from either: i) MPS being human in vitro models, or ii) new experiments enabled by fluidics (Figure 1). The current state of the art will be reviewed, and opportunities for MPS to address a range of existing and new applications will be explored. The challenges in bringing MPS to an end user community, well versed in cell culture, but with little experience of (micro)fluidics, are highlighted and potential solutions suggested. For the success of MPS, it is key they establish translational relevance in applications important to the pharmaceutical industry. The studies needed to establish translational relevance are complex and can likely only be successfully achieved through partnerships between academia, companies commercializing MPS and pharmaceutical scientists. The form of these partnerships is discussed and issues in their implementation are examined.
Figure 1.
Proposed uses of MPS in the pharmaceutical development pipeline
Modeling of the liver
The liver is a complex organ, involved in myriad processes essential to life, including protein production, drug metabolism, glucose storage, and is frequently a target organ for drug toxicity. The basic structural unit of the liver is the hepatic lobule, in which an array of hepatic sinusoids carries blood from the portal triad to the central vein. The sinusoid is composed of plates of hepatocytes, the parenchymal cell of the liver, lined with liver sinusoidal endothelial cells. Resident within the sinusoid are non-parenchymal cells, namely Kupffer cells, the liver resident macrophage, and hepatic stellate cells, which are implicated in extracellular matrix production and tissue remodeling. Hepatocytes form tight junctions separating the blood side of the sinusoid from the bile duct, which is lined with cholangiocytes. Hepatocytes are highly metabolically active cells and consequently an oxygen gradient exists down the liver sinusoid. Comprehensive reviews of the structure and anatomy of the liver are available elsewhere.1,2
The culture of primary hepatocytes has long been a target for the application of innovative cell culture systems, owing to: (i) a broad interest in these cells for drug safety and metabolism studies; (ii) a rapid loss of viability in suspension cultures; and (iii) the difficulty in maintaining differentiated state in adherent monolayer cultures. Approaches including optimization of medium conditions,3 sandwich culture,4,5 micro-patterned co-cultures,6 scaffold-supported and unsupported three dimensional (3D) culture,7,8 and MPS9,10 have all been demonstrated to improve hepatocyte viability and function in the long-term culture. Liver MPS have been demonstrated to mimic the histoarchitecture of the liver sinusoid,11,12 supply the metabolic demands of hepatocytes through the application of flow,13 and are readily incorporated into interconnected MPS. These features differentiate liver MPS from other in vitro liver culture systems, but inevitably bring with them both complexity and cost. Thus, for liver MPS to be widely adopted, the advantages must be thoroughly demonstrated, translational relevance to humans established, and costs be acceptable to the end user.
The field of in vitro liver cell culture, particularly with emphasis on toxicology, has been well reviewed elsewhere.1,2,14–16 This review will focus on technologies which are nearing, or have reached commercial maturity. Interconnected MPS, which feature as one component a liver MPS will be addressed in a later section. For simplicity, advanced in vitro liver systems can be categorized as innovative two-dimensional (2D) culture, static 3D cultures and MPS. Of the innovative 2D systems, the micro-patterned co-cultures of hepatocytes and 3T3 fibroblasts developed by Prof. S. Bhatia6 and commercialized by Ascendance (formerly Hepregen), and the Hurel random co-cultures,17 stand out as rare examples of advanced systems which have been published on by the pharmaceutical industry.18–22 Innovative 2D systems have the advantage of being in a format which is already familiar to biologists and can be easily integrated into current workflows. Workflow compatibility is also typically afforded by static 3D cultures, which are often deployed in a multi-well plate format and can either be formed through spontaneous aggregation of cells as spheroids, or organoids as in the InSphero system,7,23 or through the use of a scaffold.24,25 The use of a scaffold presents the opportunity to further control the cellular microenvironment by modulating the mechanical properties of the surfaces in contact with the cell.
The number of companies offering liver MPS, which combine some mimicry of histoarchitecture in combination with flow, has grown over the past decade, in part reflecting the increased research funding directed towards this area. CN Bio Innovations offer the mesofluidic LiverChip® system, developed by the group of Prof. L. Griffith, in which the liver vasculature capillary bed is approximated by an array of perfused liver microtissues.13,26,27 Hepatocytes and optionally non-parenchymal cells are seeded into an array of micro-channels, in which they form annular 3D structures. Cell culture medium is perfused through the scaffold, and passes through the portion of the microchannel not occluded by cells. Thus, each cell containing micro-channel can be considered a simple mimic of the liver sinusoid. Smaller scale, more microfluidic liver MPS are also available.28,29 Publications from pharmaceutical scientists on the use of Liver MPS are comparatively rare and where they do exist have typically been focused on drug metabolism.30,31
There are a variety of liver MPS and other advanced liver culture systems on the market and it is clear that many are being actively evaluated by academic and pharmaceutical scientists. However, there clearly remain issues to be addressed on the path to a more faithful in vitro recapitulation of the human liver. As MPS vendors, the authors frequently interact with academic researchers and scientists from pharmaceutical companies both large and small. The issues detailed below often feature in discussions with this community of potential end users.
Hepatic scaling
A typical culture of 375,000 primary human hepatocytes, in a single well of a 24-well plate, will be cultured in 500 µL of cell culture medium, which is changed every 1–2 days. This corresponds to 1.3 µL of medium, per 1000 hepatocytes, per day. Making a simplistic comparison, the ratio for man is circa 0.008 µL of blood, per 1000 hepatocytes, per day; assuming 5 L of blood, 50% cleaned each day, and 300 billion hepatocytes in the liver. The use of dilute systems with high medium to cell ratios has likely evolved for several reasons: (i) convenience; (ii) to ensure adequate nutrient supply and buffering capacity; and (iii) to guard against evaporation from multi-well plates placed in cell culture incubators. The frequent complete changing of medium in well plate and flask culture poorly mimics the in vivo situation, as any autocrine and paracrine factors which have accumulated are entirely removed from the system. Intuitively more appropriately scaled systems, in which the levels of extracellular factors are closer to in vivo, might be expected to be more predictive of the in vivo response. MPS systems are well placed to address the scaling issue, as microfluidic channels permit high surface area to volume ratios, giving large areas for cell attachment with constrained volumes.32
Sensing
Moving to a less dilute system, with lower medium to cell ratios, is appealing as concentrations of secreted autocrine and paracrine factors increase. This may be achieved by decreasing medium volume, increasing cell number or a combination of both. If the chosen route is decreased medium volumes, this has practical implications for measurement and sensing. A single ELISA or proteomics measurement by LC/MS will typically require tens to hundreds of microlitres of medium, which might consume all the available media from a single MPS or medium change. Thus, to achieve the maximum value from scaled MPS, online analysis tools or sampling and analysis, which can operate on micro and nanolitre volumes, need to be developed and integrated with platforms. Integration of online sensors into MPS allows continuous temporal measurement of one or more parameters. The recent review by Marx et al. gives an excellent overview of the area.33 Turning specifically to the liver, measures of metabolic function are of interest and sensors integrated with liver MPS have been demonstrated.2,34 When integrating online sensors with MPS the additional complexity associated with sensor connections, sensor stability, calibration and any risks of contamination need to be considered. An alternative approach to integrating sensors is to enable the precise withdrawal of micro or nanolitre samples for analysis, ideally with minimum disruption to the cultures. These samples might be withdrawn and stored for later analysis,35 or fed directly to the analyzer.36,37 Dependent on the number and volume of samples taken, automated methods of replacing the volume of medium lost from the system may be required. In combination with the above approaches, imaging will remain an important and widely used tool for analyzing MPS cultures. MPS are often specifically designed to enable online imaging38 as positioning the microscope objective close enough to the cells whilst still allowing for flow channels, pumps and associated tubes can be challenging.
