Abstract
Gastrointestinal diseases are a significant health care and economic burden. Prevention and treatment of these diseases have been limited by the available human biologic models. Microphysiological systems comprise organ-specific human cultures that recapitulate many structural, biological, and functional properties of the organ in smaller scale including aspects of flow, shear stress and chemical gradients. The development of intestinal microphysiological system platforms represents a critical component in improving our understanding, prevention, and treatment of gastrointestinal diseases. This minireview discusses: shortcomings of classical cell culture models of the gastrointestinal tract; human intestinal enteroids as a new model and their advantages compared to cell lines; why intestinal microphysiological systems are needed; potential functional uses of intestinal microphysiological systems in areas of drug development and modeling acute and chronic diseases; and current challenges in the development of intestinal microphysiological systems.
Impact statement
The development of a gastrointestinal MPS has the potential to facilitate the understanding of GI physiology. An ultimate goal is the integration of the intestinal MPS with other organ MPS. The development and characterization of nontransformed human intestinal cultures for use in MPS have progressed significantly since the inception of the MPS program in 2012, and these cultures are a key component of advancing MPS. Continued efforts are needed to optimize MPS to comprehensively and accurately recapitulate the complexity of the intestinal epithelium within intestinal tissue. These systems will need to include peristalsis, flow, and oxygen gradients, with incorporation of vascular, immune, and nerve cells. Regional cellular organization of crypt and villus areas will also be necessary to better model complete intestinal structure.
Keywords: Human intestinal enteroids, microphysiological systems
Introduction
Microphysiological systems (MPSs) are in vitro microfluidic platforms that use human cells and recapitulate the physiology of the tissue through the implementation of fluid flow and mechanical factors. In addition, these platforms are designed to physically couple two or more organs in a scalable fashion. The development of these systems is already advancing the understanding of organ function and diseases of lung, liver, nervous system, skin, pancreas, kidney, and intestine.1 The recent development of new models of the human intestinal epithelium2 has provided novel tools which can now be used to propel the development of the intestinal MPS forward. These new technologies are reviewed below and their potential use in furthering the development of the intestinal MPS discussed.
Intestinal disorders, including specific diarrheal diseases such as celiac disease, inflammatory bowel disease, irritable bowel syndrome, infectious diarrhea, and metabolic and genetic diseases such as cystic fibrosis and cancer, affect approximately 60–70 million Americans each year with an economic cost of close to $150 billion.3 Symptoms such as abdominal pain, gastroenteritis, diarrhea, and constipation cause an estimated 72 million ambulatory care visits and 4.6 million hospitalizations yearly.3 The gastrointestinal (GI) tract is one of the most architecturally and functionally complex organs of the body and performs many physiological functions including digestion, absorption, hormone and enzyme secretion, peristalsis, detoxification, excretion, and immunity. Alterations in these functions occur in disease and involve the dysregulation of pathways such as those that regulate metabolism, motility, and immunity. Prevention and therapy of gastrointestinal diseases are important priorities, yet development of effective drugs to treat intestinal disorders has been limited by the available human biological models, which do not fully recapitulate the human intestine, and are lacking in their effectiveness of rapid and accurate evaluations of potential compounds.
As a major component of the GI tract, the small and large intestinal epithelium functions both in the secretion and uptake of molecules, such as enzymes, defensive peptides, hormones, water, nutrients and drugs. The epithelium also functions as a barrier between the external environment, which contains a large amount of microbial species, and the rest of the body. Junctional complexes maintain a physical barrier between the body and the intestinal lumen. The intestinal epithelium consists of a wide variety of polarized cell types organized with a regional dependent architecture. Absorptive cells function in nutrient, electrolyte, and water uptake as well as in drug absorption. Secretory cells produce mucin and bicarbonate, which add to the protective barrier. Stem cells renew the epithelium, Paneth cells produce anti-microbial peptides and contribute to the stem cell niche, and enteroendocrine cells mostly affect and coordinate intestinal effects on other intestinal cells including immune and neuronal cells, and other body organs, such as the liver, kidney, and brain, through the production and secretion of chemical messengers such as hormones and neurotransmitters. Many secreted products communicate with other organs starting with the liver via the portal circulation and then are widely disseminated to other organs including the brain and kidney through the systemic circulation. The intestinal lumen contains many species of resident commensal organisms that affect the function of the epithelium through such mechanisms as maintenance of the barrier, control of the immune response, and regulation of metabolic function. Additionally, the lamina propria contains capillaries, lymph, immune and inflammatory components, and the enteric nervous system. The muscularis mucosa, which lies below the lamina propria, is composed of a three layer thick smooth muscle that gives some structure to the epithelium and its mechanical effects play a role in epithelial homeostasis.
Shortcomings of classical culture systems of the intestinal epithelium
Many models have been used to study the gastrointestinal tract including static in vitro transformed cell lines and intact animal models.4 In vitro classical culture models (e.g. immortalized human colon cancer cells) consist of a single epithelial cell population, and while they have been used for many years in GI research, they do not fully recapitulate normal tissue architecture, segment specificity, biology, molecular signaling including paracrine and autocrine cross-talk between different cell types in the intestinal epithelium, or susceptibility to human pathogens.4 In addition, it is not clear how much a transformed cell line recapitulates the function of an untransformed heterogeneous epithelium. Genetic drift, lack of selective pressure, and sub-cloning have led to many different versions of the colon cancer cell lines such as Caco-2 that are used by different laboratories. In some cases, immortalization or transformation alters the response of the cells to drugs or infectious agents as they are selected for proliferation and resistance to cell death.5 Various studies have commented on the lack of functional correlation between Caco-2 cells and human intestine. Mainly, various drug transporters are either mislocalized or have inaccurate expression in Caco-2 cells when compared to human intestine.6–8 There are some reports of co-culturing Caco-2 cells with HT-29X or HT29-5M21 cells, a mucin secreting population, or with PBMC-derived macrophages and dendritic cells in an attempt to improve the in vitro cultures to mimic a more in vivo like response.9–11 Although these cultures have additional complexity compared to Caco-2 cell cultured alone, the use of transformed epithelial (Caco-2) cells still raises questions as to the validity of recapitulating true intestinal epithelial biology with these transformed cell cultures. Computational modeling or systems biology approaches are often necessary to take in vitro responses from these cells and extrapolate predictive in vivo responses such as absorption and transport but these approaches have had limited accuracy.12–15 Animal models can recapitulate the human intestine from an anatomical and structural standpoint but exhibit limitations in recapitulating physiology and subsequent responses to drugs targeting intestinal disorders, depending on the animal.16,17 In fact, failures are common when preclinical animal testing is used to predict adverse outcomes arising from sequential, multi-organ metabolism of drugs and xenobiotics.18 Other factors that limit the use of animal models in development of intestinal therapies include the labor-intensive and expensive nature of maintaining an animal colony, the lack of genetic diversity often critical to disease development and drug treatments, the lack of suitability for high-throughput drug screening, and the inability to model human patient- and population-specific physiology and pathophysiology.17,19 These considerations highlight the interest and need for models of human intestine that recapitulate human physiology and function yet are compatible with high-throughput analyses.
