Abstract
Myelin is a lipid-rich sheath formed by the spiral wrapping of specialized glial cells around axon segments. Myelinating glia allow for rapid transmission of nerve impulses and metabolic support of axons, and the absence of or disruption to myelin results in debilitating motor, cognitive, and emotional deficits in humans. Because myelin is a jawed vertebrate innovation, zebrafish are one of the simplest vertebrate model systems to study the genetics and development of myelinating glia. The morphogenetic cellular movements and genetic program that drive myelination are conserved between zebrafish and mammals, and myelin develops rapidly in zebrafish larvae, within 3–5 days post-fertilization. Myelin ultrastructure can be visualized in the zebrafish from larval to adult stages via transmission electron microscopy, and the dynamic development of myelinating glial cells may be observed in vivo via transgenic reporter lines in zebrafish larvae. Zebrafish are amenable to genetic and pharmacological screens, and screens for myelinating glial phenotypes have revealed both genes and drugs that promote myelin development, many of which are conserved in mammalian glia. Recently, zebrafish have been employed as a model to understand the complex dynamics of myelinating glia during development and regeneration. In this chapter, we describe these key methodologies and recent insights into mechanisms that regulate myelination using the zebrafish model.
INTRODUCTION
The evolution of myelin, which surrounds and protects axons, was a critically important innovation in the jawed vertebrate lineage (Zalc, Goujet, & Colman, 2008). Myelin is a lipid-rich sheath that facilitates saltatory conduction velocity without a substantial increase in axonal diameter; thus, myelin permits higher brain complexity within a bony skeleton. In humans, the importance of the myelin sheath is most easily realized when it is lost or disrupted, such as in diseases including multiple sclerosis and Charcot-Marie-Tooth. Despite its importance, the cellular and molecular mechanisms that drive the migration, morphogenesis, terminal differentiation, and regeneration of myelinating glia are incompletely understood.
Though the myelin sheath itself is ultrastructurally and biochemically similar in the central and peripheral nervous systems (CNS and PNS, respectively), the cell types that form the myelin sheath are derived from different precursor populations, and the genetic programs and morphogenetic behaviors driving myelin formation are quite different between the CNS and PNS. Myelin is formed in the CNS by oligodendrocytes (OLs) in a multistep developmental process characterized by expression of molecular markers and dramatic changes in cell morphology. OL precursor cells (OPCs; Olig2+ and Sox10+) originate from discrete ventral regions of the neural tube during early embryonic development and in the subventricular region of the brain during adult neurogenesis (Ackerman & Monk, 2015; Emery, 2010). Once specified, OPCs proliferate and migrate remarkable distances to populate the entire CNS. Once migration ceases, a subset of OPCs transition into premyelinating OLs (Nkx2.2+), which extend filapodia-like processes and ensheath axons but do not begin myelination (Kirby et al., 2006; Kucenas et al., 2008; Mitew et al., 2014; Snaidero et al., 2014; Zhu et al., 2014). Finally, premyelinating OLs select axons, terminally differentiate to become myelinating OLs, and iteratively wrap axonal segments (Fig. 1A).
FIGURE 1. Myelinating glia in the zebrafish.
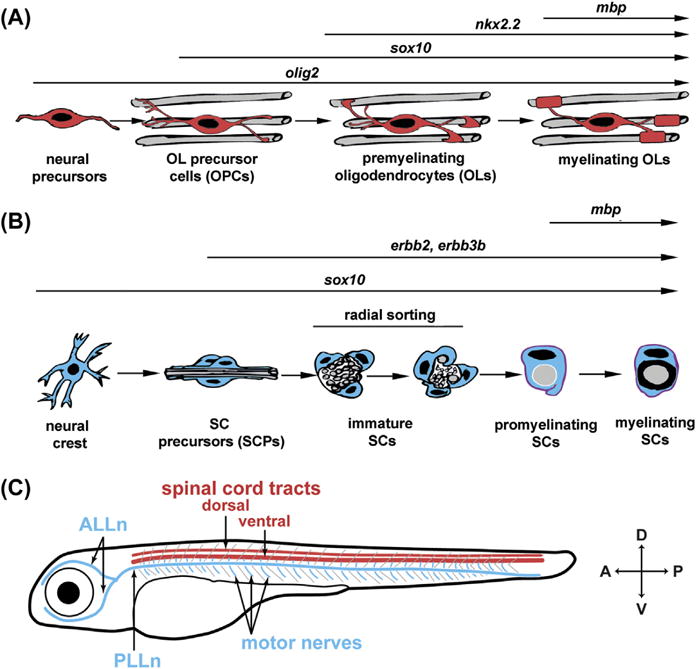
(A) Oligodendrocyte (OL) development. Neural precursor cells, depicted in red, are born within the ventral neural tube during early embryonic development. Once specified, OL precursor cells (OPCs) proliferate and migrate. Once migration ceases, a subpopulation of OPCs begin to extend protrusions, contact axons (premyelinating OLs), and terminally differentiate to become myelinating OLs. (B) Schwann cell (SC) development. Immature SCs associate with bundles of axons, begin the process of radial sorting by segregating individual axons based on diameter, and spiral their cytoplasmic membranes to form myelin. The accompanying arrowheads show the key markers for SC development discussed in this review. (C) Schematized locations of commonly studied myelinated axons in zebrafish larvae at 3–5 days postfertilization. Compass in lower right denotes dorsal (D), ventral (V), anterior (A), and posterior (P). CNS is shown in red and denotes the dorsal and ventral tracts of the spinal cord. PNS is shown in blue and marks the anterior and posterior lateral line nerves (ALLn and PLLn, respectively) and motor nerves that extend ventrally from the spinal cord.
