Abstract
Colorectal cancer (CRC) remains a major health problem worldwide and the detailed mechanisms of CRC still need further understanding. Circular RNAs (circRNAs), a special class of endogenous RNAs, have emerged recently as a new potential player in governing fundamental biological process and cancer progression. In this study, we chose circRNA0003906 as a targeted circRNA to evaluate its expression pattern and clinical value in CRC patients. circRNA0003906 expression level in 6 CRC cell lines and 122 paired CRC tissues was measured by quantitative real-time polymerase chain reaction. Then, the potential correlation between circRNA0003906 expression level and clinicopathological factors of CRC patients was analyzed. Additionally, a receiver operating characteristic curve was built to evaluate the diagnostic value of circRNA0003906. Our results showed that circRNA0003906 expression level was dramatically downregulated in both CRC tissues and cell lines. Moreover, the downregulation of circRNA0003906 level significantly correlated with lymphatic metastasis and poor differentiation. In addition, the area under the receiver operating characteristic curve of circRNA0003906 for CRC was 0.818 (P<0.001). Taking consideration of all of these results, circRNA0003906 may be potentially involved in the colorectal cancerogenesis and serve as a potential biomarker for the diagnosis and treatment of CRC.
Keywords: circular RNA, circRNA0003906, colorectal cancer, biomarker
Introduction
Colorectal cancer (CRC) is a major health problem that is the third most common malignancy and the fourth most common cause of death in the world.1 With the progress of surgical skills and more options used for CRC therapy, the long-term survival of CRC patients in the last decade has been greatly improved, but it is still unsatisfactory due to the lack of reliable diagnostic biomarkers for early cancer detection. Thus, it is crucial to identify some new molecular biomarkers to raise the efficiency of tumor diagnosis and therapeutic application.
Circular RNAs (circRNAs), a class of endogenous noncoding RNA (ncRNA), are formed mainly by back-splicing and covalent binding, thus presenting as covalently closed continuous loops.2 They are as abundant, stable, and conserved and usually exhibit tissue- or developmental-stage-specific expression.3 In the past two decades, they were considered as errors in splicing and thought to be functionless. To date, plenty of circRNAs have been identified in different cell lines and species.4 Emerging evidence has revealed that circRNAs participate in fundamental biological process and may become ideal biomarkers in the diagnosis and prognosis of cancers.5–7 However, until now little is known about their roles in human CRC.
Human circRNA0003906 is one of circRNAs with 14317 nt in spliced sequence length. Its gene is located at chr6:29989443–30003760, and it has no associated gene symbol. In this study, we selected circRNA0003906 as a targeted circRNA because it may be associated with CRC according to the circRNA bioinformatics analysis.
Based on the above information, we first verified circRNA0003906 expression level in CRC cell lines and paired CRC tissues. Then, the potential relationship between circRNA0003906 expression level and clinicopathological factors of CRC patients was analyzed. Moreover, a receiver operating characteristic (ROC) curve was built to evaluate its diagnostic value. Taken together, our results indicate that circRNA0003906 may be potentially involved in the colorectal cancerogenesis and serve as a potential biomarker for the diagnosis and treatment of CRC.
Materials and methods
Tissue samples and clinical information
A total of 122 CRC tissues and matched nontumor tissues were obtained from patients who underwent surgical resection between 2013 and 2016 without prior radiotherapy or chemotherapy in the General Surgery Department of the First Affiliated Hospital of Nanchang University, People’s Republic of China. The matched nontumor tissues were generally 5 cm away from the primary tumors. Meanwhile, 40 healthy colon tissues used for diagnostic analysis were obtained from noncancer patients. The patient cohort was typical for CRC patients: 72.9% of patients were male, 72.1% were ≥60 years old, 29.4% had rectal cancers, and 2.4% had with distant metastases. None of them had received chemotherapy or radiotherapy before surgery and were identified by pathological diagnosis. Written informed consent was obtained from each patient and this study was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University. All the specimens were immediately snap-frozen and stored at −80°C in an ultra-low temperature refrigerator until further use. Clinicopathologic data was available for each of the 122 patients.
