Abstract
Purpose
Recent progress in understanding the molecular biology of epithelial ovarian cancer has not yet translated into individualized treatment for these women or improvements in their disease outcome. Gene expression has been utilized to identify distinct molecular subtypes, but there have been no reports investigating whether or not molecular subtyping is predictive of response to bevacizumab in ovarian cancer.
Experimental Design
DASL gene expression arrays were performed on FFPE tissue from patients enrolled on the ICON7 trial. Patients were stratified into four TCGA molecular subtypes. Associations between molecular subtype and the efficacy of randomly assigned therapy with bevacizumab were assessed.
Results
Molecular subtypes were assigned as follows: 122 immunoreactive (34%), 96 proliferative (27%), 73 differentiated (20%), and 68 mesenchymal (19%). In univariate analysis patients with tumors of proliferative subtype obtained the greatest benefit from bevacizumab with a median PFS improvement of 10.1 months (HR 0.55 [95%CI 0.34–0.90], p=0.016). For the mesenchymal subtype, bevacizumab conferred a non-significant improvement in PFS 8.2 months (HR 0.78 [95%CI 0.44–1.40], p=0.41). Bevacizumab conferred modest improvements in PFS for patients with immunoreactive subtype (3.8 months; p=0.08) or differentiated subtype (3.7 months; p=0.61). Multivariate analysis demonstrated significant PFS improvement in proliferative subtype patients only (HR 0.45 [95%CI 0.27–0.74 p=0.0015]).
Conclusions
Molecular subtypes with the poorest survival (proliferative and mesenchymal) derive a comparably greater benefit from treatment that includes bevacizumab. Validation of our findings in an independent cohort could enable the use of bevacizumab for those patients most likely to benefit, thereby reducing side effects and healthcare cost.
Keywords: Ovarian cancer, molecular subtypes, bevacizumab, angiogenesis
INTRODUCTION
Ovarian cancer has the highest mortality rate of all gynecologic malignancies1. Recent progress in understanding the molecular biology of epithelial ovarian cancer has not yet translated into individualized treatment for these women or improvements in their disease outcome. Most patients initially respond to platinum-based chemotherapy but the majority relapse and die from drug-resistant disease2. This underscores the significant clinical need for more effective and refined treatment strategies. Despite classifying epithelial ovarian cancer into high grade serous, endometrioid, clear cell and mucinous histologies, the disease continues to be treated with a “one size fits all” approach. Gene expression analysis of fresh frozen ovarian cancers performed in the Australian Ovarian Cancer Study and The Cancer Genome Atlas (TCGA) has led to a molecular classification of four subtypes of high-grade serous (HGS) ovarian cancer: proliferative, mesenchymal, immunoreactive and differentiated3,4. In contrast to the original TCGA report, we recently demonstrated that these four subgroups have prognostic significance when well-annotated with complete clinical follow-up5,6. Furthermore, we demonstrated that these molecular subtypes could also be used to classify high grade, advanced stage endometrioid and clear cell ovarian cancers6. However, the clinical practice of stratifying ovarian cancer patients into different targeted treatment subgroups based on their molecular classification has not yet been adopted.
In ongoing efforts to refine treatment approaches to ovarian cancer while acknowledging the unique biological differences between these four molecular subtypes of the disease, novel targeted agents are being developed and investigated. Of particular interest is, bevacizumab, an anti-angiogenic monoclonal antibody that binds to all isoforms of the vascular endothelial growth factor (VEGF)-receptor ligand VEGF-A. The Gynecologic Cancer Inter Group (GCIG) International Collaboration on Ovarian Neoplasms (ICON7) trial and the Gynecologic Oncology Group study 218 (GOG-218) were two phase III trials in ovarian cancer which showed statistically significant improvements in median progression free survival (PFS) of 2.3 and 3.8 months, respectively, when bevacizumab was added to standard first-line chemotherapy7,8. However, neither trial showed a statistically significant improvement in overall survival (OS) in unselected patients. This modest clinical improvement has led to limited use of bevacizumab in the frontline treatment of ovarian cancer. Unfortunately, there are currently no predictive biomarkers that can help to identify patients who would derive a larger clinical benefit from frontline treatment with bevacizumab. We now have information, based on gene expression data, that the mesenchymal and proliferative ovarian cancer molecular subtypes are both defined by overexpression of genes that are relevant to angiogenesis and VEGF-A, the target of bevacizumab3–5. Thus, an improved response to bevacizumab in ovarian cancer may be expected if it is used as a targeted therapeutic for patients within the angiogenic driven mesenchymal and proliferative subgroups. In this current study, we hypothesize that these gene expression-derived molecular subtypes can serve as biomarkers to identify patients with differential sensitivity to bevacizumab in ovarian cancer. In order to identify TCGA molecular subtypes, whole genome gene expression analysis was performed using stored formalin-fixed paraffin-embedded (FFPE) tumors from ICON7 trial participants who were treated with and without bevacuzimab4,7. Clinical data for these patients was subsequently analyzed for potential sub-type specific differences in outcome when treated with and without bevacizumab. The primary objective of this study was to evaluate the therapeutic treatment impact of bevacizumab on PFS based on molecular subtyping. The secondary objective of this study was to assess the impact of treatment with bevacizumab on OS based on molecular subtyping,
MATERIAL AND METHODS
Study Subjects
Patients were derived from the AGO-OVAR11 trial, the German contribution to the ICON7 multicenter phase III trial in which patients with peritoneal, tubal or ovarian carcinoma were randomized to carboplatin and paclitaxel with or without bevacizumab7. Of 533 patients enrolled in the AGO-OVAR11 trial, paraffin-embedded tissue was available for a total of 423 patients with primary ovarian, fallopian tube or primary peritoneal cancer confirmed by expert gynecopathologic review9,10. Adequate RNA (described below) was available from 391 patients, and expression array data for 359 patients passed quality control steps (described below). This resulted in a total of 359 patients for further analysis.
