Abstract
Objective
The aim of this study was to investigate whether serum samples from the Department of Defense Serum Repository (DoDSR) are of sufficient quality to detect microRNAs (miRNAs), cytokines, immunoglobulin E (IgE), and polycyclic aromatic hydrocarbons (PAHs).
Methods
MiRNAs were isolated and quantified by polymerase chain reaction (PCR) array. Cytokines and chemokines related to inflammation were measured using multiplex immunoassays. Cotinine and IgE were detected by enzyme-linked immunoassay (ELISA) and PAHs were detected by Liquid Chromatography/Mass Spectroscopy.
Results
We detected miRNAs, cytokines, IgE, and PAHs with high sensitivity. Eleven of 30 samples tested positive for cotinine suggesting tobacco exposure. Significant associations between serum cotinine, cytokine, IgE, PAHs, and miRNA were discovered.
Conclusion
We successfully quantified over 200 potential biomarkers of occupational exposure from DoDSR samples. The stored serum samples were not affected by hemolysis and represent a powerful tool for biomarker discovery and analysis in retrospective studies.
Environmental exposures can be a major consequence of military service when personnel deploy. A number of environmental and occupational exposures, including burn pit smoke, dust storms, and diesel exhaust, may predispose personnel to long-term health issues such as chronic obstructive pulmonary disease (COPD), asthma, and bronchiolitis.1 The Department of Defense Serum Repository (DoDSR) has collected serum from service members before and after deployment.2 A major knowledge gap is whether these serum samples can be used to detect specific biomarkers that may be useful to predict and/or monitor potential health issues in postdeployment service members. Thus, it is crucial to determine whether serum samples from the DoDSR are of sufficient quality to identify putative biomarkers and signs of exposure, including changes in microRNAs (miRNA), cytokines, chemokines, polycyclic aromatic hydrocarbons (PAHs), and other potential biomolecules that are present in serum.
MiRNAs are a regulatory class of small noncoding RNAs that exhibit tightly controlled spatial and temporal expression patterns. Alterations in miRNA expression can have a profound impact on biological function, as a single miRNA can regulate expression of 100 to over 1000 predicted target genes.3 Due to their small size and resistance to nuclease mediated degradation, circulating miRNA can be detected in serum. Several recent studies show that even after long-term storage at −20°C, miRNA levels remain relatively unchanged.4,5 Several miRNA species are aberrantly expressed in disorders such as cardiovascular disease, COPD, and cancer.6–8 Further, specific miR-NAs can serve as diagnostic and prognostic biomarkers for chemical exposures or diseases.9 Furthermore, Deng et al10 have shown that environmental PAH exposures have altered plasma miRNA levels. One limitation in using miRNA expression for diagnostic and bio-marker discovery purposes is the small amount of patient material available. However, recent advances in miRNA isolation and detection have vastly improved the analysis of serum miRNA.11
In addition to the great promise that miRNAs hold as biomarkers, other factors present in serum are also potential indicators of health, disease, and environmental exposure. Cytokines and chemokines such as interleukin (IL)-6, IL-8, and IL-1β are present in serum samples and are altered by health status.12–14 The immunoglobulin E (IgE) class of antibodies is present at high levels in serum from asthmatics and people with allergy. Furthermore, serum IgE levels are readily increased by certain environmental exposures.15,16 Thus, serum IgE may be an important indicator of allergy and/or environmental exposure.
Finally, other xenobiotic compounds derived from combustion of material present in burn pits, diesel exhaust, or cigarettes can be considereddirect markersof exposure. A number of toxicants and types of smoke produce PAHs and dioxin derivatives that can accumulate in exposed personnel. Thus, the ability to detect PAHs and dioxins in DoDSR serum samples may be of great value to identify exposures and link other biomarkers directly to exposure.17,18
The current study was a proof of concept to demonstrate we could detect miRNAs, cytokines, IgE, and PAHs in DoDSR serum samples and determine whether they are of sufficient quality for use as potential biomarkers. In this study, 30 archival serum samples with no identifier information were chosen to test the viability of these putative biomarkers. In addition, we assessed whether we could integrate results from different classes of biomarkers to discover novel associations.
We found that serum samples from the DoDSR are of sufficient quality to detect over 200 putative biomarkers, including miRNA, cytokines, IgE, and PAHs. Specific biomarkers strongly correlate with each other. We also detected novel associations in this pilot study (n =30) that warranted further investigation in the larger cohort study of 200 deployed and 200 controls who did not deploy.
MATERIALS AND METHODS
Study Design
Thirty archival serum samples that lacked identifier information (unknown subject, unknown collection date) were obtained from the DoDSR for a pilot study, including targeted analysis of miRNAs, cytokines, chemokines, cotinine status, IgE levels, and PAH levels. The samples were stored by the DoDSR at −30°C until thawing, aliquoting, and shipping for analysis. The serum samples were thawed at 4°C overnight, and then aliquoted for the different extractions and assays as described below. In addition, serum was collected from three normal healthy donors at the University of Rochester for use as internal reference standard. These samples were processed and stored at −80°C for no more than 30 days before thawing and extraction with the DoDSR samples.
Isolation and Analysis of miRNA From Serum Samples
MiRNAwas extracted from 0.15 mL of serum samples using the miRCURY RNA isolation kit for biofluids (Exiqon, Boston, MA) following the manufacturer’s instructions. Importantly, 1 μg of bacteriophage MS2 RNA (Roche, Basel, CHE) was added to each sample as a carrier to increase consistency of purification. Purified miRNA (50 μL elution volume) was then subjected to single miRNA TaqMan Reverse transcription-quantitative PCR (RT-qPCR) reactions (Applied Biosystems, Foster City, CA) for quality control before further analysis. All samples passed initial quality control indicating the presence of high-quality miRNA. MiRNA from all samples (15 μL RNA per sample) were then converted into cDNA using the Universal reverse transcriptase miRNA kit (Exiqon). The subsequent cDNA was subjected to analyses on a low-density PCR array for 179 key miRNAs present in a known serum/plasma panel (Exiqon) and amplification and quantification were performed on a Roche Lightcycler 480. Each miRNA was analyzed in duplicate on a 384-well PCR plate and specific melt curves were evaluated for proper amplification. PCR quantification cycles (Cq) values after quality control were compiled and data were normalized using Normfinder software.19 In our study, the most stable normalizer was the average expression of miRNAs common to all samples (n =57, common miRNAs). After normalization, datawere converted into relative miRNA levels for each specific miRNA.
Cotinine Analysis
Tobacco use was determined by cotinine analysis. Cotinine was measured in 0.02 mL of serum using a commercial enzyme-linked immunoassay (ELISA) kit (CO096D; Calbiotech, Spring Valley, CA). Values less than 1.0 ng/mL were defined as nonusers and greater than 10 ng/mL as tobacco users. Values between 1 and 10 ng/mL would meet the criteria for exposure to second-hand tobacco smoke.20 One sample was within this range (1.6 ng/mL) and was grouped with the nonsmokers for subsequent analyses.
Cytokine and Chemokine Analysis
A multiplex panel was designed by selecting biomarkers that were expected to have a high likelihood of revealing immunological and inflammatory changes that might be associated with service-related exposures to pollutants (Luminex, Austin, TX). The cytokine panel included 22 cytokines and chemokines associated with inflammation, B and T cell function and allergy: Eotaxin, Granulocyte-colony stimulating factor, Flt-3L, Granulocyte-macrophage colony-stimulating factor, Fractalkine, Interferonγ, Growth related oncogene, IL-10, IL-12p40, IL-12-p70, IL-13, sCD40L, IL-17A, IL-1ra, IL-10, IL-1α, IL-5, IL-6, IL-8, monocyte chemoattractant protein-1, Tumor necrosis factorα, and Vascular endothelial growth factor. Each sample was assayed in duplicate, requiring a total of 0.05 mL of serum to simultaneously measure these biomarkers. Standard curves and internal controls were run to ensure accuracy and reproducibility of the multiplex immunosorbent assays.
IgE Analysis by ELISA
Serum IgE was measured in 0.01 mL of serum using a commercial ELISA kit (Bethyl Laboratories, Montgomery, TX) according to the manufacturer’s directions. Positive and negative controls as well as specific, quantitative standards were used in all assays to ensure data quality.
