Abstract
Background
The objective of our investigation is to study the relationship between the rs4939827 SNP in the SMAD7 gene, Mediterranean diet pattern and the risk of colorectal cancer.
Methods
We examined 1087 cases of colorectal cancer and 2409 population controls with available DNA samples from the MCC-Spain study, 2008–2012. Descriptive statistical analyses, and multivariate logistic mixed models were performed. The potential synergistic effect of rs4939827 and the Mediterranean diet pattern was evaluated with logistic regression in different strata of of adherence to the Mediterranean diet and the genotype.
Results
High adherence to Mediterrenean diet was statistically significantly associated with colorectal cancer risk. A decreased risk for CRC cancer was observed for the CC compared to the TT genotype (OR = 0.65 and 95% CI = 0.51–0.81) of the rs4939827 SNP Also, we could show an association between the Mediterranean diet pattern (protective factor) and rs4939827. Although the decreased risk for the CC genotype was slightly more pronounced in subjects with high adherence to Mediterrenean diet, there was no statistically significant synergistic effect between genotype CC and adherence to the Mediterranean dietary pattern factors.
Conclusion
The SMAD7 gene and specifically the allele C could be protective for colorectal cancer. An independent protective association was also observed between high adherence Mediterranean diet pattern and CRC risk. Findings form this study indicate that high adherence to Mediterranean diet pattern has a protective role for CRC cancer probably involving the Tumor Growth Factor- β pathway in this cancer.
Electronic supplementary material
The online version of this article (10.1186/s12881-017-0485-5) contains supplementary material, which is available to authorized users.
Keywords: Colorectal cancer, SMAD7, rs4939827, Mediterranean diet, Gene-environment
Background
Colorectal cancer (CRC) has the third highest incidence in men, and the second highest in women worldwide [1]. This cancer has an increasing incidence in developing countries [2] probably due to the increasing prevalence of environmental factors that contribute to the development of CRC, for instance, dietary pattern and physical activity [3]. On the other hand, mortality levels are decreasing in countries with specialized care and better screening services [4]. Gene-Environment (GxE) interactions may play an essential role in increasing the susceptibility to developing colorectal cancer [3]. Among these factors, it is important to pay attention to the Mediterranean Diet Pattern adherence, since lower adherence to this pattern is more common as a direct consequence of the general westernization of the lifestyle’s population [5–7]. Assessing the adherence to a Mediterranean dietary pattern is one of the particularly interesting approaches which may help to understand the relationship between diet and CRC [8]. However, the effect of this pattern over health depends on many individual aspects, involving genetic factors and polymorphisms. The lack of knowledge about the interactions between diet-polymorphism is a huge problem for the public health [9].
Earlier studies show a possible relationship between the diet pattern and the rs4939827 SNP in SMAD7 (SMAD family member 7), which has been associated to CRC previously. The SMAD7 gene acts as a Transforming Growth Factor Beta (TGF-β) family inhibitor by blocking the pathway signaling (Fig. 1) [10]. TGF-β encodes for a cytokines family, which are multifunctional peptides that control some process like cell proliferation. In this way, when TGF-β is inhibited by the interaction with SMAD7, cell proliferation is promoted and this could lead to the development of cancer. Another key gene in the undirected regulation of SMAD7 expression is the gene Ski (SKI proto-oncogene), which acts by blocking the TGF-β target genes, as SMAD7 [11, 12].
Fig. 1.
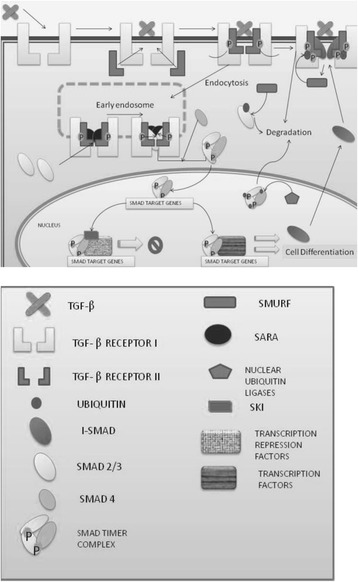
Signaling pathway TGFβ-SMAD
The interaction between the Mediterranean diet and the SMAD7 gene may be due to the action of other genes that are involved in the TGF-β pathway, in particular SMAD3 (SMAD family member 3), which is inhibited by the first one [13]. This pattern of diet is characterized by high fiber contents and before a fermentative process Na-butyrate is produced. This compound interacts with SAMD3, highlighting the signaling of this pathway [11]. In this way, we can check that the relationship between the dietary pattern and SMAD7 expression is real.
The goal of this article is to investigate the relationship between the rs4939827 SNP, the Mediterranean Diet Pattern and the risk of CRC.
Method
Study population
MCC-Spain is a multicentric case-control study with population controls aiming to evaluate the influence of environmental exposures and their interaction with genetic factors in common tumors in Spain (prostate, breast, colorectal, gastroesophageal and chronic lymphocytic leukemia). Between September 2008 and December 2013, subjects between 20 and 85 years of age (matched by 5-year age interval) with histologically-confirmed newly-diagnosed colon or rectum cancer (ICD-10: C18, C19, C20, D01.0, D01.1, D01.2) were recruited in 23 Spanish hospitals from 12 Spanish provinces. Simultaneously, population-based controls frequency-matched to cases, by age, sex and region with the join distribution of the tumors included in MCC were randomly selected from primary care centers within hospitals’ catchment areas. All participants signed an informed consent. Approval for the study was obtained from the ethical review boards of all recruiting centers. Additional information regarding the study design is provided elsewhere [14].
In our study, 3496 individuals were included from this MCC-Spain study. For this analysis, 1087 cases of colorectal cancer and 2409 controls were involved with available DNA samples for genetic analysis.