Zonation
Oxygen consumption by hepatocytes leads to a decrease in oxygen concentration from ∼100 µM to 50 µM in the blood, as it flows down the liver sinusoid from the portal triad to the central vein.13 For comparison, the saturation concentration of oxygen in cell culture medium at 37℃ is 201 µM. The sinusoid is considered to contain three zones, and it is well established that hepatocytes in different zones are functionally distinct owing to the variation in oxygen tensions.39 For example, necrotic cell death associated with acetaminophen toxicity is typically localized in Zone 3 where expression of cytochrome P450s is high.40 The ability to control oxygen tension and model zonation in vitro has been explored using a variety of systems including liver MPS.13,40,41 A liver MPS model which captured the differing hepatic function of the three zones of the liver, and enabled the response of each to be interrogated individually would likely be of interest, particularly for toxicology and drug metabolism applications. In the interests of practicality, it would be preferable if oxygen tensions in the medium could be modulated without the need to resort to hypoxic incubators or sparging of gas. It also remains to be understood what, if any role, the zone from which hepatocytes were originally isolated plays in their functioning in vitro, or their ability to become zonated.
Complex co-cultures
The culture of primary hepatocytes in all formats has advanced significantly in the past decade and this has led to the desire to increase the complexity of the model, through the incorporation of liver non-parenchymal cells. Liver MPS are well positioned to enable the formation of co-cultures of hepatocytes and non-parenchymal cells, in which cells are added in controlled ratios, in precisely defined locations within the system. Given spatial separation of cells, for example in channels on either side of a porous membrane, the potential exists at the end of the experiment to recover and analyze individual populations separately. These features should be a commercially attractive, as they will enable end users to tailor a liver model to answer their specific biological questions rather than simply aiming to develop a model which recapitulates all possible functions of the liver.
Cultures of primary hepatocytes with Kupffer cells, stellate cells, endothelial cells and mixed fractions of non-parenchymal cells, have been reported in both MPS11,17,42 and other advanced liver culture systems.10,43,44 The source of the non-parenchymal cells must be carefully considered as this is likely to have a significant bearing on the biological authenticity of the model. Primary human non-parenchymal cells can be isolated from fresh tissue or obtained cryopreserved from commercial vendors, but a donor match between hepatocytes and non-parenchymal cells can rarely be achieved. In many instances, primary hepatocytes are combined with cell lines such as EA.hy296 or HUVEC (endothelial cell lines) or LX-2, the stellate cell line12,45 as these cells are more readily available then primary non-parenchymal cells. Whether combining cells from different donors or combining cell lines, the biological and particularly immunological implications need to be considered.
The addition of non-parenchymal cells to a liver model provides additional biological functionality, which is of use in application areas including drug metabolism31,44 toxicology9,24,26 and disease modeling.43 In drug-induced liver injury, a role for underlying inflammation has been proposed.46 If primary hepatocytes are co-cultured in a liver MPS with primary Kupffer cells, the liver resident macrophage, the cell death induced by toxicants with known immune liabilities such as trovofloxacin47 is enhanced if lipopolysaccharide the model TLR4 agonist is added.7 The addition of non-parenchymal cells holds promise in the development of disease models in which multiple cell types are implicated in the progression of the disease. Diseases such as non-alcoholic steatohepatitis (NASH), the more severe manifestation of simple hepatic steatosis,48 will surely require models comprised of at least hepatocytes and stellate cells.49 It should be noted that not all disease models require complex co-culture. For example, the study of monogenetic liver diseases such as alpha-1-antitrypsin disorder can likely be very satisfactorily addressed with monocultures.50
Bile ducts
Polarization, tight junction, and bile canaliculi formation are frequently demonstrated using in vitro liver models,6 and are considered important for achieving uptake and efflux transporter function. Although bile canaliculi can be formed in vitro, no existing model has yet demonstrated a network of canaliculi leading to a bile duct. In monolayer cultures of primary hepatocytes, a network of canaliculi is formed in the plane of the hepatocytes with bile acids released into the cell culture medium.51 Devices that enable the contents of the canalicular network, namely bile acids to be collected, independent from the culture medium would more closely mimic in vivo physiology, be analytically useful, and offer opportunities for the modeling of enterohepatic circulation. Liver MPS incorporating multiple micron sized channels wherein the placing and spatial orientation of cells can be controlled, might offer the opportunity to produce a model which has bile duct-like features. Optimally these duct-like features would be lined with cholangiocytes.
Cell culture medium
The cell culture medium typically used for the culture of primary human hepatocytes is complex, typically having up to 100 components. The necessity for many of these components is difficult to establish from the literature and indeed if co-culture of multiple liver cell types is to be successful, as is typical in liver MPS, the effects of individual components on all the cell types present must be understood. For example, dexamethasone, the potent synthetic glucocorticoid, is often added to culture medium to maintain the differentiated state of primary hepatocytes. Glucocorticoids are anti-inflammatory and indeed if hepatocyte and Kupffer cell co-cultures are attempted in the presence of dexamethasone, at nanomolar concentrations the response of the Kupffer cells to inflammatory stimuli is significantly attenuated.42 A full review of medium components is beyond the scope of this work, suffice to note that in building an in vitro liver model, be it microphysiological or otherwise, if medium components are added at supra-physiological concentrations, it is important to consider the effects this may have on cell function and signaling. For example, Winnike et al. determined from metabolomics analysis that despite primary human hepatocytes being cultured in 25 mM glucose and 688 nM insulin, they appeared to be in a state of extreme starvation.52 This work points to the need for hepatocyte culture medium, which promotes a more physiologically relevant metabolic state.