Cell culture
Normal human colon epithelial cell line (NCM460) and six CRC cell lines (SW480, SW620, HCT8, HCT116, HT29, and LoVo) were purchased from Cell Bank of Type Culture Collection (Shanghai, People’s Republic of China). The cells were incubated in Roswell Park Memorial Institute-1640 medium (Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum. They were maintained in the recommended culture conditions and incubated at 37°C in a humidified environment containing 5% CO2.
RNA extraction and quantitative real-time polymerase chain reaction
About 50 mg freshly frozen tissues from paired tissue samples or 1 million cells from cultured cell lines were obtained for total RNA extraction by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instruction. RNA quality was calculated with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA with an RNA Integrity Number more than six was considered to be eligible. Then, 1 μg total RNA was used to synthesize cDNA with random primers using the GoScript Reverse Transcription System (Promega, Madison, WI, USA). The procedure for RNA reverse transcription was as follows: incubation at 42°C for 1 h, 70°C for 10 min, and 4°C for forever. Human circRNA0003906 was selected for experimental validation using quantitative real-time polymerase chain reaction (qRT-PCR). It was achieved by using ABI 7500 Real-time PCR system (Applied Biosystems, Carlsbad, CA, USA). The melt curve of circRNA0003906 showed that the primer set only had one specific peak, suggesting that the amplification was specific and no secondary targets were produced (Figure S1). Divergent primers of circRNA0003906 were designed and synthesized by Sangon Biotech Cooperation, Ltd (Shanghai, People’s Republic of China). The primers were used to ensure that circRNA0003906 was amplified through head-to-tail junction. The amplified PCR products of circRNA0003906 were verified by DNA sequencing. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize the relative amount of gene expression. The sequences of circRNA0003906 and GAPDH primers were as follows: 5′-TCTGGCAGACCAGGGGTTAC-3′ (sense) and 5′-TGCCTTGCCGCCATCTTTTA-3′ (antisense) for circRNA0003906; 5′-GTC AGC CGC ATC TTC TTT TG-3′ (sense) and 5′-GCG CCC AAT ACG ACC AAA TC-3′ (antisense) for GAPDH. The relative expression level was calculated using the 2−ΔΔCt method with GAPDH as the control. All data was expressed as the mean ± standard deviation of three independent experiments.
Statistical analysis
Differences in continuous data between pairs of groups were detected by independent Student’s t-test, whereas clinicopathological variables were assessed by either Pearson’s χ2 test or two-sided Fisher’s exact test. These statistical analyses were performed by using IBM SPSS Statistics for Windows, Version 22.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism version 5.0 (GraphPad Software, La Jolla, CA, USA). The ROC curve was also constructed by using IBM SPSS version 20.0. A value of P<0.05 was considered to indicate statistical significance.
Results
Expression of circRNA0003906 in CRC cell lines and tissues
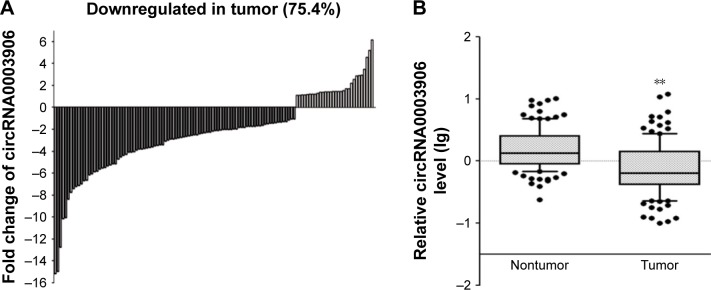
We first performed qRT-PCR to detect the expression of circRNA0003906 in six CRC cell lines. Of the six CRC cell lines, five (HCT116, SW480, SW620, HT29, and LoVo) expressed lower level of circRNA0003906 than normal colon epithelial cell line NCM460 (Figure 1). Similar to the situations in cell lines, qRT-PCR analysis of paired primary cancerous and adjacent noncancerous tissues from 122 CRC patients showed that 92 of these cases (75.4%) exhibited significant downregulation of circRNA0003906 in cancerous tissues (Figure 2A). In all the 122 tissue pairs, circRNA0003906 expression was significantly lower in tumors compared to adjacent noncancerous tissues (P<0.0001) (Figure 2B).
Figure 1.