Lifestyle information
A computerized epidemiological questionnaire including self-reported socio-demographic and anthropometric data, family history of cancer, environmental exposures, use of selected drugs, reproductive history and current and past lifestyle behaviors (including leisure time physical activity and sedentary lifestyle) was administered by trained personnel in a face-to-face interview at enrolment. Waist and hip circumferences were measured by the interviewer [14]. Subjects were provided a previously validated semi-quantitative Spanish Food Frequency Questionnaire (FFQ) [15] which was modified to include regional products. The FFQ included 140 food items, and assessed usual dietary intake during the previous year. The FFQ included the specific cooking methods for meat and some pictures to establish how thoroughly-cooked participants prefer it. The FFQ was self-administered and returned by mail or filled out face to face (global response rate 88%). Frequency data was used to derive amount (g/day, g/1000 kcal) of each of the individual food types and summary variables. The food composition table has been a compiled table from the Centre for Higher Studies in Nutrition and Dietetics (CESNID) and other specific sources [16]. Cross-check questions on food groups intakes were used to adjust the frequency of foods eaten and reduce misreporting of food groups with large numbers of items [17, 18]. Data from this questionnaire was used to obtain a score in order to know the adherence to the Mediterranean diet by Sofi F [19]. Sofi F. et al. have computed the adherence to this type of diet taking into account 9 groups of foods (vegetables, legumes, meat, fish, integral cereals, fruit, dairy products, alcohol, olive oil) [19]. The range of this score goes from zero to eighteen points. From that score, we create subsets based on individuals with low (if the score is <9), medium (from 9 to 11 points) or high (if the score is higher than eleven points) adherence to this kind of diet [12]. Since the Mediterranean diet has a high fiber content and the SMAD7 gene can interact with it, an analysis of this factor was considered essential.
For the variable of physical activity, the last 10 years were taken into account. This parameter also includes recreational physical activity. The assessed parameter was METs, a measured unit of metabolism. The responses obtained allowed us to create four subpopulations: sedentary, low physical activity, average physical activity and intense physical activity.
Samples processing
Peripheral blood (27 ml) was drawn from participants, which were aliquoted in whole blood, plasma, cellular fraction for DNA extraction, and serum and stored at −80 °C. Saliva was collected for subjects refusing to donate blood with the Oragene® DNA Kit and stored at room temperature until DNA extraction. We collected biological samples for DNA extraction for participants with interview, as well as toenail and hair samples. In 4 centers (Madrid, Cantabria, Asturias and Huelva) cases and controls also donated urine samples (60 ml) that were aliquoted and frozen at −80 °C. Fresh tumor biopsies or paraffin embedded samples are available in all participating hospitals. Standardized basic clinical and pathological information on the diagnosis and treatment of tumors was collected from hospital records by using a predefined format.
Genetic analyses were carried out within MCC-Spain and also through participation in international consortia. The InfiniumHumanExomeBeadChip from Illumina was used to genotype >200,000 coding markers plus 6000 additional custom variants on the pathways of interest [14].
SNP selection
After a literature search, different SNPs that were associated with CCR were selected. These SNPs were processed with the PLINK software in order to verify that they had been collected in the database of the MCC-Spain. This file contains the basic data needed for statistical analysis (cases and controls, SNPs, and the identifier of each individual). Finally, after an ad-hoc evaluation of the polymorphisms, rs4939827 was selected and we performed the statistical analyses described in the following section because only this SNP was considered relevant in its relation with colorectal cancer (see Additional file 1: Table S1).
Statistical methods.
First, a descriptive statistical analysis was performed to determine the characteristics of our study population. For age, mean and standard deviation were calculated. For the rest of variables, we calculate frequencies of cases and controls using STATA. Table 1 show variables used in analysis and how they were classified.
Table 1.
Descriptive analysis results. Characteristics of cases and controls
| CONTROLS | CASES | p-values | ||||
|---|---|---|---|---|---|---|
| N | % | N | % | |||
| 2409 | 1087 | – | ||||
| Age | (Mean/SD) | 63.13 | 11.5 | 66.64 | 10.4 | – |
| Sex | Men | 1300 | 54 | 708 | 65.1 | <0.0001 |
| Women | 1109 | 46 | 379 | 34.9 | ||
| First Degree Family History | No | 2081 | 90.6 | 855 | 82.1 | <0.0001 |
| Yes | 216 | 9.4 | 186 | 17.9 | ||
| Cultural level | Less than primary school | 409 | 17 | 288 | 26.5 | <0.0001 |
| Primary school | 803 | 33.3 | 458 | 42.1 | ||
| Secundary school | 698 | 29 | 221 | 20.3 | ||
| University | 499 | 20.7 | 120 | 11 | ||
| BMI | Normal weight | 759 | 31.5 | 259 | 23.8 | <0.0001 |
| Less than normal weight | 23 | 1 | 13 | 1.2 | ||
| Overweight | 873 | 36.2 | 399 | 36.7 | ||
| Obesity | 754 | 31.3 | 416 | 38.3 | ||
| Total energy (Kcal/Day) | Low | 795 | 33 | 296 | 27.2 | <0.0001 |
| Medium | 794 | 33 | 320 | 29.4 | ||
| High | 820 | 34 | 471 | 43.3 | ||
| METS | Sedentarism | 855 | 35.5 | 468 | 43.1 | <0.0001 |
| Low physical activity | 385 | 16 | 134 | 12.3 | ||
| Moderate physical activity | 313 | 13 | 109 | 10 | ||
| High physical activity | 856 | 35.5 | 376 | 34.6 | ||
N Number, BMI Body Mass Index, METS Metabolic Equivalent of Task. The cut point for category of total energy are <1586.3 Kcal/day, ≥ 1586.3 Kcal/day & < 2046.4 Kcal/day and ≤2046.4 for low, medium and high level respectively.For the physical activity, the cut points are 0 when the there is no physical activity, < 8 METS*hours/week for low physical activity, ≥ 8 and < 16 METS*hours/week for moderated physical activity and >16 METS*hours/week for high physical activity (according to American College of Sport Medicine)
Then multivariate logistic mixed models, including the study region as a random effect term, were performed to evaluate the association between genotypes of selected polymorphisms and the risk of CRC. The same analysis was made in order to show the association of the component of the Mediterranean diet with the colorectal cancer. Adjusted odds ratios and confidence intervals at 95% were calculated by reference to the homozygous genotype of the most common allele (T allele). The minimally adjusted odds ratio was calculated by adjusting by sex, age and educational background. The adjusted odds ratios were calculated taking into account the variables mentioned before and BMI, total energy intake, family history first degree, race and METS.