Polycyclic Aromatic Hydrocarbon Analysis
The concentrations of 16 PAHs [nathphalene, acenaphthylene, acenapthene, fluorene, phenanthrene, anthracene, fluoranthene, pyrene, benzo(a)anthracene, chrysene, benzo(b)fluoranthene, benzo(k)fluoranthene, benzo(a)pyrene, dibenzo(a,h)anthracene, benzo(ghi)pyrlene, and indeno(123cd)pyrene] were quantified in the serum samples. Samples of 0.10 mL of serum were subjected to liquid–liquid extraction using the approach of Sirimanne et al21 that employs cloud-point extraction to preconcentrate, extract, and cleanup the PAHs from human serum. The resulting samples were then analyzed by gas chromatography/mass spectrometry (ThermoScientific TRACE GC/DSQ MS).
The separations were performed using an Rxi-XLB-fused silica column with a low polarity proprietary phase. The column dimension was 30 mm ×0.25 mm ID ×0.25 μm df. It exhibited extremely low bleed, ideal for PAHs analysis. The inlet temperature was 220°C. Samples are injected with splitless injection. High-purity He was used as the carrier gas and kept constant at 1.2 mL/min. The GC oven initial temperature was 60°C and held for 1 minute, then the temperature was increased to 210°C at 12°C/min, then to 320°C at 8°C/min, and the final temperature was held for 10 minutes. The specific conditions for the MS were as follows: EI with Secondary Ions Mass-positive mode. The filament emission current was set at 70 μA. MS transfer line temperature was set at 300°C and ion source temperature at 200°C. Five-point calibration PAH standard mixtures were run to determine the instrument response factors. The resulting chromatograms and mass spectra were inspected to ensure proper peak integration and the peak areas were the converted to concentrations using the response factors and the known surrogate recoveries. The PAH concentration results were then reported as ng of each detected PAH per mL of serum.
Statistical Analysis
Data were tested for statistical significance using unpaired Student t-test. Correlation analysis between relative miRNA levels, cytokines, IgE, and PAHs was determined by Spearman correlation. Benjamin–Hochberg method was used wherever indicated to correct P values for multiple hypothesis testing. To test the association of miRNA/cytokines with PAH levels, a linear regression analysis was performed using cotinine as a covariate. The R package miRNApath22 was used to perform the pathway analysis. Pathway information was obtained from KEGG23,24 and Biocarta25 databases. The heatmaps were created utilizing gplots.
RESULTS
DoDSR Serum Samples Contain High Levels of Specific miRNAs
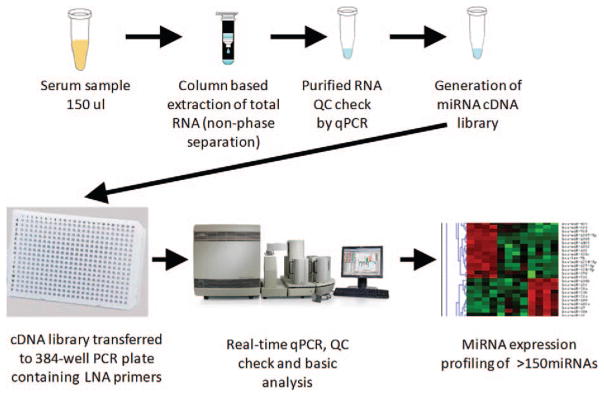
On the basis of our pilot studies of freshly isolated and stored serum samples, 0.15 mL of serum was determined to be adequate to detect individual miRNA species by quantitative real-time PCR (qPCR). To test if DoDSR serum samples were useful for detection of miRNA, we used 0.15 mL of serum from 30 different archived serum samples for RNA extraction (Fig. 1). Extraction was performed by column-based purification without phenol phase separation. After sample elution, RNA was used in single miRNA qPCR reactions. All samples were found to contain miRNA and were used for profiling of 179 miRNAs by qPCR array as described in the Materials and Methods.
FIGURE 1.
Isolation and detection of miRNAs from serum. Schematic outline of serum miRNA isolation, quality control, and profiling analysis. Briefly, 150 μL of serum from DoDSR samples was used to isolate miRNA using affinity column based chemistry. After isolation, a small portion of sample was used in single miRNA RT-qPCR assays to demonstrate successful miRNA analysis. Following successful single qPCR reactions, samples were profiled using qPCR assays for over 150 miRNAs in duplicate in a 384-well plate. Data were then processed and normalized to determine relative expression levels of each miRNA in the serum samples.
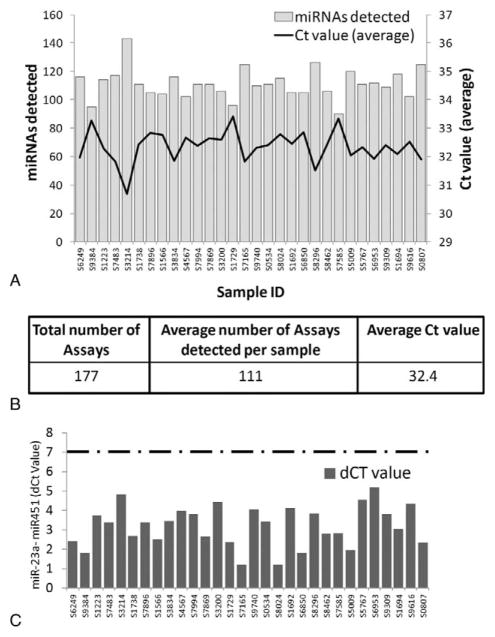
MiRNA profiling revealed that on average, 111 miRNAs out of the 179 assayed were detected per sample, with all 30 samples having 90 or more miRNAs detected (Fig. 2A and B). The average PCR cycle in which miRNA amplification reached the fluorescent threshold (Ct) value was 32.4 with a range of 30.8 to 33.4. Furthermore, 57 miRNAs were common among all 30 samples indicating robust and consistent miRNA detection in DoDSR serum samples.
FIGURE 2.
Identification and quality assessment of miRNAs present in serum. (A), 30 archival samples are listed on the x-axis with the number of miRNAs detected in each sample on the left side y-axis. The average PCR threshold cycle (Ct) for detected miRNA per sample is listed in the right side y axis. (B) Summary of miRNA profiling data. One hundred seventy-seven miRNAs were assayed with an average of 111 miRNAs detected per sample. (C) Assessment of red blood cell lysis (hemolysis) by detection of two different miRNAs. MiR-451 is present in red blood cells, while miR-23a is not. The difference in Ct (dCt) value between miR-23a and miR-451 can be used to assess hemolysis. Samples with a dCt value 7.0 or greater would be flagged for potential hemolysis (any sample with dCt above dotted line). All DoDSR samples had a dCt value of 5.5 or lower, demonstrating that hemolysis is not a concern.
Serum Samples Are Not Affected by Red Blood Cell Lysis (Hemolysis)
One potential drawback and source of variation in serum sample miRNA profiling is contamination from cellular-derived miRNA, especially from red blood cell lysis (hemolysis) during serum or plasma collection.26 To assess the possibility of hemolysis in DoDSR serum samples, the levels of two specific miRNAs were used. MiRNA-451 is present at high levels in red blood cells, while miR-23a is primarily in serum and plasma and not affected by cell lysis. Thus, the expression ratio between miR-451 and miR-23a can be used to assess potential hemolysis of serum samples.26 As in the study by Blondal et al,26 we used the difference in PCR threshold cycle value (dCt) between miR-451 and miR-23a to test for red blood cell miRNA contamination. A dCT value of 7.0 or higher suggests hemolysis and thus contaminating cellular miRNA in the serum samples, while samples with lower dCT values are not affected by hemolysis.26 Here, all samples had dCT values of 5.5 or below and thus red blood cell lysis does not appear to be an issue (Fig. 2C).
miRNA Expression Patterns Are Different in Smokers Versus Nonsmokers
As we determined that the archival DoDSR serum samples contained adequate levels of miRNA and were free of contaminating red blood cell miRNA, we looked for novel associations between specific miRNA and other detectable analytes. Cotinine, a breakdown product of nicotine, is readily detected in serum from tobacco users. Here, we measured cotinine levels in the DoDSR samples by ELISA. Serum cotinine values less than 1.0 ng/mL were defined as nontobacco users and values greater than 10.0 ng/mL were classified as tobacco users. Serum cotinine values between 1 and 10 ng/mL meet the criteria for exposure to second-hand tobacco smoke.20 One sample was within this range (1.6 ng/mL) and was grouped with the nonsmokers for subsequent analyses. Eleven of the 30 archival samples exceeded the threshold for tobacco use, although we cannot distinguish between smoking (cigarette, cigar or pipe), smokeless tobacco, or electronic cigarettes.
