SUMMARY
Interleukin 21 (IL21) plays key roles in humoral immunity and autoimmune diseases. It is known to function in mature CD4+ T follicular B cell helper (TFH) cells but its potential involvement in early T cell ontogeny is unclear. Here, we find that a significant population of newly activated thymic and peripheral CD4+ T cells functionally express IL21 soon after birth. This naturally occurring population, termed natural (n)TH21 cells, exhibits considerable similarity to mature TFH cells. nTH21 cells originating and activated in the thymus are strictly dependent on AIRE and express high levels of NUR77 consistent with a bias toward self-reactivity. Their activation/expansion in the periphery requires gut microbiota and is held in check by FoxP3+ TREG cells. nTH21 cells are the major thymic and peripheral populations of IL21+ cells to expand in an IL21-dependent humoral autoimmune disease. These studies link IL21 to T cell ontogeny, self-reactivity and humoral autoimmunity.
Graphical Abstract
INTRODUCTION
The helper T (TH) cell cytokine Interleukin 21 (IL21) acting through its broadly expressed receptor, (IL21R), is a critical driver of T-dependent humoral immune responses, supports anti-viral and anti-tumor responses but also promotes autoimmune diseases and the development of lymphomas (Davis et al., 2015; Ettinger et al., 2008; Jain et al., 2015; Spolski and Leonard, 2014). Given the importance of IL21 in health and disease there is need for a deeper understanding of its cellular ontogeny.
Current concepts concerning the cellular ontogeny of IL21 are based largely on studies of adult mice after immunization, infection, or stimulation in vitro. Such studies point to CD4+ T follicular B cell helper (TFH) cells as the major but not exclusive source of IL21. Prototypical TFH cells express high levels of CXCR5, ICOS and PD1 and localize in B cell follicles where they to help drive antigen-activated B cells to form germinal centers (GCs), affinity mature and differentiate to plasma and memory B cells. This IL21- competent stage is preceded by an IL21-negative, B cell-independent “pre-TFH” stage that is characterized by cell surface expression of ICOS and CXCR5 but not PD1 (Barnett et al., 2014; Choi et al., 2011; Crotty, 2014; Goenka et al., 2011). It is initiated when naïve cells are stimulated via their TCRs by antigens presented by DC in a milieu that includes IL6 and ICOS/ICOSL costimulatory factors (Akiba et al., 2005; Barnett et al., 2014; Choi et al., 2011; Crotty, 2014). This directs a transcriptional network governed largely by BCL6 and associated early acting factors that repress alternative TH fates by limiting expression of BLIMP1 (Choi et al., 2015; Johnston et al., 2009; Liu et al., 2014; Liu et al., 2012; Xu et al., 2015; Yu et al., 2009). This program is reinforced as pre-TFH cells re-encounter cognate antigens as they enter B cell follicles, upregulate CXCR5 and clonally expand as mature CXCR5hi PD1+ TFH and GC TFH cells expressing high levels of IL21. However, residence in B cell follicles may not be absolute as IL21 is expressed by TFH-like cells, termed extrafollicular TFH (ETFH) that help extrafollicular B cells differentiate into plasmablasts and contribute to autoimmune diseases (Bubier et al., 2009; Lee et al., 2011; Odegard et al., 2008). Moreover, natural killer T (NKT) cells, TH17 cells and intestinal CCR9+ CD4+ T cells and IFNγ memory TH cells can also be sources of IL21 (Coquet et al., 2007; Hsieh et al., 2004; Korn et al., 2009; McGuire et al., 2011; Wei et al., 2007).
As most information on the cellular ontogeny of IL21 is limited to induction and immunization paradigms, we sought here to investigate the potential involvement of IL21 in early stages of T cell development and differentiation in naïve mice. Through use of an IL21 fluorescence reporter mouse, we show that a significant population of CD4+ T cells that are activated in the thymus and periphery soon after birth functionally express IL21. This naturally occurring population, termed natural (n)TH21 cells, exhibits considerable transcriptional overlap with mature TFH cells. nTH21 cells originating in the thymus are strictly dependent on AIRE, express high levels of NUR77 and increase greatly if their TCRs are deliberately biased toward autoreactivity. Their activation/expansion in the periphery is supported by gut microbiota and is held in check by FoxP3 TREG cells. nTH21 cells are also the major thymic and peripheral populations of IL21+ cells to expand in an IL21-dependent humoral autoimmune disease. These studies link IL21 to early stages of T cell ontogeny, self-reactivity and humoral autoimmunity.
RESULTS
The IL21-VFP knock-in reliably reports IL21 expression
The IL21-VFP knock-in reporter allele contains an internal ribosomal entry site (IRES)-VFP LoxP-flanked PGK-NeoR selection cassette inserted into the non-coding portion of exon 5 of the mouse Il21 locus by targeted transgenesis in C57BL6/N (B6)-derived embryonal stem cells (Figures S1A and S1B). Founder mice were crossed to germ-line expressing cre mice to genetically excise the NeoR selection cassette. IL21-VFP heterozygotes and homozygotes lacking the selection cassette were born in expected Mendelian ratios and were healthy. There were no alterations in the proportions of splenic lymphocyte subpopulations or the frequencies of naïve cells, spontaneously activated CD4+ CD44+ T cells or CD4+ ICOS+ T cells as determined by flow cytometry, indicating that the reporter did not alter the normal immune status (Figures S1C and data not shown). No differences were noted in the percentages of VFP+ cells between heterozygous and homozygous mice (Figure S1D).
To determine if IL21-VFP reliably reports Il21 expression in vitro, we performed RT-qPCR analysis on FACS-sorted splenic VFP+ and VFP− CD4+ T cells isolated from IL21-VFP mice after stimulation in vitro with anti-CD3/CD28 antibodies. Il21 and VFP transcripts were expressed selectively and at equivalent levels by VFP+ gated cells (Figure S1E). We also found that IL21 was only in supernatants collected from VFP+ CD4+ T cells after they had been stimulated with anti-CD3/CD28 for 36 hrs, sorted, and then cultured independently for 24 hrs. IL21 was accompanied by increased expression of IL2 and IL10. In contrast, IL17 and IFNγ were most prominent in supernatants of VFP−CD4+ T cells (Figure S1F). Thus, VFP accurately reported the transcription and secretion of IL21 by activated CD4+ T cells in vitro.
To determine cellular patterns of VFP after immune challenge, IL21-VFP reporter mice were immunized with DNP-KLH. Their splenic and lymph node cells were analyzed 11 days later by flow cytometry. Consistent with VFP expression being limited to TFH and related cells, a significant fraction of CD44+ICOS+CD4+ T cells were VFP+ with many also expressing PD1 (Figure S2A) and approximately 40% of PD1+ cells being VFP+ (Figure S2B). Also notable was the lack of VFP expression by CD4+ T cells expressing the NKT cell marker NK1.1+ and other lymphocyte or leukocyte populations (Figure S2A).
To determine if IL21-VFP reliably reports intracellular IL21, we analyzed splenic CD4+ T cells from 6 wk old B6 mice that were wild type (WT) or heterozygous for IL21-VFP after stimulation with PMA/ionomycin (Figure S2C). VFP (detected by anti-GFP) and intracellular IL21 were expressed concordantly only by activated CD44+ cells with the frequencies of IL21+ cells reduced likely because of lower sensitivity of intracellular IL21 detection (Figure S2C). Similar protocols were used to compare intracellular staining of VFP and IL21 in BXSB.Yaa mice whose lupus-like disease is characterized by elevations in TFH, ETFH and IL21 (Bubier et al., 2009). Intracellular VFP and IL21 were coordinately expressed in higher intensities and frequencies by the activated CD44+ population (Figure S2D). To determine the anatomical positioning of IL21-VFP cells, we analyzed VFP+ cells in splenic sections of BXSB.Yaa IL21-VFP mice that were in an early stage of disease by immunohistochemistry. Consistent with TFH, GC TFH and ETFH as the sources of IL21, VFP+ T cells were found in B cell follicles, most strongly within PNA+ GC, as well as in T cell zones (Figure S2E). Taken together, the results show that IL21-VFP reliably reports the native patterns of IL21 expression both in vitro and in vivo.