RNA Isolation
Using FFPE tumor, three 1mm cores were obtained from areas containing >70% tumor nuclei. Total RNA was isolated from these cores using a Qiagen AllPrep DNA/RNA FFPE kit according to the manufacturer's protocol using a Qiacube robot. RNA concentration was measured with a ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA). Real time RT-PCR was performed to amplify small regions of two abundant mRNAs: the 18S rRNA and gACTB to assess RNA quality.
Whole Genome DASL Microarray Analysis
Specimens were randomly allocated to RNA extraction and assay run order. In brief, 200 ng of RNA was analyzed using the Illumina Whole-Genome DASL HT assay with the HumanRef-8 Bead Chip (Cat. No. DA-905-1096) corresponding to 29K gene transcripts or 21K unique genes according to the manufacturer's protocol.
Gene Expression Quality Control
Gene expression data quality was assessed via residual minus versus average plots, box plots, and jitter plots to view experimental artifacts such as batch effects10. In addition, numerical measures such as stress and dfbeta, measures of the magnitude of change due to normalization, were utilized11. Criteria for exclusion were median stress >1 (0 samples were excluded) and median dfbeta >1 (35 samples were excluded). Data were normalized on the log2 scale via quantile normalization. Per-probe batch effects remaining after normalization were removed by calculating residuals from per-probe linear models12.
Molecular Subtype Assignment
Molecular classification was determined blinded to demographic and clinical information. Briefly, each given sample was assigned to a subtype according to similarity between observed expression and per-subtype expression centroids learned from TCGA. De novo clustering was also performed, confirming the existence of four subtypes (Appendix 1). In both TCGA and our own de novo clustering studies, consensus clustering approach was used to ensure that only stable clustering solutions were kept after multiple re-runs.
Statistical Analysis
As defined in the original ICON7 report7, primary endpoints were PFS and OS. Kaplan Meier curves and log rank tests were used to visualize unadjusted results. As observed previously7, non-proportional hazards were evident for PFS (p<0.0001). Thus, restricted means hypothesis tests were conducted for PFS over the duration of bevacizumab treatment (18 months) and at 36 and 42 months for unadjusted models. Restricted means measure the area under the survival curve, and so more accurately measure differences in outcome in the presence of non-proportional hazards.
Covariate-adjusted testing was conducted in a two-step manner. First, similar to a propensity score13,14, a clinical risk score was calculated by fitting a Cox regression model to all patients based on high risk of progression (ICON 7 high risk group: suboptimally cytoreduced Stage III with >1.0 cm residual disease at the end of surgery, inoperable Stage III, all Stage IV)7, age (continuous), histology (serous; other), and grade (1, 2 or 3). Second, the predicted value, the Xβ̂, was used as an offset in Cox models. Given the sample size, we focused on effect size and report actual p-values, using 0.05 as an indicator of statistical significance, and utilized Bonferroni multiple testing criteria for subgroup comparisons.
RESULTS
Patient Cohort
The baseline characteristics of the 359 patients included in the analysis were well balanced between treatment groups (Table 1). 77.2% of tumors were of serous histology and one third of patients were at high risk of progression. An optimal cytoreduction (to residual disease of ≤ 1 cm) was performed in 76.3% of patients. At a median follow-up of 26.9 months (range 0 – 43.6), 226 patients (63%) had a PFS event and 91 patients (25%) had died.
Table 1.
Baseline characteristics by treatment arm
| Bevacizumab (N=189) |
Standard (N=170) |
Total (N=359) |
|
|---|---|---|---|
| Molecular subgroup | |||
| Differentiated | 36 (19.0%) | 37 (21.8%) | 73 (20.3%) |
| Immunoreactive | 69 (36.5%) | 53 (31.2%) | 122 (34.0%) |
| Mesenchymal | 37 (19.6%) | 31 (18.2%) | 68 (18.9%) |
| Proliferative | 47 (24.9%) | 49 (28.8%) | 96 (26.7%) |
| Age (years) at randomization | |||
| Mean (SD) | 58.1 (11.0) | 57.4 (11.2) | 57.8 (11.1) |
| Range | (26.0–80.0) | (21.0–80.0) | (21.0–80.0) |
| Race | |||
| White | 189 (100.0%) | 167 (98.2%) | 356 (99.2%) |
| Asian | 0 (0.0%) | 3 (1.8%) | 3 (0.8%) |
| ECOG score | |||
| 0 | 77 (40.7%) | 78 (45.9%) | 155 (43.2%) |
| 1 | 93 (49.2%) | 83 (48.8%) | 176 (49.0%) |
| 2 | 19 (10.1%) | 9 (5.3%) | 28 (7.8%) |
| Origin of cancer | |||
| Ovary | 169 (89.4%) | 150 (88.2%) | 319 (88.9%) |
| Fallopian tube | 7 (3.7%) | 7 (4.1%) | 14 (3.9%) |
| Primary peritoneum | 12 (6.3%) | 12 (7.1%) | 24 (6.7%) |
| Multiple sites | 1 (0.5%) | 1 (0.6%) | 2 (0.6%) |
| Histology | |||
| Serous | 150 (79.4%) | 127 (74.7%) | 277 (77.2%) |
| Clear cell | 7 (3.7%) | 7 (4.1%) | 14 (3.9%) |
| Endometrioid | 4 (2.1%) | 6 (3.5%) | 10 (2.8%) |
| Mucinous | 3 (1.6%) | 2 (1.2%) | 5 (1.4%) |
| Mixed | 15 (7.9%) | 21 (12.4%) | 36 (10.0%) |
| Other | 10 (5.3%) | 7 (4.1%) | 17 (4.7%) |
| FIGO stage | |||
| I/IIA | 14 (7.4%) | 12 (7.1%) | 26 (7.2%) |
| IIB/IIC | 12 (6.3%) | 11 (6.5%) | 23 (6.4%) |
| III | 134 (70.9%) | 117 (68.8%) | 251 (69.9%) |
| IV | 29 (15.3%) | 30 (17.6%) | 59 (16.4%) |
| Outcome of surgery | |||
| Optimal | 145 (76.7%) | 129 (75.9%) | 274 (76.3%) |
| Sub-Optimal | 43 (22.8%) | 40 (23.5%) | 83 (23.1%) |
| missing | 1 (0.5%) | 1 (0.6%) | 2 (0.6%) |
| High-risk of progression* | |||
| No | 130 (68.8%) | 110 (64.7%) | 240 (66.9%) |
| Yes | 59 (31.2%) | 60 (35.3%) | 119 (33.1%) |
| Grade | |||
| 1 or 2 | 46 (24.5%) | 28 (16.8%) | 74 (20.8%) |
| 3 | 142 (75.5%) | 139 (83.2%) | 281 (79.2%) |
| Missing | 1 | 3 | 4 |
High risk of progression: suboptimal debulked stage III, inoperable Stage III, all stage IV patients.