In the PNS, specialized glia called Schwann cells (SCs) form myelin, with both similar and distinct mechanisms as CNS myelination (reviewed in Jessen & Mirsky, 2005). During embryogenesis, SC precursors (SCPs) are proliferative and migrate along with growing axons. Once migration ceases, SCPs differentiate into immature SCs that are associated with many axons. At this point, immature SCs secrete extracellular matrix (ECM) molecules that constitute the basal lamina (Court, Wrabetz, & Feltri, 2006). Through a process called radial sorting, immature SCs select and segregate large caliber axons and repeatedly wrap their membranes around individual axonal segments, ultimately generating the myelin sheath (Fig. 1B).
The morphological progression of myelinating glia is well characterized in mammals, but only relatively recently have zebrafish become a premiere tool for in vivo analysis of OLs and SCs (Ackerman & Monk, 2015; Brösamle & Halpern, 2002; Monk & Talbot, 2009). OL development is easily monitored in larval zebrafish CNS, most frequently in the spinal cord, whereas the anterior and posterior lateral line nerves (ALLn and PLLn, respectively) and motor nerves are useful models for SC myelination (Fig. 1C). Here, we describe recent advances in imaging, genetic, and pharmacological tools to investigate conserved mechanisms of myelinating glial cell development and regeneration using the zebrafish.
1. VISUALIZATION OF MYELINATING GLIA IN ZEBRAFISH
A major advantage to using zebrafish as a model system is the ability to perform in vivo imaging to study myelination. Transgenic reporter lines expressing fluorescent proteins under control of glial-specific promoters are ideal tools to study the behavior of myelinating cells in a living vertebrate. Several valuable transgenic reporter lines are available to visualize myelination throughout development (Table 1). Zebrafish transgenic lines are predominantly generated using the Gateway-compatible Tol2Kit (Kwan et al., 2007). Concurrently, the Tol2 sites can also be used to generate enhancer trap lines in which a Gal4 transactivator is placed downstream of a promoter of interest and activates a reporter under Upstream Activating Sequence (UAS) control. Importantly, for the purposes of this review, several transgenic reporter lines are available to study myelination in both the CNS and PNS, at different stages of development, and the expression of these reporters are evolutionarily conserved.
Table 1.