Fetal calf serum is frequently a component of cell culture medium. Serum is a chemically undefined, non-human component and hence should be removed from human liver MPS, if suitable functional performance of the cells can be maintained. Chemically defined medium, which is serum free but will still contain some amount of protein, typically bovine serum albumin at circa 0.1 mg/mL is preferred. Extended culture of primary human hepatocyte monocultures,1 hepatocyte/Kupffer cell co-cultures,31,42,53 hepatocyte/Kupffer/stellate triple cultures (unpublished data), and hepatocyte/mixed non-parenchymal cell fraction,11 can be achieved in chemically defined, serum-free medium. The ability to sustain cultures for more than a few days in the complete absence of protein, other than that produced by the cells, remains an open question. The concentrations of protein in defined medium are far below those in blood where total protein and serum albumin are circa 70 mg/mL and 50 mg/mL, respectively. The concentration of protein in liver MPS medium will also influence pharmacokinetics, as it will lead to lower free drug concentrations, as most drugs exhibit at least some protein binding. As a minimum, protein/serum concentrations and the free, as opposed to total, drug concentrations in any experiment should be considered, particularly when comparing between culture systems, as this can radically effect the interpretation of metabolism and toxicology data. For liver MPS systems, the presence of serum or protein may lead to pumping issues particularly if concentrations are high and the pump action is harsh, resulting in protein denaturation and aggregation.54
Interconnected MPS
Following the early work of Prof. M. Schuler,55,56 the interest in interconnected MPS or multi-MPS has increased in recent years, and has been catalyzed by significant funding from the Defence Advanced Research Projects Agency (DARPA), the National Institutes of Health, and the European Union. Interconnect MPS are being developed by a rapidly expanding number of vendors, including TissUse, Hurel, Hesperos, Emulate, Nortis, Kirkstall, and CN Bio Innovations. Most of the interconnected MPS are small-scale microfluidic devices. These systems typically incorporate a liver MPS, owing to the central importance of the liver in drug metabolism and toxicity, connected to a number of other organ MPS. Access to these technologies is achieved through purchase of products, contract research services or research alliances, dependent on the vendor. This section will focus on commercially available interconnected MPS; however, given the limited published data from vendors, examples from academia will also be included.
The choice of which MPS to interconnect must be driven by the intended application. To make the platform or assay fit for purpose, at a minimum it must feature the organ or tissue mimics required to recapitulate the relevant biological processes. When considering which and how many MPS to interconnect, a balance must be struck between capturing the required biology, preserving usability, and limiting cost. The field of interconnected MPS is still in its infancy, but as the field advances, and new insights are gained into human physiology and disease, the fit for purpose question will need to be continuously revisited.
There already exist examples of interconnected MPS targeted to specific applications (Figure 1). For toxicology, interconnected MPS featuring common target organs57 such as liver, heart, and kidney have been developed,55,58,59 whilst for cosmetics testing, the linking of skin and liver has been demonstrated.60 In drug metabolism studies, modeling of first pass metabolism with an interconnected gut and liver has been exemplified in a number of different MPS configurations.60–62 In efficacy testing, the conversion of prodrug to its active form, and the subsequent study of the effects on a target tissue has been achieved through the interconnection of a liver MPS with a cancer tumor model.32,63 Looking to the future, the study of the diseases such as fatty liver disease and diabetes would be enabled by a liver, pancreas, adipose tissue and muscle interconnected MPS.
A key area in which interconnected MPS may yield new biological insights, is the role of inflammation and immune response. For example, low level chronic inflammation of the liver driven by bacterial components leaking from the gut has been implicated in increased susceptibility to drug-induced liver injury,47 whilst massive leakage drives acute liver failure. Interconnected MPS should be well suited to studies in this area with the ability to control the cell types and inflammatory stimuli, combined with time course sampling. The inclusion of an adaptive immune component as part of an interconnected MPS would expand potential applications into areas such as immune responses to viral infection, and the study of immuno-modulatory therapies. Given the highly species specific nature of immune responses, an all human interconnected MPS with immune components should be a valuable tool able to replace animal models.
The science of interconnected MPS is advancing rapidly; however, there remain many unanswered questions in the field. Many are conceptually similar to those faced in the development of liver MPS, with the obvious caveat that interconnection of MPS increases complexity and hence the magnitude of any issue. Sensing is highly relevant to interconnected MPS, but the area will not be revisited as the issues are similar to those discussed in the previous section.
Interconnected MPS scaling
The appropriate scaling of interconnected MPS remains perhaps the largest challenge in the field. As discussed, the scaling of a single MPS or organ can be challenging and this is multiplied when systems are interconnected. The key questions become on what basis should both the individual MPS and the inter-MPS interactions be scaled, and how does one account for any tissues or organs which are not present? This area has recently been extensively reviewed, with residence time, allometric and functional scaling all highlighted.64,65 As noted by Wikswo et al., matching the enormous efficiency of the human circulatory system presents a significant challenge.64 A 70 kg man has approximately 5 L of blood. Scaling this using any methodology to typical in vitro cultures, leads to the need for the whole system to contain only microliters of culture medium.
This presents practical challenges which may be at least in part, overcome through careful design of the individual MPS and the means by which fluid is transferred between the MPS. If one is constrained to working with total system volumes in the microliter range, then volumes in fluidic paths, connectors linking MPS, and dead volumes within the MPS, must be aggressively controlled. One potential approach is to develop compact systems, with on-board pumps and short interconnections, where currently individual MPS and pumps are often separate units linked with lengths of tube. Aside from scaling, the elimination of tubing offers benefits as it removes a potential source of non-specific drug binding and bubble formation. An alternative approach to scaling systems, which eliminates tubing, is to use robotic handling to transfer aliquots of medium between MPS. This approach offers great flexibility in fluidic routing, but the transfer of fluids between MPS cannot be continuous. Whichever solution is selected, transfer rates of fluid and fluid mixing will need to be carefully considered to ensure that the fluid dynamics of system do not product artifactual results.
The issue of device scaling should not be solely considered an engineering design challenge, as mathematical modeling also has a role to play. In the likely event that interconnected MPS cannot be developed, which are perfectly scaled in every facet, provided a robust physiologically based pharmacokinetic (PBPK) or systems biology type model can be created, then experimental results may still be usefully interpreted. A separate, but related topic is the scaling of in vitro results generated in single and interconnected MPS to the in vivo situation. This will be of critical importance in determining the translational relevance of the systems. Existing techniques may be applied,17,66 but likely new methodologies will be required, and synergistic development of these methodologies will occur together with MPS.
Availability of multiple cell types
The need to have several different cell types available for each interconnected MPS, and to have the individual MPS available for experimentation at the same time, poses a considerable logistical challenge. The individual MPS must either be produced locally, within the research facility, or purchased from vendors in ready to use form. The local production of MPS will require biological expertise, specific to each organ or tissue, with this requirement being more stringent if primary cells are used. This suggests that the deployment of higher order interconnected systems will be limited to larger research facilities.
To lessen the issue of cell and MPS availability, much of the early work in the field has been performed with MPS composed of carcinoma cell lines, which can be readily sub-cultured and are consequently always available. If primary cells are to be used in the interconnected MPS, the logistical requirements point to the need for either cryopreserved cells or the purchasing of ready to use MPS from a vendor. The ability to culture some or all the MPS in isolation for a period before combining to form an interconnect MPS will doubtless also be required.
Quality of individual organs
MPS systems like animals and monolayer cell cultures are simply models; approximations of a given tissue, organ, disease or system. Individual MPS will have different levels of biological authenticity. This needs to be considered when MPS are combined to form an interconnected system. For example, if one highly functional MPS, composed of primary cells, was connected to other MPS composed of carcinoma cell lines of limited functionality, how would this effect the overall functionality of the system? Using the liver to exemplify this point, Kratochwil et al. compared cell lines, stem cell-derived hepatocytes, and primary hepatocyte cultures for drug metabolizing capacity, and found the metabolic capacity of primary hepatocytes in a number of formats to be 10-fold higher than HepG2 cells.22 If primary cells are used, donor-to-donor variability should also be considered.66 Where possible, the level of functionality between interconnected MPS should be balanced, but if this is not possible the differences should at least be understood and due account taken of these when interpreting experimental results.