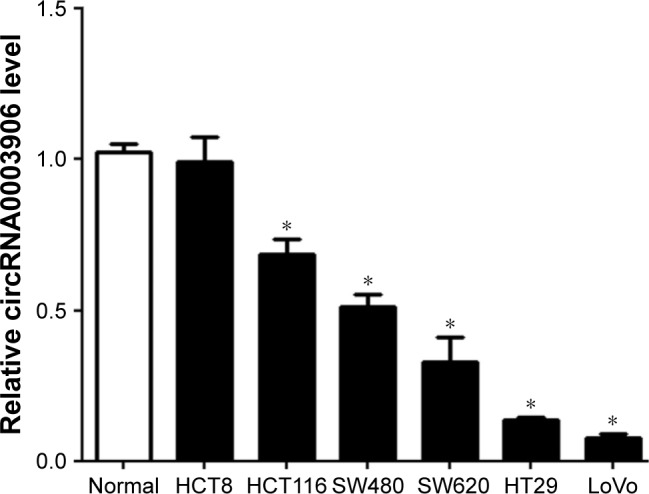
Relative circRNA0003906 expression in CRC cell lines.
Notes: circRNA0003906 expression levels in six CRC cell lines (HCT8, HCT116, SW480, SW620, HT29, and LoVo) and normal colon epithelial cell line NCM460 was determined by qRT-PCR. The values are given as mean ± SD of three independent experiments. *P<0.05.
Abbreviations: CRC, colorectal cancer; qRT-PCR, quantitative real-time polymerase chain reaction; SD, standard deviation.
Figure 2.
Relative circRNA0003906 expression in CRC tissues.
Notes: (A) Of 122 CRC patients, 92 (75.4%) exhibited significant downregulation of circRNA0003906 in cancerous tissues compared with adjacent nontumor tissues. (B) circRNA0003906 expression level in 122 CRC tissues compared with adjacent nontumor tissues. The values are mean ± SD of three independent experiments. **P<0.0001.
Abbreviations: CRC, colorectal cancer; SD, standard deviation.
Correlation between circRNA0003906 and clinicopathological factors
To verify whether circRNA0003906 could be an important molecular marker for predicting clinical outcomes in CRC patients, we assessed the correlation between circRNA0003906 expression level and clinicopathological characteristics. In clinicopathological correlation analysis, the 122 CRC patients were segregated into low and high circRNA0003906 expression groups. There were no significant correlations with age, gender, tumor diameter, T classification, TNM stage, carcinoembryonic antigen level, and carbohydrate antigen 19-9 level (Table 1). However, compared with the high circRNA0003906-expression group, the low circRNA0003906-expression group exhibited a significantly higher incidence of poor differentiation (P=0.026) and lymphatic metastasis (N classification) (P<0.001). It is well known that poor differentiation and lymphatic metastasis are both independent risk factors of CRC patients’ survival. Our result imply that circRNA0003906 may play a role as a suppressor in CRC patients.
Table 1.
Clinicopathological correlation of circRNA0003906 expression in CRC
| Variables | Tumor circRNA0003906 expression
|
P-valuea | |
|---|---|---|---|
| Low | High | ||
| Age | 0.419 | ||
| <60 years | 19 | 15 | |
| ≥60 years | 42 | 46 | |
| Gender | 0.308 | ||
| Male | 47 | 42 | |
| Female | 14 | 19 | |
| Tumor diameter | 0.560 | ||
| <5 cm | 40 | 43 | |
| ≥5 cm | 21 | 18 | |
| Differentiation | 0.026 | ||
| Well + moderate | 31 | 43 | |
| Poor | 30 | 18 | |
| T classification | 0.147 | ||
| 1–2 | 25 | 33 | |
| 3–4 | 36 | 28 | |
| N classification | <0.001 | ||
| 0 | 21 | 46 | |
| 1–2 | 40 | 15 | |
| TNM stage | 0.146 | ||
| I–II | 29 | 37 | |
| III–IV | 32 | 24 | |
| CEA | 0.389 | ||
| Negative | 45 | 49 | |
| Positive | 16 | 12 | |
| CA199 | 0.783 | ||
| Negative | 54 | 53 | |
| Positive | 7 | 8 | |
Notes: CRC patients receiving radical operation were segregated into circRNA0003906 high/low expression groups (calculation of cut-off by ROC analysis).
