Abstract
Structural dynamics of Ras proteins contribute to their activity in signal transduction cascades. Directly targeting Ras proteins with small molecules may rely on movement of a conserved structural motif, switch II. To understand Ras signaling and advance Ras targeting strategies, experimental methods to measure Ras dynamics are required. Here we demonstrate the utility of hydrogen-deuterium exchange mass spectrometry to measure Ras dynamics by studying representatives from two branches of the Ras superfamily, Ras and Rho. A comparison of differential deuterium exchange between active (GMPPNP-bound) and inactive (GDP-bound) proteins revealed differences between the families, with the most notable differences occurring in the phosphate-binding loop and switch II. The P-loop exchange signature correlated with switch II dynamics observed in molecular dynamics simulations focused on measuring main chain movement. Hydrogen-deuterium exchange provides a means of evaluating Ras protein dynamics which may be useful for understanding mechanisms of Ras signaling, including activated signaling of pathologic mutants, and for targeting strategies that rely on protein dynamics.
Keywords: Protein dynamics, Rho, signal transduction, Hydrogen-deuterium exchange mass spectrometry, guanosine diphosphate (GDP)
Graphical Abstract

INTRODUCTION
Members of the Ras superfamily play roles in numerous diseases including cardiovascular disease [1], retinal degeneration [2], neurodegenerative conditions [3], developmental disorders, and cancer [4, 5]. The Ras subfamily has attracted particular attention because a large proportion of malignant tumors contain Ras mutations, and to date, no effective Ras-targeted therapies are clinically available [6]. As investigators have searched for targetable vulnerabilities in signal transduction pathways mediated by oncogenic mutant Ras, it has become clear that protein dynamics contribute significantly to the functional output of subsets of Ras mutations, and that understanding these differences may lead to new therapeutic opportunities [7, 8]. While molecular dynamic simulations have provided mechanistic insights, experimental techniques complimentary to existing technology (e.g., NMR) by which Ras protein dynamics may be studied empirically are needed. In an effort to develop complementary methodologies we have applied hydrogen-deuterium exchange (HDX) mass spectrometry (MS) to compare the dynamics of two branches of the Ras superfamily, which may enable higher throughput analysis of protein dynamics, especially given the increasing availability of high-throughput MS instrumentation.
Over 170 members of the Ras superfamily of small GTP-binding proteins (GTPases) have been identified in eukaryotes [9]. These small, monomeric GTPases operate as molecular switches within various cellular signaling pathways and can be sub-divided into at least five subfamilies: Ras, Rho, Rab, Arf, and Ran [10]. In general, these families are each designated to a different portion of the subcellular environment, where the proteins that comprise them are responsible for promoting a distinct set of unique cellular tasks. Despite their varied roles and responsibilities, many Ras superfamily members have similar tertiary structures, regardless of diverse primary structures (FIGURE 1). The main component of the tertiary structures is the G domain, an approximately 20 kDa α,β protein with a mixed six-stranded β sheet and five helices [11]. The general structure is illustrated in FIGURE 1b using K-Ras as an example. The switch mechanism primarily consists of two dynamic structural motifs, referred to as switch I and switch II, which form a portion of the nucleotide binding pocket and are critical interfaces for downstream effector interactions that result in cellular signaling [12–15]. The GxxGxGKS/T region, or phosphate-binding loop (P-loop), and the N/TKxD region are also very important for binding nucleotides. The G domain is followed (beginning at residue 165 for Ras subfamily members) C-terminally by a hypervariable domain that is highly divergent between Ras superfamily members [16]. The hypervariable domain is responsible for proper protein trafficking and subcellular localization, and it is typically modified to enable a number of functions including membrane association [17].
FIGURE 1.
Comparison of primary and tertiary structures for the Ras superfamily proteins studied. (a) Sequences for all proteins studied, aligned using Clustal Omega [66, 67]. Residue numbering is based on N-Ras, beginning with the initiator methionine and ending at the C-terminus. Yellow boxes indicate regions of notably high dissimilarity between proteins, due either to purification tags or sequence insertions of more than one amino acid. The red box corresponds to the P-loop and the green box corresponds to the N/TKxD motif. The cyan box represents the DxxG motif, and the blue and magenta boxes correspond to switch I and switch II, respectively. (b) The crystal structure of active K-Ras (PDB: 3GFT) with the highlighted regions from the sequence alignment indicated by their respective colors. The non-hydrolysable GTP analogue is represented by sticks, with each atom colored by element. (c) Table showing how each protein compares to the others in terms of percent sequence identity and root mean square deviation of tertiary structure alignment (RMSD, in angstroms). Percent sequence identity was calculated using the entire sequence of each protein studied, excluding tags. Tertiary structural alignments were performed using the cealign function in PyMol (The PyMol Molecular Graphics System, Version 1.8 Schrödinger, LLC.), which utilizes the combinatorial extension (CE) algorithm to align backbone α-carbons. These structural alignments were performed on accepted crystal structures of the inactive variants of each protein, with PDB codes detailed in parentheses underneath protein names.
Ras superfamily proteins alternate between distinct conformational states by binding either GDP or GTP. Activation occurs when the GDP-bound protein binds with a guanine-nucleotide exchange factor (GEF). This exposes the nucleotide binding site through interactions with the switch domains, allowing GDP to escape [14]. The empty catalytic site is then free to bind GTP or rebind GDP, but the binding of GTP is heavily favored due to a ten-fold higher cellular concentration (200–400 μM) than GDP [18–29]. By comparing crystal structures, the conformational changes associated with activation are primarily found in the regions around switch I (RMSD 4.77 Å for K-Ras, comparing PDB: 4DSN and 4OBE) and switch II (RMSD 2.96 Å), the latter of which extends away from the active site when not constrained by interactions with the γ-phosphate of GTP. These changes allow the GTP-bound protein to bind downstream effector proteins that perpetuate cellular signaling. Hydrolytic activity, either intrinsic or stimulated by a GTPase activating protein (GAP), facilitates the hydrolysis and elimination of the γ-phosphate of GTP [13]. GDP remains in the nucleotide binding pocket, and the protein relaxes to its inactive state. Proteins from the Rho and Rab subfamilies are also subjected to further regulation by guanine nucleotide dissociation inhibitors (GDIs), which bind to the inactive proteins and prevent GDP exchange and subsequent localization at the membrane [30].