TCGA-defined Molecular Subtype Determination
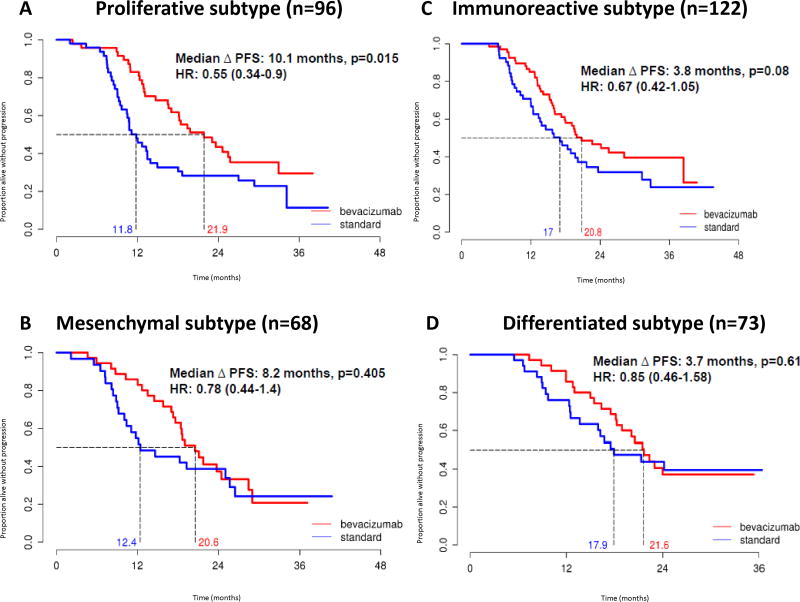
Molecular subtype assignment was as follows: 73 differentiated (20%), 122 immunoreactive (34%), 68 mesenchymal (19%), and 96 proliferative (27%). These frequencies are comparable to those found in prior studies4. When we applied unsupervised non-negative matrix factorization (NMF) clustering with k=4 groups, the average cophenetic coefficient was 0.993, indicating high reproducibility of this classification (Chi square test P<0.001) (Figure S1, 2). Baseline characteristics within molecular subgroups are shown in Table 2. Among mesenchymal and proliferative subgroups, 43% and 42% patients, respectively, met the criteria of the ICON7 high risk group, compared to 25% and 26% in the differentiated and immunoreactive subgroups, respectively4. Consistent with prior reports, patients with mesenchymal and proliferative tumors also had inferior PFS compared to the differentiated and immunoreactive subgroups (Figure 1)5,6.
Table 2.
Baseline Characteristics by Molecular Subgroups
| Differentiated (N=73) |
Immunoreactive (N=122) |
Mesenchymal (N=68) |
Proliferative (N=96) |
Total (N=359) |
|
|---|---|---|---|---|---|
| Treatment | |||||
| Bevacizumab | 36 (49.3%) | 69 (56.6%) | 37 (54.4%) | 47 (49.0%) | 189 (52.6%) |
| Standard | 37 (50.7%) | 53 (43.4%) | 31 (45.6%) | 49 (51.0%) | 170 (47.4%) |
| Age (years) at randomization | |||||
| Mean (SD) | 52.0 (12.9) | 57.0 (9.8) | 59.5 (11.3) | 62.0 (9.1) | 57.8 (11.1) |
| Range | (21.0–75.0) | (35.0–77.0) | (21.0–80.0) | (37.0–80.0) | (21.0–80.0) |
| Race | |||||
| White | 73 (100.0%) | 122 (100.0%) | 65 (95.6%) | 96 (100.0%) | 356 (99.2%) |
| Asian | 0 (0.0%) | 0 (0.0%) | 3 (4.4%) | 0 (0.0%) | 3 (0.8%) |
| ECOG score | |||||
| 0 | 29 (39.7%) | 64 (52.5%) | 23 (33.8%) | 39 (40.6%) | 155 (43.2%) |
| 1 | 38 (52.1%) | 45 (36.9%) | 42 (61.8%) | 51 (53.1%) | 176 (49.0%) |
| 2 | 6 (8.2%) | 13 (10.7%) | 3 (4.4%) | 6 (6.3%) | 28 (7.8%) |
| Origin of cancer | |||||
| Ovary | 73 (100.0%) | 105 (86.1%) | 57 (83.8%) | 84 (87.5%) | 319 (88.9%) |
| Primary peritoneal | 0 (0.0%) | 8 (6.6%) | 8 (11.8%) | 8 (8.3%) | 24 (6.7%) |
| Fallopian tube | 0 (0.0%) | 7 (5.7%) | 3 (4.4%) | 4 (4.2%) | 14 (3.9%) |
| Multiple sites | 0 (0.0%) | 2 (1.6%) | 0 (0.0%) | 0 (0.0%) | 2 (0.6%) |
| Histology | |||||
| Serous | 53 (72.6%) | 91 (74.6%) | 56 (82.4%) | 77 (80.2%) | 277 (77.2%) |
| Clear cell | 5 (6.8%) | 5 (4.1%) | 3 (4.4%) | 1 (1.0%) | 14 (3.9%) |
| Endometrioid | 3 (4.1%) | 2 (1.6%) | 1 (1.5%) | 4 (4.2%) | 10 (2.8%) |
| Mucinous | 1 (1.4%) | 1 (0.8%) | 3 (4.4%) | 0 (0.0%) | 5 (1.4%) |
| Mixed | 9 (12.3%) | 13 (10.7%) | 3 (4.4%) | 11 (11.5%) | 36 (10.0%) |
| Other | 2 (2.7%) | 10 (8.2%) | 2 (2.9%) | 3 (3.1%) | 17 (4.7%) |
| FIGO stage | |||||
| I/IIA | 8 (11.0%) | 8 (6.6%) | 3 (4.4%) | 7 (7.3%) | 26 (7.2%) |
| IIB/IIC | 10 (13.7%) | 8 (6.6%) | 1 (1.5%) | 4 (4.2%) | 23 (6.4%) |
| III | 44 (60.2%) | 91 (73.7%) | 50 (73.6%) | 66 (68.7%) | 251 (69.8%) |
| IV | 11 (15.1%) | 15 (12.3%) | 14 (20.6%) | 19 (19.8%) | 59 (16.4%) |
| Grade | |||||
| 1 or 2 | 28 (38.9%) | 16 (13.3%) | 15 (22%) | 15 (15.8%) | 74 (20.8%) |
| 3 | 44 (61.1%) | 104 (86.7%) | 53 (77.9%) | 80 (84.2%) | 281 (79.2%) |
| Missing | 1 | 2 | 0 | 1 | 4 |
| Outcome of surgery | |||||