Universal or common cell culture medium
The purpose of interconnected MPS is to enable cross talk between MPS, thereby uncovering system level biological events only previously able to be studied in animal models. This cross talk may be through either soluble factors, such as cytokines, or the migration of cells between tissues. In either case, a path for communication between MPS must be established. Intuitively, the solution is a single common culture medium which bathes all the interconnected MPS. However, it is likely that each MPS will have been developed with its own bespoke medium, optimized through an iterative process to best maintain the viability and function of a specific cell type. In selecting a common medium for several interconnected MPS, the components within each medium should be analyzed, commonality sort, and components liable to have detrimental effects on other cells or tissues identified. A balance must be struck by substituting components, or modifying concentrations, to find a common medium which keeps all MPS functional, but may not offer optimal function for each individual MPS. The challenge of finding a common medium is increased if primary cells are used, as the medium used for their culture tends to be more diverse than that for carcinoma cell lines.
There are alternatives to using a single common medium, to which all MPS are exposed for the duration of the culture. If the rate of transfer of medium between MPS can be controlled, then a scenario can be envisaged in which each MPS starts the culture in its own bespoke medium and then these mediums are mixed at a controlled rate. This can be a useful strategy, particularly for early experiments, or where the biology of the MPS may still be under development, as interconnection can be achieved without having a fully developed common medium. The disadvantage of the strategy is that the rate of interaction of the MPS is governed by medium considerations, rather than physiological relevance. An alternative approach is to endothelialize MPS, or include porous channels within the MPS, to create effectively two systems separated by an endothelial barrier. It becomes possible to have one culture medium on the lumen side of the endothelialize channels, analogous to blood, and a second for the tissue side of each MPS. Assuming the endothelial barrier maintains integrity, the media are effectively separated and can thus be optimized accordingly. This is an elegant approach being pursued by several groups; however, the endothelialization of channels is not without issues, and consideration also needs to be given to mass transfer across the endothelial barrier.
Current and future applications for MPS
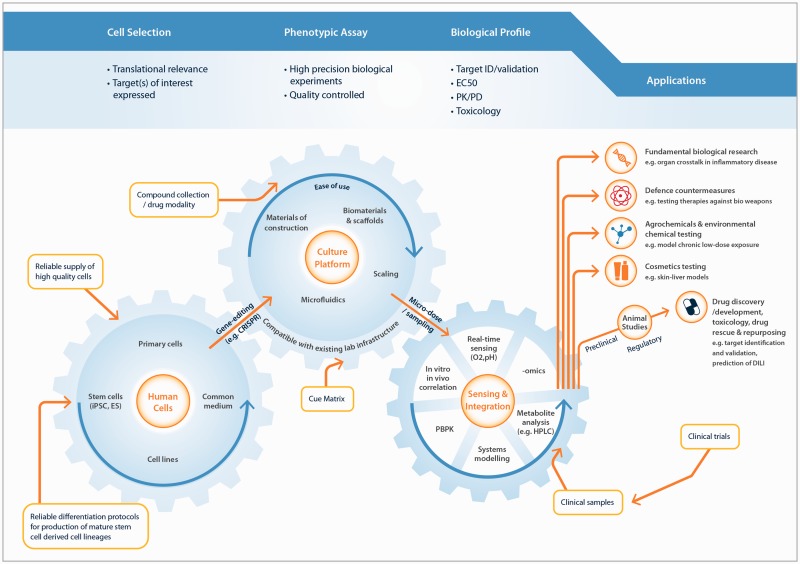
Given that MPS are currently in development for most tissues in the body, and that the interconnection of these systems represents a further level of sophistication, a wide field of possible applications is available (Figure 2). When considering the value an MPS can offer, the biological or translational relevance of the data produced is likely to be paramount. Does the MPS, or interconnected MPS, provide a more faithful recapitulation of the in vivo biology? Does it generate a result which more readily translates to the in vivo, or a unique result which cannot be produced by other models or means? The biological problems typically studied with MPS are complex and multi-faceted; for example, drug-induced liver injury, which requires multiple liver cell types to be present; or diseases with an inflammatory component, which require the trafficking of signalling molecules between organs (Figure 2). These problems are unlikely to be adequately understood through the measurement of a single end-point assay and favor comprehensive characterization of the system, through temporal sampling and -omics analysis. To establish the translational relevance of a MPS, these large data sets must be modeled and rationalized. Here, approaches, including PBPK models, quantitative systems pharmacology, and systems biology, will all be of utility for specific applications. Academic researchers in the MPS field are already pursuing this linkage of MPS with quantitative modeling,67 and in the authors’ opinion, this represents a rational approach to discovering applications in which MPS add significant value in the drug discovery process.
Figure 2.
Roadmap for MPS development
The commercialization of MPS or interconnected MPS, and their transition from a tool used by the laboratory which developed them, to products suitable for use by a wider community, is a challenging and costly step. It is conceivable that products may achieve a limited degree of success in academic, or research settings, by proving a generally improved model of human physiology. To achieve widespread uptake in the pharmaceutical industry, it is likely that specific applications in which these systems offer unique information, will need to be discovered. The success of the search for these applications of value, or so called “killer applications”, will determine the commercial success of MPS and the companies producing them.
Liver MPS applications can be broadly categorized as toxicology, drug metabolism and disease modeling. The search for applications began in toxicology and drug metabolism, but this has yet to bear significant commercial fruit. The need for more predictive models of drug-induced liver injury, or improved means to measure the metabolism of low clearance compounds, is well recognized, but the authors believe that early applications of value will be found in disease modeling. Here, MPS will be deployed for target identification and validation studies or as phenotypic screens, albeit that the size or throughput of these screens might be limited to at most a few thousand compounds. Application areas may include diseases currently poorly served by animal models, such as viral hepatitis or modeling of diseases implicating multiple cells types such as fatty liver disease, liver fibrosis, and metastatic liver cancer. The opportunity to explore new modalities, such as RNAi and viral delivery, also represents an interesting application for liver MPS. The development paradigm and key data sets required when advancing new modalities to the clinic are less well defined than for small molecules, and this presents an opportunity for new technologies, such as liver MPS to become an established standard. An often-posed question regarding liver MPS is: could a similar result be achieved or application addressed with 2D, innovative 2D, or static 3D cultures? In the authors’ experience, definitively showing a given result can only be produced using a liver MPS is challenging. As applications are developed, the community should ensure that the uniqueness and value of liver MPS over other systems, is clearly demonstrated in robust and fair comparisons.
The field of potential “killer applications” for interconnected MPS is wide, but currently poorly defined. In the near term, systems with 2, 3 or 4 MPS will provide a bridge to move interconnected systems from academic to industrial laboratories. Early applications of value may include: (i) modeling of diseases with an inflammatory component, (ii) assays that allow simultaneous assessment of toxicity and efficacy, and (iii) healthy and diseased MPS interacting with an adaptive immune model (Figure 2). The interconnection of seven, 10, or more MPS has potential applications for the study of bio-distribution, an experiment which is at present performed in animals. These platforms may also be used in the study of disease and development of treatments for which clinical trials are difficult to perform in a timely fashion, e.g. viral disease with sporadic outbreaks, or unethical e.g. bioweapons. These higher order systems are under development primarily in academia, through research consortiums, and published literature is at present rare.68
Given the diversity of human biology, it is likely that MPS will find multiple areas or applications in which they can contribute unique information. This speaks to a commercial landscape with multiple vendors each offering MPS, which address a specific set of applications. It is unlikely that a single vendor with a single technological solution will be able to address all applications of interest. Given the mix of skills required, in the search for unique applications for interconnected MPS, forming partnerships and working collaboratively will be important. The MPS community is one born out of partnerships, in the first instance biologist, (bio)engineers, and physicists coming together to design and develop MPS. The question becomes how best to forge the next generation of partnerships which move MPS from academia into wide industrial adoption.