Single amino acid mutations can activate Ras proteins leading to constant, persistent downstream signaling as seen with K-, H-, and N-Ras in cancer. Such Ras mutations are present in 20–30% of all human tumor types, and over 90% of tumors for certain diseases such as pancreatic cancer [6, 31]. Recent advances to target specific Ras mutants directly, such as K-Ras G12C with covalent inhibitors, have generated considerable optimism that direct Ras therapy may be possible [6, 32, 33]. In particular, compounds that target a pocket termed the ‘switch II pocket’ (SIIP) of Ras deserve mention from a protein dynamics perspective, because SIIP binders are observed to cause dramatic reordering of switch II into conformations not previously seen, despite the availability of many X-ray and NMR Ras structures [33, 34]. While targeting the SIIP has only been attempted for K-Ras G12C, it is possible that the SIIP strategy may be extended to other Ras proteins or oncogenic mutants. Additional techniques to identify candidates would be valuable in this regard.
In the course of developing covalent GTP-competitive K-Ras G12C inhibitors [32], it became apparent that analysis of Ras by HDX MS provided valuable information about movement and conformational change during GDP/GTP switching. As described many times in the literature, HDX MS can be used to compare the conformations of different protein states, such as native versus non-native, wild type versus mutant, and/or free versus associated with another protein or ligand [35, 36]. The results are complementary to X-ray crystallography and can provide information silenced by crystallization or give access to proteins that will not crystallize or diffract well. We applied HDX MS to five representative members of the Ras subfamily (K-Ras, H-Ras, N-Ras, R-Ras, Rap1B) and three members of the Rho subfamily (CDC42, Rac1, and RhoA) to answer questions about the extent to which conformational changes were conserved upon nucleotide switching, if changes occurred in similar places, and to group the proteins into classes according to how they respond to activation. We then correlated the data based on conservation of primary and tertiary structure.
RESULTS AND DISCUSSION
Ras family proteins have variable sequence but conserved tertiary structure
Similarity between the proteins chosen for investigation was quantified based on sequence and tertiary structure. A sequence alignment, including N- and C-terminal tags, is shown in FIGURE 1a. Each construct included the G domain and some portion of the hypervariable domain. The crystal structure of the G domain from activated K-Ras (PDB: 3GFT), chosen as a general representation of the G domain structure, is shown in FIGURE 1b, and a number of the critical structural motifs are highlighted (the hypervariable domain is not shown because its structure has not been observed in structural studies and is therefore unknown). The P-loop, also known as the Walker-A-motif, consists of sequence GxxGxGKS/T where x is any amino acid and binds to the β-phosphate of GDP and the β,γ-phosphates of GTP. This is followed by the switch I region, which is a loop and β-strand considered at minimum to contain residues 32–38. Within switch I, a threonine residue forms a hydrogen bond with an oxygen atom from the γ-phosphate of GTP. Upon binding GTP, this region changes conformation [37–41] and stabilizes the β-strand that immediately succeeds it, thereby allowing that strand to more easily bind to downstream effector proteins [12–15, 41]. Switch II consists of a helical/loop region containing residues 59–67, which changes conformation depending on which nucleotide is bound. The conserved DxxG motif immediately precedes the switch II region, and it confers specificity for guanosine and contributes to phosphate-binding. The final conserved element is a loop containing the sequence N/TKxD, and this interacts with the guanosine base.
While there is some sequence similarity, pairwise comparisons (FIGURE 1c, top numbers) quantitatively reveal the divergence of sequence among the family members. Certain sequences are very similar, for example K-Ras and H-Ras share 94.0% sequence identity; however, CDC42 and Rap1B share only 26.5% sequence identity. The proteins studied share an average of 47.4% sequence identity, with the Ras subfamily proteins sharing 67.2% sequence identity and the Rho subfamily proteins sharing 60.7%. When the tertiary structures of these proteins were compared through alignments of α-carbon atoms, the relatively wide range of sequence similarity translated into a comparatively narrow range of RMSDs (FIGURE 1c, bottom numbers in parenthesis). Overall, the average RMSD between all Ras proteins studied was 1.87 Å. The Ras subfamily proteins have an average RMSD of 1.25 Å and the Rho subfamily proteins have an average RMSD of 1.67 Å. The three most similar proteins studied were K-, H-, and N-Ras. These proteins all share greater than 90% sequence identity, and their tertiary structures align with RMSDs of 0.69–1.10 Å. In general, greater disparity in sequence between two proteins also corresponded to greater variability in tertiary structure. For example, K-Ras and N-Ras share 91.7% sequence and have an RMSD of 0.82 Å, but K-Ras and CDC42 share only 33.3% sequence and have an RMSD of 3.30 Å. Not surprisingly, sequence and especially tertiary structure seem to be more highly conserved within a given Ras subfamily compared to proteins from different subfamilies.
Deuterium incorporation of inactive Ras proteins
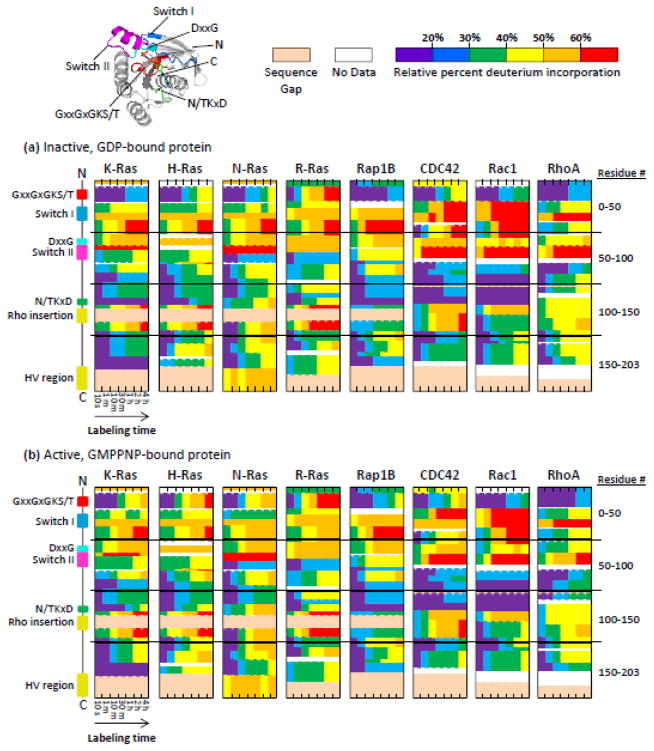