Statistical analyses were performed with χ2 test or Fisher’s exact test.
Abbreviations: CA199, carbohydrate antigen 19-9; CEA, carcinoembryonic antigen; CRC, colorectal cancer; ROC, receiver operating characteristics.
The diagnostic value of circRNA0003906 in CRC
To further estimate the diagnostic value of circRNA0003906 in CRC patients, a ROC curve was performed to distinguish 122 CRC tissues from 40 healthy controls. The ROC curve is a comprehensive index that can reflect on the specificity and sensitivity of continuous variables. Figure 3 shows that the area under the ROC curve was up to 0.818 (95% confidence interval =0.749–0.888; P<0.001). The specificity and sensitivity were 0.725 and 0.803, respectively. Hence, circRNA0003906 might be a diagnostic indicator of CRC.
Figure 3.
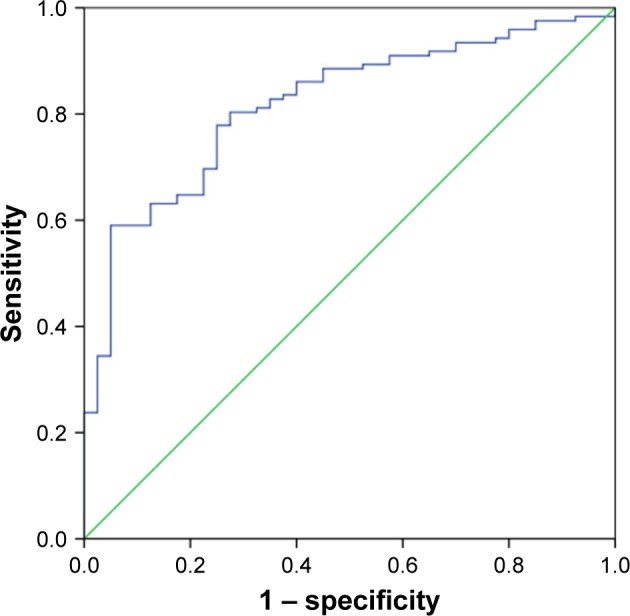
The ROC curve for differentiating CRC tissues from healthy controls.
Notes: The ROC curve was constructed using IBM SPSS Statistics for Windows, Version 22.0 (IBM Corp., Armonk, NY, USA). The area under the ROC curve was 0.818 (95% confidence interval =0.749–0.888; P<0.001). The specificity and sensitivity were 0.725 and 0.803, respectively.
Abbreviations: CRC, colorectal cancer; ROC, receiver operating characteristics.
Discussion
Accumulating evidence from the human genome sequencing projects suggests that more than 80% of the human genome is actively transcribed into RNA, even though less than 3% of the human genome codes for translated proteins.8,9 RNAs that do not code proteins are collectively called as ncRNAs. In the past two decades, many studies have deeply explored the role of ncRNAs in cell biology and carcinogenesis. In particular, the role of microRNAs in posttranscriptional regulation has been very well established, and currently their involvement in cancers has been thoroughly studied.10,11 Moreover, the involvement of specific long ncRNAs in tumor formation, progression, and metastasis in many human malignancies has also been proved.12,13 Nowadays, circRNAs are considered to be a newly rising star in governing cancer development and progression.
circRNAs, which were found more than 20 years ago from a few transcribed genes, are widely expressed in human cells.14–16 Compared with other ncRNAs, circRNAs are characterized as highly conserved sequences and a high degree of stability.17 These properties enable circRNAs to have the potential to become ideal biomarkers for the diagnosis of human diseases. It was reported that circRNAs may take part in the progression of several benign diseases, such as heart failure, atherosclerosis, and Alzheimer’s disease.18–20 Recently, the relationship between circRNAs and cancers was further investigated. Evidence revealed that circRNA100876 and circRNA0013958 were markedly upregulated in lung cancer and correlated with unfavorable clinicopathological factors,21,22 while downregulation of circRNA100269 and circRNA0001895 was observed in gastric cancer as they might act as tumor suppressor genes.23,24 To date, the most prevalent regulatory mechanism of circRNAs is that circRNAs may function by targeting miRNAs and act as miRNA sponge.18,23
In the field of CRC, a negative correlation of global reduction of circRNA abundance and proliferation is observed.25 Moreover, the dysregulation of circRNA001988, circRNA001569, circRNA0000069, and circRNA0020397 have been detected in CRC and correlated with poor clinicopathological parameters.26–29 These results strongly suggested that circRNAs might play an important role in CRC progression and have a great potential in clinical application. However, little was known about the relationship between circRNA000390 and CRC.