Finally, linear regression was used for associations between polymorphisms and the study variables. For this, the online software used was SNPstats, by the log-additive model mainly, although we also observed the results of other models (dominant, codominant, recessive and overdominant) [20]. By using STATA, the rs4939827 SNP association with the Mediterranean diet pattern was evaluated with logistic regression. For that, we stratified the variable of adherence to the Mediterranean diet and the genotype was also taken into account.
Results
The initial study population consists of 6090 individuals, distributed into 2140 cases and 3950 controls. Among them, 3496 had both genetic information and the dietary pattern available, the algorithm of missing data can be seen in Fig. 2. Table 1 shows the main characteristics of those individuals which the analysis was performed with. Analyzing the missing data, it has been observed that only men have significant differences (in both case and control groups). The rest of variables do not show significant differences (Data not shown).
Fig. 2.
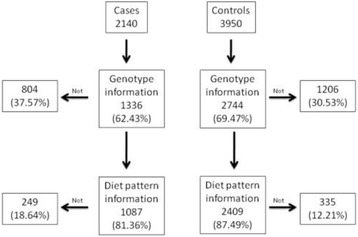
Algorithm for cases and controls for this study, selected from the population which participated in the project
Descriptive analysis
In this study 3496 participants have been included, 1087 cases (65.1% males; 34.9% females) and 2407 controls (54% males; 46% females). The characteristics of the individuals of this study are described in Table 1. Data belong to a multi case-control study and matches were performed by frequencies in all types of tumors (colorectal, mama, prostate, leukemia, stomach), so it can be the reason for the mismatch between the sex distribution between affected cases and frequency-matched controls.
In the study population, the average age is 66.6 (±10.4) in cases and 63.1 (±11.5) in controls. Physical activity habits show a sedentary population, with 43.0% in cases and 35.5% in controls who do not practice physical activity. BMI shows that only 23.8% in cases and 31.5% in controls have an adequate weight. Most of the population suffers from obesity or being overweight.
Mediterranean diet pattern and components analysis
The components of the Mediterranean diet pattern by case-control status are shown in Table 2. We observe that there are significant differences in alcohol intake in the last 10 years between cases and controls, however we have not got any clear evidence that alcohol intake in the Mediterranean diet is associated with CRC. The opposite case can be found in vegetables and legumes, where a high intake of them may be protective against colorectal cancer. On the other hand, meat and dairy products are risk factors when they are consumed in high quantities. Other components (integral cereals, fish and olive oil) do not show significant differences. Fruit cannot be analyzed because there are not enough subjects (Table 2).
Table 2.
Association between intake levels of food components in the Mediterranean diet anc case (affection) status. The cut points are based on the score criteria for Sofi’s Mediterranean diet pattern
| CONTROLS | CASES | OR minimally adjusted | CI (95%) | OR adjusted | CI (95%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||||
| 2409 | 1087 | ||||||||
| Alcohol | Moderate intake (12 g/day-24 g/day) | 612 | 25.4 | 386 | 35.5 | 1 | 1 | 1 | 1 |
| Hight intake (>24 g/day) | 1471 | 61.1 | 537 | 49.4 | 1.17 | 0.92–1.49 | 0.76 | 0.60–0.97 | |
| Low intake (<12 g/day) | 326 | 13.5 | 164 | 15.1 | 0.77 | 0.61–0.98 | 1.04 | 0.81–1.33 | |
| Vegetables | Low intake (<100 g/day) | 563 | 23.4 | 306 | 28.2 | 1 | 1 | 1 | 1 |
| Moderate intake (100 g/day-250 g/day) | 1378 | 57.2 | 624 | 57.4 | 0.82 | 0.68–0.98 | 0.85 | 0.70–1.03 | |
| Hig/dayh intake (>250) | 468 | 19.4 | 157 | 14.4 | 0.54 | 0.42–0.70 | 0.55 | 0.42–0.72 | |
| Legumes | Low intake (<70 g/day) | 1561 | 64.8 | 670 | 61.6 | 1 | 1 | 1 | 1 |
| Moderate intake (70 g/day-140 g/day) | 383 | 15.9 | 177 | 16.3 | 0.94 | 0.76–1.16 | 0.97 | 0.77–1.21 | |
| Hig/dayh intake (>140 g/day) | 465 | 19.3 | 240 | 22.1 | 0.75 | 0.60–0.92 | 0.77 | 0.61–0.96 | |
| Meat | Low intake (<80 g/day) | 185 | 7.7 | 152 | 14.0 | 1 | 1 | 1 | 1 |
| Moderate intake (80 g/day-120 g/day) | 442 | 18.3 | 236 | 21.7 | 1.4 | 1.15–1.70 | 1.29 | 1.05–1.60 | |
| Hig/dayh intake (>120 g/day) | 1782 | 74.0 | 699 | 64.3 | 1.98 | 1.54–2.55 | 1.74 | 1.32–2.30 | |
| Dairy products | Low intake (<180 g/day) | 1541 | 64.0 | 693 | 63.8 | 1 | 1 | 1 | 1 |
| Moderate intake (180 g/day-270 g/day) | 557 | 23.1 | 250 | 23.0 | 0.95 | 0.74–1.23 | 0.97 | 0.74–1.28 | |
| Hig/dayh intake (>270 g/day) | 311 | 12.9 | 144 | 13.2 | 1.02 | 0.82–1.29 | 1.01 | 0.79–1.30 | |