| Optimal (< 1cm residual tumor) | 59 (80.8%) | 99 (81.1%) | 48 (70.6%) | 68 (70.8%) | 274 (76.3%) |
| Sub-Optimal (>1cm residual tumor) | 13 (17.8%) | 22 (18.0%) | 20 (29.4%) | 28 (29.2%) | 83 (23.1%) |
| Inoperable | 1 (1.4%) | 1 (0.8%) | 0 (0.0%) | 0 (0.0%) | 2 (0.6%) |
| High-risk of progression* | |||||
| No | 55 (75.3%) | 90 (73.8%) | 39 (57.4%) | 56 (58.3%) | 240 (66.9%) |
| Yes | 18 (24.7%) | 32 (26.2%) | 29 (42.6%) | 40 (41.7%) | 119 (33.1%) |
High risk of progression suboptimal debulked stage III, inoperable Stage III, all stage IV.
Figure 1.
Kaplan Meier analysis of progression free survival for bevacizumab vs. standard treatment stratified by TCGA subtype.
Bevacizumab Effects on PFS by Molecular Subtype
Univariate analysis was performed for PFS between treatment arms stratified by molecular subtypes using standard Kaplan-Meier. Multivariate analysis was then performed using Cox model analyses, adjusting for high risk of progression (suboptimal stage III, inoperable Stage III, and all stage IV patients), age, grade, and histology. Patients with proliferative and mesenchymal tumors obtained greater benefit from bevacizumab than did the immunoreactive or differentiated subgroups. Among the proliferative subgroup, median PFS improved by 10.1 months and was statistically significant (21.9 versus 11.8 months, unadjusted HR 0.55 [95% CI 0.34–0.90], p=0.016; adjusted HR 0.45 [95% CI 0.27–0.74], p=0.0015)) (Figure 1A, Table 3). In the mesenchymal subtype, non-significant prolongations in PFS of 8.2 months (20.6 versus 12.4 months, unadjusted HR 0.78 [95% CI 0.44–1.40], p=0.41). The immunoreactive subtype demonstrated a non-significant prolongation in PFS of 3.8 months (20.8 versus 17 months; unadjusted HR 0.67 [95% CI 0.42–1.05], p=0.08) (Figure 1C, Table 3). The prolongation in PFS in the differentiated subtype was 3.7 months (21.6 versus 17.9 months, unadjusted HR 0.85 [95% CI 0.46–1.85], p=0.61) (Figure 1D, Table 3). Changes in PFS for the mesenchymal, immunoreactive, and differentiated groups remained non-significant in multivariate analysis (Table 3).
Table 3.
Unadjusted and adjusted progression free and overall survival in patients treated with bevacizumab vs. standard treatment, stratified by TCGA molecular subtype
| Progression Free Survival | Overall Survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Unadjusted | Adjusted | Unadjusted | Adjusted | |||||
| HR (95% CI) |
p- value |
HR (95% CI) |
p-value | HR (95% CI) |
p- value |
HR (95% CI) |
p-value | |
| All (n=359) | 0.68 (0.53–0.89) | 0.005 | 0.64 (0.49–0.83) | 0.0.0008 | 0.68 (0.45–1.03) | 0.07 | 0.66 (0.44–1.00) | 0.05 |
| Mesenchymal (n=68) | 0.78 (0.44–1.40) | 0.41 | 0.80 (0.45–1.43) | 0.45 | 0.56 (0.23–1.34) | 0.19 | 0.55 (0.23–1.32) | 0.18 |
| Proliferative (n=96) | 0.55 (0.34–0.90) | 0.016 | 0.45 (0.27–0.74) | 0.0015* | 0.52 (0.25–1.08) | 0.08 | 0.50 (0.24–1.03) | 0.06 |
| Immunoreactive (n=122) | 0.67 (0.42–1.05) | 0.08 | 0.66 (0.42–1.05) | 0.08 | 0.76 (0.33–1.76) | 0.52 | 0.72 (0.31–1.67) | 0.45 |
| Differentiated (n=73) | 0.85 (0.46–1.58) | 0.61 | 0.75 (0.40–1.39) | 0.36 | 1.41 (0.53–3.71) | 0.49 | 1.55 (0.59–4.10) | 0.38 |
P value for subgroup comparison significant after Bonferroni correction. Adjusted analyses included high risk of progression (suboptimal stage III, inoperable Stage III, and all stage IV patients), age, grade, and histology.