Precocious expression of IL21 is a property of an appreciable population of activated peripheral CD4+ T cells that arise postnatally in naïve mice
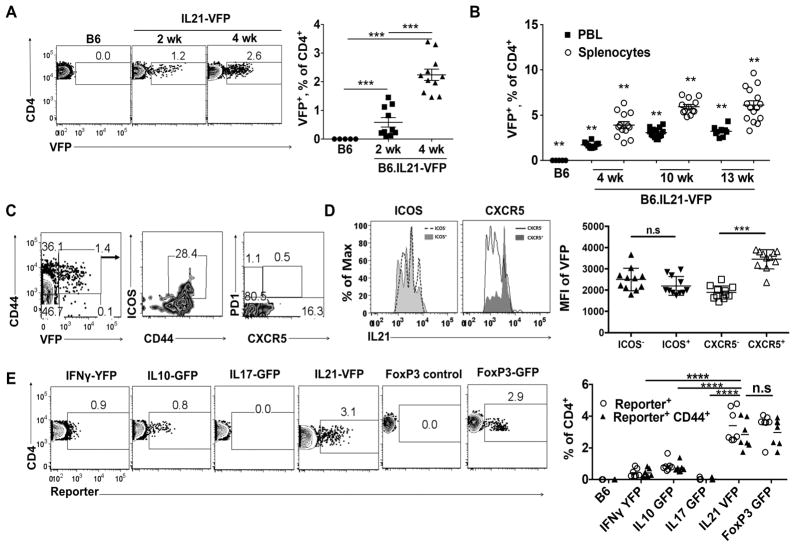
We then used the IL21-VFP reporter to investigate the patterns of IL21 expression in young naive mice. FACS analysis of spleen cells from 2–4 wk old B6.IL21-VFP mice consistently revealed small populations of VFP+ CD4+ T cells (Figure 1A). Splenic frequencies increased to ~6–7% at 13 wk, paralleled by lower percentages amongst CD4+ T cells in blood at each age (Figure 1B). Essentially all VFP+ cells expressed high levels of the pan-activation marker, CD44, while expression of ICOS and CXCR5 were limited to subpopulations of these cells (Figure 1C). Expression of VFP as measured by mean fluorescence intensity (MFI) did not differ between ICOS+ and ICOS− cells, but VFP intensities were increased in CXCR5+ cells (Figure 1D). A genetic deficiency in Cd1d1, the MHC-like selecting molecule for NKT cells, did not impact the frequencies of VFP+ CD4 T cells indicating that NKT cells were not an appreciable source of IL21 in naive mice (data not shown). We conclude that IL21 expression by CD4+CD44+ CXCR5− PD1− cells is a property of a significant fraction of spontaneously activated, conventional CD4+ T cells that accrue in the periphery of naïve mice within weeks of birth and we term them natural IL21-expressing T helper (nTH21) cells.
Figure 1. IL21-expressing CD4 T cells are the major population of activated CD4 T cells that arises in young naïve mice.
(A) Example FACS plots of splenic VFP+ CD4+ T cells from 2 and 4 wk old naïve IL21- VFP and reporter-negative control mice (B6:n=5; 2 wk:n=10; 4 wk: n=11). (B) Summary of splenic and PBL VFP+ CD4 T cell frequencies from naïve IL21-VFP mice of indicated ages. Increase over time is significant by P ≤0.02 (Kruskal-Wallis) (B6:n=5; 4wk:n=10,12; 10wk:n=13,14; 13wk: n=9,15) (C) Representative analyses of CD44, ICOS, CXCR5 and PD1 expression by VFP+ splenic CD4+ T cells from 4 wk old naïve IL21-VFP mice (n=15). (D) Left: Representative MFI of VFP in CD4 T cells based on ICOS and CXCR5 expression. Right: combined data (n=11). (E) Left: Representative FACS plots comparing reporter expression of CD4 T cells from splenocytes of 4 to 6 wk old IL21-VFP, IL17-GFP, IL10-GFP, IFNγ-YFP and FoxP3-GFP heterozygous reporter mice on a B6 background. Right: Summarized percentages of reporter-positive CD4 T cells (circle) and gated as CD44hi (triangle) (B6:n=4; IFNγ:n=7; IL10:n=8; IL17:n=4; IL21:n=7; FoxP3:n=6). n, number of mice/cohort. Representative of at least four independent experiments. ** P ≤0.01, *** P≤0.001; *** P ≤0.0001; n.s, not significant. Mean ± SD is indicated. See also Figure S1 and S2.
We next compared month old reporter mice for the frequencies of nTH21 cells vs. frequencies of cells expressing alternative T-helper cytokines, IFNγ, IL10, and IL17a, as revealed by age-matched cytokine reporters. In all cases, reporter expression was restricted to splenic CD4+ T cells showing an activated CD44+ phenotype (Figure 1E). VFP+ cells were considerably increased over cells expressing alternative cytokine reporters. Naturally activated (n) FoxP3+ CD44+ TREG cells develop in appreciable frequencies in young naïve mice (Wyss et al., 2016; Yang et al., 2015). Comparisons of splenic CD4+ T cells from naïve FoxP3-GFP reporter and IL21-VFP mice showed that the frequencies of activated CD44+ GFP+ and VFP+ populations were comparable (Figure 1E). Thus, nTH21 cells are found in the periphery of young, naive mice at frequencies rivaled only by FoxP3+ nTREG cells.
Cellular, chemokine and cytokine requirements of nTH21 cells
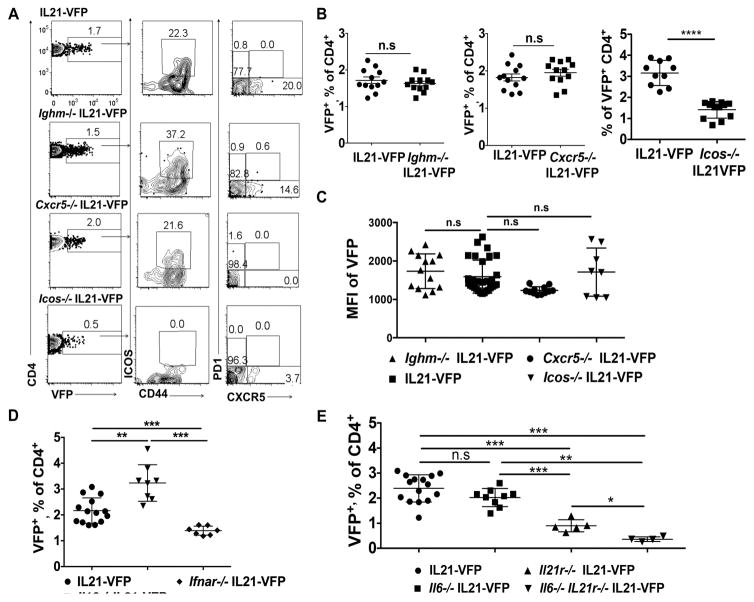
Signals from B cells, ICOS and CXCR5 contribute to the maturation of IL21-competent TFH. To determine if such signals are required for nTH21 cells we compared the frequencies of splenic VFP+ cells from naïve Ighm−/−, Cxcr5−/−, Icos−/− and WT IL21-VFP mice. The lack of B cells or CXCR5 had no significant effect on the frequencies of VFP+ cells (Figures 2A and 2B). However, the MFIs of VFP in mice lacking CXCR5 were lower (Figure 2C). A deficiency in ICOS caused a ~50% reduction in frequency (Figure 2B) but did not affect the VFP MFIs (Figure 2C). Thus, unlike prototypic TFH, B cells, ICOS and CXCR5 are not essential for the generation of nTH21 but ICOS increases their frequencies and CXCR5 enhances their expression of IL21.
Figure 2. Cellular, chemokine and cytokine requirements of IL21+ CD4 T cells.
(A) FACS plots of VFP+CD4 T cells in PBL from 4 wk old WT, Ighm−/−, Cxcr5−/− and Icos−/− IL21-VFP mice. Also shows co-expression of CD44, ICOS, CXCR5 and PD1 (n=12). (B) Frequencies of VFP+CD4 T cells in PBL from Ighm−/−, Cxcr5−/−, and Icos−/− and WT IL21- VFP mice (n=12). (C) MFI of VFP by CD4+ T cells; pooled data from PBL of 4 wk old Ighm−/− (n=13), WT (n=28) Cxcr5−/− (n=12), and Icos−/− (n=8) IL21-VFP mice. (D) Frequencies of VFP+ CD4 T cells in 5–6 wk old WT (n=14), Il10−/− IL21-VFP (n=8) and Ifnar−/− IL21-VFP mice. (E) Frequencies of VFP+ CD4 T cells in 4–6 wk old WT (n=15), Il6−/− IL21-VFP (n=9), Il21r−/− IL21-VFP (n=5) and Il6−/− Il21r−/− (n=4) IL21-VFP mice. Representative of two to four independent experiments. * P ≤0.05; ** P ≤0.01; *** P≤0.001; n.s, not significant. Mean ± SD is indicated.
We then examined the cytokine requirements of nTH21 cells. After crossing the IL21-VFP reporter onto mice homozygous for knockout alleles of Il6, Il10, Ifnar1 and Il21r, we quantified percentages of splenic nTH21 cells. Mice deficient in IL10 had significantly increased frequencies in nTH21 while a deficiency in IFNAR1 resulted in significant reductions (Figure 2D). Mice lacking IL6 showed minimal reductions, a deficiency in IL21R caused significant reductions, and mice doubly deficient in IL6 and IL21R had the greatest reductions in nTH21 (Figure 2E). Thus, nTH21 are supported by IFN1 and IL21 individually and by IL6 in combination with IL21.
Comparative RNAseq-based transcriptomics of nTH21 and other naturally arising CD4+ T cell populations
To gain insight into the molecular processes that distinguish nTH21 cells, we performed paired-end mRNAseq on FACS-purified splenic CD4+ T cells from 4 wk old IL21-VFP reporter mice based on the following criteria: naïve VFP− ICOSlocells (N); activated VFP−ICOShi cells (ACT); and VFP+ ICOSint/hi (nTH21) (Figure S3A). Pre-sort analysis indicated that the great majority of VFP+ ICOSint/hi cells were CXCR5−/PD1− negative by flow cytometry (Figure S3A–B) but transcriptional elevations of Cxcr5 and less so for Pdcd1 (gene for PD1) were detected (Figure S3C). Icos and Cd44 transcripts were similarly upregulated by ACT and nTH21 populations, whereas Il21 and VFP were expressed coordinately and almost exclusively by the gated nTH21 cells (Figure S3C).