New models (organoids and human intestinal enteroids) and their advantages compared to cell lines
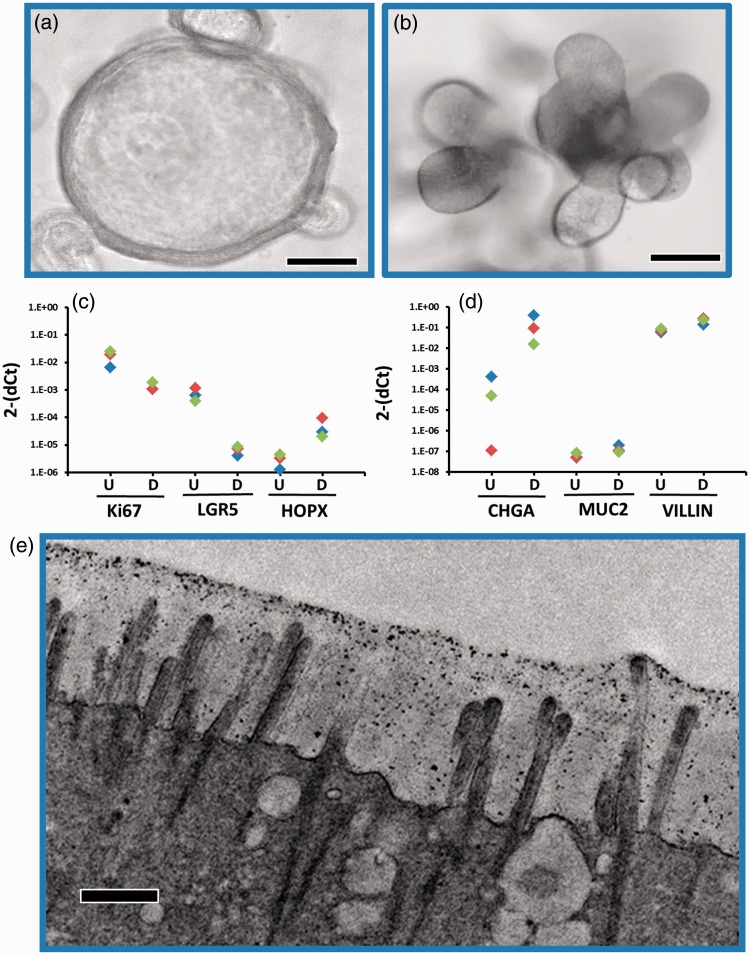
Two new models of the human GI epithelium have recently been developed: induced pluripotent stem cell (iPSC)-derived intestinal organoid cultures20 and ex vivo intestinal enteroid cultures derived from either a single LGR5+ stem cell/Paneth cell unit or multi-cell human intestinal crypts2,21 obtained from human intestinal tissue. These cultures have emerged as a potentially transformative and biologically relevant model of intestinal biology.2,22–24 The iPSC-derived cultures have a more immature, fetal tissue phenotype making them valuable to study developmental biology,25,26 and much current effort is focused on understanding the signals required to drive their maturation. The ex vivo intestinal enteroids are derived from either biopsies or surgical specimens from both the large and small intestine. Cell cultures are established from stem cells isolated from the crypts along with Paneth cells and are propagated as three-dimensional cultures in Matrigel® with media containing appropriate growth factors (Wnt3a, Noggin, R-spondin) to stimulate stem cell growth. These 3D enteroids, which express stem cell and proliferative markers, can be passaged indefinitely once they are established and retain intestinal regional-specific properties as well as multiple host phenotypic properties (Figure 1(a) and (b)). Removal of the growth factors stops the stem cell proliferation and the cells differentiate into mature intestinal cell types found in the epithelium (Figure 1(c) and (d)). Thus, these multicellular cultures recapitulate the central features of normal intestinal epithelial structure and function including cell polarization, the presence of the brush border with microvilli (Figure 1(e)), and appropriate physiologic responsiveness.27,28 These multicellular cultures represent a step towards returning to the whole organ with the retention of host genetic and phenotypic properties. In addition, these cultures maintain specific gene expression profiles reflecting their region of origin (duodenum, jejunum, ileum, and colon).29 These features will be useful in assessing the influence of genetic heterogeneity and regional specificity on multiple conditions in the intestine including infection, injury, and drug transport, metabolism and toxicity.
Figure 1.
Properties of human intestinal enteroids (HIEs). Jejunal crypts were propagated in vitro for 6–10 passages in Matrigel® and media containing WNT3A. WNT was removed from the cultures resulting in differentiation and the presence of the mature cell types that line the villi in the small intestine. (a, b) Light micrograph of one individual line illustrating the 3D structure in the proliferative (A) or differentiated (B) state, scale bar 50 µm. (c, d) Expression levels of stem cell (LGR5, HOPX), proliferative markers (Ki67), and differentiation markers (CHGA, MUC2, VILLIN) were assessed in three individual lines by RT-qPCR in the proliferative (U) or differentiated (D) state. Each color represents an individual enteroid line. (e) Electron microscopy was used to confirm the presence of the brush border with microvilli on the apical surface of the differentiated jejunal HIEs, scale bar 500 nm.