It is common to see pharmaceutical companies engaging in collaborative research with academic laboratories. Engagement with MPS vendors more typically occurs through technology evaluation programs. The evaluation program will typically compare the performance of different MPS, against each other, and an already established in-house assay. Whilst the rationale for evaluation programs is clear, when viewed from the perspective of the customer, namely a pharmaceutical company, it is the authors’ experience that they are of limited value in discovering new or unique applications for technologies, such as MPS. Many focus on determining if MPS #1 is better than MPS #2 or the existing assay, where precisely what “better” means is difficult to define. The authors’ experience of working with pharmaceutical companies is that those who take the time to engage collaboratively with MPS vendors and seek to answer specific and well-defined questions in their evaluation, have an improved chance of a successful outcome. MPS are currently promising, but relatively immature. They will benefit from being developed collaboratively between MPS vendors and pharmaceutical scientists, to achieve systems which are usable, affordable, and offer unique insights. If one accepts that development through partnerships offers the highest chance of success then MPS vendors, pharmaceutical companies and government funders need to determine how the costs and benefits of such partnerships are divided.
Challenges limiting wider adoption
MPS may enable superior modeling of human biology and responses to endogenous and exogenous agents. However, adoption of MPS will only accelerate if the additional value gained from the new systems is sufficient to compensate users for the costs associated with purchasing and embedding the new technology within their organization. These costs may come in many forms including: reconfiguration of laboratories, capital expenditure on equipment, changes to workflow, retraining of personnel and payments to access new CRO services. In these regards, there remains a considerable gap between user expectations and what is currently delivered by MPS. This can, in part, be attributed to MPS being in the early stages of product development and commercialization, meaning that innovations and efficiencies that will ultimately lead to best-in-class products and lower cost of goods have not yet been achieved. However, user expectations are also shaped by contextual factors, such as personal knowledge, technical, organizational and policy factors, and in the case of MPS, these factors currently favor incumbent technologies.
The pharmaceutical industry possesses enormous infrastructure, both physical and intellectual, dedicated to 2D monolayer cell culture and animal experimentation. These have been their primary tools for half a century or more. Whilst it is hoped MPS can be a disruptive technology, to achieve initial uptake, MPS must find value integrating with existing well-established infrastructure, technical expertise, and workflows.
Materials of construction
A key challenge for the field is how to achieve quantifiable results when many devices include elastomers such polydimethylsiloxane (PDMS), which are known to absorb small molecules.69,70 The high surface-area-to-volume ratio of micro-channels71 and the increased development of highly lipophilic drugs, exacerbate this issue. Another concern with elastomeric materials, and PDMS in particular, is the leaching of uncured monomers from the bulk material to the cell culture fluid.70 Given that elastomeric materials are pervasive in microfluidics, either as materials of construction for whole devices, seals, tubes or pump components, it is unlikely they can be completely eliminated. Three approaches to this problem can be taken: (i) reduce the quantity of elastomer contacting drug containing medium; (ii) reduce the tendency of common elastomers like PDMS to absorb through chemical or surface modification72; (iii) select elastomeric materials with inherently lower sorption.69,73 Finding practical solutions to the problem of drug sorption, and incorporating these into commercially available devices will be key to offering end users precision in vitro models.
Infrastructure
There is a world of equipment dedicated to the set-up, manipulation, imaging and analysis of loose lid multi-well plates and flasks. The adoption of MPS will undoubtedly be more rapid if methodologies can be found that allow MPS to interface with these already existing technologies, thereby removing the need to additionally purchase ancillary equipment to support any new MPS introduced to the laboratory. However, care should be taken to ensure that changes to MPS to facilitate alignment with current infrastructure do not compromise the function and quality of the biology of the MPS.
Technical expertise
The end users for commercial MPS will be biologists, toxicologists and pharmacologists, whilst those primarily engaged in their development are engineers and physicists. The technical skills and working practices of these groups are different. The challenge comes in presenting MPS to the end user in an integrated format, or package. This will enable the user to focus on their research goals, rather than having to learn new techniques and skills to operate a device which is unfamiliar or counter-intuitive. For example, biologists typically use open-well plates or flasks in which cells can readily by accessed and dissociate from the surface for analysis, whilst many MPS house cells in closed micro-channels, where access to the cells is restricted. Solutions are required that maintain the advantages of culture in micro-channels, whilst giving biologists the access to cells/tissues enabling them to perform a full range of analytical techniques.
The need for movement of fluid in MPS, particularly interconnected MPS, makes it inevitable MPS will be more complex than simple monolayer cultures. Thus, a key to increasing adoption of MPS is to find ways to produce fluid movement or flow systems which are easy to use and intuitive for end users without a background in fluidics. The requirement to connect MPS to ancillary pumps with lengths of tubing, whilst tolerated in the academic environment is often a frustration and concern for pharmaceutical scientists. The ability to produce integrated systems with either pumpless flow, on-board pumps or simple sterile connection to off-board pumps will be of importance.
Workflows
To be incorporated into pharmaceutical workflows, even for investigational type applications, MPS must achieve a minimum throughput. In the near term, MPS will not compete directly with high throughput formats such as 384- or 1536-well plates, but conversely the ability to culture only one or two MPS at any one time, seriously limits utility. The authors' believe the optimal throughput for MPS lies in systems able to culture simultaneously between 6 and 96 replicates. This range is achievable with current technologies.27,29,74 As MPS are scaled up to include more replicates the challenge will be to retain usability and robustness.
Cell sourcing
MPS often use primary human cells. The sourcing, quality control, general fragility, and donor-to-donor variability of primary cells restricts their use to a specialist community. For MPS to achieve wide adoption, detailed handling protocols or equipment which automates and simplifies the manipulation of primary cells will be required. Mature lineages derived from stem cells, be they embryonic or iPSC, represent a promising alternative to primary cells, and have already been cultured in MPS.75 The opportunity to derive a potentially limitless supply of all cells used in an MPS, from an autologous source, is appealing as it eliminates concerns over lot-to-lot variability and immune compatibility.58 The relative immaturity and consequent lack of function of some stem cell-derived lineages when compared to primary cells remains an issue.22 The opportunity to use MPS to drive the differentiated state of stem cell-derived lineages, closer to adult primary cells, is a promising avenue and likely maturation of both technologies will be mutually beneficial for wider adoption.
Summary
MPS have the potential to be a disruptive technology, providing unique insights into human biology, valuable to both basic research and the pharmaceutical industry. There remain key challenges to be overcome in commercializing MPS and making them accessible to a wide group of users. In the search for the most valuable applications in which to deploy these technologies, the multidisciplinary nature of MPS and pharmaceutical research makes development through partnerships the logical strategy.