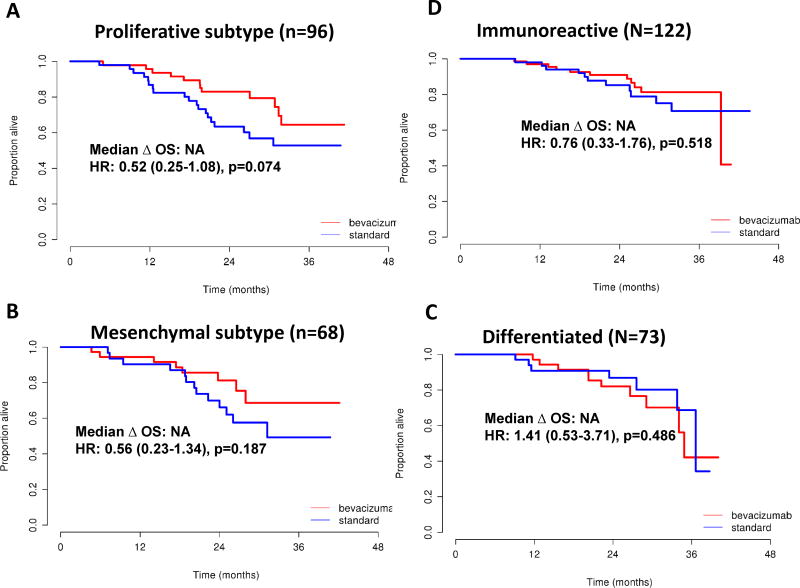
Bevacizumab Effects on OS by Molecular Subtype
The overall survival data was not mature at the time of analysis, but preliminary analysis was performed using OS between treatment arms in molecular subtypes in a univariate analysis by standard Kaplan-Meier and in a multivariate analysis using Cox model analyses, adjusting for high risk of progression (suboptimal stage III, inoperable Stage III, and all stage IV patients), age, grade, and histology. The proliferative subtype showed a trend towards improved OS (median not reached, unadjusted HR 0.52 [95% CI 0.25–1.08], p=0.08; adjusted HR 0.50 [95% CI 0.24–1.03], p=0.06) (Figure 2A, Table 3). In the mesenchymal subtype OS (unadjusted HR of 0.56] 95% CI 0.23–1.34], p=0.19) were observed (Figure 2B, Table 3). The immunoreactive subtype demonstrated an OS unadjusted HR of 0.76 [95% CI 0.33–1.76], p=0.52) (Figure 2C, Table 3). The differentiated subtype OS was unadjusted HR of 1.41 [95% CI 0.53–3.71], p=0.49) (Figure 2D, Table 3).
Figure 2.
Kaplan Meier analysis of overall survival for bevacizumab vs. standard treatment in patients, stratified by TCGA subtype.
Changes in OS for the mesenchymal, immunoreactive, and differentiated groups remained non-significant in multivariate analysis (Table 3).
Restricted Means Analysis for PFS by Molecular Subtype
As reported in the primary results for the clinical trial, there was a significant violation of the assumption of proportional hazards. Thus, we also assessed the effect of bevacizumab on PFS using a restricted means approach, as performed in the parent trial. During the first 18 months of therapy, which corresponds to the treatment duration of bevacizumab, the strongest bevacizumab treatment effect was observed in the proliferative group (p=0.0004) followed by the mesenchymal subgroup (p=0.0121); this treatment effect persisted within the proliferative subgroup only, as seen at 36 months (p=0.010) and 42 months (p=0.011) respectively (Table S1).
Bevacizumab Treatment Effects on PFS and OS
For all patients, there was an improvement in median PFS for the bevacizumab arm compared to the standard arm of 6.5 months (21.1 vs. 14.6, unadjusted HR 0.68 [95% CI 0.53–0.89], p=0.005) (Figure S3A, Table 3). When adjusted for high risk of progression, age, grade, and histology, Cox regression analysis demonstrated that bevacizumab conferred significant improvements in PFS (adjusted HR 0.64 [95% CI 0.49–0.83], p=0.0008), but OS was of borderline significance (unadjusted HR 0.68, [95% CI 0.45–1.03], p=0.07; adjusted HR 0.66, [95% CI 0.44–1.00], p=0.05) (Table 3).
CONCLUSION
The precise role of anti-angiogenic therapy of ovarian cancer continues to evolve. While both GOG 218 and ICON7 showed improvement in progression free survival with the addition of bevacizumab to standard platinum based chemotherapy in the treatment of ovarian cancer, predicting which patients will derive the greatest benefit from this anti-angiogenic therapy has remained challenging. Through work performed by the Australian Ovarian Cancer Study, the TCGA and our own group, gene expression analysis has identified four distinct molecular subtypes of high grade serous ovarian cancer with prognostic significance. Of these four types, the mesenchymal and proliferative subtype share an angiogenic gene expression signature and may respond to anti-angiogenic therapy.