| Fruits | Low intake (<150 g/day) | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Moderate intake (150 g/day-300 g/day) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Hig/dayh intake (>300 g/day) | 2409 | 100 | 1087 | 100 | omitted | omitted | |||
| Cereals | Low intake (<130 g/day) | 2327 | 96.6 | 1047 | 96.3 | 1 | 1 | 1 | 1 |
| Moderate intake (130 g/day-195 g/day) | 71 | 2.9 | 32 | 2.9 | 0.97 | 0.62–1.51 | 1.08 | 0.68–1.74 | |
| Hig/dayh intake (>195 g/day) | 11 | 0.5 | 8 | 0.7 | 1.57 | 0.62–4.01 | 1.42 | 0.50–4.02 | |
| Fish | Low intake (<100 g/day) | 169 | 7.0 | 91 | 8.4 | 1 | 1 | 1 | 1 |
| Moderate intake (100 g/day-250 g/day) | 663 | 27.5 | 304 | 28.0 | 0.91 | 0.67–1.22 | 0.9 | 0.65–1.25 | |
| Hig/dayh intake (>250 g/day) | 1577 | 65.5 | 692 | 63.7 | 0.82 | 0.62–1.09 | 0.84 | 0.62–1.16 | |
| Olive oil | Low intake (<0.1 g/day) | 115 | 10.6 | 245 | 10.2 | 1 | 1 | 1 | 1 |
| Moderate intake (0.1 g/day-0.99 g/day) | 206 | 19.0 | 513 | 21.3 | 0.88 | 0.72–1.07 | 0.85 | 0.69–1.05 | |
| Hig/dayh intake (>1 g/day) | 766 | 70.5 | 1651 | 68.5 | 0.85 | 0.67–1.10 | 0.91 | 0.70–1.21 | |
The minimally adjusted odds ratio (OR) was calculated adjusting by sex, age (as continuous variable) and educational level from a multivariate logistic mixed model.Area was used as random variable. The adjusted odds ratios were calculating taking into account the variables mentioned before and BMI, total energy intake, family history fist degree, race and METS. (N: Number; CI: Confidence Interval). Cut-offs are based on score criteria of Sofi’s Mediterranean Pattern [19]
Taking into account the adherence to the Mediterranean dietary pattern, 68.6% of cases and in 75.3% of controls have a high or medium adherence to the Mediterranean dietary pattern. All these variables show significant differences between cases and controls (Table 3).
Table 3.
Association between SNP rs4939827 and the Mediterranean dietary pattern adherence in relation with colorectal cancer taking into account the genotype
| Cases | Controls | Adjusted OR | ||||
|---|---|---|---|---|---|---|
| n | % | n | % | OR | CI 95% | |
| Low | 341 | 31.4 | 594 | 24.7 | 1 | – |
| TT | 110 | 32.2 | 171 | 28.8 | 1 | – |
| CT | 169 | 49.6 | 302 | 50.8 | 0.83 | 0.59–1.17 |
| CC | 62 | 18.2 | 121 | 20.4 | 0.70 | 0.44–1.08 |
| Medium | 431 | 39.7 | 1010 | 41.9 | 0.85 | 0.70–1.03 |
| TT | 154 | 35.7 | 316 | 31.29 | 1 | – |
| CT | 211 | 49 | 478 | 47.2 | 0.87 | 0.65–1.15 |
| CC | 66 | 15.3 | 216 | 21.38 | 0.64 | 0.44–0.93 |
| High | 315 | 29.0 | 805 | 33.4 | 0.64 | 0.51–0.79 |
| TT | 123 | 39.1 | 242 | 30.1 | 1 | – |
| CT | 144 | 45.7 | 405 | 50.3 | 0.75 | 0.55–1.02 |
| CC | 48 | 15.2 | 158 | 19.6 | 0.63 | 0.42–0.96 |
CT vs TT: Adjusted Odd Ratio (OR) = 0.82 and 95% Confidence Interval (CI) = 0.69–0.98
CC vs TT: OR = 0.65 and 95% CI = 0.51–0.81
The adjusted odds ratios (OR) was calculating taking into account the variables sex, age, educational level, BMI, total energy intake, family history fist degree, METS. Area was used as random variable (N: Number; CI: Confidence Interval)
Associations of rs4939827 with CRC and other factors
The rs4939827 SNP was statistically significantly associated with colorectal cancer according to several models. The most important result is referred to the Log-Additive model (p-value = 1.00E-04). The association of this polymorphism with CRC can also be observed in other models in a significant way: codominant model (p-value = 2e-04); dominant model (p-value = 6e-04); recessive model (p-value = 8e-04). The analysis of association of this polymorphism with risk factors is also shown in the summary table.
rs4939827 associations with risk factors of this study
After analyzing the association between rs4939827 and risk factors, statistically significant results can be observed in the case of the degree of adherence to the Mediterranean dietary pattern. Other factors assessed did not show a significant association with the SNP (data not shown). We conducted an analysis for rs4939827 with an interaction term to evaluate the independent main effect of the Mediterranean diet and main effect of the protective rs4939827 allele (see Additional file 2: Table S2). However, none of the results provide a significant p-value although the overdominant model is close to p-value = 0.05 (p-value = 0.07), determining that C allele it the dominant one.
The CC genotype (16.1% in cases and 20.5% in controls) seems to be protective compared to the TT (35.6% in cases and 30.3% in controls) genotype (ORa TTvsCT = 0.82 IC (95%) = 0.69–0.98; ORa TTvsCC = 0.65 IC (95%) = 0.51–0.81). When we check the association with adherence pattern to the Mediterranean diet (only statistically significant association), it can be observed that high adherence is a protective factor (Fig. 3 and Table 3). However, we could not see a synergistic effect between both genotype CC and adherence to the Mediterranean dietary pattern factors.