To our knowledge, this study is the first to identify the role of circRNA000390 in CRC. Our findings showed that circRNA0003906 expression level was significantly downregulated in both CRC tissues and CRC cell lines. It is generally known that among all the clinical factors affecting CRC survival, tumor cell differentiation is one of predominant causes for CRC metastasis and recurrence.30,31 Meanwhile, it has been well identified that lymphatic metastasis is an independent predictor of CRC survival and local recurrence.32,33 Our data indicated that the downregulation of circRNA0003906 level significantly correlated with poor differentiation (P=0.026) and lymphatic metastasis (P<0.001). Thus, it seems possible that circRNA0003906 plays an important role in the prediction of CRC survival and recurrence.
Besides detecting the role of circRNA0003906 in CRC survival and recurrence, we also explored the value of circRNA0003906 in CRC diagnosis. In this study, a ROC curve was constructed to differentiate CRC tissues from healthy controls. The area under ROC curve of circRNA0003906 was up to 0.818 (P<0.001). The specificity and sensitivity was up to 0.725 and 0.803, respectively. Hence, circRNA0003906 may be used as a potential biomarker in the diagnosis of CRC.
It should be noted that in this study, we examined the role of circRNA0003906 only by clinical analysis, and future studies should be performed in vitro and vivo to further confirm the exact role of circRNA0003906 in CRC.
Conclusion
To sum up, the present study for the first time suggests that circRNA0003906 was significantly downregulated in CRC and correlated with poor differentiation and lymphatic metastasis. Our results indicated that circRNA0003906 may play a crucial role in colorectal cancerogenesis and is a potential biomarker for CRC diagnosis and treatment.
Supplementary material
The melt curve of circRNA0003906.
Acknowledgments
This study was supported by Project Fund of Technology Department of Jiangxi Province (number 3202710) and we thank the surgeons and nurses who kindly facilitated patient information collection.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6(6):1167–1176. [PMC free article] [PubMed] [Google Scholar]
- 4.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 5.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35:1751–1756. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancermetastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 15.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 16.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 18.Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 19.Chang HY, Burd CE, Jeck WR, et al. Expression of linear and novel circular Forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genetics. 2010;6(12):e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukiw WJ. Circular RNA (circRNA) in Alzheim-er’s disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao JT, Zhao SH, Liu QP, et al. Over-expression of circRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi: 10.1111/febs.14132. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Liu H, Li W, Yu J, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY) 2017;9(6):1585–1594. doi: 10.18632/aging.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao Y, Chen L, Lu R, et al. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39(4):1010428317699125. doi: 10.1177/1010428317699125. [DOI] [PubMed] [Google Scholar]
- 25.Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057–8067. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 28.Guo JN, Li J, Zhu CL, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. doi: 10.2147/OTT.S123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis, and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41(9):1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 30.Barresi V, Reggiani Bonetti L, Ieni A, Caruso RA, Tuccari G. Histological grading in colorectal cancer: new insights and perspectives. Histol Histopathol. 2015;30(9):1059–1067. doi: 10.14670/HH-11-633. [DOI] [PubMed] [Google Scholar]
- 31.Lee JL, Yu CS, Kim M, Hong SM, Lim SB, Kim JC. Prognostic impact of diagnosing colorectal neuroendocrine carcinoma using the World Health Organization 2010 classification. Surgery. 2014;155(4):650–658. doi: 10.1016/j.surg.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 32.O’Connor ES, Greenblatt DY, LoConte NK, et al. Adjuvant chemotherapy for stage II colon cancer with poor prognostic features. J Clin Oncol. 2011;29(25):3381–3388. doi: 10.1200/JCO.2010.34.3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwitalla S, Ziegler PK, Horst D, et al. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell. 2013;23(1):93–106. doi: 10.1016/j.ccr.2012.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The melt curve of circRNA0003906.