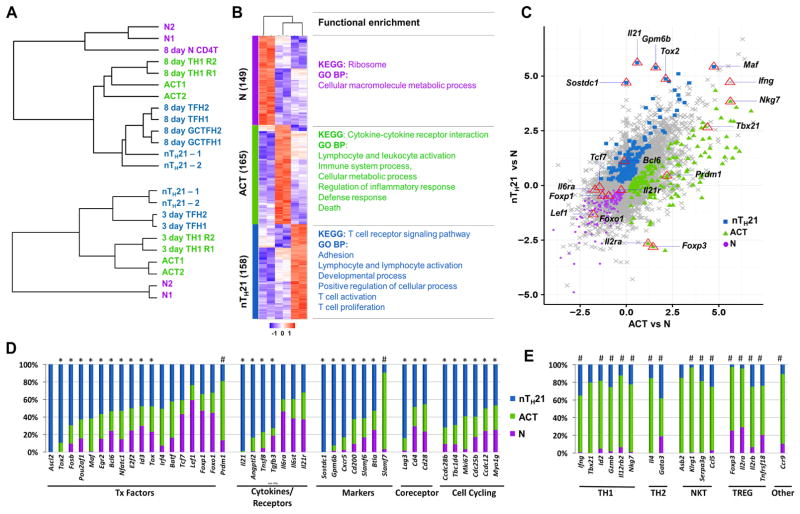
We then used hierarchical clustering to evaluate these populations in the context of published microarray datasets of polyclonal mature TFH, GC TFH, TH1 and naïve cells isolated from mice 8 days after acute infection with lymphocytic choriomeningitis virus (LCMV) (Yusuf et al., 2010) (Figure 3A top), and mRNAseq datasets derived from ‘early’ IL2Rα−Blimp1− TFH and IL2R α−Blimp1+ TH1 cells generated from naïve SMARTA TCR transgenic CD4 T cells adoptively transferred to B6 mice and harvested 3 days after infection with the Armstrong strain of LCMV (Choi et al., 2015) (Figure 3A bottom). nTH21 cells clustered closely with TFH generated 3 and 8 days following infection, whereas ACT cells clustered closely with 3 or 8 day TH1 cells.
Figure 3. RNAseq-based transcriptomic analysis of natural CD4 T cell populations.
(A) Comparisons of RNAseq profiles of N (CD4+ VFP− ICOSlo CD44−), ACT (CD4+ VFP−ICOShi CD44+) and nTH21 (CD4+ VFP+ ICOShi/int CD44+) populations with microarray-based datasets with. Top: naïve, TH1, TFH and GC TFH 8 days after an infection with LCMV (Yusuf et al., 2010). Bottom: RNAseq datasets of early TFH (IL-2Rα− Blimp1− CXCR5+ PD1lo) and early TH1 (IL-2Rα +Blimp1+CXCR5−) cells isolated 3 days after LCMV infection (Choi et al., 2015). (B) Heat map of the 471 genes that best discriminate N, ACT and nTH21 with representative KEGG and GO BP functional enrichments (P<1 × 106 and 1×10−8, respectively. (C) Scatterplot of the Log2 difference between each ACT sample and the N sample with the most discriminating N, ACT, and nTH21 genes colored. Reference genes are indicated. (D) Expression patterns of N, ACT and nTH21 for selected TFH-associated genes. Data are shown as % representation of each transcript in the specified population. *, Selectively enriched in nTH21; and #, transcripts significantly different at ≤0.05 by nTH21 in comparison to N and ACT. (E) Expression patterns of genes of interest for TH1, TH2, NKT and TREG. See also Figure S3, Table S1, Table S2 and Table S3.
We then took advantage of our N, ACT and nTH21 datasets to identify groups of genes that most reliably discriminate these populations. We minimized technical noise inherent to low read counts by including only genes having >20 transcripts per million (TPM) in at least one population. Of the 6996 genes meeting these criteria, 471 showed significantly higher expression levels in one subpopulation (Table S1). These included 148 for N; 165 for ACT; and 158 for nTH21 (Figure 3B). Functional gene enrichment analysis of these gene sets using DAVID (https://david.ncifcrf.gov/) showed that nTH21 cells were most enriched for genes associated with T cell development, differentiation, activation, proliferation and adhesion (Figure 3B, Table S2). Thus, nTH21 cells stood out for their transcriptional engagement in T cell activation and signaling processes.
As presented by a 3-way scatterplot (Figure 3C) and gene-selective comparisons (Figures 3D and 3E; Table S3), nTH21 cells were uniquely distinguished by their heightened expression of a number of transcription factors prototypic of TFH cells, including Ascl2, Fosb, Pouaf1, Tox2, and Bcl6, and low expression of the BLIMP1-encoding gene, Prdm1, comparable with N cells. High expression of archetypal TFH markers, including but not limited to Sostdc1, Btla, Cd200, Slamf6, and Gpm6b, further supported TFH relatedness. Consistent with a high activation state, nTH21 cells were distinguished by their elevated expression of TCR-related signaling genes, Lag3, Cd4 and Cd28, and a number of cell cycling genes (Figure 3D). nTH21 cells were surprisingly restricted in their effector cytokines; in addition to Il21, only Ifng was expressed at appreciable levels that were still lower than for the ACT population (Figure 3E). ACT cells, in contrast, were readily distinguished not only by their TH1-relatedness but also by expression patterns consistent with the inclusion of NKT cells, FoxP3 TREG, CCR9+ TH, and some TH2 cells (Figure 3E). Expression of key TH17 signature genes, Rorc, Il17a, and Il17f, was minimal in the nTH21 or ACT populations, arguing that few TH17 cells were present (Table S3).
We then mined our RNAseq data to evaluate Trav and Trbv usage patterns of the naïve (N), activated (ACT) and IL21-ACT (nTH21) populations. ACT showed significant bias, including substantially increased usages of Trav11 and Trav11d, while usages by nTH21 cells did not differ from those of naïve cells (Figure S3C). Taken together, the results show that IL21 expression marks an actively signaling population with diverse TCR usage that is readily distinguished from alternative CD4+ lineages and is equipped with many of the same central transcriptional factors and cell surface receptors of prototypical TFH cells.
IL6/IL21/BCL6-dependent nTH21 are the major natural T-helper population to develop in the neonatal thymus
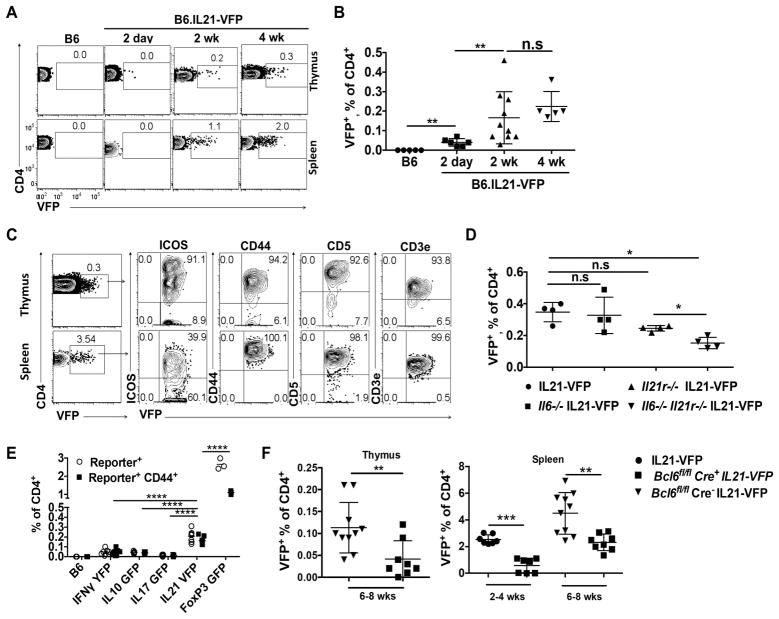
The preceding results focused on nTH21 in the periphery. To address the potential thymic ontogeny of nTH21, we performed parallel analyses of thymocytes and splenocytes from WT and naïve IL21-VFP mice at 2 d, 2 wk and 4 wk of age. VFP+ CD4+ CD8− thymocytes were detected by 2 d and increased to 0.2–0.3% at 2–4 wk with frequencies 5- to 10-fold lower than for their splenic counterparts (Figures 4A and 4B). To determine if the VFP+ thymocytes share cell surface markers with peripheral nTH21, we analyzed both populations for expression of ICOS, CD44, CD5 and CD3e. Only expression of ICOS was notably different between the thymic and splenic nTH21, with the great majority of thymic nTH21 expressing unusually high levels (Figure 4C). It should be noted that all thymic VFP+ cells are CD44+CXCR5− PD1− and are thus all nTH21 (data not shown). We then addressed their requirements for IL6 and IL21 by comparisons of nTH21 in IL21-VFP mice that were WT or deficient in IL6 and/or IL21R. Only combined deficiencies in IL6 and IL21R resulted in significant reductions of nTH21, consistent with thymic nTH21 being supported by the synergistic effects of IL6 and IL21 (Figure 4D)
Figure 4. IL6/IL21/BCL6-dependent nTH21 cells are a major natural T-helper population to develop in the neonatal thymus.