Why intestinal MPSs are needed
MPSs are biomimetic systems consisting of organ-specific human cultures that recapitulate many structural, biological, and functional properties of the organ in small scale (micro refers to 10−6 that of the actual size). These systems have emerged based on advances in bioengineering, microchip technology, microfluidics, and tissue engineering.30 MPSs replicate several aspects of human organs that have been lacking in tissue culture models and have the potential to bridge the gap between in vitro static culture of transformed cells and the complexity of the human system in vivo. They model the three-dimensional architecture of the organ, involve simultaneous formation of two or more tissues, and replicate many aspects of the organ microenvironment.31 The terminology “organs-on-a-chip” or “human-on-a-chip” is currently being used to describe systems in which multiple modular organs in an MPS are linked and integrated using microfluidic technology.32 Microfluidics provide critical physical cues such as flow rate and direction, shear stress, and chemical gradients that are important at both the tissue and organ level.33 Morphology and architecture of the epithelium depend in part on the extracellular microenvironment. Several materials including membranes, PDMS, and hydrogels are currently being used in MPS. PDMS has been used widely; however, it does not appear to be an optimal model for the in vivo extracellular matrix because it absorbs small hydrophobic molecules affecting physiological and pharmacological studies.34–36 Synthetic hydrogels may provide a more physiologically relevant matrix.37–39 Hydrogels overcome the limitations of PDMS and closely mimic the physical properties of the extracellular matrix. Gjorevski et al.40 have recently defined the extra-cellular matrix parameters required for intestinal stem cell expansion by the utilization of modular synthetic hydrogels, while others have employed microchambers or droplets to control the cellular microenvironment.41,42 Since MPS represent a more complex culture system that can mimic the whole human, they are developed and evaluated to determine if the efficiency of the drug development process can be improved with their use. These systems have great potential to facilitate new drug screening; their use in pharmacokinetic and toxicology assays are expected to provide more accurate predictions of clinical trial outcome. The ideal MPS will have the ability to obtain absorption, distribution, metabolism, excretion, and toxicity (ADMETOX) profiles along the timeline of a repeated dose substance test in coupled human organs.
There are several critical features that are important to develop a physiologically relevant model of the human intestine in vitro. The epithelium must contain a heterogeneous population of cells that is spatially organized, mimicking the crypt-villus axis where the proliferating cells are located in the crypt and the differentiated cells in the villus. The epithelium must be supported by matrices that mimic the lamina propria and the muscular layers that form a concentric tube along with the epithelium. The epithelium also experiences luminal unidirectional sheer stress on its apical surface while the basolateral surface is exposed to blood flow. The intestine is further exposed to periodic squeezing by the muscular layers (peristalsis) that propels the luminal contents from the mouth to the anus. Additionally, the intestine has a unique relationship to the liver in that the basolateral outflow from the intestinal epithelium is directly transported to the liver through the portal system. Conversely, liver produced products are secreted into the lumen of the duodenum and act on the apical side of the intestinal epithelium. Thus, enterohepatic signaling is critical to understand the biology of both tissues. Previously available in vitro models of the GI epithelium consist of transformed cell lines that do not reflect the heterogeneity and special organization present in vivo. New models using enteroids or iPS-derived organoids are being developed to overcome these limitations. The glass and plastic tissue culture supports used to culture these cells do not reflect the actual mechanical forces the epithelium would experience in vivo, and static culture conditions do not incorporate fluid flow.
Current platforms are focusing on introducing fluid sheer stress, flow direction and rate, and chemical gradients to well characterized gastrointestinal transformed cell lines and are being used to examine whether these changes in cellular microenvironment, polarization, spatial organization, and/or differentiation of the cultures better resemble that found in vivo. There have been several reports linking the addition of dynamic flow to static transformed cells with increased cell growth, differentiation, and polarization.43 Kim et al.44 have examined the response of fluidic cultures of Caco-2 cells and reported villi-like folds, increasing surface area, and cell reorganization that they compared to in vivo properties. However, there has been much discussion over whether these structures truly represent villi. Other work has focused on using a “human-on-a-chip”-based approach to link multiple transformed cell lines from different tissues such as the intestinal Caco-2/HT29-MTX and liver HepG2,C3A cell lines45 using microfluidic engineering to examine aspects of the enterohepatic communication pathways. However, these systems are still developed using transformed cell lines causing questions as to whether the findings have biological relevance. Wang et al.46 have used an air–liquid interface (ALI) culture to propagate enteroid cultures and report that they form a highly uniform 3D serpentine pattern that exhibits all the characteristics of a differentiated intestinal epithelium.46 The continued development of an MPS using iPSC-derived intestinal organoids or crypt stem cell-derived enteroids will increase the complexity of current MPS comprising transformed cells and providing a more biological relevant cell culture system in which new knowledge can be learned and the heterogeneous nature of the human population can be modeled.
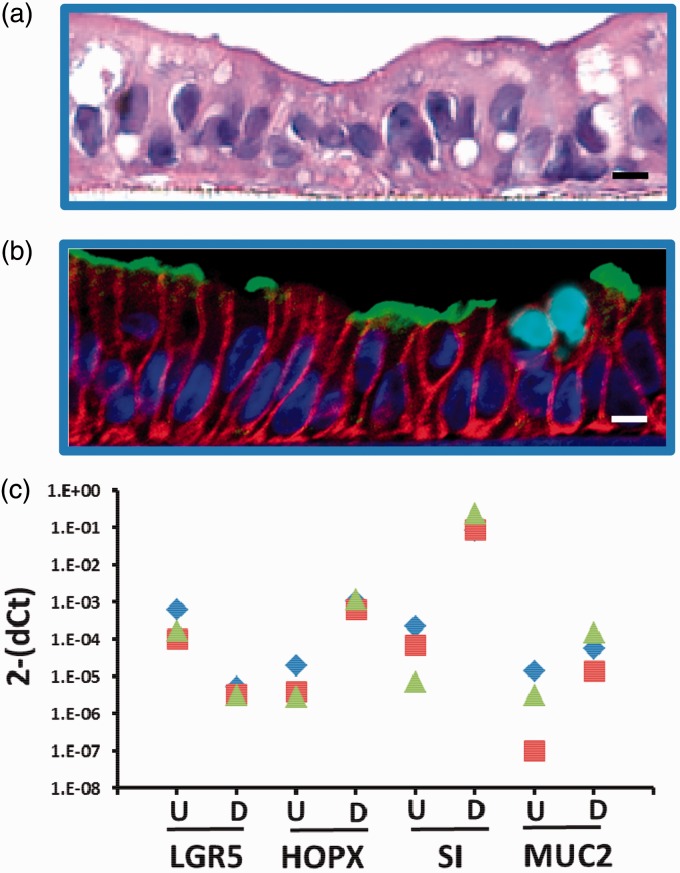
With increasing knowledge about stem cell differentiation and better characterization of organoids and enteroids, these cell lines are becoming invaluable to further intestinal MPS platforms for uses in personalized medicine and establishing multi-organ system “human-on-a-chip.” The ability to disperse 3D enteroid cultures and then grow them as 2D monolayers on Matrigel®, collagen, or other extracellular matrices represents an important advance by providing separate access to apical and basolateral surfaces of the epithelium.47–49 These monolayers appear to be identical to their 3D counterparts in terms of composition and physiology; the monolayers remain multi-cellular and express differentiated cell type specific markers expected in the epithelium (Figure 2). The monolayer format now allows the enteroid cultures to be adapted to MPS platforms. The MPS can be seeded with proliferating intestinal stem cells to form the monolayers, differentiated by the withdrawal of growth factors, and then used for drug testing or other biological investigation. It is unknown how MPS factors, such as fluid shear stress, mechanical forces that mimic peristalsis, and spatial organization of cells, will affect the growth and differentiation of these cultures. Progress is underway to develop and validate these cultures for adaptation to a number of MPS platforms.