Next, we assessed whether the miRNA levels were different in cotinine positive and cotinine negative samples. We identified several miRNAs that had differential expression between the two groups. As this was a pilot study with limited sample size, we found miRNAs with significant differential expression with a cut off of P value less than 0.1. The 19 different miRNAs that met this criterion are listed in Table 1. Some of the miRNAs such as miR-133a and miR-133b are from the same miRNA family and contain similar seed sequences. However, many of these miRNAs are from different miRNA families and contain distinct seed sequences. We wondered whether any of these miRNAs are predicted to target similar biological pathways. To assess similarities, we grouped predicted miRNA target genes by cognate KEGG pathways using miRNApath (Fig. 3).22,27 Interestingly, multiple cellular pathways are regulated by multiple miRNAs that are impacted by smoke exposure (Fig. 3). For example, multiple miRNAs target pathways related to cell proliferation and differentiation, including the cell cycle, Transforming Growth Factor Beta, mammalian target of rapamycin, ErbB, and focal adhesion pathways. Immune cell function is also multiply targeted, including toll-like receptors, B- and T-cell receptors, Fc receptors, Jak-Signal transducer and activator of transcription signaling, and chemokine signaling. These data suggest that strong relationships between tobacco use and regulation of key cellular and immunological pathways are mediated by miRNAs.
TABLE 1.
Top 5 miRNAs That Are Differentially Expressed in Cotinine-Positive vs Cotinine-Negative Serum Samples
| miRNA | Fold Change | P |
|---|---|---|
| miR-95 | 9.7 | 0.02 |
| miR-2110 | 3.9 | 0.09 |
| miR-133a | 3.1 | 0.08 |
| miR-30e-3p | 2.9 | 0.05 |
| miR-133b | 2.2 | 0.06 |
| miR-146b-5p | 1.9 | 0.09 |
| miR-152 | 1.8 | 0.06 |
| miR-139-5p | 1.8 | 0.02 |
| miR-22-5p | 1.5 | 0.30 |
| miR-660-5p | 1.5 | 0.08 |
| miR-10b-5p | 1.5 | 0.06 |
| miR-126-3p | 1.4 | 0.03 |
| miR-23b-3p | 1.3 | 0.01 |
| miR-26a-5p | 1.3 | 0.06 |
| miR-20a-5p | 0.8 | 0.07 |
| miR-19b-3p | 0.7 | 0.05 |
| miR-19a-3p | 0.7 | 0.09 |
| miR-17-5p | 0.3 | 0.05 |
| miR-551b-3p | 0.1 | 0.03 |
FIGURE 3.
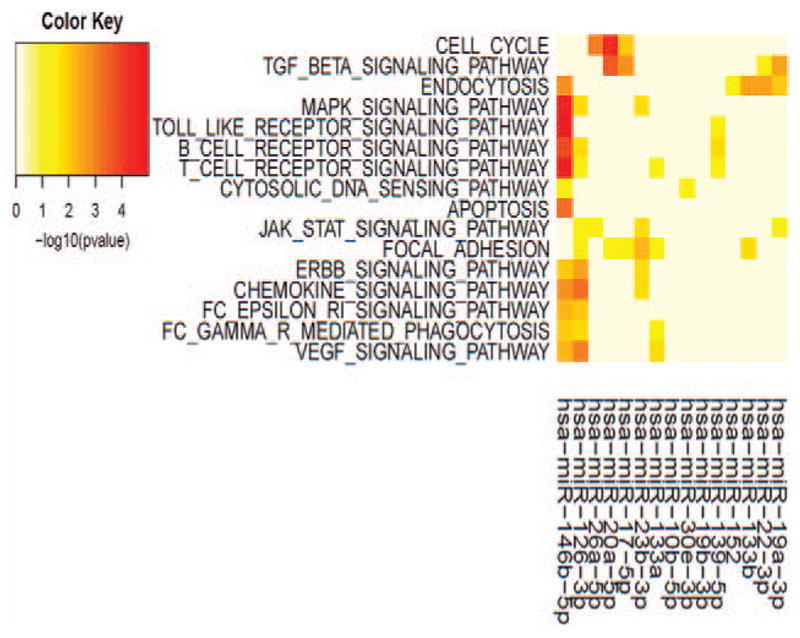
Pathway analysis of miRNAs differentially expressed in cotinine-positive samples. Pathway analysis (see Materials and Methods) of differentially expressed miR-NAs in cotinine-positive samples based on ranking according to P values was performed in R (R package miRNApath1 for pathway analysis). Serum cotinine levels were used to group the samples into tobacco users (n =11) compared with non-tobacco users (n =19) as described in the Materials and Methods section. The color map P values indicate the strength of the association between the miRNA and the relevant pathway. Pathways targeted by fewer than two miRNAs and miRNAs that targeted fewer than two pathways were omitted for clarity.
Cytokines, Chemokines, and IgE Are Detectable in DoDSR Serum Samples
Although novel associations were identified between cotinine status and miRNA levels, we continued to test the 30 DoDSR serum samples for other biomolecules including 22 different cytokines/chemokines and IgE. Cytokines and chemokines were detected using specific, quantitative antibody-based multiplex assay panels, while IgE was measured by specific ELISA. The results of these assays are listed in Table 2. To ensure that stored DoDSR samples were not affected by sample degradation and are of sufficient quality to measure these analytes, we used freshly isolated serum from three healthy control donors as reference (mean values listed in Table 2, far right column). We detected serum cytokines/chemokines in similar ranges between DoDSR samples and the control samples demonstrating that the DoDSR serum samples are of sufficient quality to reliably measure these putative biomarkers.
TABLE 2.
Cytokine, Chemokine, and IgE Analysis of 30 DoDSR Serum Samples
| Analyte | Mean ± SD (pg/mL) | Median | Lower Quartile | Upper Quartile | Ref Sample* Mean (pg/mL) |
|---|---|---|---|---|---|
| Eotaxin | 115.6 ± 46.1 | 83.4 | 83.4 | 147.1 | 115.5 |
| G-CSF | 41.9 ± 30.3 | 21.8 | 21.8 | 49.9 | 43.5 |
| Flt3-L | 12.4 ± 20.3 | 0.3 | 0.3 | 15.6 | 55.7 |
| GM-CSF | 7.7 ± 10.0 | 1.0 | 1.0 | 11.9 | 15.1 |
| Fractalkine | 73.4 ± 71.0 | 28.7 | 28.7 | 94.5 | 135.2 |
| IFNγ | 13.4 ± 19.3 | 2.1 | 2.1 | 12.3 | 28.8 |
| GRO | 896.3 ± 810.9 | 517.8 | 517.8 | 833.2 | 1170.1 |
| IL-10 | 10.4 ± 15.1 | 0.3 | 0.3 | 15.0 | 5.2 |
| IL-12p40 | 33.8 ± 53.2 | 0.3 | 0.3 | 50.2 | 18.1 |
| IL-12p70 | 2.3 ± 2.9 | 0.3 | 0.3 | 3.0 | 2.5 |
| IL-13 | 8.9 ± 16.3 | 0.3 | 0.3 | 13.7 | 13.1 |
| sCD40L | 1061.9 ± 856.0 | 330.9 | 330.9 | 1393.2 | 3934.8 |
| IL-17A | 10.7 ± 29.0 | 1.9 | 1.9 | 7.4 | 11.9 |
| IL-1ra | 35.7 ± 51.0 | 3.8 | 3.8 | 50.6 | 41.6 |
| IL-1α | 15.5 ± 24.9 | 0.3 | 0.3 | 24.0 | 22.2 |
| IL-1β | 4.4 ± 5.7 | 0.3 | 0.3 | 8.0 | 2.8 |
| IL-5 | 1.6 ± 2.9 | 0.3 | 0.3 | 1.0 | 1.0 |
| IL-6 | 5.1 ± 7.3 | 0.3 | 0.3 | 7.2 | 7.6 |
| IL-8 | 79.0 ± 70.0 | 37.7 | 37.7 | 89.9 | 14.0 |
| MCP-1 | 646.3 ± 258.1 | 463.9 | 463.9 | 756.9 | 504.6 |
| TNFα | 7.2 ± 3.4 | 5.0 | 5.0 | 9.9 | 5.8 |
| VEGF | 197.0 ± 218.3 | 68.0 | 68.0 | 236.5 | 239.9 |
| IgE | 482.1 ± 893.6 | 196.8 | 49.9 | 518.1 | N/A |
Cytokine and chemokine levels were determined by a multiplex immunosorbent assay. IgE levels were determined by ELISA. The mean amount of each serum component detected ±the standard deviation across all samples is shown in column 2. Then, columns 3 to 6 show median value, lower, and upper quartiles.