Each of the eight proteins was overexpressed, purified, and then independently exposed to deuterium. Using standard and well-established methodology (see Experimental Procedures), the exchange reaction was quenched, labeled proteins were digested into pieces with pepsin, and the location and magnitude of deuteration measured using MS (representative mass spectra are shown in Supplemental FIGURE S1). The experiments were performed for both the GDP-bound (inactive) form and the non-hydrolysable GTP analogue GMPPNP-bound (active) form after nucleotide exchange, as described in the next section. Of note, GMPPNP affinity for Ras has been measured at approximately 20 times below the affinity for GTP. Nevertheless, the affinity remains high in the sub- to low-nanomolar range, so this was not expected to impact the relevance of results obtained from our system [42, 43]. The percent deuteration in different portions of the inactive form of each protein is shown in FIGURE 2a, and the same is shown for each active form in FIGURE 2b. The results indicate differences in the deuterium incorporation of both the inactive and active states of these eight proteins; however, such data must be interpreted carefully [44]. Variations in protein sequence produce differences in the loss of deuterium during mass analysis, and changes to digestion patterns as a result of sequence difference make comparisons between proteins complicated. Calculations of the theoretical back-exchange at each backbone amide position in each protein analyzed in this study (Supplemental FIGURE S2) show just how diverse the back-exchange can be due to differences in sequence. The end result is that data such as those in FIGURE 2 cannot be used to make a comparison of the differences between proteins (meaning comparing horizontally across proteins in FIGURE 2) without a correction for deuterium back-exchange during analysis. No back-exchange correction was performed in this work for two reasons: first, in general it can be difficult to prepare maximally deuterated reference controls [45] for proteins and we were not certain that in such controls all positions would have become labeled such that a correction would be valid; and second, the main goal was to measure the differences between the GMPPNP- and GDP-bound forms (comparing vertically for a given protein in FIGURE 2). For comparisons of exchange of the same protein in two different bound states, a back-exchange correction is not necessary and a relative comparison [46] can be performed. The peptide fragments produced during pepsin digestion were, as expected, different for these eight proteins (Supplemental FIGURE S3). The highly-conserved regions often generated similar peptides for most proteins, but the less conserved regions did not. Therefore, comparison of exchange across regions of diverse sequence can be challenging. However, this issue is nearly eliminated by considering the differences in deuteration between the GMPPNP- and GDP-bound states of a given protein, since the peptic peptides produced upon digestion of the two states of a single protein were identical, including in sequence. Considerations about differences in back- and forward-exchange resulting from sequence variation are no longer relevant. HDX differences measured in these comparison experiments report on how changes to the bound nucleotide influence deuteration. Sequence variation between the proteins could certainly change the response to the nucleotides during the labeling step, and therefore affect the amount of deuterium. Our comparison experiments mainly focused on monitoring the differences between active and inactive forms of the proteins, and then relating these differences to structural comparisons.
FIGURE 2.
Relative percent deuterium incorporation of (a) inactive, GDP-bound and (b) active, GMPPNP-bound Ras proteins. Peptic peptides where deuterium incorporation was measured run vertically from N- to C-terminus, with horizontal lines delineating blocks of 50 residues. Labeling time increases from left to right, as shown. Each box is colored according to the deuterium scale at the top right. The locations of key structural motifs are shown at the left (aligned to each box) and highlighted on the crystal structure of active K-Ras (PDB: 3GFT) in the upper left corner. Regions of a given protein represented in light beige indicate sequence gaps, or locations in the sequence where at least one other protein studied (besides the given protein of interest) contains an insertion of one or more amino acid residues.
Preparing GDP- and GTP-loaded forms
Overexpression and purification of Ras proteins from bacteria results in primarily the GDP-bound form. To generate the active form, GDP must be exchanged for GTP (or in this case a non-hydrolysable analogue of GTP, GMPPNP). As GDP is bound extremely tightly (Kd = 0.6 – 60 nM) [43, 47, 48], it can be difficult to achieve 100% GMPPNP loading and nucleotide exchange efforts frequently result in a mixture of protein bound to GDP and GMPPNP. We therefore optimized nucleotide exchange and analyzed nucleotide content by HPLC using an ion exchange column for nucleotide separation. The measured GMPPNP and GDP content for the samples analyzed here is shown in Supplemental FIGURE S4a. To characterize the spread of the HDX MS data as a result of different levels of nucleotide loading, we performed multiple biological replicates [49, 50] wherein each replicate was composed of an independent expression of protein followed by purification, nucleotide switching and characterization by HPLC, and finally HDX MS analysis. The variability of the measured deuterium levels in these replicates (Supplemental FIGURE S4b) was relatively low for the GDP-bound form, but was high in some regions for the GMPPNP-bound forms. Unequal GMPPNP content was the primary cause for localized deuterium-level variability in the GMPPNP-bound forms. The locations of differences in deuteration between GMPPNP- and GDP-bound forms remained consistent between biological replicates of the same protein while the magnitude did not. We could reliably determine which peptides, or which parts of the structure, underwent changes between the two nucleotide-bound forms. The magnitude of the difference in deuteration between the forms was much more difficult to quantify, but did not detract from the ability to say with some confidence which proteins behave the same and which behave differently. Subtle changes in deuterium levels between proteins as a result of differences in the amount of GDP removed seem to us to be nearly impossible to measure without an absolutely 100% GDP-free form.
Ras family proteins activate differentially
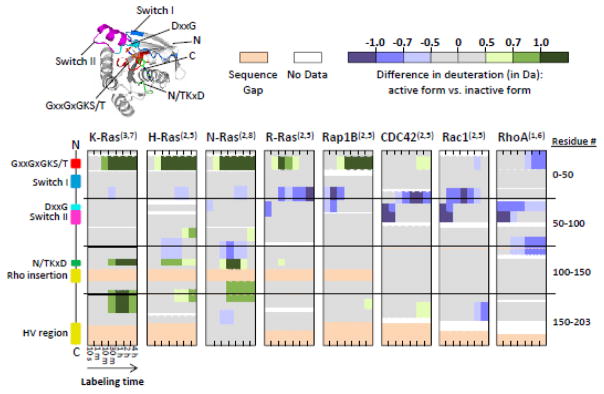
To compare the GDP- or GMPPNP-bound forms of the multiple proteins in the study, differences in deuterium levels for each of the peptic peptides were calculated by subtracting the deuterium level of the GDP-bound form from the deuterium level of the GMPPNP-bound form (representative deuterium uptake curves shown in Supplemental FIGURE S5). The resulting numerical values were color-coded (FIGURE 3) with positive values (more deuterium in the GMPPNP-bound form) colored green and negative values (less deuterium in the GMPPNP-bound form) colored purple. These results highlight regions of the proteins that undergo changes in backbone amide hydrogen exchange when the proteins switch between states, and indicate that not all proteins respond similarly to nucleotide switching.
FIGURE 3.