(A) Representative thymic and splenic VFP expression patterns of CD4+8− T cells from naïve IL21-VFP mice at varied ages. (B) Combined thymus data from A (see figure 1A for spleen) (B6:n=5; 2d:n=6; 2wk:n=10; 4wk:n=5) (C) Expression of ICOS, CD44, CD5 and CD3 by splenic and thymic nTH21 (VFP+ CD4+) from 6 wk old mice (n=6). (D) Comparisons of thymic nTH21 frequencies from WT, Il6−/−, Il21r−/− and Il6−/−Il21r−/− IL21-VFP mice (n=4). (E) 4–6 wk old heterozygous reporter mice were analyzed for thymic levels of IFNγ-YFP (n=6), IL10-GFP (n=5), IL17-GFP (n=5), IL21-VFP (n=6) and FoxP3-GFP (n=3). Total percentages of reporter-positive CD4+8− T cells and of those that are also CD44+ are shown. Statistics shown are comparing the CD44+ subsets. (F) Frequency comparisons of thymic and splenic nTH21 cells (VFP+ CD4+8−) from IL21-VFP heterozygous CD4-Cre+ Bcl6fl/fl, CD4-Cre− Bcl6fl/fl and Bcl6 WT mice at indicated ages (n=7–10). Representative of at least two independent experiments. * P ≤0.05, ** P ≤0.01, **** P≤0.0001; n.s, not significant. Mean ± SD is indicated. See also Figure S4.
We next used fluorescence reporters to compare the proportions of IL21+ CD4+ CD8− thymocytes with those reporting alternative cytokines (IFN, IL10, IL17) or FoxP3 (Figure 4E). nTH21 cells were the only population found in appreciable frequency by the cytokine reporters, but in lower frequencies than Foxp3+ cells. Thus, and in agreement with patterns found in the periphery, thymic nTH21 cells are a major naturally-activated population exceeded only by thymic nTREG cells.
Given the central role for BCL6 in directing antigen-induced TFH cells (Johnston et al., 2009; Nurieva et al., 2009, Yu et al., 2009; Liu et al., 2012), we used CD4-cre to conditionally delete a Bcl6fl allele in IL21-VFP mice. When analyzed at 2 and 6–8 wks of age, Cre+ Bcl6fl/fl IL21-VFP mice showed substantial reductions in thymic and splenic frequencies of nTH21 cells compared with CD4-cre− Bcl6fl/lfl littermate and IL21-VFP controls (Figure 4F). Floxing of Bcl6 did not alter the proportions of double negative, single negative and double positive or activated (CD44+) CD4+ thymocytes (Figure S4). The appreciable reductions of thymic and peripheral nTH21 cells in Bcl6-floxed mice are thus consistent with a role for BCL6 in the lineage determination of thymic and peripheral nTH21 cells. Whether the residual VFP+ cells arise from incomplete floxing or BCL6-independence as has been described for IL21+ IFNγ memory TH cells generated after infection with Plasmodium chabaudi (Carpio et al., 2015) remains to determined.
Thymic but not peripheral nTH21 require AIRE and express high levels of Nur77
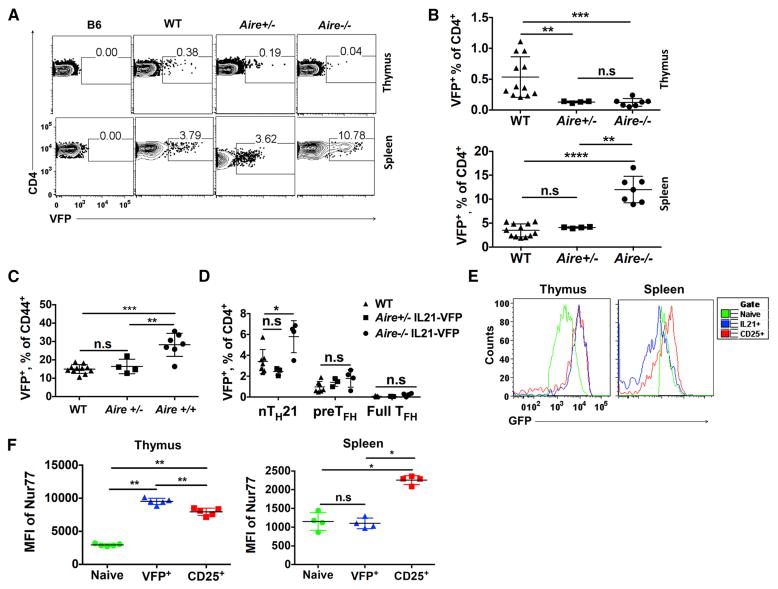
A primary function of the autoimmune regular, AIRE, is to force the ectopic expression of self-p/MHC molecules not usually encountered in the thymus, including peripheral tissue antigens (PTA), to drive negative selection (Liston et al., 2003). However, recent studies have also uncovered a more nuanced role in which AIRE also safeguards from autoimmunity by diverting the most highly self-reactive thymocytes from conventional T cells to TREG (Kieback et al., 2016; Malchow et al., 2016; Malhotra and Jenkins, 2016). Deficiency in AIRE thus results in the diminution of nTREG capacity and the emergence of self-reactive conventional CD4+ T cells that can inflict systemic autoimmune disease. As thymic nTH21 develop with comparable timing and frequencies to nTREG cells, we determined the effects of AIRE deficiency on their development. The frequencies of nTH21 cells were greatly and equivalently reduced in Aire−/− and +/− compared with WT mice suggesting that AIRE controls the thymic development of nTH21 cells in a highly dosage sensitive manner (Figures 5A and 5B). By comparison, in the periphery, AIRE deficiency resulted in a substantial increase in the overall percentages of splenic VFP+ cells and the proportion of activated CD44+ CD4+ cells that were VFP+ (Figures 5B–C). These increases were predominately in the nTH21 population (Figures 5D). Thus, AIRE supports the thymic development of nTH21 cells but it represses their expansion or generation de novo in the periphery.
Figure 5. Thymic but not peripheral nTH21 cells require AIRE and express high levels of Nur77.
(A–B) Thymic and splenic VFP+ CD4+ T cell frequencies in 7 wk old WT (n=12), Aire+/− (n= 4) and Aire−/− (n=7) IL21-VFP mice. (C) Effect of Aire copy number on frequencies of CD44+ splenic VFP+ CD4+ T cells (WT:n=11; Aire+/−:n=4; Aire−/−:n=7). (D) Splenic TH sources (CXCR5lo PD1− nTH21; CXCR5hi PD1− pre-TFH; CXCR5hi PD1+ full TFH) in WT (n=7), Aire+/− (n=4), Aire−/− (n=4) IL21-VFP mice. (E) Histograms of Nur77-GFP expression by thymic and splenic VFP+, CD25+ and naïve CD4+ T cells (n=5). (F) Summary data from E. Representative of at least three independent experiments.* P ≤0.05, ** P ≤0.01, *** P ≤0.001, **** P≤0.0001; n.s, not significant. Mean ± SD is indicated.
The strong dependence on AIRE raised the possibility that thymic nTH21 cells are positively selected by strong TCR signals in response to self-p/MHCII ligands, as has been described for thymic nTREG cells (Kieback et al., 2016; Malchow et al., 2016; Malhotra and Jenkins, 2016). Nur77 (encoded by Nr4a1) is upregulated immediately after TCR stimulation. Thus, Nur77-GFP transgenic mice provide a convenient means to proxy the strength of TCR/self-p/MHC signaling independent of cytokine induction (Hogquist and Jameson, 2014; Moran and Hogquist, 2012; Qiao et al., 2012). To test if thymic nTH21 cells are actively signaling through their TCRs, we produced Nur77-GFP, IL21-VFP dual reporter mice and used flow cytometry to analyze Nur77-GFP expression by VFP+ and CD25+ CD4+ T cells. Comparison of MFIs of GFP showed that Nur77 was highly expressed by thymic nTH21 cells almost identically to thymic CD25+ TREG cells (Figures 5E and 5 F). However, the MFI of NUR77-GFP for splenic nTH21 cells was reduced to levels comparable to those of naïve cells (Figures 5E and 5F). These results suggest that thymic nTH21 cells are responding strongly to self-p/MHCII signals and to a similar degree as thymic nTREG cells. The reduced expression of Nur77 by nTH21 in the periphery could be due to a dampened need for TCR signaling after thymic exit or by nTH21 that arise de novo in the periphery.