Figure 2.
Properties of human intestinal enteroids after culturing in monolayer format. Jejunal enteroid lines were cultured in monolayer format followed by withdrawal of WNT to induce differentiation. (a) H&E stain of a jejunal monolayer. Scale bar 10 µm. (b) Immunofluorescent staining of markers of differentiated intestinal epithelium (Blue, nuclear; Red, E-cadherin; Green, Sucrase Isomaltase, Teal, Muc2). Scale bar 10 µm. (c) RT-qPCR was performed to assess expression levels of stem cell (LGR5, HOPX) and differentiation markers (Sucrase Isomaltase [SI], mucin [MUC2]) in proliferative (U) or differentiated (D) enteroid lines. Each color (blue, green, red) represents an individual enteroid line.
Potential functional uses of intestinal MPS
Drug development
The development of optimized “gut-on-a-chip” modules represents a critical goal, as oral administration of therapeutic agents is often the preferred route for clinical use. Therefore, intestinal MPS are being developed to study intestinal behavior related to drugs or xenobiotics. The intestine is the largest organ in the body and absorption, metabolism, transport and clearance of drugs in the intestine can affect downstream availability, toxicity, and efficacy in particular to the liver. There is evidence that for some drugs, there are segment-specific preferential absorption and metabolism rates.50 Defining effects regulated by specific regions of the GI tract on bioavailability of compounds to the liver are an important goal and challenge of the drug development community. Recapitulation of drug metabolism pathways, such as that of acetaminophen’s first pass metabolism in intestinal cells and the production of liver toxic metabolites, has already been successfully performed using first iterations of these platforms.51 An intestinal MPS could be used initially to determine the kinetics of absorption, transport, and metabolism of compounds by the intestinal epithelium. Once the output from the intestine has been characterized, the modular unit can be plugged into the multiple organ system, which can then be configured for unidirectional flow (i.e. intestine->liver->[target organ(s)]->kidney) or a looped flow with feedback from the other organ modules in the system. In these linked systems, scaling will be a major concern and the multi-MPS platform must be designed to accurately reflect in vivo physiology as well as relative quantitative contributions of each organ to modeled blood flow.52 These considerations are important in pre-clinical and clinical drug testing to determine the bioavailable fraction of an orally administered compound that enters the systemic circulation and ultimately affects dosing regimens and drug effectiveness. Currently, uptake and metabolism transformation of many orally administered drugs by the GI tract is often not considered. Organoids and enteroids combined with an MPS platform offer a complex model system in which key pharmacologic responses could be defined and documented in a patient-specific way.
Microfluidic platforms using multiple Transwell® permeable supports seeded with the Caco-2 transformed cell line are already being used to predict drug bioavailability provided by the intestine and subsequent liver metabolism and provide proof-of-principle data that MPS platforms can be used to model pharmaceutical testing.53–55 There are several limitations to the use of Caco-2 cells in these studies, as they are a good model of drug absorption56 but lack the required factors necessary for metabolic pathways that model in vivo responses.57 The response of the colon carcinoma line to several drugs differs dramatically from the response in enteroid cultures58 suggesting that the enteroids might be more suitable and reflective of the in vivo metabolic response when studying drug toxicity. Using the human intestinal enteroids (HIEs) to assess anticipated toxicities as well as discovery of unexpected adverse drug effects will be a great strength of this model. Initial studies indicate HIE cultures exhibit expected toxicity when treated with indomethacin (Figure 3). Caco-2 cells are also being used in microfluidic cultures to study real time intestinal permeability to model substances such as curcumin to study pharmacokinetic processes that regulate bioavailability, drug metabolism, absorption, and excretion.59 As HIEs are adapted to the microfluidic platforms, they have the potential to better predict the response of the human intestinal epithelium to novel drugs. Drug toxicity can be measured in HIEs using a variety of techniques as has been done previously in cancer cell lines. Enteroid cell death can be monitored in real time by microscopy directly on some platforms; by LDH assay of the effluent from the monolayer; and by terminal analysis using confocal microscopy, flow cytometry, and western blotting (Figure 3(a) to (f)).
Figure 3.

Indomethicin treatment of HIEs results in cell toxicity. (a–b) 3D jejunal HIEs were treated overnight with indomethicin or vehicle and dead cells imaged following overnight incubation with 0.1 ug/ml propidium iodide. (c, d) HIEs were treated with either vehicle or indomethicin. Dead cells marked by propidium iodide uptake were quantified by flow cytometry. (e) LDH was quantified in the HIE supernatants by LDH ELISA assay after HIE treatment with increasing concentrations of indomethicin. (f) Cell death also can be analyzed by detection of cleaved caspase three by Western blotting and quantified densitometrically following vehicle or indomethicin treatment and normalized to GAPDH.