Acknowledgement
The authors thank the M.I.T PhysioMimetics project team for discussions on microphysiological systems.
Authors’ contributions
All authors wrote, reviewed and edited the manuscript.
Declaration of conflicting interests
All authors are full time employees of and hold stock or options in CN Bio Innovations Ltd, vendor of the LiverChip® microphysiological system.
References
- 1.Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Bottger J, Braeuning A, Budinsky RA, Burkhardt B, Cameron NR, Camussi G, Cho CS, Choi YJ, Craig Rowlands J, Dahmen U, Damm G, Dirsch O, Donato MT, Dong J, Dooley S, Drasdo D, Eakins R, Ferreira KS, Fonsato V, Fraczek J, Gebhardt R, Gibson A, Glanemann M, Goldring CE, Gomez-Lechon MJ, Groothuis GM, Gustavsson L, Guyot C, Hallifax D, Hammad S, Hayward A, Haussinger D, Hellerbrand C, Hewitt P, Hoehme S, Holzhutter HG, Houston JB, Hrach J, Ito K, Jaeschke H, Keitel V, Kelm JM, Kevin Park B, Kordes C, Kullak-Ublick GA, LeCluyse EL, Lu P, Luebke-Wheeler J, Lutz A, Maltman DJ, Matz-Soja M, McMullen P, Merfort I, Messner S, Meyer C, Mwinyi J, Naisbitt DJ, Nussler AK, Olinga P, Pampaloni F, Pi J, Pluta L, Przyborski SA, Ramachandran A, Rogiers V, Rowe C, Schelcher C, Schmich K, Schwarz M, Singh B, Stelzer EH, Stieger B, Stober R, Sugiyama Y, Tetta C, Thasler WE, Vanhaecke T, Vinken M, Weiss TS, Widera A, Woods CG, Xu JJ, Yarborough KM, Hengstler JG. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 2013; 87: 1315–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LeCluyse EL, Witek RP, Andersen ME, Powers MJ. Organotypic liver culture models: meeting current challenges in toxicity testing. Crit Rev Toxicol 2012; 42: 501–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ullrich A, Berg C, Hengstler JG, Runge D. Use of a standardised and validated long-term human hepatocyte culture system for repetitive analyses of drugs: repeated administrations of acetaminophen reduces albumin and urea secretion. ALTEX 2007; 24: 35–40. [DOI] [PubMed] [Google Scholar]
- 4.Ryan CM, Carter EA, Jenkins RL, Sterling LM, Yarmush ML, Malt RA, Tompkins RG. Isolation and long-term culture of human hepatocytes. Surgery 1993; 113: 48–54. [PubMed] [Google Scholar]
- 5.Kono Y, Yang S, Roberts EA. Extended primary culture of human hepatocytes in a collagen gel sandwich system. In Vitro Cell Dev Biol Anim 1997; 33: 467–72. [DOI] [PubMed] [Google Scholar]
- 6.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol 2008; 26: 120–6. [DOI] [PubMed] [Google Scholar]
- 7.Messner S, Agarkova I, Moritz W, Kelm JM. Multi-cell type human liver microtissues for hepatotoxicity testing. Arch Toxicol 2013; 87: 209–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell CC, Hendriks DF, Moro SM, Ellis E, Walsh J, Renblom A, Fredriksson Puigvert L, Dankers AC, Jacobs F, Snoeys J, Sison-Young RL, Jenkins RE, Nordling A, Mkrtchian S, Park BK, Kitteringham NR, Goldring CE, Lauschke VM, Ingelman-Sundberg M. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci Rep 2016; 6: 25187–25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivaraman A, Leach JK, Townsend S, Iida T, Hogan BJ, Stolz DB, Fry R, Samson LD, Tannenbaum SR, Griffith LG. A microscale in vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction. Curr Drug Metab 2005; 6: 569–91. [DOI] [PubMed] [Google Scholar]
- 10.Maher SP, Crouse RB, Conway AJ, Bannister EC, Achyuta AK, Clark AY, Sinatra FL, Cuiffi JD, Adams JH, Kyle DE, Saadi WM. Microphysical space of a liver sinusoid device enables simplified long-term maintenance of chimeric mouse-expanded human hepatocytes. Biomed Microdevices 2014; 16: 727–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheeler SE, Clark AM, Taylor DP, Young CL, Pillai VC, Stolz DB, Venkataramanan R, Lauffenburger D, Griffith L, Wells A. Spontaneous dormancy of metastatic breast cancer cells in an all human liver microphysiologic system. Br J Cancer 2014; 111: 2342–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prodanov L, Jindal R, Bale SS, Hegde M, McCarty WJ, Golberg I, Bhushan A, Yarmush ML, Usta OB. Long-term maintenance of a microfluidic 3D human liver sinusoid. Biotechnol Bioeng 2016; 113: 241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domansky K, Inman W, Serdy J, Dash A, Lim MH, Griffith LG. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip 2010; 10: 51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebrahimkhani MR, Neiman JA, Raredon MS, Hughes DJ, Griffith LG. Bioreactor technologies to support liver function in vitro. Adv Drug Deliv Rev 2014; 69-70: 132–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bale SS, Vernetti L, Senutovitch N, Jindal R, Hegde M, Gough A, McCarty WJ, Bakan A, Bhushan A, Shun TY, Golberg I, DeBiasio R, Usta OB, Taylor DL, Yarmush ML. In vitro platforms for evaluating liver toxicity. Exp Biol Med (Maywood) 2014; 239: 1180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauschke VM, Hendriks DF, Bell CC, Andersson TB, Ingelman-Sundberg M. Novel 3D culture systems for studies of human liver function and assessments of the hepatotoxicity of drugs and drug candidates. Chem Res Toxicol 2016; 29: 1936–55. [DOI] [PubMed] [Google Scholar]
- 17.Novik E, Maguire TJ, Chao P, Cheng KC, Yarmush ML. A microfluidic hepatic coculture platform for cell-based drug metabolism studies. Biochem Pharmacol 2010; 79: 1036–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khetani SR, Kanchagar C, Ukairo O, Krzyzewski S, Moore A, Shi J, Aoyama S, Aleo M, Will Y. Use of micropatterned cocultures to detect compounds that cause drug-induced liver injury in humans. Toxicol Sci 2013; 132: 107–17. [DOI] [PubMed] [Google Scholar]
- 19.Wang WW, Khetani SR, Krzyzewski S, Duignan DB, Obach RS. Assessment of a micropatterned hepatocyte coculture system to generate major human excretory and circulating drug metabolites. Drug Metab Dispos 2010; 38: 1900–5. [DOI] [PubMed] [Google Scholar]
- 20.Chan TS, Yu H, Moore A, Khetani SR, Tweedie D. Meeting the challenge of predicting hepatic clearance of compounds slowly metabolized by cytochrome P450 using a novel hepatocyte model, HepatoPac. Drug Metab Dispos 2013; 41: 2024–32. [DOI] [PubMed] [Google Scholar]
- 21.Atienzar FA, Novik EI, Gerets HH, Parekh A, Delatour C, Cardenas A, MacDonald J, Yarmush ML, Dhalluin S. Predictivity of dog co-culture model, primary human hepatocytes and HepG2 cells for the detection of hepatotoxic drugs in humans. Toxicol Appl Pharmacol 2014; 275: 44–61. [DOI] [PubMed] [Google Scholar]