FoxP3+ CD4 T cells and microbial depletion restricts the expansion and maturation of peripheral but not thymic nTH21
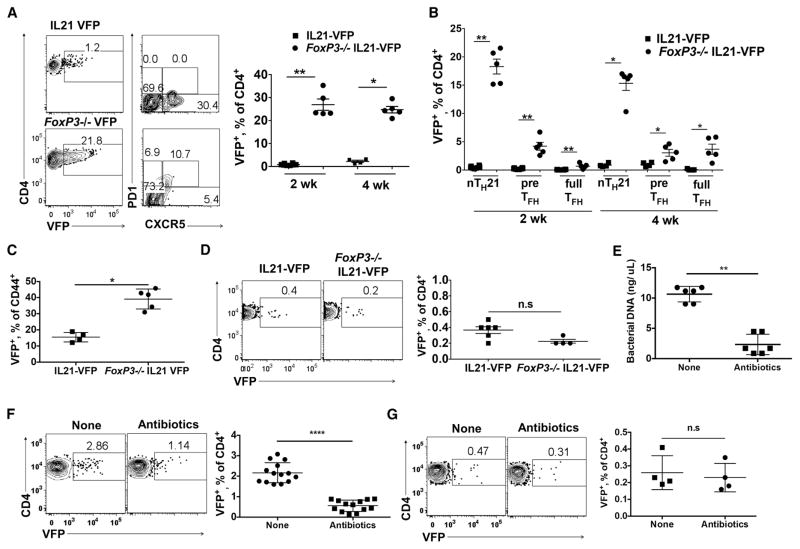
As nTREG are potent inhibitors of autoimmunity and are found in young naïve mice at frequencies comparable to thymic and peripheral nTH21 cells, we asked if they restrain nTH21 cells. Foxp3-deficient (scurfy) mice are deficient in nTREG cells and consequentially develop severe autoimmune disease within 3–4 wk of birth (Ramsdell and Ziegler, 2014). We generated Foxp3− IL21-VFP reporter mice and analyzed their male progeny at 2 wk of age, when they display few signs of disease, and at 4 wk when they display overt disease. FoxP3 deficiency caused substantial increases in splenic VFP+ cells (Figure 6A) that were primarily nTH21 rather than CXCR5+ PD1− pre- and CXCR5+ PD1+ full-TFH cells (Figure 6B). The increases in FoxP3-deficient mice were not simply due to global increases in all T-helper subsets because about 40% of CD44+ T cells were VFP+ positive (Figure 6C). However, deficiency in FoxP3 had no significant effect on thymic nTH21 frequencies (Figure 6D). Thus, nTH21 cells that develop in the thymus are insensitive to thymic nTREG cells but are greatly restrained in the periphery by this potent regulatory population.
Figure 6. FoxP3 and depletion of gut microbiota restrict the expansion and maturation of peripheral but not thymic nTH21 cells.
(A) (Left); Representative plots of VFP+ gated splenic T cells from 4 wk old Foxp3− (n=5) and WT (n=4) IL21-VFP male mice. (Right); Summary data showing 2 and 4 wks. (B) VFP+ CD4+ T cells from A at 2 weeks further distinguished as nTH21, pre-TFH or full TFH cells (VFP:n=7; FoxP3:n=5). (C) Effects of FoxP3-deficiency on frequencies of activated CD44+ splenic VFP+ CD4+ T cells (VFP:n=4; FoxP3:n=5). (D) Analysis of thymic VFP+ CD4+8− T cells from 4 wk old Foxp3− (n=4) and WT (n=5) IL21-VFP male mice. (E) Levels of fecal bacterial DNA detected by 16s RNA pyrosequencing from IL21-VFP mice untreated or treated for 4 wks with ampicillin/sulfatrim (n=6). (F) Analyses of splenic (n=13–14) and (G) thymic VFP+ CD4+ T cells (n=4) from antibiotic-treated and untreated mice. Representative of at least two independent experiments. * P ≤0.05, ** P ≤0.01, **** P≤0.0001; n.s, not significant. Mean ± SD is indicated.
It has been established that enteric microbiota support the activation and expansion of TH subsets including TFH (Block et al., 2016; Teng et al., 2016). We therefore addressed the possibility that the expansion of nTH21 in the periphery was influenced by signals from gut microbiota. Cohorts of IL21-VFP mice were treated with ampicillin and sulfatrim in their drinking water starting at 3 wk of age or were left untreated. This treatment resulted in the substantial reduction of fecal bacterial as measured by bacterial16S DNA (Figure 6E). Flow cytometric analyses indicated that antibiotic treatment dramatically reduced the frequencies of splenic VFP+ T cells while not altering the frequencies of thymic VFP+ cells (Figure 6F and 6G). Thus, stimulatory signals from the gut microbiome support nTH21 cells in the periphery while not affecting those developing in the thymus.
Peripheral and thymic nTH21 develop in unusually high frequencies in the IL21-dependent K/BXN model of autoimmune arthritis
Given that thymic development of nTH21 cells requires AIRE and constitutes the major IL21+ population to develop in normal and more so in FoxP3 deficient mice, we considered the possibility that heightening their positive selection to self-p/MHC ligands would have disease consequences. TCR transgenic KRN x NOD F1 (K/BXN) mice spontaneously develop autoantibodies against the ubiquitous glycolytic enzyme, glucose 6 phosphate isomerase (GPI), that cause severe arthritis (Korganow et al., 1999; Kouskoff et al., 1996). This humoral autoimmune disease critically requires transgenic CD4+ T cells that are positively selected by the NOD MHC class II molecule Ag7 to respond strongly to self-GPI/MHCII ligands, and is strictly dependent on IL21 (Block and Huang, 2013; Jang et al., 2009). Its severity also depends on stimulatory cues from enteric bacteria (Block et al., 2016; Wu et al., 2010)
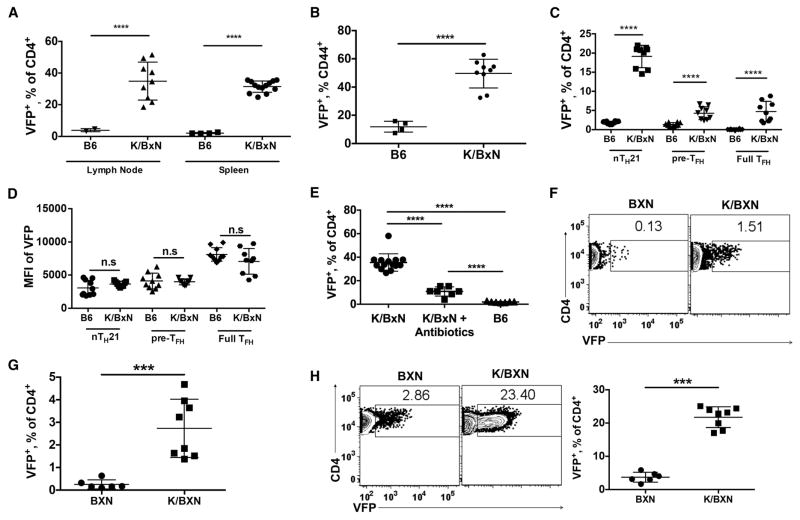
To investigate the possible involvement of nTH21 cells in this strictly IL21-dependent disorder, we produced K/BXN mice carrying the IL21-VFP reporter and characterized their IL21-expressing T cells. Spleens and mesenteric lymph nodes of 10 wk old K/BXN mice showed marked increases in VFP+ cells compared with age-matched IL21-VFP controls, averaging over 37% of CD4+ T cells and 50% of activated CD44+ cells (Figure 7A and 7B). The most striking increase was in nTH21 cells (Figure 7C), although full TFH expressed elevated IL21 as indicated by higher MFIs (Figure 7D). These increases in VFP+ cells were largely negated by antibiotic treatment indicating a key role for gut microbiota (Figure 7E).
Figure 7. Thymic and peripheral nTH21 develop in unusually high frequencies in the IL21-dependent K/BXN model of autoimmune arthritis.
(A) Analysis of VFP+ CD4+ T cells from mesenteric lymph nodes (B6:n=2; K/BxN:n=9) and spleens (B6:n=4; K/BxN:n=13) of 10 wk old B6 IL21-VFP and K/BXN IL21-VFP mice. (B) Percentages of VFP+ CD44hi splenic CD4+ T cells of B6 (n=4) and K/BxN (n=9) IL21-VFP mice. (C) VFP+ CD4 T cells further distinguished as nTH21, pre-TFH or full TFH cells (n=9). (D) MFI of VFP expression by CD4+ T cells from C. (E) Comparison of CD4+ VFP+ T cells from K/BxN (n=14), antibiotic treated K/BxN (n=7) and B6 (n=7) IL21-VFP mice. Analysis of VFP+ CD4+ T cells from (F & G) thymi and (H) spleens of 6 wk old BXN (n=6) and K/BXN (n=8) IL21-VFP mice. Representative of at least two independent experiments. *** P ≤0.001, **** P≤0.0001; n.s, not significant. Mean ± SD is indicated.
We then addressed whether these dynamics were limited to the periphery. Comparison of thymic VFP+ CD4 T cells from K/BXN with control KRN TCR transgene-negative B6xNOD F1 (BXN) IL21-VFP mice showed that K/BXN mice had striking (~10-fold) elevations in thymic TH21 cells as compared with BXN controls in parallel to similar elevations of VFP+ cells in the periphery (Figure 7F–H). Thus, skewing the T cell repertoire toward increased self-reactivity substantially enhanced the thymic selection of nTH21 resulting in abnormally high frequencies of nTH21 and matured TFH populations that depend on microbial stimuli.
DISCUSSION
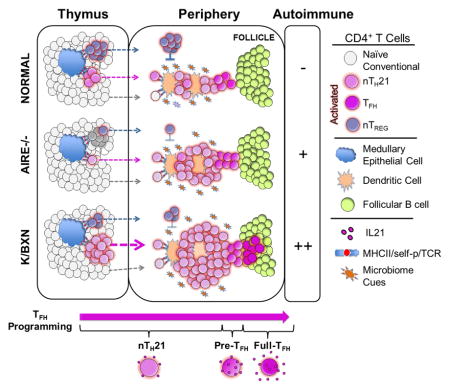
Classically, thymocytes with high TCR affinity for self-p/MHC ligands are eliminated, while those with weak but sufficient affinities are positively selected and maintained in the periphery in the naïve state through continued basal self-p/MHC signaling to await immune challenge (Hogquist and Jameson, 2014). However, this paradigm does not account for an assortment of naturally activated α/β CD4+ T cell subpopulations found in naïve mice at early age. These include nTREG, NKT, nTH17, and intestinal CCR9+ TH cells and can be further distinguished by their thymic and/or peripheral l origin (Jenkinson et al., 2015; Marks et al., 2009; McGuire et al., 2011; Wyss et al., 2016; Yadav et al., 2013). Through use of a rigorously validated IL21 reporter mouse, we describe a distinct naturally activated population in which IL21 is functionally expressed and found in surprisingly high frequencies in the thymus and periphery of mice shortly after birth and without deliberate immune challenge.