Other approaches seek to optimize integration of HIEs in MPS platforms with other organ modules to contribute to drug evaluation. The feasibility of such studies on an MPS platform was recently demonstrated by the functional coupling of small intestine (jejunum), liver, kidney (proximal convoluted tubules), blood–brain barrier, and skeletal muscle through the sequential organ-to-organ transfer of media between MPS platforms.60 In this work, linked human tissues replicated drug/toxin metabolism and novel tissue responses were observed to terfenadine, vitamin D3, and trimethylamine (TMA). Metabolic activity of each tissue was preserved as expected with sequential drug intermediates quantifiable.60 This study represents an advance in integrating various organs to screen drugs and chemicals for toxicity. After successful integration, scaling becomes an important barrier to modeling accurate physiological responses. The challenge lies in the technology and expertise required to integrate or directly physically couple each organ system in the right scale and the design of a universal “one size fits all” MPS platform to accommodate the unique features of each organ. Platforms that incorporate individual inserts with permeable supports or chambers with apical and basolateral flow seem to meet the needs of most models. However, currently there is no single device available that is able to achieve this successfully and is one of the future challenges that remains to be solved.
Modeling diseases: GI and the role of other organ systems
The study of acute gastrointestinal diseases may benefit from using MPS platforms to probe the pathogenesis of these diseases in a system that better replicates the gastrointestinal environment. Many acute diseases have infectious etiologies and although much has been learned about pathogenesis from static transformed cell lines, the use of the organoid and enteroid primary human epithelial lines in an MPS platform will provide a more physiological environment with flow and sheer stress to study how pathogens such as human rotavirus and norovirus48,61 infect and potentially alter epithelial responses. In addition, regional contributions of specific intestinal segments to pathogenesis can be examined. MPS platforms will also help identify global metabolic changes that occur following infection that affect other organs besides the primary target organ. The response of these “non-infected” organs to the presence of the pathogen or to pathogen-induced signals can then be investigated. The development of intestinal MPS platforms also will be of potential value to rapidly assess the impact of emerging biologics, toxins, and pathogens. With recent bioengineered attacks and the continual emergence of infectious diseases that rapidly spread world-wide, there is a tremendous need to bolster our capabilities to rapidly gain information to assist in combating these threats. MPS offer the opportunity to dramatically reduce the time and the cost of development of new treatments as well as assessment of their safety and efficacy. Studying emerging pathogenic infections of the GI tract in the context of a multiple organ MPS platform can contribute to the implementation of global health plans to combat both known and other emerging infectious pathogens.24
The study of chronic gastrointestinal diseases such as IBD, IBS, cystic fibrosis, infectious or chronic diarrhea, celiac disease, and cancer may also benefit from using MPS platforms that better mimic the physiological environment of the GI tract to understand the role that intra-organ communication contributes to these disease processes in ways that have been previously inaccessible. Many of these diseases are not modeled adequately in small animals due to species differences such as immune responses and microbiota.62,63 GI diseases can also affect multiple organs and the response at the organ level may have global effects that cannot be discovered using cells from a single organ. MPS platforms containing enteroids can be made from individuals that have mutations that are linked to specific disease, such as the TTC7 gene that is linked to infantile IBD that results in epithelial barrier defects, and can be used to assess therapies that can reverse the effects of the mutation on the epithelium.64,65 High-throughput screening methods are being developed to test patient-derived cultures from patients with chronic intestinal illnesses for drug discovery.66 Notable examples include the screening of iPSC-derived cholangiocytes from cystic fibrosis patients with a mutation in CFTR for drugs that could correct the CF phenotype through reductions in misfolding and stabilization of the CFTR protein.67 Once effective drugs are identified, their efficacy, toxicity, and effects on downstream organs can be pursued on MPS platforms.
Tumor metastasis is a major factor predicting poor clinical outcomes and increased mortality. Metastasis occurs when cells that originate from the primary tumor migrate through the lymphatic and vascular systems to remote organs where they ultimately implant, establish lesions, and compromise the function of the invaded organ. Little is known about the mechanics by which a disseminated cancer cell seeds and overtakes organs. This process has been difficult to study due to a lack of model systems. Cancer metastasis in animals has questionable relevance to human metastasis16,18 and although studies in conventional transformed cultures have provided much of our understanding of tumor growth, they are not able to provide information about the complex interactions that occur between the cancer cell and its microenvironment, nor are they able to model the specific steps of metastasis. The MPS has the potential to overcome several of these limitations. Recently, a metastasis-on-a-chip was described that allowed real-time tracking of labeled colon cancer cells from a gut model to a downstream liver module using circulatory flow.68 CRISPR-CAS9-mediated gene editing of healthy human enteroids has allowed the evaluation of candidate genes in carcinogenesis69,70 and examination of these lines in a multi-organ MPS will provide important information about the genes that might mediate metastasis. Systems such as these have the potential to unlock much information concerning the process of tumor spread.