- 22.Kratochwil NA, Meille C, Fowler S, Klammers F, Ekiciler A, Molitor B, Simon S, Walter I, McGinnis C, Walther J, Leonard B, Triyatni M, Javanbakht H, Funk C, Schuler F, Lave T, Parrott NJ. Metabolic profiling of human long-term liver models and hepatic clearance predictions from in vitro data using nonlinear mixed-effects modeling. AAPS J 2017; 19: 534–50. [DOI] [PubMed] [Google Scholar]
- 23.Hendriks DF, Fredriksson Puigvert L, Messner S, Mortiz W, Ingelman-Sundberg M. Hepatic 3D spheroid models for the detection and study of compounds with cholestatic liability. Sci Rep 2016; 6: 35434–35434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostadinova R, Boess F, Applegate D, Suter L, Weiser T, Singer T, Naughton B, Roth A. A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol Appl Pharmacol 2013; 268: 1–16. [DOI] [PubMed] [Google Scholar]
- 25.Schutte M, Fox B, Baradez MO, Devonshire A, Minguez J, Bokhari M, Przyborski S, Marshall D. Rat primary hepatocytes show enhanced performance and sensitivity to acetaminophen during three-dimensional culture on a polystyrene scaffold designed for routine use. Assay Drug Dev Technol 2011; 9: 475–86. [DOI] [PubMed] [Google Scholar]
- 26.Dash A, Inman W, Hoffmaster K, Sevidal S, Kelly J, Obach RS, Griffith LG, Tannenbaum SR. Liver tissue engineering in the evaluation of drug safety. Expert Opin Drug Metab Toxicol 2009; 5: 1159–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kostrzewski T, Cornforth T, Snow SA, Ouro-Gnao L, Rowe C, Large EM, Hughes DJ. Three-dimensional perfused human in vitro model of non-alcoholic fatty liver disease. World J Gastroenterol 2017; 23: 204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao P, Maguire T, Novik E, Cheng KC, Yarmush ML. Evaluation of a microfluidic based cell culture platform with primary human hepatocytes for the prediction of hepatic clearance in human. Biochem Pharmacol 2009; 78: 625–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang M, Neuzil P, Volk T, Manz A, Kleber A. On-chip three-dimensional cell culture in phaseguides improves hepatocyte functions in vitro. Biomicrofluidics 2015; 9: 034113–034113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vivares A, Salle-Lefort S, Arabeyre-Fabre C, Ngo R, Penarier G, Bremond M, Moliner P, Gallas JF, Fabre G, Klieber S. Morphological behaviour and metabolic capacity of cryopreserved human primary hepatocytes cultivated in a perfused multiwell device. Xenobiotica 2015; 45: 29–44. [DOI] [PubMed] [Google Scholar]
- 31.Long TJ, Cosgrove PA, Dunn RT, 2nd, Stolz DB, Hamadeh H, Afshari C, McBride H, Griffith LG. Modeling therapeutic antibody-small molecule drug-drug interactions using a three-dimensional perfusable human liver coculture platform. Drug Metab Dispos 2016; 44: 1940–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bale SS, Sridharan GV, Golberg I, Prodanov L, McCarty WJ, Usta OB, Jindal R, Yarmush ML. A novel low-volume two-chamber microfabricated platform for evaluating drug metabolism and toxicity. Technology (Singap World Sci) 2015; 3: 155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marx U, Andersson TB, Bahinski A, Beilmann M, Beken S, Cassee FR, Cirit M, Daneshian M, Fitzpatrick S, Frey O, Gaertner C, Giese C, Griffith L, Hartung T, Heringa MB, Hoeng J, de Jong WH, Kojima H, Kuehnl J, Leist M, Luch A, Maschmeyer I, Sakharov D, Sips AJ, Steger-Hartmann T, Tagle DA, Tonevitsky A, Tralau T, Tsyb S, van de Stolpe A, Vandebriel R, Vulto P, Wang J, Wiest J, Rodenburg M, Roth A. Biology-inspired microphysiological system approaches to solve the prediction dilemma of substance testing. ALTEX 2016; 33: 272–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bavli D, Prill S, Ezra E, Levy G, Cohen M, Vinken M, Vanfleteren J, Jaeger M, Nahmias Y. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc Natl Acad Sci U S A 2016; 113: E2231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark AM, Wheeler SE, Taylor DP, Pillai VC, Young CL, Prantil-Baun R, Nguyen T, Stolz DB, Borenstein JT, Lauffenburger DA, Venkataramanan R, Griffith LG, Wells A. A microphysiological system model of therapy for liver micrometastases. Exp Biol Med (Maywood) 2014; 239: 1170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wikswo JP, Block FE, 3rd, Cliffel DE, Goodwin CR, Marasco CC, Markov DA, McLean DL, McLean JA, McKenzie JR, Reiserer RS, Samson PC, Schaffer DK, Seale KT, Sherrod SD. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans Biomed Eng 2013; 60: 682–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marasco CC, Enders JR, Seale KT, McLean JA, Wikswo JP. Real-time cellular exometabolome analysis with a microfluidic-mass spectrometry platform. PLoS One 2015; 10: e0117685–e0117685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA, Ingber DE. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med 2012; 4: 159ra47–159ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jungermann K, Kietzmann T. Oxygen: modulator of metabolic zonation and disease of the liver. Hepatology 2000; 31: 255–60. [DOI] [PubMed] [Google Scholar]
- 40.Allen JW, Khetani SR, Bhatia SN. In vitro zonation and toxicity in a hepatocyte bioreactor. Toxicol Sci 2005; 84: 110–9. [DOI] [PubMed] [Google Scholar]
- 41.Rotem A, Toner M, Tompkins RG, Yarmush ML. Oxygen uptake rates in cultured rat hepatocytes. Biotechnol Bioeng 1992; 40: 1286–91. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar U, Rivera-Burgos D, Large EM, Hughes DJ, Ravindra KC, Dyer RL, Ebrahimkhani MR, Wishnok JS, Griffith LG, Tannenbaum SR. Metabolite profiling and pharmacokinetic evaluation of hydrocortisone in a perfused three-dimensional human liver bioreactor. Drug Metab Dispos 2015; 43: 1091–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bale SS, Geerts S, Jindal R, Yarmush ML. Isolation and co-culture of rat parenchymal and non-parenchymal liver cells to evaluate cellular interactions and response. Sci Rep 2016; 6: 25329–25329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen TV, Ukairo O, Khetani SR, McVay M, Kanchagar C, Seghezzi W, Ayanoglu G, Irrechukwu O, Evers R. Establishment of a hepatocyte-kupffer cell coculture model for assessment of proinflammatory cytokine effects on metabolizing enzymes and drug transporters. Drug Metab Dispos 2015; 43: 774–85. [DOI] [PubMed] [Google Scholar]
- 45.Rennert K, Steinborn S, Groger M, Ungerbock B, Jank AM, Ehgartner J, Nietzsche S, Dinger J, Kiehntopf M, Funke H, Peters FT, Lupp A, Gartner C, Mayr T, Bauer M, Huber O, Mosig AS. A microfluidically perfused three dimensional human liver model. Biomaterials 2015; 71: 119–31. [DOI] [PubMed] [Google Scholar]