Raw difference (in Da) between active, GMPPNP-bound and inactive, GDP-bound Ras proteins. From top to bottom, each visual shows the protein of interest from N- to C-terminus, with labeling time increasing from left to right. The superscript numbers in parentheses to the right of each protein name indicate the number of biological replicates (uniquely expressed and purified protein preparations) and the total number of labeling replicates analyzed, respectively, for that particular protein. The visual on the far left indicates the locations of key structural motifs, which are also highlighted on the crystal structure of active K-Ras (PDB: 3GFT) in the upper left corner. Gaps in coverage indicate regions of protein for which there was no peptide coverage and therefore no data were generated. Sequence gaps, represented in light beige, indicate regions where at least one protein had an insertion of one or more amino acids that other proteins did not share.
We expected the most significant differences would occur in switches I and II given multiple observations from various structural methods that the conformation of these regions are different when comparing the GDP and GTP-bound states, or analogues thereof. No differences in HDX were detected in switch I for any proteins, which we attribute to the nature of the HDX measurement. While HDX MS is sensitive to changes in the exchange of backbone amide hydrogens it may not be influenced by interactions involving primarily sidechains [35]. In switch I, which is well-exposed to solvent and mostly not involved in hydrogen bonding due to secondary structure, changes to the nucleotide would be expected to primarily affect side chain interactions and therefore not necessarily alter HDX. In switch II, in contrast, there was a pattern of significantly less deuterium exchange at the early time points for the GMPPNP- vs. GDP-bound states for CDC42, Rac1, and RhoA. This is consistent with Ras structures showing that the GTP-bound form is ‘closed’ and the elements of switch II less solvent accessible relative to the GDP-bound form. Surprisingly, the same deuteration signature was not evident in Ras subfamily members.
Striking patterns were seen in the P-loop (GxxGxGKS/T, FIGURE 3), where K-, H-, and N-Ras showed more deuterium in the GMPPNP-bound (active) state compared to the GDP-bound (inactive) state, represented by the band of dark green near the N-terminus of each of these three proteins. Rap1B showed the same general trend as K-, H-, and N-Ras of increased deuteration in the active form. R-Ras differed from the other Ras subfamily proteins in that it was less affected in the P-loop by nucleotide switching. Interestingly, the Rho subfamily proteins CDC42, Rac1, and RhoA, which showed decreases in deuteration at early time points as discussed above, did not show the same degree of an increase in deuteration upon activation in the P-loop as the Ras subfamily proteins, and Rac1 and RhoA actually incorporated less deuterium in the active state.
In the region between switch I and switch II, R-Ras, Rap1B, CDC42, and Rac1 showed areas of reduced deuterium incorporation. Between switch II and the N/TKxD motif there was disparity between the proteins, with some showing areas of more deuterium upon activation and some showing less. In the N/TKxD motif, all Ras subfamily proteins except Rap1B showed more deuterium exchange upon activation while none of the Rho subfamily proteins showed any significant differences upon activation. Following the Rho insertion (Rho subfamily proteins contain an additional α-helix that Ras subfamily proteins do not) was another area of disparity between proteins. This is partly to be expected given that this region leads into and contains the C-terminal hypervariable domain, thusly named because it is used for protein trafficking and subcellular localization, and as such the protein sequences are more highly divergent here than anywhere else.
To determine where the changes upon activation were located in the proteins, and if tertiary features could be correlated with dynamics, differences in deuteration upon activation were mapped onto the tertiary structures of each protein. Because it is difficult to show every subtle difference detailed in FIGURE 3 on a single structure for each protein, differences in deuteration at each deuterium exchange time point were summed to yield a “net deuteration” value for each peptide (see Experimental Procedures). These net deuteration values were then color-coded and mapped onto the respective crystal structures (FIGURE 4). The crystal structures were organized around a phylogenetic tree based on sequence identity. One thing that was immediately apparent was that similar proteins did not necessarily have identical deuterium incorporation differences between states. For example, the HDX MS data reporting how these proteins respond to nucleotide switching do not indicate that K-, H-, and N-Ras are similar in terms of activation-induced dynamics despite the fact that K-, H-, and N-Ras are similar in terms of primary and tertiary structure.
FIGURE 4.
The total differences in deuterium incorporation for each protein highlighted on the respective crystal structure of that protein (PDB codes as indicated). For a given region, the total difference in deuterium incorporation is the sum of the individual differences at each time point for that region. The crystal structures of these proteins are organized using a phylogenetic tree, generated using Clustal Omega in conjunction with DrawTree [64, 65], which is shown in the middle. At the top, key conserved structural motifs are shown using this linear representation as well as highlighted on the crystal structure of active K-Ras (PDB: 3GFT).
FIGURE 4 reveals some interesting correlations, including the broad differences between the Ras and Rho subfamilies. Proteins at one end of the phylogenetic tree (K-Ras, H-Ras, N-Ras, R-Ras, and Rap1B) show strong increases in deuteration upon activation in the P-loop but do not show significant differences in the switch II region, whereas proteins at the other end of the tree (CDC42, Rac1, and RhoA) show decreases in deuteration upon activation in switch II but do not show strong differences in the P-loop. The exclusivity between the P-loop and switch II suggests an interaction between the two elements. This argument is also based on the fact that switch II interacts directly with the P-loop in certain functional states of Ras. Furthermore, switch II is one of the most mobile elements of Ras as demonstrated by comparing structures of Ras across the superfamily and by comparing GTP- vs. GDP-bound structures of Ras isoforms. The ability to experimentally measure switch II mobility also has important implications for therapeutic approaches which directly target Ras, especially in light of the recently discovered switch II pocket (SIIP) inhibitors of K-Ras G12C that rely on movement of switch II for binding [33, 34, 51]. If increased deuteration in the P-loop or decreased deuteration in switch II correlates with switch II mobility, HDX MS may provide an experimental means of determining which other Ras proteins might be targetable by SIIP-binding molecules.