Current challenges for the development of intestinal MPS
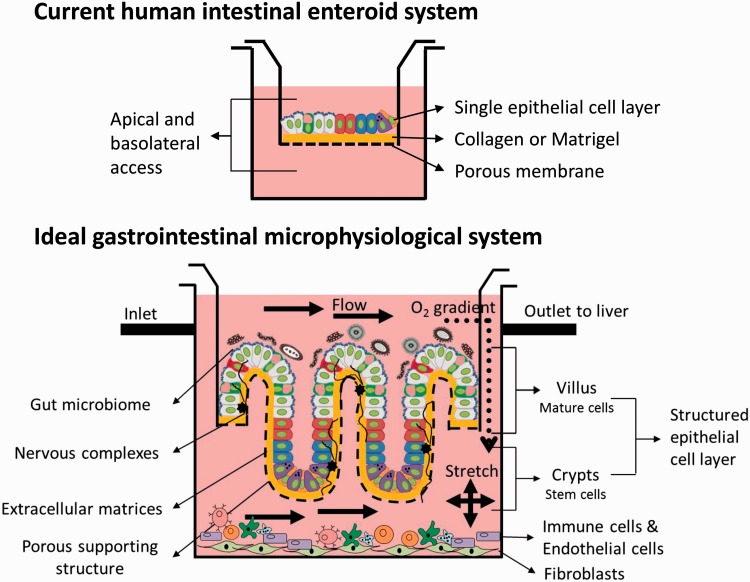
The development of gastrointestinal MPS will facilitate the understanding of physiologically relevant interactions with the ultimate goal being integration with the other organs. Current MPS have limitations that need to be overcome (Table 1). First, current cultures do not fully represent the complexity of the intestine but only the intestinal epithelium. Optimized future MPS intestinal platforms will need to incorporate the addition of immune cells, vasculature, and nerves.71,72 Recently, aspects of the enteric nervous system have been integrated into iPSC-derived cultures and formed neuroglial structures similar to a myenteric and submucosal plexus.73 Other groups are working with incorporating macrophages and myofibroblasts and assessing changes in growth and differentiation of intestinal progenitor cells.72 In addition, incorporation of the microbiome will be an important aspect to develop in these systems as it makes major contributions to human health and disease. The microbiome is a previously unappreciated “complementary organ” that metabolizes nutrients and drugs, regulates the immune system, and maintains intestinal homeostasis. Attempts at integration of these two organs have been modeled. The probiotic, Lactobacillus rhamnosus, was introduced into an MPS containing transformed cells resulting in colonization of the MPS with the organism and positive effects on the cells such as increased barrier function.44,74 Others have looked at the effects of microbial communities on intestinal enteroid growth and differentiation.72 Two features are emerging as critical to the integration of the microbiome in the GI MPS. First, the presence of flow is important to replace used nutrients, remove waste, and prevent toxic bacterial build-up (Figure 4). The development of a “blood mimic” will be essential to advancement in this area. Second, regulation of oxygen levels is essential to maintain the balance of aerobic and anaerobic species that represent the true intestinal microbial flora. Another critical aspect of developing a GI MPS will be the incorporation of the immune system (Figure 4). The GI tract is the largest immune organ in the body; therefore, studying biological responses of an intestinal MPS without this important aspect of the GI tract will be incomplete. In addition to the microbiome and immune system, a fully relevant intestinal MPS will have a differentiated multicellular composition and exhibit cellular organization including crypts and villi (Figure 4). Attayek et al.75 are designing chemical gradients consisting of stem cell signaling factors that result in basal luminal patterning of colonic cells in vitro.75 Research such as this will contribute to the design and development of gut MPS platforms. With these considerations, the human intestinal MPS will provide a new in vitro model system in which diseases can be studied, drugs can be tested, and mechanisms can be dissected.
Table 1.
Limitations and current challenges for the development of intestinal MPS
| Limitations | Current challenges |
|---|---|
| HIEs are in flat monolayer format that lacks the intestinal architecture | Engineer crypt-villus like structures and integrate chemical gradient for the establishment of stem cell and differentiated cell zones |
| HIEs only contain intestinal epithelium | Incorporated immune, vasculature and nervous systems |
| HIEs lack a microbiome | Flow and oxygen gradients are needed for the colonization of the microbiome |
MPS: Microphysiological systems; HIEs: human intestinal enteroids.
Figure 4.
Ideal gut MPS platform with primary cells and other important GI components integrated with basolateral flow to the liver and apical access.
Acknowledgments
This work was supported by National Institutes of Health grants NCATS U18- TR000552 and UH3-TR00003, U19-AI116497, RO1-AI080656, P30-DK089502, P30 DK56338 and Howard Hughes Medical Institute grant 570076890.
Authors’ contributions
SB, JB, WZ, MD and MKE wrote the manuscript; SB, JB, XZ, UK, JI, NZ, OK performed the experiments; all authors reviewed, revised and approved the manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Benam KH, Dauth S, Hassell B, Herland A, Jain A, Jang KJ, Karalis K, Kim HJ, MacQueen L, Mahmoodian R, et al. Engineered in vitro disease models. Ann Rev Pathol 2015; 10: 195–262. [DOI] [PubMed] [Google Scholar]
- 2.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009; 459: 262–5. [DOI] [PubMed] [Google Scholar]
- 3.Peery AF, Dellon ES, Lund J, Crockett SD, McGowan CE, Bulsiewicz WJ, Gangarosa LM, Thiny MT, Stizenberg K, Morgan DR, et al. Burden of gastrointestinal disease in the United States: 2012 update. Gastroenterology 2012; 143: 1179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le FE, Chesne C, Artusson P, Brayden D, Fabre G, Gires P, Guillou F, Rousset M, Rubas W, Scarino ML. In vitro models of the intestinal barrier. The report and recommendations of ECVAM Workshop 46. European Centre for the Validation of Alternative methods. Altern Lab Anim 2001; 29: 649–68. [DOI] [PubMed] [Google Scholar]
- 5.Kutuk O, Letai A. Alteration of the mitochondrial apoptotic pathway is key to acquired paclitaxel resistance and can be reversed by ABT-737. Cancer Res 2008; 68: 7985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awortwe C, Fasinu PS, Rosenkranz B. Application of Caco-2 cell line in herb-drug interaction studies: current approaches and challenges. J Pharm Pharm Sci 2014; 17: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larregieu CA, Benet LZ. Drug discovery and regulatory considerations for improving in silico and in vitro predictions that use Caco-2 as a surrogate for human intestinal permeability measurements. AAPS J 2013; 15: 483–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun D, Lennernas H, Welage LS, Barnett JL, Landowski CP, Foster D, Fleisher D, Lee KD, Amidon GL. Comparison of human duodenum and Caco-2 gene expression profiles for 12,000 gene sequences tags and correlation with permeability of 26 drugs. Pharm Res 2002; 19: 1400–16. [DOI] [PubMed] [Google Scholar]
- 9.Leonard F, Collnot EM, Lehr CM. A three-dimensional coculture of enterocytes, monocytes and dendritic cells to model inflamed intestinal mucosa in vitro. Mol Pharm 2010; 67: 2103–19. [DOI] [PubMed] [Google Scholar]