Fig. 3.
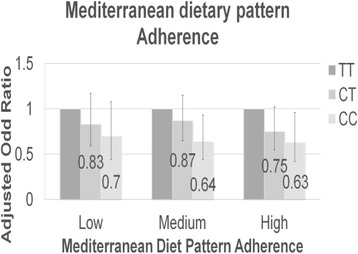
Association between SNP rs4939827 and the Mediterranean dietary pattern adherence in relation with colorectal cancer taking into account the genotype. Reference: TT genotype. Vertical axis: Adjusted Odd Ratio (The adjusted odds ratios (OR) was calculating taking into account the variables sex, age, socioeconomic level, BMI, total energy intake, family history fist degree, METS and area was used as random variable). Abscissa axis: Mediterranean Diet Pattern Adherence
Discussion
SMAD7 is a target gene of the signaling pathway TGFβ-SMAD. For the TGFβ expression, it must be joined with its receptor type II. This binding stimulates the receptor type I, which binds to the above complex and, the complete assembly, is able to regulate the formation and activation (by phosphorylation) of the heterodimeric complex SMAD2/3. After the activation, this complex joins SMAD4 (SMAD family member 4) protein, which helps the complex translocation to the nucleus. In this organelle, the activation of the expression of target genes of TGFβ occurs, including the SMAD7 gene and other genes responsible for proliferation inhibition and cell division. This happens when the SKI protein is not in the nucleus, which is capable of binding with the heterotrimeric complex and inhibit its function, inhibiting the expression of the SMAD7 gene in this way (Fig. 1) [6, 7].
In this study of 1087 cases of CRC and 2409 controls, the rs4939827 SNP with the CC genotype was associated with a reduction of the risk for colorectal cancer. This may be due to SMAD7 participation modulating the TGFβ pathway. SMAD7 protein is capable of binding with TGFβ, causing its ubiquitination and destruction. In this way, the action of TGFβ is inhibited and cell proliferation can occur. To clarify the role of SMAD7 in cell growth in the CRC, Halder et al. [21], overexpressed this gene. This fact produces a higher cell growth than in normal conditions (21). At the same time, it has been observed that inhibition of SMAD7 with a specific oligonucleotide reduces cell growth in CRC [22]. This data leads to the hypothesis that rs4939827 with genotype CC is a protective genotype because the correct expression of the SMAD7 gene is inhibited, preventing cell proliferation and reducing the susceptibility to colorectal cancer.
This hypothesis is supported by B. Zhang et al. (2014) [23] who share the idea that the gene variant rs4939827 may be associated with an increase of CRC survivorship [23]. Similarly, Slattery M. et al. (2010) [24] also agree with our results as they show that the CC genotype is inversely associated with the risk of colon cancer. Thus, they conclude that individuals containing the homozygous recessive gene variant rs4939827 show a reduced risk of colon cancer by a 27% [24]. Thompson C.L. et al. (2009) [25] performed an analysis of the SNP taking into account the dominant model, stratifying the population according to gender. They noticed that women with the C allele were associated with a decreased risk of CRC. In contrast, they did not obtain significant results in men [25].
The CC genotype of this polymorphism was also associated with survival of other cancers. For example, according to Geng, T.T. et al. (2015) [26], the dominant model showed that the rs4939827 polymorphism was significantly associated with a decreased risk of esophageal cancer by 0.67 fold due to a possible inhibition of the SMAD7 gene [26].
The T allele must also be taken into account. Although T allele does not show a significant risk in our results, many authors support the idea by Jung KJ et al. (2015) [27], which defended the hypothesis that the T allele of this SNP was a risk factor for colorectal cancer and rectal cancer [27]. Furthermore, according to analysis made by Baert-Desurmont S. et al. (2015) [28] an increased risk is observed depending on the number of T alleles present in genotype [28]. Ho J. W. et al. (2011) [29] analyzed the risk of TC heterozygous and TT homozygous genotypes. They identified both as risk genotypes, but only the homozygous genotype showed a significant result [29]. Tenesa A. et al. (2008) [30], after taking into account the location of the tumor to perform the analysis, noted that the T allele is a risk factor mainly for rectal cancer and not for the colon cancer [30]. In our results, we cannot verify that information, since we did not identify the T allele as a risk allele, only the C allele could be identified as a protective allele. Tenesa A. et al. (2010) [31] also associated the T allele of the rs4939827 SNP with an increase of mortality [31]. Passarelli M.N. et al. (2011) [32] found that SMAD7 variants that inhibit TGFβ completely may reduce its tumor suppressor activity (resulting in an increased risk), but can also reduce their ability to promote their metastatic promoter activity (resulting in a slower progression of the tumor and improved survival) [32]. Finally, Yao et al. (2015) [33] support that the T allele increases the risk of CRC in Caucasian population [33].
If we analyze the SNP association with diet, our results are statistically significant. However, few authors refer to the association of this factor with the rs4939827 polymorphism. As it is mentioned before, association with the diet is due to the TGFβ signaling pathway genes that interact with SMAD7. There is evidence that this association may be due to the ingestion of high amounts of fiber in the Mediterranean diet. Vegetables and fruit, for example, are highly fermentable fiber sources, while products such as wheat bran are low fermentable. The difference is the amount of short-chain fatty acid formed after fermentation of the fiber by the colon’s bacteria in this organ, as described by Nguyen K.A. et al. (2006) [11]. So, food containing fermentable fiber (fruit and vegetables) will produce higher amount of fatty acids than that containing short chain poorly fermentable fiber. Therefore, there is a positive relationship between the consumption of large amounts of fiber and a lower risk of colon cancer. This may be due to the fact that the Na-butyrate (Na-B, the main product of the fermentation of fiber) is able to induce cell cycle arrest, cellular differentiation and, even, apoptosis. It seems that the Na-B is capable of inducing, selectively, Smad3 phosphorylation, so pathway activity of TGF-β is enhanced [6, 7]. Despite this, we have not found a statistically significant interaction in our results. It may be due it has not got enough statistical power or the actual interaction is carried out with SMAD3. This produces that TGF-β is not ever ubiquitined and degraded, so cyclins jogging cell proliferation (p15, p21 and p27) are expressed [34]. Besides, one of SMAD7 functions is to produce cell cycle arrest in G1 phase. However, because of this mutation, this activity cannot be carried out by this process. Na-B intake helps this action be covered, further minimizing the risk of uncontrolled cell proliferation.