To further assess correlations between this increased P-loop deuteration and switch II mobility we conducted molecular dynamic simulations extending over 50 ns using either X-ray crystal structures or homology models of the Ras proteins studied by HDX MS. Simulations for both the GTP- and GDP-bound forms were performed. RMSD of the protein mainchains over the courses of simulations are shown (Supplemental FIGURE S6) and show that equilibrium was achieved prior to 30 ns. Switch II mobility was assessed by monitoring the variability of main chain positions over the course of the simulation. All protein frames were aligned on the starting reference frame and the root mean square fluctuation (RMSF) was calculated to measure the local changes along the protein backbone chain. Larger numbers for RMSF indicate significant movement in the associated region of the protein backbone during a simulation run. Main chain RMSF is plotted across the protein sequence in FIGURE 5. We note that the RMSF for K-Ras is higher in switch II compared to other Ras family members, including H-Ras. While surprising given the high sequence similarity between H-Ras and K-Ras, this is consistent with prior simulations of K-Ras vs. H-Ras A59G [52]. GTP -bound proteins showing a significant P-loop exposure signature (H-Ras, N-Ras, K-Ras, R-Ras, Rap1B, and CDC42) all demonstrate an RMSF peak greater than 2 units in the switch II region, whereas those showing no P-loop signature (Rac1 and RhoA) do not. We reasoned that a difference in mobility between GTP- vs. GDP-bound states might translate into increased P-loop deuteration as seen in the HDX MS data. To evaluate this possibility, we calculated the difference in RMSF between GTP- and GDP-bound forms (FIGURE 5, right). While in some cases this did yield the expected significant positive difference in favor of GTP for proteins demonstrating the P-loop signature (K-Ras, R-Ras, CDC42), there were other P-loop proteins for which the positive difference was positive but small (H-Ras, N-Ras, Rap1B). For proteins showing no HDX MS P-loop signature (Rac1 and RhoA), there was minimal difference between GTP- and GDP-bound simulations. A likely explanation for the association between a high P-loop signature and increased switch II dynamics is that a dynamic switch II leaves the adjacent P-loop more exposed and therefore subject to more rapid exchange.
FIGURE 5.
Analysis of Ras proteins by MD Simulation. The root mean square fluctuation (RMSF) is used to characterize local changes along protein backbone residues whose positions are shown on the x axis. Changes in RMSF (ΔRMSF, in Å) are calculated from left to right to indicate the difference between GTP- and GDP-bound states for each protein. The switch II region is highlighted in red for plots showing RMSF and green in ΔRMSF.
Taken together these data suggest that GTP- and GDP-associated switch II mobility may be a key functional distinction between different Ras subfamilies. Specifically, by these data we would predict that for Ras subfamily members, a relatively hyperdynamic switch II confers specific features that could influence interactions with effectors or the membrane, rates of nucleotide hydrolysis, or rates of nucleotide exchange. In turn, these features may also translate into higher order functions such as increased signaling plasticity. For example, the importance of switch II engagement at the time of GTP hydrolysis [53] suggests that a hyperdynamic switch II would lead to lower intrinsic GTP hydrolysis rates and an increased dependence on GAPs for inactivation of the GTPase, or that in the cell, Ras proteins might remain in an activated state longer than Rho proteins. While measurements of GTP hydrolysis for both RAS [7] and RHO [54] proteins have been measured, and both are greatly stimulated by GAPs, to our knowledge these rates have never been directly compared to one another. Similarly, the proportion of cellular GTP-bound Ras or Rho proteins is also technically difficult to measure and has not been directly compared. While it is tempting to speculate how protein dynamics or resulting functional differences might subserve the known functions of Ras vs. Rho proteins, such as regulation of transcription or cytoskeletal reorganization [55, 56], general or specific conclusions are difficult to reach without additional dedicated study. As a first step, it would be interesting to subject members of other Ras subfamilies not included in this study (e.g., Rab, Ran, and Arf) to similar analyses to determine if dynamic features reliably correlate with primary structure.
Given that HDX MS is capable of characterizing aspects of switch II dynamics, another exciting possibility is that this technique will be a useful method for evaluating compounds which occupy the switch II binding site, as has been done for the activating K-Ras G12C mutant [32]. Indeed, these compounds place switch II in a conformation not previously observed, but at the same time are expected to stabilize switch II by virtue of interactions between these compounds and switch II residues. It is therefore likely that switch II binders will have a particular HDX MS signature that could be useful not only for studying the impact of these compounds on K-Ras G12C, but also for exploring the potential of this class of compounds on other K-Ras mutants or other Ras superfamily members.
CONCLUSIONS
HDX MS provides a method by which dynamic characteristics of Ras proteins may be classified, as in this work where distinguishing hallmarks were found between eight members of the Ras and Rho subfamilies. In particular, the five Ras subfamily proteins displayed a striking HDX MS signature in the P-loop that is exclusive of a pattern for switch II found in Rho family proteins. These two signatures correlate with switch II dynamics as measured by MD simulations. Measurements of Ras protein dynamics by HDX MS may be useful for studying the functional characteristics of Ras subfamilies. Additionally, HDX MS may prove useful for understanding functional effects of oncogenic mutations, the development of direct Ras inhibitors such as switch II pocket binders, or other targetable vulnerabilities that depend upon Ras protein dynamics.
EXPERIMENTAL PROCEDURES
Purification of Ras proteins
DNA encoding K, N, H-Ras fused with N-terminal multi-histidine tags (or GST in the case of H-Ras) were cloned into pJExpress vectors (DNA 2.0, Menlo Park, CA). R-Ras and Rap1B were cloned into modified pET28a vectors with N-terminal histidine tags. Rac1, RhoA, and CDC42 were cloned into pET24a vectors with C-terminal histidine tags. All proteins were overexpressed in the E coli. B21 strain. Cultures for each were grown in Luria broth at 37 °C with shaking at 225 rpm until the OD600 reached 0.8. The temperature was reduced to 16 °C and expression was induced with 0.5 mM IPTG. After 8–12 hours, bacteria were harvested by centrifugation for 15 minutes at 4 °C. The pellet was resuspended in lysis buffer (20 mM NaH2PO4, 500 mM NaCl, 5% glycerol, 20 mM imidazole) containing 1 mM PMSF, 1 mM benzamidine, and 1 mg/ml lysozyme then flash frozen. Cells were lysed to completion using sonication and the lysates clarified by centrifugation for 35 minutes. Supernatants were filtered using 0.22 μm syringe filters, and applied to Ni-NTA resin (Biorad, Hercules, CA). Elution was accomplished with a step gradient to buffer containing 20 mM NaH2PO4, 1 M NaCl, 5 % glycerol, and 100 mM imidazole. Proteins were immediately buffer exchanged using a desalting column into Buffer A (20 mM Tris-HCl, 100 mM NaCl, pH 8.0).
Nucleotide exchange
Proteins were diluted to 45 μM using Buffer A and incubated with either 5 mM GDP or 0.5 mM GMPPNP. For reactions containing GMPPNP, 4 units alkaline phosphatase per μg of protein was also included similar to methods described previously [57]. Reactions were incubated at 25 °C for 2 hours with gentle agitation, then buffer exchanged to remove unbound nucleotides using Buffer A and Amicon Ultra 0.5 mL spin columns (10,000 Da MWCO) wherein proteins were subjected to 3 cycles of concentrating from 500 μL to 100 μL, then adding 400 μL Buffer A to the protein solution. The final protein concentration was assessed using Bradford reagent (Biorad). Protein concentrations were adjusted to 90 μM using Buffer A and flash frozen in liquid nitrogen.