- 10.Nollevaux G, Deville C, El MB, Zorzi W, Deloyer P, Schneider YJ, Peulen O, Dandrifosse G. Development of a serum-free co-culture of human intestinal epithelium cell-lines (Caco-2/HT29-5M21). BMC Cell Biol 2006; 7: 20–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilgendorf C, Spahn-Langguth H, Regardh CG, Lipka E, Amidon GL, Langguth P. Caco-2 versus Caco-2/HT29-MTX co-cultured cell lines: permeabilities via diffusion, inside- and outside-directed carrier-mediated transport. J Pharm Sci 2000; 89: 63–75. [DOI] [PubMed] [Google Scholar]
- 12.Egan WJ, Lauri G. Prediction of intestinal permeability. Adv Drug Deliv Rev 2002; 54: 273–89. [DOI] [PubMed] [Google Scholar]
- 13.Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov 2015; 14: 248–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed SS, Ramakrishnan V. Systems biological approach of molecular descriptors connectivity: optimal descriptors for oral bioavailability prediction. PLoS One 2012; 7: e40654–e40654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heikkinen AT, Korjamo T, Monkkonen J. Modelling of drug disposition kinetics in in vitro intestinal absorption cell models. Basic Clin Pharmacol Toxicol 2010; 106: 180–8. [DOI] [PubMed] [Google Scholar]
- 16.Hackam DG, Redelmeier DA. Translation of research evidence from animals to humans. JAMA 2006; 296: 1731–2. [DOI] [PubMed] [Google Scholar]
- 17.Martic-Kehl MI, Schibli R, Schubiger PA. Can animal data predict human outcome? Problems and pitfalls of translational animal research. Eur J Nucl Med Mol Imag 2012; 39: 1492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol 2004; 172: 2731–8. [DOI] [PubMed] [Google Scholar]
- 19.Warren HS, Tompkins RG, Moldawer LL, Seok J, Xu W, Mindrinos MN, Maier RV, Xiao W, Davis RW. Mice are not men. Proc Natl Acad Sci U S A 2015; 112: E345–E345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011; 470: 105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato T, Stange DE, Ferrante M, Vries RG, van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van GJ, Siersema PD, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011; 141: 1762–72. [DOI] [PubMed] [Google Scholar]
- 22.Zachos NC, Kovbasnjuk O, Foulke-Abel J, In J, Blutt SE, de Jonge HR, Estes MK, Donowitz M. Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J Biol Chem 2016; 291: 3759–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.In JG, Foulke-Abel J, Estes MK, Zachos NC, Kovbasnjuk O, Donowitz M. Human mini-guts: new insights into intestinal physiology and host-pathogen interactions. Nat Rev Gastroenterol Hepatol 2016; 13: 633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mills M, Estes MK. Physiologically relevant human tissue models for infectious diseases. Drug Discov Today 2016; 21: 1540–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aurora M, Spence JR. hPSC-derived lung and intestinal organoids as models of human fetal tissue. Dev Biol 2016; 420: 230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finkbeiner SR, Hill DR, Altheim CH, Dedhia PH, Taylor MJ, Tsai YH, Chin AM, Mahe MM, Watson CL, Freeman JJ, et al. Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Rep 2015; 4: 1140–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 2013; 340: 1190–4. [DOI] [PubMed] [Google Scholar]
- 28.Foulke-Abel J, In J, Yin J, Zachos NC, Kovbasnjuk O, Estes MK, de JH, Donowitz M. Human enteroids as a model of upper small intestinal ion transport physiology and pathophysiology. Gastroenterology 2016; 150: 638–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Middendorp S, Schneeberger K, Wiegerinck CL, Mokry M, Akkerman RD, van WS, Clevers H, Nieuwenhuis EE. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 2014; 32: 1083–91. [DOI] [PubMed] [Google Scholar]
- 30.Ingber DE. Reverse engineering human pathophysiology with organs-on-chips. Cell 2016; 164: 1105–9. [DOI] [PubMed] [Google Scholar]
- 31.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science 2010; 328: 1662–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huh D, Kim HJ, Fraser JP, Shea DE, Khan M, Bahinski A, Hamilton GA, Ingber DE. Microfabrication of human organs-on-chips. Nat Protoc 2013; 8: 2135–57. [DOI] [PubMed] [Google Scholar]
- 33.Mehling M, Tay S. Microfluidic cell culture. Curr Opin Biotechnol 2014; 25: 95–102. [DOI] [PubMed] [Google Scholar]
- 34.van der Meer AD, van den Berg A. Organs-on-chips: breaking the in vitro impasse. Integr Biol 2012; 4: 461–70. [DOI] [PubMed] [Google Scholar]
- 35.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Meth 2005; 2: 119–25. [DOI] [PubMed] [Google Scholar]
- 36.Berthier E, Young EW, Beebe D. Engineers are from PDMS-land, Biologists are from Polystyrenia. LabChip 2012; 12: 1224–37. [DOI] [PubMed] [Google Scholar]
- 37.Chung BG, Lee KH, Khademhosseini A, Lee SH. Microfluidic fabrication of microengineered hydrogels and their application in tissue engineering. Lab Chip 2012; 12: 45–59. [DOI] [PubMed] [Google Scholar]
- 38.Seliktar D. Designing cell-compatible hydrogels for biomedical applications. Science 2012; 336: 1124–8. [DOI] [PubMed] [Google Scholar]
- 39.Huang GY, Zhou LH, Zhang QC, Chen YM, Sun W, Xu F, Lu TJ. Microfluidic hydrogels for tissue engineering. Biofabrication 2011; 3: 012001–012001. [DOI] [PubMed] [Google Scholar]
- 40.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordonez-Moran P, Clevers H, Lutolf MP. Designer matrices for intestinal stem cell and organoid culture. Nature 2016; 539: 560–4. [DOI] [PubMed] [Google Scholar]
- 41.Kwapiszewska K, Michalczuk A, Rybka M, Kwapiszewski R, Brzozka Z. A microfluidic-based platform for tumour spheroid culture, monitoring and drug screening. Lab Chip 2014; 14: 2096–104. [DOI] [PubMed] [Google Scholar]
- 42.Ziolkowska K, Stelmachowska A, Kwapiszewski R, Chudy M, Dybko A, Brzozka Z. Long-term three-dimensional cell culture and anticancer drug activity evaluation in a microfluidic chip. Biosens Bioelectron 2013; 40: 68–74. [DOI] [PubMed] [Google Scholar]
- 43.Sakolish CM, Esch MB, Hickman JJ, Shuler ML, Mahler GJ. Modeling barrier tissues in vitro: methods, achievements, and challenges. EBioMedicine 2016; 5: 30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HJ, Ingber DE. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol 2013; 5: 1130–40. [DOI] [PubMed] [Google Scholar]