Therefore, through this work, we investigated whether molecular subtyping by gene expression analysis could identify ovarian cancer patients who would preferentially benefit, in terms of PFS and OS, from the addition of bevacizumab to standard initial systemic therapy. Using archival tumor samples from a subset of women with ovarian cancer treated on the randomized ICON7 trial, we were able to reclassify these patients into four different molecular subtypes and analyze clinical response with and without the addition of bevacizumab. Overall, we were able to demonstrate that the addition of bevacizumab to carboplatin and paclitaxel in all patients resulted in a significant improvement in PFS of 6.5 months, but OS was not prolonged, reflecting the original findings from the ICON7 trial7. When assessing clinical response between all four TCGA molecular subtypes, the patients with proliferative tumors who received bevacizumab had a prolongation in PFS of 10.1 months compared to the standard control arm (no bevacizumab). This improvement in PFS for the proliferative group remained statistically significant even after adjusting for age, grade, histology, and high risk of progression (HR 0.45, p=0.0015). In addition, bevacizumab exerted a strong effect in the mesenchymal subtype with the largest benefit over the period of therapy (18 months, restricted means analysis). The immunoreactive subtype was the largest subtype (34%), yet showed a modest PFS effect (median increase in PFS of 3.8 months, p=0.08) when treated with bevacizumab but this was not accompanied by a survival benefit. Finally, patients with the differentiated molecular subtype showed the weakest treatment effect (increase in median PFS of 3.7 months, p=0.61) from bevacizumab and no survival benefit (OS HR=1.41, 95% CI: 0.53–3.71, p=0.49).
Based on findings in four large phase III trials (GOG 218, ICON7, OCEANS, AURELIA), bevacizumab was approved for first-line treatment of patients with newly diagnosed, advanced ovarian carcinoma by the European Medicines Agency. However, considering its lack of impact on OS, bevacizumab has not been granted approval by the United States Food and Drug Administration for the treatment of primary ovarian cancer. Realizing these limitations in recommending treatment for all patients with primary ovarian cancer with bevacizumab, our results may provide support for treatment of a subset of patients with ovarian cancer, thereby improving benefit while reducing risk for adverse events and treatment cost. To our knowledge, this report is the first to investigate possible molecular predictors of response to bevacizumab utilizing a randomized, placebo-controlled trial of patients with primary ovarian cancer. Others have used retrospective approaches to study genetic alterations associated with anti-angiogenic response, or mechanisms that may contribute to resistance. For example, in an analysis of 84 ovarian cancer samples, expression of Notch/Delta-like ligand 4 was shown to be lower in tumors from patients who responded to bevacizumab15. Usingtranscriptional profiling on 129 ovarian cancers, an “angiogenesis signature” was described and validated in ten gene expression datasets and associated with improved OS16, but an unknown number of patients were treated with bevacizumab. For the current study, we hypothesized that bevacizumab may be more effective in those subtypes (the mesenchymal and proliferative) with upregulation of proangiogenic genes and/or upregulation of stromal components that may secrete proangiogenic factors. For example, overexpression of SOX11, a defining alteration in the proliferative subtype, is associated with increased microvessel density17. Mesenchymal tumors are characterized by HOX upregulation, an important promoter of capillary morphogenesis and angiogenesis through VEGF18,19. Interestingly, the mesenchymal subtype of glioblastoma, a tumor which can also be divided into four unique molecular subtypes based on gene expression profling20, has the worst overall prognosis but appears to have an improved response to bevacuzimab21 and is currently the subject of ongoing clinical investigation [http://clinicaltrials.gov/show/NCT01392209]. In addition, a recent study from Sandmann et al. used a NanoString based gene expression molecular classifier to stratify patients treated in the phase III frontline glioblastoma AVAglio trial into established molecular subtypes. All patients in this study who were treated with bevacizumab had an improvement of PFS but without an OS benefit. However, when stratified by molecular subtype, the proneural group showed a significant improvement in PFS and OS with the addition of bevacuzimab22. As in our current study, the work by Sandmann et al. demonstrated the feasibility of clinical application of gene expression based classifiers to select patients benefiting most from antiangiogenic treatment22. Additional studies in breast cancer and lymphoma have implemented clinically reliable and reproducible subtyping assays in other cancers23,24.
Our study sought to establish the validity of stratifying ovarian cancer patients into molecular subtypes based on gene expression data to predict response to bevacizumab. Our results are strengthened by the use of samples from a large, randomized controlled clinical trial comparing the addition of bevacizumab to standard treatment versus standard treatment alone, with high quality robust clinical follow-up. Importantly, we were able to demonstrate the ability to obtain high quality gene expression data from FFPE tissues in 85% of cases. The Australian Ovarian Cancer Study Group has recently evaluated the feasibility of using FFPE tissue in comparison to fresh frozen tissue from the same tumor and were able to correctly classify 80% of the FFPE samples25. In addition, they compared 1) PCR-based assays (low density arrays and Fluidigm), 2) the fluorescent oligonucleotide array NanoString and 3) a targeted RNA sequencing assay (Illumina). The NanoString assay emerged as the best clinically applicable platform and can be used in individual patients for molecular subtype assignment25,26. The utility of molecular subtype assignment using the NanoString platform is currently being validated in 3000 retrospective ovarian cancer samples, by the Ovarian Tumor Tissue Analysis (OTTA) consortium. The use of FFPE has important clinical practicality in contrast to the TCGA’s use of fresh frozen material which is less readily available and inconvenient to transport. Finally, an additional strength of our data lies in the fact that rather than creating another de novo molecular classification, we used the four gene signatures previously described by the Australian Ovarian Cancer Study3 and validated by TCGA4. In contrast to the original TCGA report, we recently demonstrated that these four subgroups have prognostic significance when well-annotated with complete clinical follow-up5,6. The proliferative and mesenchymal signatures had shorter survival when compared to the immunoreactive group (adjusted OR 1.52, 1.84, respectively). It is encouraging that bevacizumab appears to confer the greatest benefit for the two molecular subgroups (proliferative and mesenchymal) with the worst prognosis. Interestingly, these two subtypes also showed the greatest benefit over the duration of bevacizumab therapy, which rapidly diminished following cessation of treatment. This suggests merit in investigating prolongation of bevacizumab therapy in patients with proliferative or mesenchymal tumors.
In this current work, we analyzed only a subset of the entire population of women treated on ICON7 but could be further strengthened by analysis of a replication cohort which was not available.