The extent of nucleotide exchange was verified by extracting nucleotides and analyzing the extracts by HPLC using an anion exchange column. Protein samples were buffer exchanged into Buffer A immediately before processing using 0.5 mL Zeba™ (ThermoFisher Scientific) desalting cartridges (7,000 Da MWCO) to remove unbound nucleotides. The protein concentration was adjusted to 45 μM with Buffer A and protein was denatured by adding 12 M urea in a 1:1 volumetric ratio to release bound ligands. Samples were centrifuged at 21,000 ×g for 5 minutes to pellet any insoluble material. The supernatant was applied to a BioSAX NP-10 column (Agilent Technologies, Santa Clara, CA) and nucleotides were separated by a sodium acetate gradient ranging from 0 to 1 M sodium acetate over 25 minutes. Nucleotides were detected by UV absorbance at 260 nm. Peak assignments were made based on retention times of GDP and GMPPNP standards. Percent loading was calculated using the area under the curve obtained from the UV traces.
Hydrogen-deuterium exchange mass spectrometry (HDX MS)
HDX MS experiments were performed in a similar manner to those described previously [32, 58]. Both the GDP- and GMPPNP-bound forms of each protein (K-Ras, H-Ras, N-Ras, R-Ras, Rap1B, CDC42, Rac1, and RhoA) were independently labeled with deuterium, using identical experimental conditions to allow for comparison between each set of bound forms. Preliminary deuterium labeling experiments were performed in duplicate, each time with an independent protein preparation, and at least one subsequent deuterium labeling experiment was performed in triplicate on each protein to confirm initial results.
Hydrogen/deuterium exchange was initiated by diluting 2.0 μL of nucleotide-bound protein (75–100 μM protein in 20 mM Tris-HCl, 100 mM NaCl, pH 8.0) 15-fold in the identical buffer containing 99% deuterium oxide at room temperature. The labeling reaction was quenched to approximately pH 2.5 at seven predetermined time points (10 s, 1 m, 10 m, 30 m, 1 h, 2 h, and 4 h) through the addition of 32.0 μL of quench buffer (2.0 M guanidinium chloride, 0.8% formic acid, pH 2.4) at 0 °C. Quenched samples were immediately flash frozen using dry ice and were stored for less than one week at −80 °C prior to analysis. For each deuterium labeling time-course, triplicate samples were prepared so that mass determination could be performed three times for each time point and for each protein:nucleotide combination. To measure extent of deuterium incorporation, each sample was rapidly thawed and injected into a Waters nanoACQUITY UPLC equipped with HDX technology [59]. Online pepsin digestion [60] was performed at 15 °C as described previously [58], and resulting peptic peptides were trapped using a Waters VanGuard™ BEH C18 1.7 μm guard column followed by separation using a 5–35% acetonitrile gradient over six minutes at a flow rate of 60 μL/min and a Waters ACQUITY UPLCR® HSS T3 1.8 μm 1.0 × 50 mm analytical column. All mobile phases contained 0.1% formic acid. After each sample, wash cycles were performed to minimize carry-over between analytical injections [61]. Mass spectral analyses were performed using a Waters Synapt G1 mass spectrometer equipped with a standard ESI source in V Mode. Peptic peptides were identified in undeuterated control samples using Waters MSE and Waters ProteinLynx Global Server (PLGS) 2.5 on the same UPLC/QTOF system used for HDX MS experiments [62]. Peptic peptide maps were generated for each protein using DynamX 2.0 software (Waters), which was also used to filter peptides and generate deuterium incorporation graphs. These deuterium incorporation graphs were created by plotting the relative amount of deuterium in each peptic peptide at each exchange time, or by subtracting the centroid of the isotopic distribution for each labeling time point from the centroid of the isotopic distribution of the undeuterated control species. Since the data were not corrected for back-exchange, the plots show the relative deuterium level at each time point for each peptide [46]. HDX MS analysis on this type of experimental system has been well-characterized previously, and the error of each data point generally does not exceed ±0.15 Da [63]. For this series of experiments, the average error for a single data point was ±0.09 Da within a single biological replicate. As such, differences in the relative deuterium level that exceeded 0.50 Da between the two protein states were considered to be meaningful.
To calculate the net difference in deuteration in FIGURE 4, for each peptide, deuteration differences at each labeling time point were summed. Values ranged from −5.37 Da to 12.34 Da, and an uncapped linear scale covering <-4 Da to >8 Da was created (FIGURE 4) to indicate relative magnitudes of the summed differences. Given there were 7 time points and a threshold of 0.5 Da difference was considered meaningful (FIGURE 3, see also above), a net difference of 3.5 Da would appear to indicate a meaningful difference between the nucleotide-bound states. We elected to use a threshold of 2.0 Da to represent meaningful net differences as it captured the vast majority of peptides showing a meaningful difference (>0.5 Da) at one or more time points in the time course, although a few peptides with minor (albeit reproducible) differences (e.g., perhaps just near 0.5 Da at 1–3 time points) were excluded. There was only a single peptide (found in R-Ras) that did not exceed the 0.5 Da threshold at any time point but did exceed the threshold of 2.0 Da representing a meaningful net difference. The crystal structures are organized around a phylogenetic tree based on a comparison of sequence identity, which was created using DrawTree [64, 65]. Comparisons of sequence identity were generated by using Clustal Omega [66, 67] to compare the protein constructs analyzed in this study, excluding purification tags.
Molecular dynamics (MD) simulations
The Schrödinger Maestro package (Schrödinger Release 2016-2: Maestro, version 10.6, Schrödinger, LLC, New York, NY, 2016.) was used to perform molecular dynamics. Systems were prepared from high-resolution crystal structures of Ras superfamily members available in the PDB: Rac1, CDC42, RhoA, H-Ras, N-Ras, K-Ras, Rap1B, and R-Ras. Each system was simulated with the active state (GTP analogue-bound) and inactive state (GDP-bound). The N-Ras and R-Ras in active state, which are not available in PDB, were generated by using the Homology Modeling module with GMPPNP. The complete PDB ID (GDP-bound, GTP analogue-bound) list was: Rac1 (2P2L, 1I4T), CDC42 (1A4R, 1E0A), RhoA (1FTN, 3TVD), H-Ras (4Q21, 1JAH), K-Ras (4DSU, 3GFT), Rap1B (4M8N, 4HDO), N-Ras (3CON), and R-Ras (2FN4). The Protein Preparation module was used for model construction, including adding missing atoms, H-bond assignments, and restrained minimization. All systems were neutralized by adding charge-neutralizing counter ions with 10 Å buffering distance in SPC solvent model. No ion-excluded region was included. The 50 ns simulations were carried out by Desmond Molecular Dynamics module with constant temperature (300 K) and pressure (1.0 bar) in NPT ensemble class.