Column 7 is the mean values obtained from internal, freshly isolated serum samples (n =3).
G-CSF, Granulocyte-colony stimulating factor; GM-CSF, Granulocyte-macrophage colony-stimulating factor; GRO, Growth related oncogene; IFN, Interferon; MCP, monocyte chemoattractant protein; TNF, Tumor necrosis factor; VEGF, Vascular endothelial growth factor.
MiRNAs, Cytokines, and IgE Levels Correlate in Archival DoDSR Samples
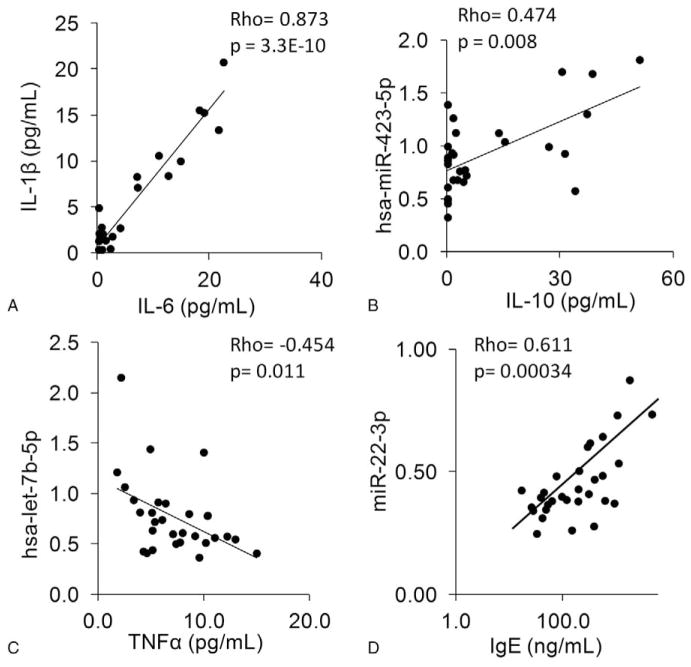
To make further use of the accumulated data from this initial pilot study, correlations were tested to determine whether specific cytokines and/or miRNA levels are associated. The first correlation tested was between the inflammatory cytokines IL-1β and IL-6, which are often detectable at high levels in many disease states.28 The correlation between IL-1β and IL-6 was highly significant with a Spearman’s rank correlation of 0.87, P <0.001 (Fig. 4A).
FIGURE 4.
Individual Spearman rank correlations between specific serum components, including cytokines, miRNAs, and IgE. Different serum components were analyzed for correlation using Spearman rank method. Each correlation used all 30 DoDSR samples and these analyses were done independently of cotinine status. (A) IL-1β and IL-6 levels are highly correlated in DoDSR serum samples. (B) The serum expression of miR-423-5p correlates with IL-10 levels. (C) There is a negative correlation between let-7b-5p and Tumor necrosis factorα levels. (D) Serum IgE levels correlate with expression of miR-22-3p.
We also looked for potential novel associations with specific miRNA and cytokines within the data set (n =30 samples). In serum containing high levels of IL-10, there was a correlation with miR-423-5p levels, rho =0.47, P =0.008 (Fig. 4B). The serum level of Tumor necrosis factor α was found to be negatively associated with the miRNA, let7b-5p, rho =−0.45, P =0.012 (Fig. 4C). In addition, serum IgE levels were significantly associated with levels of miR-22-3p, rho =0.61, P =0.00034 (Fig. 4D). As several associations between miRNA and cytokines were identified, we graphically illustrated multiple correlations simultaneously (Fig. 5). The top 25 miRNAs that were positively associated with three or more different cytokines, IgE, or cotinine and their correlation P values are represented in this heat map. This mapping reveals many novel associations and clusters of associations within the data. For example, the lower left portion of the heat map identifies significant associations with the three cytokines: Interferonγ, Vascular endothelial growth factor, IL-17A, and the six miRNAs: miR-605, miR-375, miR-2110, mir-365-3p, miR-424-5p, and miR-328. Several other clusters of cytokines and miRNAs are also revealed through heat mapping. Taken together, these results show clear relationships between cytokines and miRNA in DoDSR samples.
FIGURE 5.
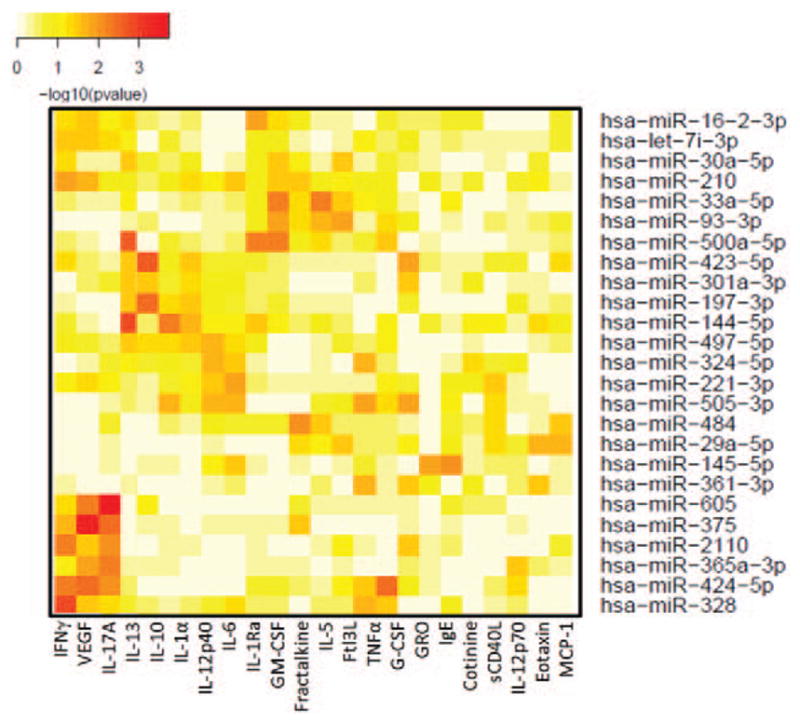
Cluster analysis of correlations between miRNA and cytokines. The miRNAs (y-axis) with significant P values for >3 cytokines (x-axis) are plotted. The complete set of correlations is provided as supplemental table 1 (http://links.lww.com/JOM/A269). The color map from yellow to red indicate lower to high –log10 (P values).
Serum Samples Can Be Used to Detect Environmental Exposure to PAHs
PAHs are an important set of molecules to measure environmental exposures. PAHs are a class of multiple compounds present in smoke from the combustion of tobacco, diesel exhaust, and JP 8 combustion as accelerant in open burn pit operations.17,29,30 A portion of the archival DoDSR serum samples was used to detect 16 different PAHs using liquid chromatography-mass spectroscopy (LC-MS). The results listed in Table 3 show that multiple different PAHs were detected in the DoDSR samples (note that in Table 3, the shaded sample IDs correspond to cotinine positive samples). Three PAHs, naphthalene, anthracene, and Benzo[a]pyrene, were detected in all 30 samples. Several other PAHs were detected in at least two of the 30 samples showing that PAH accumulation can be measured in DoDSR serum.
TABLE 3.