Transcriptomic analyses provided a glimpse into lineage bifurcations that are established at an early age without immune challenge. Whole transcriptome comparisons of naïve, activated IL21− and IL21+ nTH21 splenic cells with published data from purified early and mature TFH, GC TFH and TH1 cell populations induced after viral infection clearly established that nTH21 cells were uniquely correlated with early and mature TFH and GC TFH and antithetically to TH1 cells. In contrast, the IL21− activated population not only correlated highly with prototypic TH1 but also showed transcriptional evidence for alternative naturally-activated populations, including nTREG, NKT, and CCR9+ TH cells.
nTH21 cells stood out in their high expression of an assemblage of markers and cytokines prototypic of TFH and their low expression of genes, including Prdm1, indicative of alternative lineages. A TFH-like ontogeny was further supported by transcription factors typical of TFH that were specifically increased in nTH21 cells, including Ascl2, Tox2, Fosb, Pou2af1, Egr2, Maf and E2f2 and Bcl6. However, some TFH-associated transcription factors, including Lef1, Ifr4, Batf, were unchanged or minimally discriminated nTH21 from activated IL21− cells. Moreover, the expression patterns of FoxO1 and FoxP3, that are considered to act early on to repress antigen-induced TFH cells by negative regulation of BCL6 and IL21, respectively (Stone et al., 2015; Wang et al., 2014; Weber et al., 2015), were not consistent with their repression of nTH21 development or IL21 expression. We suggest that nTH21 is an early stage in TFH ontology that is distinguished from alternative naturally activated populations by much of the transcriptional machinery that specifies prototypical TFH cells.
nTH21 being an early stage of TFH ontology is further supported by gene knockout studies. According to concepts based on immunization paradigms in adult mice, TFH programming is initiated by encounter with antigen-presenting DC. This pre-TFH stage is characterized by the upregulation of BCL6 and cell surface expression of ICOS and CXCR5 without expression of IL21 (Akiba et al., 2005; Barnett et al., 2014; Choi et al., 2011; Crotty, 2014; Goenka et al., 2011). Similar to previously described pre-TFH, we found that nTH21 cell development is largely dependent on BCL6 and is unimpaired in B cell-deficient mice. However, nTH21 cells differed in that ICOS and CXCR5 were not required for IL21 expression. A genetic deficiency in ICOS resulted in a 50% reduction in nTH21 but not lower IL21 expression. A deficiency in CXCR5, in contrast, lowered IL21 expression but did not affect the overall frequencies of nTH21 cells. Thus, programming of nTH21 cells likely initiates when naïve cells are first activated by DC and receive early inductive signals from BCL6 and to a lesser extent ICOS. This results in nTH21 cells expressing IL21 independent of ICOS or CXCR5 signals but with heightened expression in those that have matured to CXCR5+ pre-TFH cells.
Our study of mice lacking IFN1, IL10, IL6 and IL21 signals is also consistent with TFH-relatedness of nTH21 cells while also documenting the tonic effects of these cytokines in young naïve mice. IFN1 signaling in DC stimulates TFH (Cucak et al., 2009) although IFN1 represses TFH cells and IL21 expression after viral infections (Nakayamada et al., 2014; Ray et al., 2014). We found that tonic IFN1 supports nTH21 which may suggest its role in DC conditioning. We also found that nTH21 cells are suppressed by IL10, which is in agreement with its suppression of TFH (Cai et al., 2012). IL6 supports prototypical TFH cells throughout their development and maturation and acts synergistically for IL21 at later stages (Eto et al., 2011). Consistent with synergy of IL6 and IL21, we found that a double deficiency in IL21R and IL6 substantially lowered the frequencies of nTH21 cells. Moreover, the fact that an IL21R deficiency reduced thymic and peripheral nTH21 cells provides strong evidence that IL21 is secreted and functionally consequential in both anatomical locations. Whether IL21 supports nTH21 directly through autocrine feedback (Nurieva et al., 2007) or by paracrine effects on DC and other APC, even during thymic development, remains to be determined.
In which sites do nTH21 cells originate? Our results are consistent with both thymic and peripheral origins in a similar manner to that described for naturally-activated FoxP3 TREG cells. Natural TREG can arise as a direct result of thymic selection (referred to as thymic TREG and most recently as triplehi FoxP3 TREG) or by the induction of naïve cells in the periphery (pTREG) (Wyss et al., 2016; Yadav et al., 2013). It is quite unlikely that thymic nTH21 derive from the peripheral pool because thymic nTH21 cells rarely exceed 0.4% of CD4+ CD8− thymocytes despite 5- to 10-fold higher frequencies in the periphery that accrue as normal mice age and 100-fold higher frequencies in FoxP3-deficient mice. Moreover, nTH21 adoptively transferred into the periphery of T cell-deficient mice do not access thymi despite having appreciable peripheral frequencies (data not shown). Conversely, peripheral nTH21 cells were found in normal frequencies in neonatally thymectomized mice (data not shown). Moreover, we found that peripheral nTH21 cells are greatly reduced by antibiotic treatment while thymic frequencies were not affected. Our results are thus consistent with thymic and peripheral origins of nTH21 cells with the former being insulated from peripheral stimuli until they merge with those induced in the periphery.
The diametrically opposed effects of AIRE provide further insights into differential modes of thymic and peripheral nTH21 cell induction. Recent findings showed that AIRE channels a significant fraction of most highly self-reactive T cell clones that escape negative thymic selection into a TREG lineage (Kieback et al., 2016; Malchow et al., 2016; Malhotra and Jenkins, 2016). Similarly, we found that thymic development of nTH21 is dependent on AIRE in a dose-dependent manner. Our results thus support an integral role for AIRE-dependent self-antigens in the positive thymic selection of nTH21, similarly to that described for thymic nTREG cells.
In contrast, we found that mice deficient in AIRE had substantial increases in peripheral nTH21 cells despite their reduction in the thymus. A deficiency in AIRE mistakenly diverts highly self-reactive thymocytes from TREG cells to aggressive self-reactive conventional T cells that “go rogue” upon escape into the periphery (Kieback et al., 2016; Malchow et al., 2016; Malhotra and Jenkins, 2016). Still to be determined is whether this increase can be attributed to the selective peripheral expansion of already activated nTH21 cells surviving thymic selection or by naïve self-reactive thymic emigrants freshly activated on encounter with new antigens and/or microbial cues in the periphery.
Other features shared by nTH21 and nTREG cells are their emergence in early post-natal life in frequencies that exceed other naturally activated thymic and peripheral CD4+ T cell populations. Consistent with positive selection and activation by strong avidity TCR/self-p/MHC signals, thymic nTH21 and nTREG cells both express high levels of Nur77 and are heavily dependent on AIRE. TCR transgenic mice have been instrumental in establishing the importance of strong TCR/self-p/MHC reactivity in the preferential positive selection of TREG cells (Hsieh et al., 2004; Kieback et al., 2016; Malchow et al., 2016). In a similar manner, the marked elevations of thymic and peripheral nTH21 cells that we found in KRN TCR transgenic K/BXN mice, whose development of pathogenic TFH is strictly dependent on the NOD H2g7 MHC, suggests that an alternative outcome of selection for strong TCR/self-p/MHC reactivity is the enhanced positive selection of potentially dangerous nTH21 cells. We also found that the absence of FoxP3 results in substantial increases in peripheral nTH21, findings consistent with reports showing that IL21+ T cells are increased in scurfy mice (Iwamoto et al., 2014; Wing et al., 2014). Thus, nTH21 and nTREG cells may develop self-reactivity through similar selection processes and at comparable frequencies but with opposing regulatory and autoimmune capabilities.
Our identification of nTH21 cells that arise in neonatal life may shed light on the origins of TFH and related IL21-expressing extrafollicular TH cell populations that contribute to autoimmune diseases. A feature shared by Aire−/−, Foxp3−/− and K/BxN mice is substantial increases in activated peripheral IL21+ CD4 T cells. Among these IL21+ populations, the largest increase was consistently seen in nTH21 rather than mature TFH. This points to the probable importance of nTH21 as the stage at which failure to control self-reactivity leads to the development of autoimmune disease. Thus, failure to limit the activation, expansion and further maturation of this intrinsically self-reactive population may underlie autoimmune disorders in which IL21, TFH, and autoantibodies are important pathogenic drivers.
EXPERIMENTAL PROCEDURES
Generation of IL21-VFP reporter mice
The bicistronic IL21-VFP knock-in allele (Il21tm1.1Hm) was generated in C57BL/6N ES cells by Ozgene Pty. Ltd. (Bentley WA) under the direction of HCM part of a contract with NIAID, NIH. Both hemizygous and homozygous IL21-VFP mice were used with zygosity consistent within each experiment. B6.IL21-VFP mice are available on request (D.C.R., H.C.M.) See Supplemental Experimental Procedures for reporter design and genotyping information.