One limitation in this work is that genotype x diet effect could be due to fiber intake rather than adherence to the Mediterranean diet, although both concepts are related.
Conclusions
There is an important role of SMAD7 gene in colorectal cancer as the allele C is protective against this disease. Also, we can see a protective association between rs4939827 SNP and high adherence to the Mediterranean diet pattern and we suppose that high adherence to Mediterranean diet pattern probably has a protective role. So they may participate together in the TGFβ pathway in colorectal cancer.
Additional files
Analysis of SNPs related to CRC. The minimally adjusted odds ratio (OR) was computed adjusting by sex, age, educational level. Area, places where cases and controls were recruited, was used as random variable. Association is evaluated for carrying 1 or 2 SNP minor frequency alleles relative to a reference of zero SNP minor frequency allele. (DOCX 22 kb)
Associations between polymorphisms and the Mediterranean Diet Pattern of Sofi considering different genetic models. Genetic models: codominant, dominant, recessive, overdominant, log-additive. (DOCX 16 kb)
Acknowledgements
We thank all the subjects who participated in the study and all MCC-Spain collaborators.
Funding
The study was partially funded by the “Accion Transversal del Cancer”, approved on the Spanish Ministry Council on the 11th October 2007, by the Instituto de Salud Carlos III-FEDER (PI08/1770, PI08/0533, PI08/1359, PI09/00773-Cantabria, PI09/01286-León, PI09/01903-Valencia, PI09/02078-Huelva, PI09/01662-Granada, PI11/01403, PI11/01889-FEDER, PI11/00226, PI11/01810, PI11/02213, PI12/00488, PI12/00265, PI12/01270, PI12/00715, PI12/00150, PI14/01219, PI14/0613), by the Fundación Marqués de Valdecilla (API 10/09), by the ICGC International Cancer Genome Consortium CLL (The ICGC CLL-Genome Project is funded by Spanish Ministerio de Economía y Competitividad (MINECO) through the Instituto de Salud Carlos III (ISCIII) and Red Temática de Investigación del Cáncer (RTICC) del ISCIII (RD12/0036/0036)), by the Junta de Castilla y León (LE22A10–2), by the Consejería de Salud of the Junta de Andalucía (2009-S0143), by the Conselleria de Sanitat of the Generalitat Valenciana (AP_061/10), by the Recercaixa (2010ACUP 00310), by the Regional Government of the Basque Country, by the Consejería de Sanidad de la Región de Murcia, by the European Commission grants FOOD-CT-2006-036224-HIWATE, by the Spanish Association Against Cancer (AECC) Scientific Foundation, by the Catalan Government DURSI grant 2014SGR647, by the Fundación Caja de Ahorros de Asturias and by the University of Oviedo.
Availability of data and materials
Samples: Biological samples were stored at the biobanks supported by Instituto de Salud Carlos III- FEDER: Parc de Salut MAR Biobank (MARBiobanc) (RD09/0076/00036), “Biobanco La Fe” (RD 09 0076/00021) and FISABIO Biobank (RD09 0076/00058). Also at the Public Health Laboratory from Gipuzkoa, the Basque Biobank, the ICOBIOBANC (sponsored by the Catalan Institute of Oncology), the IUOPA Biobank from the University of Oviedo and the ISCIII Biobank.
Genotyping: SNP genotyping services were provided by the Spanish “Centro Nacional de Genotipado” (CEGEN-ISCIII).
Abbreviations
- CESNID
Centre for Higher Studies in Nutrition and Dietetics
- CRC
Colorectal cancer
- FFQ
Spanish Food Frequency Questionnaire
- GxE
Gene-Environment
- Na-B
Na-butyrate
- ORa
Odd Ratio adjusted
- Ski
SKI proto-oncogene
- SMAD3
SMAD family member 3
- SMAD4
SMAD family member 4
- SMAD7
SMAD family member 7
- SNP
Single Nucleotide Polymorphism
- TGF-β
Transforming Growth Factor Beta
Authors’ contributions
VMo and VMa have made substantial contributions to conception and design; JAM and CGD have made substantial contributions to conception and design, analysis and interpretation of data and have been involved in drafting the manuscript or revising it critically for important intellectual content. CP, TFV, ER, MP, NA, JL, MHA, AT, PA, JJJM, RPP, RC, AJM, IGA, MG, BPG, VL, JMH, GCV, MK acquired data and given final approval of the version to be published. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The protocol of MCC-Spain was approved by the Ethics committees of the participating institutions. All participants were informed about the study objectives and signed an informed consent. Confidentiality of data is secured removing personal identifiers in the datasets. The database was registered in the Spanish Agency for Data Protection, number 2102672171. Permission to use the study database will be granted to researchers outside the study group, after revision and approval of each request by the Steering Committee.
More details on the organization of the project can be found online at http://www.mccspain.org.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12881-017-0485-5) contains supplementary material, which is available to authorized users.
Contributor Information
Jéssica Alonso-Molero, Email: jalonm00@estudiantes.unileon.es.
Carmen González-Donquiles, Email: cgonzd00@estudiantes.unileon.es.