Polyaromatic Hydrocarbon (PAH) Analysis Using Liquid Chromatography/Mass Spectroscopy From 30 Archival DoDSR Samples
| Sample ID | Total PAH | Naphthalene | Fluorene | Phenanthrene | Anthracene | Fluor-anthene | Pyrene | Benz (a) Anthracene | Chrysene | Benzo (a) Pyrene | Benzo (ghi) Perylene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S053405966 | 3.5674 | 0.1412 | N/F | N/F | 0.1487 | N/F | N/F | N/F | N/F | 3.2776 | N/F |
| S080712445 | 0.9787 | 0.1565 | N/F | N/F | 0.1309 | N/F | N/F | N/F | N/F | 0.6913 | N/F |
| S122353014 | 3.2746 | 0.1925 | 0.0655 | 0.0567 | 0.2847 | N/F | N/F | N/F | N/F | 2.6752 | N/F |
| S156644116 | 0.3928 | 0.1367 | N/F | N/F | 0.1304 | N/F | N/F | N/F | N/F | 0.1256 | N/F |
| S169239333 | 3.9103 | 0.2103 | N/F | N/F | 0.0912 | 0.0153 | N/F | N/F | N/F | 3.5934 | N/F |
| S169424056 | 3.6422 | 0.1833 | N/F | N/F | 0.2327 | 0.0119 | N/F | N/F | N/F | 3.2143 | N/F |
| S172938259 | 2.7362 | 0.1644 | N/F | N/F | 0.054 | N/F | N/F | N/F | N/F | 2.4849 | N/F |
| S173820505 | 2.6796 | 0.1781 | N/F | N/F | 0.2454 | N/F | N/F | N/F | N/F | 2.2561 | N/F |
| S320055488 | 2.1464 | 0.1609 | N/F | N/F | 0.0761 | N/F | N/F | N/F | N/F | 1.9094 | N/F |
| S321462178 | 1.1959 | 0.1854 | N/F | N/F | 0.284 | N/F | N/F | N/F | N/F | 0.7265 | N/F |
| S383498374 | 2.2817 | 0.1627 | N/F | N/F | 0.1111 | N/F | N/F | N/F | N/F | 1.9368 | 0.0711 |
| S456732067 | 1.1899 | 0.1352 | N/F | N/F | 0.1708 | N/F | N/F | N/F | N/F | 0.8839 | N/F |
| S500933196 | 158.358 | 0.1133 | 0.0224 | 0.0145 | 0.1759 | 0.0562 | 0.0085 | N/F | N/F | 37.2643 | 0.2594 |
| S576739980 | 1.9407 | 0.1914 | N/F | N/F | 0.2136 | N/F | N/F | N/F | N/F | 1.5357 | N/F |
| S624913446 | 2.3831 | 0.1366 | 0.0075 | 0.0055 | 0.1455 | 0.0075 | 0.0115 | N/F | N/F | 2.069 | N/F |
| S685083601 | 1.248 | 0.1667 | N/F | N/F | 0.1175 | N/F | N/F | 0.1333 | 0.1455 | 0.685 | N/F |
| S695344016 | 2.6218 | 0.1625 | 0.0326 | 0.0249 | 0.2434 | 0.0083 | N/F | N/F | N/F | 2.1084 | 0.042 |
| S716543256 | 1.8196 | 0.1803 | N/F | N/F | 0.1134 | N/F | N/F | N/F | N/F | 1.526 | N/F |
| S748323523 | 3.9861 | 0.1193 | N/F | N/F | 0.1526 | 0.018 | 0.0293 | 0.0379 | 0.0413 | 3.5876 | N/F |
| S758598913 | 7.3575 | 0.1541 | 0.0073 | 0.0071 | 0.1292 | 0.0273 | 0.0143 | N/F | N/F | 6.0341 | N/F |
| S786911942 | 1.043 | 0.1203 | N/F | N/F | 0.1925 | N/F | N/F | N/F | N/F | 0.7303 | N/F |
| S789635792 | 1.1289 | 0.2359 | N/F | N/F | 0.1576 | N/F | N/F | N/F | N/F | 0.7355 | N/F |
| S799412327 | 1.7397 | 0.1623 | N/F | N/F | 0.1571 | N/F | N/F | N/F | N/F | 1.4203 | N/F |
| S802483786 | 3.8096 | 0.1241 | N/F | N/F | 0.2445 | 0.0519 | 0.0704 | 0.0786 | 0.0537 | 3.0602 | 0.1262 |
| S829657159 | 5.899 | 0.2207 | N/F | N/F | 0.1694 | N/F | N/F | N/F | N/F | 5.5089 | N/F |
| S846210194 | 2.7672 | 0.1062 | 0.0203 | 0.0115 | 0.1987 | 0.0055 | N/F | 0.0319 | 0.0348 | 2.3583 | N/F |
| S930920676 | 2.8547 | 0.1354 | 0.0261 | 0.0161 | 0.2182 | 0.0111 | N/F | N/F | N/F | 2.4478 | N/F |
| S938451005 | 2.2668 | 0.1237 | N/F | N/F | 0.2258 | N/F | N/F | N/F | N/F | 1.9174 | N/F |
| S961642526 | 1.9535 | 0.1748 | N/F | N/F | 0.0653 | N/F | N/F | N/F | N/F | 1.7133 | N/F |
| S974027464 | 3.88 | 0.1623 | 0.0883 | 0.074 | 0.2198 | 0.0115 | 0.0112 | N/F | N/F | 3.3129 | N/F |
Total and specific PAHs were detected in DoDSR samples (highlighted samples are cotinine positive). Naphthalene, anthracene, and Benzo[a]pyrene were detected in all samples, whereas acenaphthylene, Benzo(b)fluoranthene, Benzo(k)fluoranthene, Indeno(123cd)ppyrene were assayed but not detected.
Polycyclic Aromatic Hydrocarbon (PAH) Levels Correlate With miRNAs, IL-8, and IgE
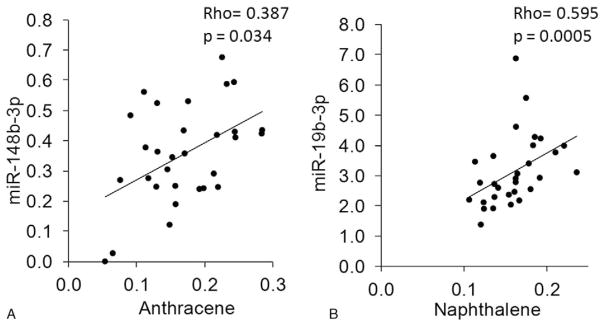
In order to find surrogate biomarkers of environmental exposure, it is important to identify associations with environmental compounds such as PAHs. We examined the associations between the three PAHs present in all 30 samples and the miRNAs that were detected. Excitingly, two novel association between PAH levels were identified (Fig. 6). Anthracene levels significantly correlated with miR-148b-3p levels, rho =0.387, P =0.034 (Fig. 6A) and napthalene levels strongly correlated with miR-19b-3p levels, rho =0.595, P =0.0005 (Fig. 6B). These results represent novel relationships between PAH levels and miRNA in human serum samples.
FIGURE 6.
Spearman rank correlation between miR-142-5p and anthracene. Significant correlations between PAH levels and specific miRNAs were identified. (A) Serum anthracene levels and miR-148b-3p expression were analyzed for correlation by Spearman rank method. (B) Serum naphthalene levels and miR-19b-3p expression were analyzed for correlation by Spearman rank method.
The presence of cotinine in serum samples indicates some tobacco use and thus may confound the effects of other environmental exposures. To avoid this problem, we controlled for cotinine status in the analysis. After cotinine levels were accounted for, other biomarkers were identified that showed an association with total serum PAH levels. The serum markers that correlated with PAH levels after using cotinine status as a covariate were four miRNAs (miR-10a-5p, miR-140-3p, miR-186-5p, miR-193b-3p), the cytokine IL-8, and the immunoglobulin, IgE, q <0.25 (Table 4).
TABLE 4.
Covariate Analysis of miRNAs, Cytokines, and IgE That Correlate With Selected PAHs
| miR-10a-5p |
| miR-140-3p |
| miR-186-5p |
| miR-193b-3p |
| IL-8 |
| IgE |
Total PAH levels were used to test for correlations with specific miRNAs, cytokines, and IgE after utilizing cotinine status as a covariate. The analysis revealed significant correlations with total PAH levels, miRNAs, IL-8, and IgE levels.
DISCUSSION
Deployed personnel have the potential to be exposed to numerous toxicants, including diesel and jet fuel exhaust, burn pit smoke, and other occupational hazards depending on deployment site.2 Environmental and occupational exposures may predispose personnel to health problems such as COPD, asthma, The top 19 differentially expressed miRNAs that were found to have P values of 0.1 or less are expressed as fold change in cotinine-positive vs negative serum (ie, in people with tobacco use or not). P values were calculated using Student t test. bronchiolitis, other cardiopulmonary illnesses, and even certain types of cancers later in life.31–33 In this pilot study, we used 30 serum samples from the DoDSR that lack any identifier information to show the feasibility of using small amounts of serum to discover the presence of biomarkers and their associations with other constituents including cytokines, miRNAs, and PAHs in the serum. We show that the DoDSR serum samples were not affected by hemolysis and that we could detect a large number of putative biomarkers, including miRNAs, cotinine status, cytokines, chemokines, and PAHs within the same sample. We identified novel associations between miRNA, cytokines, and PAHs that may be useful to predict various environmental exposures. Taken together, these results highlight the quality and usefulness of DoDSR serum for future, larger cohort studies on novel biomarker discovery.