Mice
All mouse work was conducted according to national guidelines and approved by the Institutional Animal Care and Use Committee of The Jackson Laboratory (AUS 01022). All animals were maintained in a specific pathogen-free mouse colony and experiments were performed using C57BL/6-background mice except where otherwise indicated. Experiments involving the IL21-VFP reporter were either hemizygous and homozygous for IL21-VFP with zygosity consistent within each experiment. Cohorts were age matched within one wk of age with similar numbers of male and female mice in each experiment unless otherwise noted. Ages of mice were between two and thirteen weeks of age. Details on the mouse strains, crosses, genotyping and breeding are described in Supplemental Experimental Procedures.
Flow Cytometry
Analytical flow cytometry was performed by established procedures after FMO gating of viable cells (McPhee et al., 2013) using antibodies listed in Supplemental Experimental Procedures. Samples were run on a four-laser/13-color BD LSRII cytometer (BD Biosciences) and analyzed with FlowJo software version 8 or 9 (Tree Star). For spectral discrimination of VFP and GFP, a 510/20 filter was used in combination with the 505LP filter for detection of GFP in the FITC channel, and a 542/27 filter was used in combination with a 525L filter for detection of VFP/YFP in the PE channel. In each experiment, compensation and gating were based on cells single positive for GFP or VFP.
T Cell Purification
For RNAseq splenic CD4+ T cells were isolated using Milltenyi streptavidin microbeads (Milltenyil; BD Bioscience) coupled with CD11b, CD11c, B220 and CD8 antibodies followed by a pass through the autoMACS pro separator (Milltenyi) according to the manufacturer’s protocols. Isolation of N, ACT and nTH21 cells for RNAseq was then performed by FACS based on VFP, anti-CD4 and anti-ICOS detection using a FACSAria (BD Biosciences).
RNAseq
Total RNA from purified N, ACT and nTH21 spleen cell pools were extracted with Qiagen RNeasy Mini Kits (Qiagen, Hilden, Germany). Poly-A-enriched mRNA was reverse transcribed and amplified using the NuGEN Ovation Kit (NuGEN, San Carlos, CA, USA). Paired-end cDNA was sequenced with an Illumina MiSeq at 106 base pair length (Illumina, San Diego, CA, USA). Reads were checked with FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit), trimmed with Trimmomatic, and aligned to the GRCm38.73 assembly transcriptome with Bowtie. Transcript expression levels were estimated in transcripts per million (TPM) using RSEM (Li and Dewey, 2011). Differential expression analysis across the sorted populations was conducted with EBSeq (Leng et al., 2013). Detailed analytical procedures are described in Supplemental Experimental Procedures
Immunization and cell culture
Mice were immunized intraperitoneally with 200μL Dinitrophenyl conjugated keyhole limpet hemocyanin (DNP-KLH) emulsified in complete Freund’s adjuvant and analyzed 7–11 days after immunization. For in vitro stimulation, total splenocytes from naïve B6.IL21-VFP mice depleted of red blood cells by exposure to ACK buffer were cultured in 6 well plates with 5 μg/ml of anti-CD3 and 1 μg/ml of anti-CD28 for 24 or 36hrs in 5% FBS-supplemented DMEM. Cells were then stained and sorted for being CD4+VFP− and CD4+VFP+. Gene expression analysis by RT-qPCR and serum cytokine analysis after in vitro stimulation are described in Supplemental Experimental Procedures.
Immunohistochemistry
Detailed protocols and reagents are described in Supplemental Experimental Procedures.
Statistical analyses
Statistical analyses were performed in GraphPad Prism v6 using non-parametric tests that do not assume a normal distribution. The Mann-Whitney U test was used for two-way comparisons and Kruskal-Wallis was used for multigroup comparisons. “n” refers to the number of individual mice sampled per cohort with Mean ± SD indicated. Comparisons of p ≤ 0.05 were considered to be significant. All experiments were repeated at least twice to ensure reproducibility and reliable interpretations. RNAseq analysis is described under “RNAseq” and Supplemental Experimental Procedures.
Supplementary Material
Document S1: Supplemental Experimental Procedures and Figures S1–S4.
Related to Figure 3. Excel spreadsheet contains expression and statistical data for significant genes enriched in the N, ACT and nTH21 populations.
Related to Figure 3. Excel spreadsheet contains the KEGG and GO pathway enrichment terms and statistics for the N, ACT and nTH21 populations. Tab 1 contains KEGG terms. Tab 2 contains GO terms.
Related to Figure 3. Excel spreadsheet contains the gene expression and statistical information for genes that are hallmarks of TFH cells or other CD4+ T helper subsets.
Acknowledgments
We thank J. Ward (Global VetPathology) for assistance with IHC images; D. Serreze (The Jackson Laboratory) for manuscript review; Gene Expression Service (The Jackson Laboratory) for RNA sequencing; John Wilson and Porcia Mandanhar for technical assistance. Supported in part by the Alliance for Lupus Research (D.C.R.), New England Biolabs (D.C.R), The Gina M. Finzi memorial student summer fellowship from the Lupus Foundation of America (E.A.M.), The Jackson Laboratory summer student program (S.L.K.), the Intramural Research Program of the NIH, NIAID (H.C.M., S.J., and H.W.) and The Pyewacket Foundation (X.W. and G.W.C.).
Footnotes
Accession Numbers
Author Contributions
Conceptualization, D.C.R., H.C.M., G.W.C., and E.A.M.; Methodology, D.C.R., H.C.M., G.W.C., E.A.M and X.W.; Software, G.W.C., and X.W.; Formal Analysis, E.A.M., and X.W.; Investigation, E.A.M, X.W, G.W.C, T.J.S., G.P., G.J.C., S.L.K., T.D. and S.J.; Resources, H.C.M., D.C.R., G.W.C., H.W., and T.D.; Data Curation, X.W., and G.W.C.; Writing – original draft, D.C.R., H.C.M., G.W.C., and E.A.M.; Writing - Review and editing, D.C.R., H.C.M., G.W.C., X.W., H.W., and E.A.M.; Visualization, E.A.M., S.J., and X.W.; Supervision, D.C.R., H.C.M., and G.W.C.; Funding Acquisition, D.C.R., H.C.M., and G.W.C.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiba H, Takeda K, Kojima Y, Usui Y, Harada N, Yamazaki T, Ma J, Tezuka K, Yagita H, Okumura K. The role of ICOS in the CXCR5+ follicular B helper T cell maintenance in vivo. J Immunol. 2005;175:2340–2348. doi: 10.4049/jimmunol.175.4.2340. [DOI] [PubMed] [Google Scholar]
- Barnett LG, Simkins HM, Barnett BE, Korn LL, Johnson AL, Wherry EJ, Wu GF, Laufer TM. B cell antigen presentation in the initiation of follicular helper T cell and germinal center differentiation. Journal of immunology. 2014;192:3607–3617. doi: 10.4049/jimmunol.1301284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block KE, Huang H. The cellular source and target of IL-21 in K/BxN autoimmune arthritis. J Immunol. 2013;191:2948–2955. doi: 10.4049/jimmunol.1301173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block KE, Zheng Z, Dent AL, Kee BL, Huang H. Gut Microbiota Regulates K/BxN Autoimmune Arthritis through Follicular Helper T but Not Th17 Cells. J Immunol. 2016;196:1550–1557. doi: 10.4049/jimmunol.1501904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubier JA, Sproule TJ, Foreman O, Spolski R, Shaffer DJ, Morse HC, 3rd, Leonard WJ, Roopenian DC. A critical role for IL-21 receptor signaling in the pathogenesis of systemic lupus erythematosus in BXSB-Yaa mice. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1518–1523. doi: 10.1073/pnas.0807309106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Nie X, Zhang W, Wu B, Lin J, Wang H, Jiang C, Shen Q. A regulatory role for IL-10 receptor signaling in development and B cell help of T follicular helper cells in mice. J Immunol. 2012;189:1294–1302. doi: 10.4049/jimmunol.1102948. [DOI] [PubMed] [Google Scholar]
- Carpio VH, Opata MM, Montanez ME, Banerjee PP, Dent AL, Stephens R. IFN-gamma and IL-21 Double Producing T Cells Are Bcl6-Independent and Survive into the Memory Phase in Plasmodium chabaudi Infection. PloS one. 2015;10:e0144654. doi: 10.1371/journal.pone.0144654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, Crotty S. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquet JM, Kyparissoudis K, Pellicci DG, Besra G, Berzins SP, Smyth MJ, Godfrey DI. IL-21 is produced by NKT cells and modulates NKT cell activation and cytokine production. J Immunol. 2007;178:2827–2834. doi: 10.4049/jimmunol.178.5.2827. [DOI] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucak H, Yrlid U, Reizis B, Kalinke U, Johansson-Lindbom B. Type I interferon signaling in dendritic cells stimulates the development of lymph-node-resident T follicular helper cells. Immunity. 2009;31:491–501. doi: 10.1016/j.immuni.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Davis MR, Zhu Z, Hansen DM, Bai Q, Fang Y. The role of IL-21 in immunity and cancer. Cancer Lett. 2015;358:107–114. doi: 10.1016/j.canlet.2014.12.047. [DOI] [PubMed] [Google Scholar]
- Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PloS one. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger R, Kuchen S, Lipsky PE. The role of IL-21 in regulating B-cell function in health and disease. Immunological reviews. 2008;223:60–86. doi: 10.1111/j.1600-065X.2008.00631.x. [DOI] [PubMed] [Google Scholar]