Camilo Palazuelos, Email: camilo.palazuelos@unican.es.
Tania Fernández-Villa, Email: tferv@unileon.es.
Elena Ramos, Email: lnagarciaramos@gmail.com.
Marina Pollán, Email: mpollan@isciii.es.
Nuria Aragonés, Email: naragones@isciii.es.
Javier Llorca, Email: javier.llorca@unican.es.
M. Henar Alonso, Email: mhalonso@iconcologia.net
Adonina Tardón, Email: atardon@uniovi.es.
Pilar Amiano, Email: epicss-san@euskadi.eus.
José Juan Jiménez Moleon, Email: jjmoleon@ugr.es.
Rosana Peiró Pérez, Email: peiro_ros@gva.es.
Rocío Capelo, Email: rocio.capelo@dbasp.uhu.es.
Antonio J. Molina, Email: ajmolt@unileon.es
Inés Gómez Acebo, Email: ines.gomez@unican.es.
Marcela Guevara, Email: mp.guevara.eslava@navarra.es.
Beatriz Pérez Gómez, Email: bperez@isciii.es.
Virginia Lope, Email: vicarvajal@isciii.es.
José María Huerta, Email: jmhuerta.carm@gmail.com.
Gemma Castaño-Vinyals, Email: gcastano@creal.cat.
Manolis Kogevinas, Email: kogevinas@creal.cat.
Victor Moreno, Email: v.moreno@iconcologia.net.
Vicente Martín, Email: vicente.martin@unileon.es.
References
- 1.Ferlay J, Ervik M, Dikshit R, Eser S, Mathers C, Al.E.GLOBOCAN. 2015. Available from: http://globocan.iarc.fr/old/burden.asp?selection_pop=62968&Text-p=Europe&selection_cancer=5060&Text-c=Colorectal+cancer&pYear=8&type=0&window=1&submit= Execute.
- 2.Center MM, Jemal A, Ward E. International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomark Prev. 2009;18(6):1688–1694. doi: 10.1158/1055-9965.EPI-09-0090. [DOI] [PubMed] [Google Scholar]
- 3.Hu X, Feng F, Li X, Yuan P, Luan R, Yan J, et al. Gene polymorphisms related to insulin resistance and gene-environment interaction in colorectal cancer risk. Ann Hum Biol. 2015;42(6):560–568. doi: 10.3109/03014460.2014.1002532. [DOI] [PubMed] [Google Scholar]
- 4.Ait Ouakrim D, Pizot C, Boniol M, Malvezzi M, Boniol M, Negri E, et al. Trends in colorectal cancer mortality in Europe: retrospective analysis of the WHO mortality database. BMJ. BMJ Publishing Group; 2015 Oct 6;351:h4970. [DOI] [PMC free article] [PubMed]
- 5.Pérez-Jiménez J, Elena Díaz-Rubio M, Saura-Calixto F. Contribution of macromolecular antioxidants to dietary antioxidant capacity: a study in the Spanish Mediterranean diet. Plant Foods Hum Nutr. 2015;70(4):365–370. doi: 10.1007/s11130-015-0513-6. [DOI] [PubMed] [Google Scholar]
- 6.Schwingshackl L, Hoffmann G. Adherence to Mediterranean diet and risk of cancer: an updated systematic review and meta-analysis of observational studies. Cancer Med. 2015;4(12):1933–1947. doi: 10.1002/cam4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotelli MT, Bocale D, De Fazio M, Ancona P, Scalera I, Memeo R, et al. IN-VITRO evidence for the protective properties of the main components of the Mediterranean diet against colorectal cancer: a systematic review. Surg Oncol Elsevier Ltd. 2015;24(3):145–152. doi: 10.1016/j.suronc.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Fung TT, Brown LS. Dietary patterns and the risk of colorectal cancer. Curr Nutr Rep. 2013;2(1):48–55. doi: 10.1007/s13668-012-0031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frazier-Wood AC. Dietary patterns, genes, and health: challenges and obstacles to be overcome. Curr Nutr Rep. 2015;4(1):82–87. doi: 10.1007/s13668-014-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.PB | Signaling by TGF-beta Receptor Complex. [cited 2016 May 22]. Available from: http://www.reactome.org/PathwayBrowser/#/R-HSA-170834
- 11.Nguyen KA, Cao Y, Chen JR, Townsend Jr. CM, Ko TC. Dietary fiber enhances a tumor suppressor signaling pathway in the gut. Ann Surg. 2006;243(5):617–619. doi: 10.1097/01.sla.0000216783.85214.c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YS. Milner J a. Dietary modulation of colon cancer risk. J Nutr. 2007;137:2576S–2579S. doi: 10.1093/jn/137.11.2576S. [DOI] [PubMed] [Google Scholar]
- 13.Yan X, Liao H, Cheng M, Shi X, Lin X, Feng XH, et al. Smad7 protein interacts with receptor-regulated Smads (R-Smads) to inhibit transforming growth factor-B (TGF-B)/Smad signaling. J Biol Chem. 2016;291(1):382–392. doi: 10.1074/jbc.M115.694281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castaño-Vinyals G, Aragonés N, Pérez-Gómez B, Martín V, Llorca J, Moreno V, et al. Population-based multicase-control study in common tumors in Spain (MCC-Spain): rationale and study design. Gac Sanit. Elsevier. 2015;29(4):308–315. doi: 10.1016/j.gaceta.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 15.García-Closas R, García-Closas M, Kogevinas M, Malats N, Silverman D, Serra C, et al. Food, nutrient and heterocyclic amine intake and the risk of bladder cancer. Eur J Cancer. 2007;43(11):1731–1740. doi: 10.1016/j.ejca.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Cervera P, Farran A, Zamora-Ros R. Tablas de composición de alimentos CESNID = Taules de composició d’aliments del CESNID: Universitat de Barcelona; 2004. 247 p
- 17.Nöthlings U, Hoffman K, Boeing H. Do cross-check questions improve food frequency questionnaire data? IARC Sci Publ. 2002:23–5. [PubMed]
- 18.Calvert C, Cade J, Barrett JH, Woodhouse A. Using cross-check questions to address the problem of mis-reporting of specific food groups on food frequency questionnaires. UKWCS steering group. United Kingdom Women’s cohort study steering group. Eur J Clin Nutr. 1997;51(10):708–712. doi: 10.1038/sj.ejcn.1600480. [DOI] [PubMed] [Google Scholar]