Supplementary Material
HIGHLIGHTS.
HDX MS, MD simulations of K-Ras, H-Ras, N-Ras, R-Ras, Rap1B, CDC42, Rac1, and RhoA
Ras subfamily member HDX MS is distinct from Rho subfamily member HDX MS
Dynamics and primary/tertiary structure conservation do not correlate
Identified a subfamily-dependent correlation between switch II and P-loop
Acknowledgments
We would like to thank Nathanael Gray for support and helpful guidance with this project, and Thomas Wales for technical assistance with HDX MS. Funding was provided by the Cancer Prevention Research Institute of Texas R1207 (KDW), ACS IRG-02-196-07 (KDW), Welch Foundation I-1829 (KDW), NIH R01-GM101135 (JRE), a research collaboration with the Waters Corporation (JRE), and the Dana Farber Cancer Institute/Northeastern University Joint Program in Cancer Drug Development (JRE).
ABBREVIATIONS
- GDP
guanosine diphosphate
- GTP
guanosine triphosphate
- GMPPNP
guanylyl imidodiphosphate
- P-loop
phosphate-binding loop
- HDX
hydrogen-deuterium exchange
- MS
mass spectrometry
- MD
molecular dynamics
- RMSF
root mean square fluctuation
- NMR
nuclear magnetic resonance
- SIIP
switch II pocket
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Laufs U, Liao JK. Targeting Rho in cardiovascular disease. Circ Res. 2000;87:526–8. doi: 10.1161/01.res.87.7.526. [DOI] [PubMed] [Google Scholar]
- 2.Seabra MC, Mules EH, Hume AN. Rab GTPases, intracellular traffic and disease. Trends Mol Med. 2002;8:23–30. doi: 10.1016/s1471-4914(01)02227-4. [DOI] [PubMed] [Google Scholar]
- 3.Linseman DA, Loucks FA. Diverse roles of Rho family GTPases in neuronal development, survival, and death. Front Biosci. 2007;13:657–76. doi: 10.2741/2710. [DOI] [PubMed] [Google Scholar]
- 4.Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–9. [PubMed] [Google Scholar]
- 5.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 6.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–51. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol Cancer Res. 2015;13:1325–35. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 8.Lu S, Jang H, Gu S, Zhang J, Nussinov R. Drugging Ras GTPase: a comprehensive mechanistic and signaling structural view. Chem Soc Rev. 2016 doi: 10.1039/c5cs00911a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colicelli J. Human RAS superfamily proteins and related GTPases. Science’s STKE: signal transduction knowledge environment. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiological reviews. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 11.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 12.Stieglitz B, Bee C, Schwarz D, Yildiz O, Moshnikova A, Khokhlatchev A, et al. Novel type of Ras effector interaction established between tumour suppressor NORE1A and Ras switch II. EMBO J. 2008;27:1995–2005. doi: 10.1038/emboj.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–8. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 14.Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–43. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 15.Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–43. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 16.Barbacid M. ras genes. Annual review of biochemistry. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 17.Cox AD, Der CJ. Protein prenylation: more than just glue? Curr Opin Cell Biol. 1992;4:1008–16. doi: 10.1016/0955-0674(92)90133-w. [DOI] [PubMed] [Google Scholar]
- 18.Traut TW. Physiological concentrations of purines and pyrimidines. Molecular and cellular biochemistry. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 19.Werner A, Siems W, Schmidt H, Rapoport I, Gerber G, Toguzov RT, et al. Determination of nucleotides, nucleosides and nucleobases in cells of different complexity by reversed-phase and ion-pair high-performance liquid chromatography. J Chromatogr B. 1987;421:257–65. doi: 10.1016/0378-4347(87)80406-1. [DOI] [PubMed] [Google Scholar]
- 20.Fürst W, Hallström S. Simultaneous determination of myocardial nucleotides, nucleosides, purine bases and creatine phosphate by ion-pair high-performance liquid chromatography. J Chromatogr B. 1992;578:39–44. doi: 10.1016/0378-4347(92)80222-c. [DOI] [PubMed] [Google Scholar]
- 21.Pilz R, Willis R, Boss G. The influence of ribose 5-phosphate availability on purine synthesis of cultured human lymphoblasts and mitogen-stimulated lymphocytes. J Biol Chem. 1984;259:2927–35. [PubMed] [Google Scholar]
- 22.Hauschka PV. Analysis of nucleotide pools in animal cells. Methods Cell Biol. 1973;7:361–462. doi: 10.1016/s0091-679x(08)61787-2. [DOI] [PubMed] [Google Scholar]
- 23.Snyder FF, Cruikshank MK, Seegmiller JE. A comparison of purine metabolism and nucleotide pools in normal and hypoxanthine-guanine phosphoribosyltransferase-deficient neuroblastoma cells. Biochim Biophys Acta. 1978;543:556–69. doi: 10.1016/0304-4165(78)90310-0. [DOI] [PubMed] [Google Scholar]
- 24.Jackson RC, Boritzki TJ, Morris HP, Weber G. Purine and pyrimidine ribonucleotide contents of rat liver and hepatoma 3924A and the effect of ischemia. Life Sci. 1976;19:1531–6. doi: 10.1016/0024-3205(76)90098-9. [DOI] [PubMed] [Google Scholar]
- 25.Weber G, Lui MS, Jayaram HN, Pillwein K, Natsumeda Y, Faderan MA, et al. Regulation of purine and pyrimidine metabolism by insulin and by resistance to tiazofurin. Adv Enzyme Regul. 1985;23:81–99. doi: 10.1016/0065-2571(85)90041-x. [DOI] [PubMed] [Google Scholar]
- 26.Keppler DO, Pausch J, Decker K. Selective Uridine Triphosphate Deficiency Induced by d-Galactosamine in Liver and Reversed by Pyrimidine Nucleotide Precursors EFFECT ON RIBONUCLEIC ACID SYNTHESIS. J Biol Chem. 1974;249:211–6. [PubMed] [Google Scholar]
- 27.de Korte D, Haverkort WA, van Gennip AH, Roos D. Nucleotide profiles of normal human blood cells determined by high-performance liquid chromatography. Anal Biochem. 1985;147:197–209. doi: 10.1016/0003-2697(85)90028-4. [DOI] [PubMed] [Google Scholar]
- 28.Wright DG. A role for guanine ribonucleotides in the regulation of myeloid cell maturation. Blood. 1987;69:334–7. [PubMed] [Google Scholar]
- 29.Pilz RB, Huvar I, Scheele JS, Van den Berghe G, Boss GR. A decrease in the intracellular guanosine 5′-triphosphate concentration is necessary for granulocytic differentiation of HL-60 cells, but growth cessation and differentiation are not associated with a change in the activation state of Ras, the transforming principle of HL-60 cells. Cell growth & differentiation: the molecular biology journal of the American Association for Cancer Research. 1997;8:53–9. [PubMed] [Google Scholar]