The ability to reliably detect miRNA in stored serum from the DoDSR represents a major advance in that it allows for use of miRNA as novel biomarkers of health and disease status. We found that miRNAs are closely associated with PAHs that are common environmental contaminants during deployments. This finding should increase the potential utility of the repository for novel miRNA biomarker discovery.2,34 Indeed, recent studies highlight the use of circulating miRNAs as indicators of health status in acute heart failure35, lung cancer,36 and COPD.37 While the use of circulating miRNA as novel biomarkers holds great potential, there are important technical challenges that need to be addressed that include having sufficient sample volume, suitable qualities of serum/plasma, and acquiring a reproducible assay. Several groups have isolated miRNAs from serum/plasma samples using 0.1 to 1.0 mL of sample using either column-based and/or phase separation isolation methods.4,5,38 A common limitation in accurately detecting miRNA from biofluids is phenol contamination after organic phase extraction. Here, we used 0.15 mL of serum for miRNA isolation without phase separation to increase PCR reproducibility and robustness. Using this method, we detected over 100 distinct miRNAs out of the over 150 miRNA qPCR assays performed per sample. Although other methods for miRNA profiling exist such as hybridization arrays and small RNAseq, PCR-based methods are still the most widely used, accepted, and reproducible for miRNA detection as shown in the miRQC study.39 Thus, we believe that this method holds great promise for future DoDSR miRNA biomarker studies.
Hemolysis in serum samples represents a major limitation in biomarker discovery, as red blood cell miRNA, metabolites, and other released proteins can drastically alter serum/plasma measurements.40–42 By comparing the ratio of miRNA species known to be absent in RBCs or present in high amounts, we were able to determine that the 30 pilot serum samples from the DoDSR were not affected by hemolysis. This is a key finding for subsequent biomarker studies, as it indicates that not only miRNAs but also cytokines, proteins, and other metabolites are likely unaffected by hemolysis at the time of collection. It will be important to gauge potential hemolysis in future studies using serum samples from the DoDSR as well as other repositories.
Tobacco use (primarily smoking, but also smokeless or electronic cigarettes) is an important environmental exposure that has known health impacts, especially on the respiratory and cardiovascular systems. Furthermore, tobacco smoke produces various PAHs such as benzo[a]pyrene that can confound environmental exposure from fuel combustion or burn pit smoke.43,44 Any attempt to use DoDSR samples to identify health effects of service-related exposures may be limited if reliable information about smoking status is not obtained. Serum cotinine levels have been used to distinguish tobacco users from nonusers independently of self-reporting or demographic information.45,46 Therefore, we assessed serum cotinine in our pilot samples by ELISA. We identified 11 samples that met the criteria for a smoker or regular tobacco user, one sample that met the criteria for exposure to secondhand tobacco smoke, while 18 samples had cotinine in the nonsmoker/nonuser range. Our cotinine detection was in good agreement with cotinine and other nicotine metabolites determined by mass spectrometry of the same samples (Walker et al, in this supplement). This demonstrates that serum cotinine measurements can be used to retrospectively identify tobacco users as part of future exposure studies.
After being able to discriminate tobacco users from nonusers, we were next interested in determining whether we could detect miRNAs specifically affected by tobacco use. We detected several miRNAs that were differentially expressed in tobacco users compared with nonusers. MiR-23b-3p was increased in cotinine-positive serum and is considered an oncogenic miRNA in renal cancer47 and has been suggested as a lung cancer biomarker.48 We also found increased levels of miR-139-5p and miR-126-3p in cotinine-positive serum samples. MiR-139 levels are increased in malignant endothelial cells49 and miR-126 expression has been implicated in angiogenesis50. Although there are currently no signaling pathways associated with miR-95 or miR-551b-3p in the current KEGG database, these miRNAs are also reported to be tumor suppressors.51 With the exception of miR-126-3p, none of these miRNAs has previously been reported as associated with tobacco use. A further 14 miRNAs were differentially expressed with P values less than 0.1. Pathway analysis found that many of these miRNAs target the same or related molecular pathways, and that individual pathways are targeted by multiple miRNAs that are differentially expressed in tobacco users. The most interesting of these may be pathways related to cell cycle, proliferation, and differentiation, and pathways related to immune cell function. As tobacco smoke is independently known to cause cancer and to alter B and T cell function and promote proinflammatory signaling and cytokine production, the fact that we can detect these same associations via miRNA analysis in this small sample size is further demonstration of the utility of these methods in analyzing similar samples for exposure-related biological outcomes. MiRNAs appear to be robust indicators of tobacco exposure, and the pathway analyses can be used to generate novel hypotheses for further testing.
We performed multiplex immunoassays on cytokine/chemokine levels and importantly, found that the levels of these factors are in similar ranges to that of freshly isolated serum samples. Although there was a wide range of cytokinevalues in these 30 DoDSR samples, we cannot further delineate these cytokines with age or health status, as demographic information is unavailable. However, a literature search revealed that the levels of IL-6, IL-8, IL-10, Tumor necrosis factorα, IL-12, monocyte chemoattractant protein-1, eotaxin, Vascular endothelial growth factor, and other biomolecules we detected in DoDSR serum samples are similar to previously reported values.52–55 Thus, using a limited serum volume, we can accurately detect multiple cytokines in DoDSR serum samples for biomarker discovery and for correlations with miRNA and PAHs.
Serum IgE is a sentinel signal of allergy and asthma. It has been reported that exposure to environmental toxicants including diesel exhaust and other hydrocarbon pollutants promote increased allergic sensitization in animal models and humans.56,57 Assessment of serum IgE along with PAH data may help us determine whether service-related environmental exposures contribute to allergic sensitization. Here, we were able to detect circulating IgE levels that correspond to reported serum levels of IgE.58 Without knowing classifier and allergic information about the personnel, we found a strong correlation between miR-22-3p and IgE. To date, few publications have described a relationship between miRNA and IgE. As miR-22-3p has ~660 predicted target sequences to human mRNAs, it is possible that miR-22-3p regulates genes involved in allergen response. One pathway that miR-22-3p is predicted to regulate is the Transforming Growth Factor Beta signaling pathway (P <0.05). Interestingly, others have shown that Transforming Growth Factor Beta signaling is involved in suppressing IgE production.59,60
We were able to detect several species of PAHs directly in DoDSR samples. All 30 samples contained B[a]P, anthracene, and naphthalene, three PAHs that can be derived from cigarette smoke, grilling meats, diesel exhaust, and burn pits. We identified a correlation between anthraces and naphthalene with specific miRNAs. To date, very few, if any miRNAs have been associated with PAH levels in human serum. Anthracene and napthalene exposure are most probably a result of cigarette smoking or from grilled meat consumption. Interestingly, miR-19b-3p, which was associated with naphthalene levels in this study, is part of the miR-17~92 polycistronic miRNA cluster. The miR-17~92 miRNA cluster is composed of six miRNAs that are linked to cell proliferation, inhibition of apoptosis, cancer, and metastasis.61–63 Recent studies linked an increase in the expression of the miR-17~92 cluster with lung and immune cell cancers.64,65 Future study should assess whether naphthalene itself can drive expression of miR-19b and the miR-17~92 miRNA cluster or whether this regulation is through an indirect pathway involved in cell proliferation or other pathways as described above.
Tobacco smoke is a known insult that can lead to PAH exposure and accumulation. To account for this, we performed another analysis using cotinine, a metabolite of nicotine from tobacco use, as a covariate for biomarker status. The serum markers that correlated with PAH levels after controlling for tobacco use were four miRNAs (miR-10a-5p, miR-140-3p, miR-186-5p, miR-193b-3p), the cytokine IL-8, and the immunoglobulin, IgE. At present, little is known about the role of these four miRNAs in relation to PAH exposure and IL-8 and IgE levels. Interestingly, others have shown that diesel exhaust, another source of PAH exposure, increases IL-8 levels in human cells.66 Likewise, IgE levels were associated with urine PAH metabolite levels in a cohort of pediatric allergy and asthma samples.67 Interestingly, miR-140-3p was shown to regulate CD38 expression in human airway smooth muscle cells.68 CD38 expression is also induced by IL-869 and may contribute to airway hyperresponsiveness and allergy.70 Thus, although more studies are need to understand the relationship between these biomolecules and nontobacco-related PAH exposures, associations exist that could underlie our novel findings.