One limitation of our study was that it was limited to the German AGO samples. The number of missing data, due to missing available FFPE tissue, limited tumor tissue or purity, could skewthe remaining data to larger tumors that had inherently more available FFPE tissue. Different institutional protocols in preparation of the FFPE tumor tissue used in this study could also have had influence on the RNA quality and subsequent successful DASL array data in this study. However the samples were obtained from 98 different participating study sites in Germany, reducing the risk of site specific sample preparation. Importantly, our set mirrors the stages and histologies of all women enrolled, and like the parent trial7, a statistically significant improvement in PFS, but not OS, was observed as a result of treatment with bevacizumab. This feature of the trial, as well as the proportional hazards violation of the PFS Cox model, led us to examine outcomes using multiple methods. We have provided all results and our conclusions reflect inference across the analytical techniques. Reported p values have not been penalized for multiple comparisons, but we have indicated when a p-value has met the Bonferroni multiple comparison cutoff in the tables; actual p-values have been reported so that the reader can impose multiple comparison penalties if they wish Interaction p-values are not reported due to severe lack of power. Finally, because of our limited cohort size, we could not demonstrate a statistically significant impact on OS in the proliferative subgroup. Similarly, this study was not powered to show statistically significant differences in PFS improvement between molecular subgroups. We nevertheless consider differences in the magnitude of PFS improvement within each subtype to be clinically relevant.
In order to move towards clinical application of gene expression based subtyping in ovarian cancer, our findings need to be validated retrospectively in a validation cohort. Despite significant differences between the ICON7 and the GOG218 trial (placebo controlled, did not include low stage patients and used a higher dose of bevacizumab), it might be the best possible validation cohort to date7,8. Furthermore, newer gene expression assays, like the NanoString platform, which can be used in a Clinical Laboratory Improvement Amendments (CLIA) certified laboratory to molecular subtype ovarian cancer patients, open the possibility of prospective clinical trials to enrich for specific molecular subtypes25,26. Furthermore, our group is currently initiating an investigator initiated Phase II, open-label, single-arm, multi-center study to evaluate efficacy and safety of pembrolizumab monotherapy in subjects with advanced epithelial ovarian cancer whose tumor specimens demonstrate an immunoreactive gene expression signature. We will utilize the NanoString platform in a CLIA certified laboratory to screen and classify patients into one of the four molecular subtypes using FFPE tumor tissue. The same approach could be used to conduct prospective trials enriching for patients with molecular subtypes deriving more possible benefit from treatment with bevacizumab.
In summary, the present investigation is the first to examine the correlation of molecular subtype with outcome after treatment with bevacizumab in a randomized controlled phase III trial of primary ovarian cancer. We showed that women with the proliferative and mesenchymal molecular subtypes appear to benefit most, with a prolongation in PFS and a trend toward greater OS. Validation of our findings in an independent cohort, like GOG 218 could enable the targeted use of bevacizumab for selected patients, based on their molecular subtype.
Supplementary Material
Translational Relevance.
The majority of patients with epithelial ovarian cancer have advanced stage disease at the time of diagnosis. More than half will relapse and die within five years. While anti-angiogenic treatment plays a crucial role in ovarian cancer, no biomarker has been established to identify patients benefiting most from this treatment. In this study, we test for a correlation between molecular subtype and outcome after treatment with bevacizumab in a randomized controlled phase III trial of primary ovarian cancer. Importantly, rather than create a de novo molecular classification, we utilized molecular subtypes previously described by the Australian Ovarian Cancer Study and validated by The Cancer Genome Atlas. We demonstrate that molecular subtypes with the poorest survival (proliferative and mesenchymal) derive a comparably greater benefit from treatment which includes bevacizumab. Taken together, this data indicates that stratifying patients by molecular subtype could be an effective therapeutic strategy for ovarian cancer.
Acknowledgments
The authors would like to thank all collaborating AGO study centers and their affiliated pathologists without whose support this project would not have been possible.
Financial Support: Mayo Clinic SPORE in ovarian cancer (P50 CA136393); Mayo Clinic Comprehensive Cancer Center (P30 CA015083). Wallace and Evelyn Simmers Career Development Award; Department of Defense Ovarian Cancer Academy (W81XWH-10-1-0386).
Footnotes
Conflict of interest:
Stefan Kommoss (SK): Roche - consultancy, honoraria; AstraZeneca - honoraria.
Boris Winterhoff (BW): No conflicts of interest.
Ann L. Oberg (ALO): No conflicts of interest.
Gottfried E. Konecny (GEK): Genentech, Consulting; Amgen, Novartis, Research funding
Chen Wang (CW): No conflicts of interest.
Shaun M. Riska (SMR): No conflicts of interest.
Matthew J. Maurer (MJM): No conflicts of interest.
Jian-Bing Fan (JBF): No conflicts of interest.
Craig April (CA): I am an employee and shareholder of Illumina Inc.
Viji Shridhar (VS): No conflicts of interest.
Friedrich Kommoss (FK): No conflicts of interest.
Andreas du Bois (ADB): Personal fees from Roche, MSD, Astra Zeneca, Pharmamar and Amgen.
Felix Hilpert (FH): No conflicts of interest.
Sven Mahner (SM): Roche: Research support, Advisory Board, Honoraria, Travel Support; AstraZeneca: Research support, Advisory Board, Honoraria, Travel Support; Boehringer Ingelheim: Research support, Advisory Board, Travel Support; GlaxoSmithKline: Research support, Advisory Board, Honoraria, Travel Support
Klaus Baumann (KB): Tesaro – advisory board honoraria.
Willibald Schroeder (WS): No conflicts of interest.
Alexander Burges (AB): No conflicts of interest.
Ulrich Canzler (UC): Roche – honoraria.
Jeremy Chien (JC): No conflicts of interest.
Andrew C. Embleton (ACE): No conflicts of interest.
Mahesh Parmar (MP): No conflicts of interest.
Richard Kaplan (RK): No conflicts of interest.
Timothy Perren (TP): No conflicts of interest.
Lynn C. Hartmann (LCH): No conflicts of interest.