- 30.Bollag G, McCormick F. Regulators and effectors of ras proteins. Annual review of cell biology. 1991;7:601–32. doi: 10.1146/annurev.cb.07.110191.003125. [DOI] [PubMed] [Google Scholar]
- 31.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–67. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim SM, Westover KD, Ficarro SB, Harrison RA, Choi HG, Pacold ME, et al. Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew Chem Int Ed Engl. 2014;53:199–204. doi: 10.1002/anie.201307387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patricelli MP, Janes MR, Li L-S, Hansen R, Peters U, Kessler LV, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016 doi: 10.1158/2159-8290.CD-15-1105. CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 35.Engen JR. Analysis of protein complexes with hydrogen exchange and mass spectrometry. The Analyst. 2003;128:623–8. doi: 10.1039/b212800b. [DOI] [PubMed] [Google Scholar]
- 36.Pirrone GF, Iacob RE, Engen JR. Applications of hydrogen/deuterium exchange MS from 2012 to 2014. Anal Chem. 2015;87:99–118. doi: 10.1021/ac5040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall BE, Bar-Sagi D, Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc Natl Acad Sci U S A. 2002;99:12138–42. doi: 10.1073/pnas.192453199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341:209–14. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- 39.Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, et al. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–45. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 40.Scheidig AJ, Burmester C, Goody RS. The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure. 1999;7:1311–24. doi: 10.1016/s0969-2126(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 41.Spoerner M, Herrmann C, Vetter IR, Kalbitzer HR, Wittinghofer A. Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc Natl Acad Sci U S A. 2001;98:4944–9. doi: 10.1073/pnas.081441398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scherer A, John J, Linke R, Goody RS, Wittinghofer A, Pai EF, et al. Crystallization and preliminary X-ray analysis of the human c-H-ras-oncogene product p21 complexed with GTP analogues. J Mol Biol. 1989;206:257–9. doi: 10.1016/0022-2836(89)90540-8. [DOI] [PubMed] [Google Scholar]
- 43.Feuerstein J, Goody RS, Wittinghofer A. Preparation and characterization of nucleotide-free and metal ion-free p21 “apoprotein”. J Biol Chem. 1987;262:8455–8. [PubMed] [Google Scholar]
- 44.Wales TE, Poe JA, Emert-Sedlak L, Morgan CR, Smithgall TE, Engen JR. Hydrogen Exchange Mass Spectrometry of Related Proteins with Divergent Sequences: A Comparative Study of HIV-1 Nef Allelic Variants. J Am Soc Mass Spectrom. 2016;27:1048–61. doi: 10.1007/s13361-016-1365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–31. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass spectrometry reviews. 2006;25:158–70. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 47.Hattori S, Ulsh LS, Halliday K, Shih TY. Biochemical properties of a highly purified v-rasH p21 protein overproduced in Escherichia coli and inhibition of its activities by a monoclonal antibody. Mol Cell Biol. 1985;5:1449–55. doi: 10.1128/mcb.5.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manne V, Yamazaki S, Kung HF. Guanosine nucleotide binding by highly purified Ha-ras-encoded p21 protein produced in Escherichia coli. Proc Natl Acad Sci U S A. 1984;81:6953–7. doi: 10.1073/pnas.81.22.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engen JR, Wales TE. Analytical Aspects of Hydrogen Exchange Mass Spectrometry. Annual review of analytical chemistry. 2015;8:127–48. doi: 10.1146/annurev-anchem-062011-143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moroco JA, Engen JR. Replication in bioanalytical studies with HDX MS: aim as high as possible. Bioanalysis. 2015;7:1065–7. doi: 10.4155/bio.15.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lito P, Solomon M, Li L-S, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016:aad6204. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lukman S, Grant BJ, Gorfe AA, Grant GH, McCammon JA. The distinct conformational dynamics of K-Ras and H-Ras A59G. PLoS computational biology. 2010;6 doi: 10.1371/journal.pcbi.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khrenova MG, Grigorenko BL, Kolomeisky AB, Nemukhin AV. Hydrolysis of Guanosine Triphosphate (GTP) by the Ras· GAP Protein Complex: Reaction Mechanism and Kinetic Scheme. The Journal of Physical Chemistry B. 2015;119:12838–45. doi: 10.1021/acs.jpcb.5b07238. [DOI] [PubMed] [Google Scholar]
- 54.Zhang B, Zhang Y, Wang Z-x, Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions of Rho family GTP-binding proteins. Journal of Biological Chemistry. 2000;275:25299–307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]
- 55.Reuther GW, Der CJ. The Ras branch of small GTPases: Ras family members don’t fall far from the tree. Curr Opin Cell Biol. 2000;12:157–65. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 56.Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annual review of biochemistry. 1999;68:459–86. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- 57.Herrmann C, Horn G, Spaargaren M, Wittinghofer A. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J Biol Chem. 1996;271:6794–800. doi: 10.1074/jbc.271.12.6794. [DOI] [PubMed] [Google Scholar]
- 58.Iacob RE, Zhang J, Gray NS, Engen JR. Allosteric interactions between the myristate- and ATP-site of the Abl kinase. PloS one. 2011;6:e15929. doi: 10.1371/journal.pone.0015929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–20. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Molecular & cellular proteomics: MCP. 2002;1:132–8. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 61.Fang J, Rand KD, Beuning PJ, Engen JR. False EX1 signatures caused by sample carryover during HX MS analyses. International journal of mass spectrometry. 2011;302:19–25. doi: 10.1016/j.ijms.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, et al. UPLC/MS(E); a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid communications in mass spectrometry: RCM. 2006;20:1989–94. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- 63.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. Journal of pharmaceutical sciences. 2011;100:2071–86. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–6. [Google Scholar]
- 66.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–9. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.