The DoDSR contains over 55 million serum samples, approximately 45 million of which can be linked to individual service records and demographic information. This represents a powerful but currently underutilized resource for monitoring the health status of service personnel and especially changes in health status with time in service or deployment to specific theaters of operation. Our results demonstrate that DoDSR serum samples are of high quality and that a large amount of biological data can be extracted from a small sample volume. We could detect both indicators of environmental exposure (cotinine, PAHs) and bio-markers linked to potential biological outcomes of exposure (miRNA, cytokines, IgE). Even limited by a relatively small sample size, we were able to identify novel associations between tobacco and PAH exposure and biological outcomes. Our overall results show that samples from the DoDSR are fit for uses such as pre- and postdeployment health surveillance of military personnel.
Acknowledgments
This work was supported by The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. grant number HT9404-13-1-0030, and the National Institute of Environmental Health Sciences Grant # P30-ES01247.
Footnotes
There are no conflicts of interest.
Supplemental digital contents are available for this article. Direct URL citation appears in the printed text and is provided in the HTML and PDF versions of this article on the journal’s Web site (www.joem.org).
References
- 1.Smith B, Wong CA, Boyko EJ, Phillips CJ, Gackstetter GD, Ryan MA, et al. The effects of exposure to documented open-air burn pits on respiratory health among deployers of the Millennium Cohort Study. J Occup Environ Med. 2012;54:708–716. doi: 10.1097/JOM.0b013e31825107f9. [DOI] [PubMed] [Google Scholar]
- 2.Mancuso JD, Mallon TM, Gaydos JC. Maximizing the capabilities of the DoD serum repository to meet current and future needs, report of the needs panel. Mil Med. 2015;180(10 Suppl):13–24. doi: 10.7205/MILMED-D-14-00732. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grasedieck S, Scholer N, Bommer M, Niess JH, Tumani H, Rouhi A, et al. Impact of serum storage conditions on microRNA stability. Leukemia. 2012;26:2414–2416. doi: 10.1038/leu.2012.106. [DOI] [PubMed] [Google Scholar]
- 5.Mraz M, Malinova K, Mayer J, Pospisilova S. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 2009;390:1–4. doi: 10.1016/j.bbrc.2009.09.061. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebrahimi A, Sadroddiny E. MicroRNAs in lung diseases: recent findings and their pathophysiological implications. Pulm Pharmacol Ther. 2015;34:55–63. doi: 10.1016/j.pupt.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Cho Y, Song MK, Jeong SC, Lee K, Heo Y, Kim TS, et al. MicroRNA response of inhalation exposure to hexanal in lung tissues from Fischer 344 rats. Environ Toxicol. 2015 doi: 10.1002/tox.22192. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng Q, Huang S, Zhang X, Zhang W, Feng J, Wang T, et al. Plasma microRNA expression and micronuclei frequency in workers exposed to polycyclic aromatic hydrocarbons. Environ Health Perspect. 2014;122:719–725. doi: 10.1289/ehp.1307080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen SG, Lamy P, Rasmussen MH, Ostenfeld MS, Dyrskjot L, Orntoft TF, et al. Evaluation of two commercial global miRNA expression profiling platforms for detection of less abundant miRNAs. BMC Genomics. 2011;12:435. doi: 10.1186/1471-2164-12-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arjomandi M, Wong H, Donde A, Frelinger J, Dalton S, Ching W, et al. Exposure to medium and high ambient levels of ozone causes adverse systemic inflammatory and cardiac autonomic effects. Am J Physiol Heart Circ Physiol. 2015;308:H1499–H1509. doi: 10.1152/ajpheart.00849.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swiston JR, Davidson W, Attridge S, Li GT, Brauer M, van Eeden SF. Wood smoke exposure induces a pulmonary and systemic inflammatory response in firefighters. Eur Respir J. 2008;32:129–138. doi: 10.1183/09031936.00097707. [DOI] [PubMed] [Google Scholar]
- 14.Cormier Y, Israel-Assayag E. Adiposity affects human response to inhaled organic dust. Am J Ind Med. 2006;49:281–285. doi: 10.1002/ajim.20265. [DOI] [PubMed] [Google Scholar]
- 15.Quirce S. IgE antibodies in occupational asthma: are they causative or an associated phenomenon? Curr Opin Allergy Clin Immunol. 2014;14:100–105. doi: 10.1097/ACI.0000000000000041. [DOI] [PubMed] [Google Scholar]
- 16.Eckl-Dorna J, Niederberger V. What is the source of serum allergen-specific IgE? Curr Allergy Asthma Rep. 2013;13:281–287. doi: 10.1007/s11882-013-0348-x. [DOI] [PubMed] [Google Scholar]
- 17.Kamal A, Cincinelli A, Martellini T, Malik RN. A review of PAH exposure from the combustion of biomass fuel and their less surveyed effect on the blood parameters. Environ Sci Pollut Res Int. 2015;22:4076–4098. doi: 10.1007/s11356-014-3748-0. [DOI] [PubMed] [Google Scholar]
- 18.Klingbeil EC, Hew KM, Nygaard UC, Nadeau KC. Polycyclic aromatic hydrocarbons, tobacco smoke, and epigenetic remodeling in asthma. Immunol Res. 2014;58:369–373. doi: 10.1007/s12026-014-8508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 20.Max W, Sung HY, Shi Y. Who is exposed to secondhand smoke? Self-reported and serum cotinine measured exposure in the U.S., 1999–2006. Int J Environ Res Public Health. 2009;6:1633–1648. doi: 10.3390/ijerph6051633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirimanne SR, Barr JR, Patterson DG, Jr, Ma L. Quantification of polycyclic aromatic hydrocarbons and polychlorinated dibenzo-p-dioxins in human serum by combined micelle-mediated extraction (cloud-point extraction) and HPLC. Anal Chem. 1996;68:1556–1560. doi: 10.1021/ac951028+. [DOI] [PubMed] [Google Scholar]
- 22.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database issue):D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37(Database issue):D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59:S1–S6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Thakar J, Hartmann BM, Marjanovic N, Sealfon SC, Kleinstein SH. Comparative analysis of anti-viral transcriptomics reveals novel effects of influenza immune antagonism. BMC Immunol. 2015;16:46. doi: 10.1186/s12865-015-0107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panteli KE, Hatzimichael EC, Bouranta PK, Katsaraki A, Seferiadis K, Stebbing J. Serum interleukin (IL)-1, IL-2, sIL-2Ra, IL-6 and thrombopoietin levels in patients with chronic myeloproliferative diseases. Br J Haematol. 2005;130:709–715. doi: 10.1111/j.1365-2141.2005.05674.x. [DOI] [PubMed] [Google Scholar]
- 29.Kim KH, Jahan SA, Kabir E, Brown RJ. A review of airborne polycyclic aromatic hydrocarbons (PAHs) and their human health effects. Environ Int. 2013;60:71–80. doi: 10.1016/j.envint.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 30.Farmer PB, Singh R, Kaur B, Sram RJ, Binkova B, Kalina I, et al. Molecular epidemiology studies of carcinogenic environmental pollutants. Effects of polycyclic aromatic hydrocarbons (PAHs) in environmental pollution on exogenous and oxidative DNA damage. Mutat Res. 2003;544:397–402. doi: 10.1016/j.mrrev.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Abraham JH, DeBakey SF, Reid L, Zhou J, Baird CP. Does deployment to Iraq and Afghanistan affect respiratory health of US military personnel? J Occup Environ Med. 2012;54:740–745. doi: 10.1097/JOM.0b013e318252969a. [DOI] [PubMed] [Google Scholar]
- 32.Falvo MJ, Osinubi OY, Sotolongo AM, Helmer DA. Airborne hazards exposure and respiratory health of Iraq and Afghanistan veterans. Epidemiol Rev. 2015;37:116–130. doi: 10.1093/epirev/mxu009. [DOI] [PubMed] [Google Scholar]
- 33.Kerr KJ. Gulf War illness: an overview of events, most prevalent health outcomes, exposures, and clues as to pathogenesis. Rev Environ Health. 2015;30:273–286. doi: 10.1515/reveh-2015-0032. [DOI] [PubMed] [Google Scholar]