- 46.Roth RA, Ganey PE. Intrinsic versus idiosyncratic drug-induced hepatotoxicity–two villains or one? J Pharmacol Exp Ther 2010; 332: 692–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaw PJ, Ganey PE, Roth RA. Idiosyncratic drug-induced liver injury and the role of inflammatory stress with an emphasis on an animal model of trovafloxacin hepatotoxicity. Toxicol Sci 2010; 118: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Than NN, Newsome PN. A concise review of non-alcoholic fatty liver disease. Atherosclerosis 2015; 239: 192–202. [DOI] [PubMed] [Google Scholar]
- 49.Willebrords J, Pereira IV, Maes M, Crespo Yanguas S, Colle I, Van Den Bossche B, Da Silva TC, de Oliveira CP, Andraus W, Alves VA, Cogliati B, Vinken M. Strategies, models and biomarkers in experimental non-alcoholic fatty liver disease research. Prog Lipid Res 2015; 59: 106–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, Ahrlund-Richter L, Skepper J, Semple R, Weber A, Lomas DA, Vallier L. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest 2010; 120: 3127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang K, Pfeifer ND, Kock K, Brouwer KL. Species differences in hepatobiliary disposition of taurocholic acid in human and rat sandwich-cultured hepatocytes: implications for drug-induced liver injury. J Pharmacol Exp Ther 2015; 353: 415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winnike JH, Pediaditakis P, Wolak JE, McClelland RE, Watkins PB, Macdonald JM. Stable isotope resolved metabolomics of primary human hepatocytes reveals a stressed phenotype. Metabolomics 2012; 8: 34–49. [Google Scholar]
- 53.Sunman JA, Hawke RL, LeCluyse EL, Kashuba AD. Kupffer cell-mediated IL-2 suppression of CYP3A activity in human hepatocytes. Drug Metab Dispos 2004; 32: 359–63. [DOI] [PubMed] [Google Scholar]
- 54.Hughes DJ, Cui Z, Field RW, Tirlapur UK. In situ three-dimensional characterization of membrane fouling by protein suspensions using multiphoton microscopy. Langmuir 2006; 22: 6266–72. [DOI] [PubMed] [Google Scholar]
- 55.Ghanem A, Shuler ML. Combining cell culture analogue reactor designs and PBPK models to probe mechanisms of naphthalene toxicity. Biotechnol Prog 2000; 16: 334–45. [DOI] [PubMed] [Google Scholar]
- 56.Viravaidya K, Sin A, Shuler ML. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol Prog 2004; 20: 316–23. [DOI] [PubMed] [Google Scholar]
- 57.Lasser KE, Allen PD, Woolhandler SJ, Himmelstein DU, Wolfe SM, Bor DH. Timing of new black box warnings and withdrawals for prescription medications. JAMA 2002; 287: 2215–20. [DOI] [PubMed] [Google Scholar]
- 58.Vunjak-Novakovic G, Bhatia S, Chen C, Hirschi K. HeLiVa platform: integrated heart-liver-vascular systems for drug testing in human health and disease. Stem Cell Res Ther 2013; 1: S8–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oleaga C, Bernabini C, Smith AS, Srinivasan B, Jackson M, McLamb W, Platt V, Bridges R, Cai Y, Santhanam N, Berry B, Najjar S, Akanda N, Guo X, Martin C, Ekman G, Esch MB, Langer J, Ouedraogo G, Cotovio J, Breton L, Shuler ML, Hickman JJ. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep 2016; 6: 20030–20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maschmeyer I, Hasenberg T, Jaenicke A, Lindner M, Lorenz AK, Zech J, Garbe LA, Sonntag F, Hayden P, Ayehunie S, Lauster R, Marx U, Materne EM. Chip-based human liver-intestine and liver-skin co-cultures – a first step toward systemic repeated dose substance testing in vitro. Eur J Pharm Biopharm 2015; 95: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Imura Y, Sato K, Yoshimura E. Micro total bioassay system for ingested substances: assessment of intestinal absorption, hepatic metabolism, and bioactivity. Anal Chem 2010; 82: 9983–8. [DOI] [PubMed] [Google Scholar]
- 62.Esch MB, Ueno H, Applegate DR, Shuler ML. Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab Chip 2016; 16: 2719–29. [DOI] [PubMed] [Google Scholar]
- 63.Kim JY, Fluri DA, Marchan R, Boonen K, Mohanty S, Singh P, Hammad S, Landuyt B, Hengstler JG, Kelm JM, Hierlemann A, Frey O. 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis. J Biotechnol 2015; 205: 24–35. [DOI] [PubMed] [Google Scholar]
- 64.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip 2013; 13: 3496–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abaci HE, Shuler ML. Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Integr Biol (Camb) 2015; 7: 383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsamandouras N, Kostrzewski T, Stokes CL, Griffith LG, Hughes DJ, Cirit M. Quantitative assessment of population variability in hepatic drug metabolism using a perfused three-dimensional human liver microphysiological system. J Pharmacol Exp Ther 2017; 360: 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cosgrove BD, Griffith LG, Lauffenburger DA. Fusing tissue engineering and systems biology toward fulfilling their promise. Cell Mol Bioeng 2008; 1: 33–41. [Google Scholar]
- 68.Miller PG, Shuler ML. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol Bioeng 2016; 113: 2213–27. [DOI] [PubMed] [Google Scholar]
- 69.van Midwoud PM, Janse A, Merema MT, Groothuis GM, Verpoorte E. Comparison of biocompatibility and adsorption properties of different plastics for advanced microfluidic cell and tissue culture models. Anal Chem 2012; 84: 3938–44. [DOI] [PubMed] [Google Scholar]
- 70.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET, Beebe DJ. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip 2009; 9: 2132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang JD, Douville NJ, Takayama S, ElSayed M. Quantitative analysis of molecular absorption into PDMS microfluidic channels. Ann Biomed Eng 2012; 40: 1862–73. [DOI] [PubMed] [Google Scholar]
- 72.Zhou J, Ellis AV, Voelcker NH. Recent developments in PDMS surface modification for microfluidic devices. Electrophoresis 2010; 31: 2–16. [DOI] [PubMed] [Google Scholar]
- 73.Domansky K, Leslie DC, McKinney J, Fraser JP, Sliz JD, Hamkins-Indik T, Hamilton GA, Bahinski A, Ingber DE. Clear castable polyurethane elastomer for fabrication of microfluidic devices. Lab Chip 2013; 13: 3956–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim JY, Fluri DA, Kelm JM, Hierlemann A, Frey O. 96-well format-based microfluidic platform for parallel interconnection of multiple multicellular spheroids. J Lab Autom 2015; 20: 274–82. [DOI] [PubMed] [Google Scholar]
- 75.Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks N, Mandegar M, Conklin BR, Lee LP, Healy KE. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep 2015; 5: 8883–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]