- 19.Sofi F, Macchi C, Abbate R, Gensini GF, Casini A. Mediterranean diet and health status: an updated meta-analysis and a proposal for a literature-based adherence score. Public Health Nutr. 2014;17(12):2769–2782. doi: 10.1017/S1368980013003169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics Oxford University Press. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 21.Halder SK, Beauchamp RD, Datta PK. Smad7 induces tumorigenicity by blocking TGF-β-induced growth inhibition and apoptosis. Exp Cell Res. 2005;307(1):231–246. doi: 10.1016/j.yexcr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Li Q, Zou C, Zou C, Han Z, Xiao H, Wei H, et al. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. Elsevier Ireland Ltd. 2013;335(1):168–174. doi: 10.1016/j.canlet.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B, Jia W, Matsuo K, Shin A, Xiang Y, Matsuda K, et al. Genome-wide association study identifies a new SMAD7 risk variant associated with colorectal cancer risk in east Asians. Int J Cancer. 2014;135(4):948–955. doi: 10.1002/ijc.28733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slattery ML, Herrick J, Curtin K, Samowitz W, Wolff RK, Caan BJ, et al. Increased risk of colon cancer associated with a genetic polymorphism of SMAD7. Cancer Res. 2010;70(4):1479–1485. doi: 10.1158/0008-5472.CAN-08-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson CL, Plummer SJ, Acheson LS, Tucker TC, Casey G, Li L. Association of common genetic variants in SMAD7 and risk of colon cancer. Carcinogenesis. 2009;30(6):982–986. doi: 10.1093/carcin/bgp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geng T-T, Xun X-J, Li S, Feng T, Wang L-P, Jin T-B, et al. Association of colorectal cancer susceptibility variants with esophageal cancer in a Chinese population. World J Gastroenterol. 2015;21(22):6898–6904. doi: 10.3748/wjg.v21.i22.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung KJ, Won D, Jeon C, Kim S, Kim T. Il, Jee SH, et al. a colorectal cancer prediction model using traditional and genetic risk scores in Koreans. BMC Genet. 2015;16(49):1–7. doi: 10.1186/s12863-015-0207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baert-Desurmont S, Charbonnier F, Houivet E, Ippolito L, Mauillon J, Bougeard M, et al. Clinical relevance of 8q23, 15q13 and 18q21 SNP genotyping to evaluate colorectal cancer risk. Eur J Hum Genet. 2016;24(1):99–105. doi: 10.1038/ejhg.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho JW, Choi S, Lee Y, Hui TC, Cherny SS, Garcia-Barceló M-M, et al. Replication study of SNP associations for colorectal cancer in Hong Kong Chinese. Br J Cancer. 2011;104(2):369–375. doi: 10.1038/sj.bjc.6605977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenesa A, Farrington SM, Prendergast JGD, Porteous ME, Walker M, Haq N, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40(5):631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenesa A, Theodoratou E, Din FVN, Farrington SM, Cetnarskyj R, Barnetson RA, et al. Ten common genetic variants associated with colorectal cancer risk are not associated with survival after diagnosis. Clin Cancer Res. 2010;16(14):3754–3759. doi: 10.1158/1078-0432.CCR-10-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passarelli MN, Coghill AE, Hutter CM, Zheng Y, Makar KW, Potter JD, et al. Common colorectal cancer risk variants in SMAD7 are associated with survival among pre-diagnostic non-steroidal anti-inflammatory drug users: a population-based study of post-menopausal women. Genes Chromosom Cancer. 2011;50(11):875–886. doi: 10.1002/gcc.20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao K, Hua L, Wei L, Meng J, Correlation HJ. Between CASC8, SMAD7 polymorphisms and the susceptibility to colorectal cancer. Medicine (Baltimore) 2015;94(46):e1884. doi: 10.1097/MD.0000000000001884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dancea HC, Shareef MM, Ahmed MM. Role of radiation-induced TGF-beta signaling in cancer therapy. Mol Cell Pharmacol. 2009;1(1):44–56. doi: 10.4255/mcpharmacol.09.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of SNPs related to CRC. The minimally adjusted odds ratio (OR) was computed adjusting by sex, age, educational level. Area, places where cases and controls were recruited, was used as random variable. Association is evaluated for carrying 1 or 2 SNP minor frequency alleles relative to a reference of zero SNP minor frequency allele. (DOCX 22 kb)
Associations between polymorphisms and the Mediterranean Diet Pattern of Sofi considering different genetic models. Genetic models: codominant, dominant, recessive, overdominant, log-additive. (DOCX 16 kb)
Data Availability Statement
Samples: Biological samples were stored at the biobanks supported by Instituto de Salud Carlos III- FEDER: Parc de Salut MAR Biobank (MARBiobanc) (RD09/0076/00036), “Biobanco La Fe” (RD 09 0076/00021) and FISABIO Biobank (RD09 0076/00058). Also at the Public Health Laboratory from Gipuzkoa, the Basque Biobank, the ICOBIOBANC (sponsored by the Catalan Institute of Oncology), the IUOPA Biobank from the University of Oviedo and the ISCIII Biobank.
Genotyping: SNP genotyping services were provided by the Spanish “Centro Nacional de Genotipado” (CEGEN-ISCIII).