- Goenka R, Barnett LG, Silver JS, O’Neill PJ, Hunter CA, Cancro MP, Laufer TM. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. Journal of immunology. 2011;187:1091–1095. doi: 10.4049/jimmunol.1100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC. The self-obsession of T cells: how TCR signaling thresholds affect fate ‘decisions’ and effector function. Nature immunology. 2014;15:815–823. doi: 10.1038/ni.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Iwamoto T, Suto A, Tanaka S, Takatori H, Suzuki K, Iwamoto I, Nakajima H. Interleukin-21-producing c-Maf-expressing CD4+ T cells induce effector CD8+ T cells and enhance autoimmune inflammation in scurfy mice. Arthritis & rheumatology (Hoboken, NJ) 2014;66:2079–2090. doi: 10.1002/art.38658. [DOI] [PubMed] [Google Scholar]
- Jain S, Chen J, Nicolae A, Wang H, Shin DM, Adkins EB, Sproule TJ, Leeth CM, Sakai T, Kovalchuk AL, et al. IL-21-driven neoplasms in SJL mice mimic some key features of human angioimmunoblastic T-cell lymphoma. The American Journal of Pathology. 2015;185:3102–3114. doi: 10.1016/j.ajpath.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang E, Cho SH, Park H, Paik DJ, Kim JM, Youn J. A positive feedback loop of IL-21 signaling provoked by homeostatic CD4+CD25- T cell expansion is essential for the development of arthritis in autoimmune K/BxN mice. J Immunol. 2009;182:4649–4656. doi: 10.4049/jimmunol.0804350. [DOI] [PubMed] [Google Scholar]
- Jenkinson WE, McCarthy NI, Dutton EE, Cowan JE, Parnell SM, White AJ, Anderson G. Natural Th17 cells are critically regulated by functional medullary thymic microenvironments. Journal of autoimmunity. 2015;63:13–22. doi: 10.1016/j.jaut.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science (New York, NY) 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieback E, Hilgenberg E, Stervbo U, Lampropoulou V, Shen P, Bunse M, Jaimes Y, Boudinot P, Radbruch A, Klemm U, et al. Thymus-Derived Regulatory T Cells Are Positively Selected on Natural Self-Antigen through Cognate Interactions of High Functional Avidity. Immunity. 2016;44:1114–1126. doi: 10.1016/j.immuni.2016.04.018. [DOI] [PubMed] [Google Scholar]
- Korganow AS, Ji H, Mangialaio S, Duchatelle V, Pelanda R, Martin T, Degott C, Kikutani H, Rajewsky K, Pasquali JL, et al. From systemic T cell self-reactivity to organ-specific autoimmune disease via immunoglobulins. Immunity. 1999;10:451–461. doi: 10.1016/s1074-7613(00)80045-x. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annual review of immunology. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Kouskoff V, Korganow AS, Duchatelle V, Degott C, Benoist C, Mathis D. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- Lee SK, Rigby RJ, Zotos D, Tsai LM, Kawamoto S, Marshall JL, Ramiscal RR, Chan TD, Gatto D, Brink R, et al. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. The Journal of experimental medicine. 2011;208:1377–1388. doi: 10.1084/jem.20102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. doi:1010.1093/bioinformatics/btt1087. Epub 2013 Feb 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. The Journal of experimental medicine. 2012;209:1841–1852. S1841–1824. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malchow S, Leventhal DS, Lee V, Nishi S, Socci ND, Savage PA. Aire Enforces Immune Tolerance by Directing Autoreactive T Cells into the Regulatory T Cell Lineage. Immunity. 2016;44:1102–1113. doi: 10.1016/j.immuni.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Jenkins MK. Regulatory T Cells: A Crisis Averted. Immunity. 2016;44:1079–1081. doi: 10.1016/j.immuni.2016.04.025. [DOI] [PubMed] [Google Scholar]
- Marks BR, Nowyhed HN, Choi JY, Poholek AC, Odegard JM, Flavell RA, Craft J. Thymic self-reactivity selects natural interleukin 17-producing T cells that can regulate peripheral inflammation. Nat Immunol. 2009;10:1125–1132. doi: 10.1038/ni.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire HM, Vogelzang A, Ma CS, Hughes WE, Silveira PA, Tangye SG, Christ D, Fulcher D, Falcone M, King C. A subset of interleukin-21+ chemokine receptor CCR9+ T helper cells target accessory organs of the digestive system in autoimmunity. Immunity. 2011;34:602–615. doi: 10.1016/j.immuni.2011.01.021. [DOI] [PubMed] [Google Scholar]
- McPhee CG, Bubier JA, Sproule TJ, Park G, Steinbuck MP, Schott WH, Christianson GJ, Morse HC, 3rd, Roopenian DC. IL-21 is a double-edged sword in the systemic lupus erythematosus-like disease of BXSB.Yaa mice. Journal of immunology. 2013;191:4581–4588. doi: 10.4049/jimmunol.1300439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran AE, Hogquist KA. T-cell receptor affinity in thymic development. Immunology. 2012;135:261–267. doi: 10.1111/j.1365-2567.2011.03547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayamada S, Poholek AC, Lu KT, Takahashi H, Kato M, Iwata S, Hirahara K, Cannons JL, Schwartzberg PL, Vahedi G, et al. Type I IFN induces binding of STAT1 to Bcl6: divergent roles of STAT family transcription factors in the T follicular helper cell genetic program. Journal of immunology. 2014;192:2156–2166. doi: 10.4049/jimmunol.1300675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- Odegard JM, Marks BR, DiPlacido LD, Poholek AC, Kono DH, Dong C, Flavell RA, Craft J. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. The Journal of experimental medicine. 2008;205:2873–2886. doi: 10.1084/jem.20080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Zhu L, Sofi H, Lapinski PE, Horai R, Mueller K, Stritesky GL, He X, Teh HS, Wiest DL, et al. Development of promyelocytic leukemia zinc finger-expressing innate CD4 T cells requires stronger T-cell receptor signals than conventional CD4 T cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16264–16269. doi: 10.1073/pnas.1207528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdell F, Ziegler SF. FOXP3 and scurfy: how it all began. Nature reviews Immunology. 2014;14:343–349. doi: 10.1038/nri3650. [DOI] [PubMed] [Google Scholar]
- Ray JP, Marshall HD, Laidlaw BJ, Staron MM, Kaech SM, Craft J. Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity. 2014;40:367–377. doi: 10.1016/j.immuni.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolski R, Leonard WJ. Interleukin-21: a double-edged sword with therapeutic potential. Nat Rev Drug Discov. 2014;13:379–395. doi: 10.1038/nrd4296. [DOI] [PubMed] [Google Scholar]
- Stone EL, Pepper M, Katayama CD, Kerdiles YM, Lai CY, Emslie E, Lin YC, Yang E, Goldrath AW, Li MO, et al. ICOS Coreceptor Signaling Inactivates the Transcription Factor FOXO1 to Promote Tfh Cell Differentiation. Immunity. 2015;42:239–251. doi: 10.1016/j.immuni.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, Umesaki Y, Wu HJ. Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer’s Patch T Follicular Helper Cells. Immunity. 2016;44:875–888. doi: 10.1016/j.immuni.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Geng J, Wen X, Bi E, Kossenkov AV, Wolf AI, Tas J, Choi YS, Takata H, Day TJ, et al. The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat Immunol. 2014;15:667–675. doi: 10.1038/ni.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JP, Fuhrmann F, Feist RK, Lahmann A, Al Baz MS, Gentz LJ, Vu Van D, Mages HW, Haftmann C, Riedel R, et al. ICOS maintains the T follicular helper cell phenotype by down-regulating Kruppel-like factor 2. The Journal of Experimental Medicine. 2015;212:217–233. doi: 10.1084/jem.20141432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Laurence A, Elias KM, O’Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007;282:34605–34610. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing JB, Ise W, Kurosaki T, Sakaguchi S. Regulatory T cells control antigen-specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA-4. Immunity. 2014;41:1013–1025. doi: 10.1016/j.immuni.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss L, Stadinski BD, King CG, Schallenberg S, McCarthy NI, Lee JY, Kretschmer K, Terracciano LM, Anderson G, Surh CD, et al. Affinity for self antigen selects Treg cells with distinct functional properties. Nat Immunol. 2016;17:1093–1101. doi: 10.1038/ni.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X, He R, Hao Y, Wang H, Zhao T, et al. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat Immunol. 2015;16:991–999. doi: 10.1038/ni.3229. [DOI] [PubMed] [Google Scholar]
- Yadav M, Stephan S, Bluestone JA. Peripherally induced tregs - role in immune homeostasis and autoimmunity. Front Immunol. 2013;4:232. doi: 10.3389/fimmu.2013.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science (New York, NY) 2015;348:589–594. doi: 10.1126/science.aaa7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1: Supplemental Experimental Procedures and Figures S1–S4.
Related to Figure 3. Excel spreadsheet contains expression and statistical data for significant genes enriched in the N, ACT and nTH21 populations.
Related to Figure 3. Excel spreadsheet contains the KEGG and GO pathway enrichment terms and statistics for the N, ACT and nTH21 populations. Tab 1 contains KEGG terms. Tab 2 contains GO terms.
Related to Figure 3. Excel spreadsheet contains the gene expression and statistical information for genes that are hallmarks of TFH cells or other CD4+ T helper subsets.