- 34.Perdue CL, Cost AA, Rubertone MV, Lindler LE, Ludwig SL. Description and utilization of the United States department of defense serum repository: a review of published studies, 1985–2012. PLoS One. 2015;10:e0114857. doi: 10.1371/journal.pone.0114857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ovchinnikova ES, Schmitter D, Vegter EL, Ter Maaten JM, Valente MA, Liu LC, et al. Signature of circulating microRNAs in patients with acute heart failure. Eur J Heart Fail. 2015 doi: 10.1002/ejhf.332. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 36.Sanfiorenzo C, Ilie MI, Belaid A, Barlesi F, Mouroux J, Marquette CH. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8:e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leidinger P, Keller A, Borries A, Huwer H, Rohling M, Huebers J, et al. Specific peripheral miRNA profiles for distinguishing lung cancer from COPD. Lung Cancer. 2011;74:41–47. doi: 10.1016/j.lungcan.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Kowdley KV. Method for microRNA isolation from clinical serum samples. Anal Biochem. 2012;431:69–75. doi: 10.1016/j.ab.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, et al. Evaluation of quantitative miRNA expression platforms in the micro-RNA quality control (miRQC) study. Nat Methods. 2014;11:809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 40.Yamada A, Cox MA, Gaffney KA, Moreland A, Boland CR, Goel A. Technical factors involved in the measurement of circulating microRNA biomarkers for the detection of colorectal neoplasia. PLoS One. 2014;9:e112481. doi: 10.1371/journal.pone.0112481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacLellan SA, MacAulay C, Lam S, Garnis C. Pre-profiling factors influencing serum microRNA levels. BMC Clin Pathol. 2014;14:27. doi: 10.1186/1472-6890-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powell TM, Smith TC, Jacobson IG, Boyko EJ, Hooper TI, Gackstetter GD, et al. Prospective assessment of chronic multisymptom illness reporting possibly associated with open-air burn pit smoke exposure in Iraq. J Occup Environ Med. 2012;54:682–688. doi: 10.1097/JOM.0b013e318255ba39. [DOI] [PubMed] [Google Scholar]
- 44.Jones KA, Smith B, Granado NS, Boyko EJ, Gackstetter GD, Ryan MA. Newly reported lupus and rheumatoid arthritis in relation to deployment within proximity to a documented open-air burn pit in Iraq. J Occup Environ Med. 2012;54:698–707. doi: 10.1097/JOM.0b013e3182529799. [DOI] [PubMed] [Google Scholar]
- 45.Maska LB, Sayles HR, O’Dell JR, Curtis JR, Bridges SL, Jr, Moreland LW, et al. Serum cotinine as a biomarker of tobacco exposure and the association with treatment response in early rheumatoid arthritis. Arthritis Care Res (Hoboken) 2012;64:1804–1810. doi: 10.1002/acr.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benowitz NL, Bernert JT, Caraballo RS, Holiday DB, Wang J. Optimal serum cotinine levels for distinguishing cigarette smokers and nonsmokers within different racial/ethnic groups in the United States between 1999 and 2004. Am J Epidemiol. 2009;169:236–248. doi: 10.1093/aje/kwn301. [DOI] [PubMed] [Google Scholar]
- 47.Zaman MS, Thamminana S, Shahryari V, Chiyomaru T, Deng G, Saini S, et al. Inhibition of PTEN gene expression by oncogenic miR-23b-3p in renal cancer. PLoS One. 2012;7:e50203. doi: 10.1371/journal.pone.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Begum S, Hayashi M, Ogawa T, Jabboure FJ, Brait M, Izumchenko E, et al. An integrated genome-wide approach to discover deregulated microRNAs in non-small cell lung cancer: clinical significance of miR-23b-3p deregulation. Sci Rep. 2015;5:13236. doi: 10.1038/srep13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L, Li B, Chen D, Liu L, Huang C, Lu Z, et al. miR-139 and miR-200c regulate pancreatic cancer endothelial cell migration and angiogenesis. Oncol Rep. 2015;34:51–58. doi: 10.3892/or.2015.3945. [DOI] [PubMed] [Google Scholar]
- 50.Ebrahimi F, Gopalan V, Smith RA, Lam AK. miR-126 in human cancers: clinical roles and current perspectives. Exp Mol Pathol. 2014;96:98–107. doi: 10.1016/j.yexmp.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Chen Z, Liu X, Hu Z, Wang Y, Liu M, Liu X, et al. Identification and characterization of tumor suppressor and oncogenic miRNAs in gastric cancer. Oncol Lett. 2015;10:329–336. doi: 10.3892/ol.2015.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arican O, Aral M, Sasmaz S, Ciragil P. Serum levels of TNF-alpha, IFN-gamma, IL-6, IL-8, IL-12, IL-17, and IL-18 in patients with active psoriasis and correlation with disease severity. Mediators Inflamm. 2005;2005:273–279. doi: 10.1155/MI.2005.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bostrom EA, Kindstedt E, Sulniute R, Palmqvist P, Majster M, Holm CK, et al. Increased eotaxin and MCP-1 levels in serum from individuals with periodontitis and in human gingival fibroblasts exposed to pro-inflammatory cytokines. PLoS One. 2015;10:e0134608. doi: 10.1371/journal.pone.0134608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.George ML, Eccles SA, Tutton MG, Abulafi AM, Swift RI. Correlation of plasma and serum vascular endothelial growth factor levels with platelet count in colorectal cancer: clinical evidence of platelet scavenging? Clin Cancer Res. 2000;6:3147–3152. [PubMed] [Google Scholar]
- 55.Stelmach I, Jerzynska J, Kuna P. A randomized, double-blind trial of the effect of glucocorticoid, antileukotriene and beta-agonist treatment on IL-10 serum levels in children with asthma. Clin Exp Allergy. 2002;32:264–269. doi: 10.1046/j.1365-2222.2002.01286.x. [DOI] [PubMed] [Google Scholar]
- 56.Mastrangelo G, Clonfero E, Pavanello S, Fedeli U, Fadda E, Turato A. Exposure to diesel exhaust enhances total IgE in non-atopic dockers. Int Arch Occup Environ Health. 2003;76:63–68. doi: 10.1007/s00420-002-0373-x. [DOI] [PubMed] [Google Scholar]
- 57.Samuelsen M, Nygaard UC, Lovik M. Allergy adjuvant effect of particles from wood smoke and road traffic. Toxicology. 2008;246:124–131. doi: 10.1016/j.tox.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 58.Kabesch M, Schedel M, Carr D, Woitsch B, Fritzsch C, Weiland SK, et al. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. 2006;117:269–274. doi: 10.1016/j.jaci.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 59.Joseph CL, Havstad S, Bobbitt K, Woodcroft K, Zoratti EM, Nageotte C, et al. Transforming growth factor beta (TGFbeta1) in breast milk and indicators of infant atopy in a birth cohort. Pediatr Allergy Immunol. 2014;25:257–263. doi: 10.1111/pai.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kypriotou M, Rivero D, Haller S, Mariotto A, Huber M, Acha-Orbea H, et al. Activin a inhibits antigen-induced allergy in murine epicutaneous sensitization. Front Immunol. 2013;4:246. doi: 10.3389/fimmu.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Danielson LS, Reavie L, Coussens M, Davalos V, Castillo-Martin M, Guijarro MV. Limited miR-17-92 overexpression drives hematologic malignancies. Leuk Res. 2015;39:335–341. doi: 10.1016/j.leukres.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 66.Bommel H, Haake M, Luft P, Horejs-Hoeck J, Hein H, Bartels J, et al. The diesel exhaust component pyrene induces expression of IL-8 but not of eotaxin. Int Immunopharmacol. 2003;3:1371–1379. doi: 10.1016/S1567-5769(03)00135-8. [DOI] [PubMed] [Google Scholar]
- 67.Miller RL, Garfinkel R, Lendor C, Hoepner L, Li Z, Romanoff L, et al. Polycyclic aromatic hydrocarbon metabolite levels and pediatric allergy and asthma in an inner-city cohort. Pediatr Allergy Immunol. 2010;21(2 Pt 1):260–267. doi: 10.1111/j.1399-3038.2009.00980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jude JA, Dileepan M, Subramanian S, Solway J, Panettieri RA, Jr, Walseth TF. miR-140-3p regulation of TNF-alpha-induced CD38 expression in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2012;303:L460–L468. doi: 10.1152/ajplung.00041.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rah SY, Park KH, Han MK, Im MJ, Kim UH. Activation of CD38 by interleukin-8 signaling regulates intracellular Ca2+ level and motility of lymphokine-activated killer cells. J Biol Chem. 2005;280:2888–2895. doi: 10.1074/jbc.M409592200. [DOI] [PubMed] [Google Scholar]
- 70.Guedes AG, Jude JA, Paulin J, Kita H, Lund FE, Kannan MS. Role of CD38 in TNF-alpha-induced airway hyperresponsiveness. Am J Physiol Lung Cell Mol Physiol. 2008;294:L290–L299. doi: 10.1152/ajplung.00367.2007. [DOI] [PubMed] [Google Scholar]