Ellen L. Goode (ELG): No conflicts of interest.
Sean C. Dowdy (SCD): No conflicts of interest.
Jacobus Pfisterer (JP): Speakers honoraria from Roche.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015 CA Cancer. J. Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Gavalas NG, Liontos M, Trachana SP, Bagratuna T, Arapinis C, Liacos C, et al. Angiogenesis-related pathways in the pathogenesis of ovarian cancer. Int J Mol Sci. 2013;14(8):15885–15909. doi: 10.3390/ijms140815885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clinical cancer research. 2008 Aug 15;14(16):5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011 Jun 30;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konecny GE, Wang C, Hamidi H, Winterhoff B, Kalli KR, Dering J, et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst. 2014 Oct;106(10) doi: 10.1093/jnci/dju249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winterhoff B, Hamidi H, Wang C, Kalli KR, Fridley BL, Dering J, et al. Molecular Classification of High Grade Endometrioid and Clear Cell Ovarian Cancer using TCGA Gene Expression Signatures. Gynecol Oncol. doi: 10.1016/j.ygyno.2016.02.023. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perren TJ, Swart AM, Pfisterer J, Ledermann JA, Pujade-Lauraine E, Kristensen G, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011 Dec 29;365(26):2484–2496. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- 8.Burger RA, Brady MF, Bookman MA, Fleming GF, Monk BJ, Huang H, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011 Dec 29;365(26):2473–2483. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 9.Sfakianos GP, Iversen ES, Whitaker R, Akushevich L, Schildkraut JM, Murphy SK, et al. Validation of ovarian cancer gene expression signatures for survival and subtype in formalin fixed paraffin embedded tissues. Gynecol Oncol. 2013 Apr;129(1):159–164. doi: 10.1016/j.ygyno.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kommoss S, Pfisterer J, Reuss A, Diebold J, Hauptmann S, Schmidt C, et al. Specialized pathology review in patients with ovarian cancer: results from a prospective study. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society. 2013 Oct;23(8):1376–1382. doi: 10.1097/IGC.0b013e3182a01813. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham JM, Oberg AL, Borralho PM, Kren BT, French AJ, Wang L, et al. Evaluation of a new high-dimensional miRNA profiling platform. BMC Med Genomics. 2009;2:57. doi: 10.1186/1755-8794-2-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahoney DW, Therneau TM, Anderson SK, Jen J, Kocher JP, Reinholz MM, et al. Quality assessment metrics for whole genome gene expression profiling of paraffin embedded samples. BMC Res Notes. 2013;6:33. doi: 10.1186/1756-0500-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007 Jan;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 14.Rosenbaum P, Rubin D. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. [Google Scholar]
- 15.Hu W, Lu C, Dong HH, Huang J, Shen DY, Stone RL. Biological roles of the delta family notch ligand Dll4 in tumor and endothelial cells in ovarian cancer. Cancer Res. 2011 Sep 15;71(18):6030–9. doi: 10.1158/0008-5472.CAN-10-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bentink S, Haibe-Kains B, Risch T, Fan JB, Hirsch MS, Holton K, et al. Angiogenic mRNA and microRNA gene expression signature predicts a novel subtype of serous ovarian cancer. PLoS One. 2012;7(2):e30269. doi: 10.1371/journal.pone.0030269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palomero J, Vegliante MC, Rodriguez ML, Equileor A, Castellano G, Planas-Rigol E, et al. SOX11 promotes tumor angiogenesis through transcriptional regulation of PDGFA in mantle cell lymphoma. Blood. 2014 Oct 2;124(14):2235–2247. doi: 10.1182/blood-2014-04-569566. [DOI] [PubMed] [Google Scholar]
- 18.Care A, Felicetti F, Meccia E, Bottero L, Parenza M, Stoppacciaro A, et al. HOXB7: a key factor for tumor-associated angiogenic switch. Cancer Res. 2001 Sep 1;61(17):6532–6539. [PubMed] [Google Scholar]
- 19.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010 May;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 20.Lin N, Yan W, Gao K, Wang Y, Zhang J, You Y. Prevalence and clinicopathologic characteristics of the molecular subtypes in malignant glioma: a multi-institutional analysis of 941 cases. PLoS One. 2014;9(4):e94871. doi: 10.1371/journal.pone.0094871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert M, Dignam J, Armstrong T, Wefel J, Blumenthal D, Vogelbaum M. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014 Feb 20;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandmann T, Bourgon R, Garcia J, Li C, Cloughesy T, Chinot OL, et al. Patients With Proneural Glioblastoma May Derive Overall Survival Benefit From the Addition of Bevacizumab to First-Line Radiotherapy and Temozolomide: Retrospective Analysis of the AVAglio Trial. J Clin Oncol. 2015 Sep 1;33(25):2735–44. doi: 10.1200/JCO.2015.61.5005. Epub 2015 Jun 29. Erratum in: J Clin Oncol. 2016 Sep 1;34(25):3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott DW, Wright GW, Williams PM, Lih CJ, Walsh W, Jaffe ES, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123(8) doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leong HS, Galletta L, Etemadmoghadam D, George J, Australian Ovarian Cancer Study. Köbel M, Ramus SJ, Bowtell D. Efficient molecular subtype classification of high-grade serous ovarian cancer. J. Pathol. 2015;236(3):272–277. doi: 10.1002/path.4536. [DOI] [PubMed] [Google Scholar]
- 26.Talhouk A, Kommoss S, Mackenzie R, Cheung M, Leung S, Chiu DS, Kalloger SE, Huntsman DG, Chen S, Intermaggio M, Gronwald J, Chan FC, Ramus SJ, Steidl C, Scott DW, Anglesio MS. Single-Patient Molecular Testing with NanoString nCounter Data Using a Reference-Based Strategy for Batch Effect Correction. PLoS One. 2016 Apr 20;11(4):e0153844. doi: 10.1371/journal.pone.0153844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.