- 45.Esch MB, Mahler GJ, Stokol T, Shuler ML. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip 2014; 14: 3081–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Yamamoto Y, Wilson LH, Zhang T, Howitt BE, Farrow MA, Kern F, Ning G, Hong Y, Khor CC, et al. Cloning and variation of ground state intestinal stem cells. Nature 2015; 522: 173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, Ciorba MA, Stappenbeck TS. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 2015; 64: 911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016; 353: 1387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.In J, Foulke-Abel J, Zachos NC, Hansen AM, Kaper JB, Bernstein HD, Halushka M, Blutt S, Estes MK, Donowitz M, et al. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol Gastroenterol Hepatol 2016; 2: 48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tam D, Tirona RG, Pang KS. Segmental intestinal transporters and metabolic enzymes on intestinal drug absorption. Drug Metab Dispos 2003; 31: 373–83. [DOI] [PubMed] [Google Scholar]
- 51.Mahler GJ, Esch MB, Glahn RP, Shuler ML. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol Bioeng 2009; 104: 193–205. [DOI] [PubMed] [Google Scholar]
- 52.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip 2013; 13: 3496–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imura Y, Sato K, Yoshimura E. Micro total bioassay system for ingested substances: assessment of intestinal absorption, hepatic metabolism, and bioactivity. Anal Chem 2010; 82: 9983–8. [DOI] [PubMed] [Google Scholar]
- 54.Imura Y, Yoshimura E, Sato K. Micro total bioassay system for oral drugs: evaluation of gastrointestinal degradation, intestinal absorption, hepatic metabolism, and bioactivity. Anal Sci 2012; 28: 197–9. [DOI] [PubMed] [Google Scholar]
- 55.Prot JM, Maciel L, Bricks T, Merlier F, Cotton J, Paullier P, Bois FY, Leclerc E. First pass intestinal and liver metabolism of paracetamol in a microfluidic platform coupled with a mathematical modeling as a means of evaluating ADME processes in humans. Biotechnol Bioeng 2014; 111: 2027–40. [DOI] [PubMed] [Google Scholar]
- 56.Hubatsch I, Ragnarsson EG, Artursson P. Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers. Nat Protoc 2007; 2: 2111–9. [DOI] [PubMed] [Google Scholar]
- 57.Taipalensuu J, Tornblom H, Lindberg G, Einarsson C, Sjoqvist F, Melhus H, Garberg P, Sjostrom B, Lundgren B, Artursson P. Correlation of gene expression of ten drug efflux proteins of the ATP-binding cassette transporter family in normal human jejunum and in human intestinal epithelial Caco-2 cell monolayers. J Pharmacol Exp Ther 2001; 299: 164–70. [PubMed] [Google Scholar]
- 58.Grabinger T, Luks L, Kostadinova F, Zimberlin C, Medema JP, Leist M, Brunner T. Ex vivo culture of intestinal crypt organoids as a model system for assessing cell death induction in intestinal epithelial cells and enteropathy. Cell Death Dis 2014; 5: e1228–e1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao D, Liu H, Lin JM, Wang Y, Jiang Y. Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device. Lab Chip 2013; 13: 978–85. [DOI] [PubMed] [Google Scholar]
- 60.Vernetti L, Gough A, Baetz N, Blutt SE, Broughman JR., Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci Rep 2017; 7: 42296–42296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saxena K, Blutt SE, Ettayebi K, Zeng XL, Broughman JR, Crawford SE, Karandikar UC, Sastri NP, Conner ME, Opekun AR, et al. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J Virol 2015; 90: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 2013; 110: 3507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiminez JA, Uwiera TC, Douglas IG, Uwiera RR. Animal models to study acute and chronic intestinal inflammation in mammals. Gut Pathog 2015; 7: 29–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avitzur Y, Guo C, Mastropaolo LA, Bahrami E, Chen H, Zhao Z, Elkadri A, Dhillon S, Murchie R, Fattouh R, et al. Mutations in tetratricopeptide repeat domain 7A result in a severe form of very early onset inflammatory bowel disease. Gastroenterology 2014; 146: 1028–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cutler DJ, Zwick ME, Okou DT, Prahalad S, Walters T, Guthery SL, Dubinsky M, Baldassano R, Crandall WV, Rosh J, et al. Dissecting allele architecture of early onset ibd using high-density genotyping. PLoS One 2015; 10: e0128074–e0128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gracz AD, Williamson IA, Roche KC, Johnston MJ, Wang F, Wang Y, Attayek PJ, Balowski J, Liu XF, Laurenza RJ, et al. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat Cell Biol 2015; 17: 340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogawa M, Ogawa S, Bear CE, Ahmadi S, Chin S, Li B, Grompe M, Keller G, Kamath BM, Ghanekar A. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol 2015; 33: 853–61. [DOI] [PubMed] [Google Scholar]
- 68.Skardal A, Devarasetty M, Forsythe S, Atala A, Soker S. A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol Bioeng 2016; 113: 2020–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 2015; 21: 256–62. [DOI] [PubMed] [Google Scholar]
- 70.Li M, Suzuki K, Kim NY, Liu GH, Izpisua Belmonte JC. A cut above the rest: targeted genome editing technologies in human pluripotent stem cells. J Biol Chem 2014; 289: 4594–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rogoz A, Reis BS, Karssemeijer RA, Mucida D. A 3-D enteroid-based model to study T-cell and epithelial cell interaction. J Immunol Meth 2015; 421: 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shaffiey SA, Jia H, Keane T, Costello C, Wasserman D, Quidgley M, Dziki J, Badylak S, Sodhi CP, March JC, et al. Intestinal stem cell growth and differentiation on a tubular scaffold with evaluation in small and large animals. Regen Med 2016; 11: 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Workman MJ, Mahe MM, Trisno S, Poling HM, Watson CL, Sundaram N, Chang CF, Schiesser J, Aubert P, Stanley EG, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med 2017; 23: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012; 12: 2165–74. [DOI] [PubMed] [Google Scholar]
- 75.Attayek PJ, Ahmad AA, Wang Y, Williamson I, Sims CE, Magness ST, Allbritton NL. In vitro polarization of colonoids to create an intestinal stem cell compartment. PLoS One 2016; 11: e0153795–e0153795. [DOI] [PMC free article] [PubMed] [Google Scholar]