Available Transgenic Reporters to Visualize Myelinating Glia in Zebrafish
| Gene | Labeled Cell Types | Transgenic Lines; References |
|---|---|---|
|
| ||
| cldnk | Myelinating OL and SCs | Tg(cldnk:Gal4)ue101Tg; Münzel et al. (2012) |
| foxd3 | Immature SCs through myelinating SCs | Tg(zFoxd3GFP); Gilmour, Maischein, and Nüsslein-Volhard (2002) |
| nkx2.2a | Subset of OPCs and early myelinating oligodendrocytes | Tg(nkx2.2a:mEGFP)vu16Tg; Kirby et al. (2006) |
| mbp | Myelinating OLs and myelinating SCs |
Tg(mbp:EGFP)ck1Tg; Jung et al. (2010) Tg(mbp:EGFP)ue1Tg; Almeida, Czopka, Ffrench-Constant, and Lyons (2011) Tg(mbp:EGFP-CAAX)ue2Tg; Almeida et al. (2011) Tg(mbp:GAL4-VP16)co20Tg; Hines et al. (2015) |
| mpz/p0 | Myelinating OLs | Tg(mpz[10kb]:EGFP)pt408Tg; Bai, Parris, and Burton (2014) |
| nkx2.2a | Subset of OPCs and early myelinating oligodendrocytes | Tg(nkx2.2a:mEGFP)vu16Tg; Kirby et al. (2006) |
| oligl | OL lineage cells | Tg(olig1:mEGFP)nv150Tg; Schebesta and Serluca (2009) |
| olig2 | PMN-derived progenitors, OL lineage, and motor neurons |
Tg(olig2:EGFP)vu12Tg; Shin, Park, Topczewska, Mawdsley, and Appel (2003) Tg(olig2:dsRed)vu19Tg; Kucenas et al. (2008) Tg(olig2:Kaeda)vu85Tg; Zannino and Appel (2009) |
| plp | OL lineage cells | Tg(Mmu.Plp1:EGFP)cc1Tg; Yoshida and Macklin (2005) |
| sox10 | OL lineage cells, interneurons, neural crest, and SC |
Tg(sox10:mRFP)vu234Tg; Kirby et al. (2006) Tg(sox10:EGFP)ba4Tg; Dutton et al. (2001) Tg(sox10:nls-Eos)w18Tg; Prendergast et al. (2012) Tg(sox10:Gal4-VP16)co19Tg; Das and Crump (2012) Tg(sox10:KalTA4GI); Almeida and Lyons (2015) |
In addition, myelin defects may be scored in fixed tissue with in situ hybridization (ISH) (Thisse, Thisse, Halpern, & Postlethwait, 1994) (Table 2). Whole mount ISH is widely used to describe expression patterns of genes of interest and interrogate myelin gene expression of mutants with defects in myelination. During ISH, an antisense messenger RNA (mRNA) probe tagged with either digoxigenin or fluorescein uridine-5′-triphosphate binds to the endogenous transcript. Following hybridization, the transcript is detected using an antibody specific to the tag and visualized under a light microscope.
Table 2.
Available In Situ Hybridization Probes for Myelinating Glia in Zebrafish
| Gene | Cell-Type Expression | References |
|---|---|---|
|
| ||
| 36K | Myelinating OLs | Morris et al. (2004) |
| cldnk | Myelinating OL and SCs | Takada, Kucenas, and Appel (2010) |
| ctnnd2 | Myelinating OL and SCs | Takada and Appel (2010) |
| erbb3 | Immature SCs through myelinating SCs | Lyons et al. (2005) |
| foxd3 | Immature SCs through myelinating SCs | Gilmour et al. (2002) |
| gpr126 | SCs | Monk et al. (2009) |
| krox20 | Promyelinating SCs | Lyons et al. (2005) |
| mbp | Myelinating OLs and myelinating SCs | Brosamle and Halpern (2002) and Lyons et al. (2005) |
| mid1ip1b | Myelinating OLs and myelinating SCs | Takada and Appel (2010) |
| nkx2.2a | Subset of OPCs and early myelinating oligodendrocytes | Kirby et al. (2006) |
| oct6 | Promyelinating SCs | Levavasseur et al. (1998) |
| olig1 | OL lineage cells | Schebesta and Serluca (2009) |
| olig2 | PMN-derived progenitors, OL lineage and motor neurons | Park, Mehta, Richardson, and Appel (2002) |
| mpz/p0 | Myelinating OLs | Brosamle and Halpern (2002) |
| plp1a | Myelinating OLs | Brosamle and Halpern (2002) |
| plp1b | Myelinating OLs | Brosamle and Halpern (2002) |
| sox10 | OL lineage cells, interneurons, neural crest, and SC | Dutton et al. (2001) |
| Zwilling-A and -B | Myelinating OLs and myelinating SCs | Schaefer and Brosamle (2009) |
The optical transparency and myriad transgenic lines make zebrafish a powerful and tractable system to study myelination. Zebrafish live-cell imaging has opened up new avenues for investigation of the cellular basis of myelination. One of the most exciting advances is the ability to see and study the fine details of individual cells and peer into the cellular and molecular events of migration, process extension, and internode development. During early stages of CNS myelination, there is a great deal of motility as the pro-OL extends actin-rich plasma membrane protrusions to contact multiple axonal segments, and as they mature, OLs myelinate several axon segments. Thus, a number of challenges must be met for OLs to establish and maintain their morphology and function. Meeting these challenges is the work of sophisticated architectural rearrangements within the OL that allow for process extension and wrapping of the plasma membrane and tightly regulated mechanisms of transport and motility. Increasingly, in vivo imaging in zebrafish has begun to add crucial missing pieces to the puzzle of myelin dynamics that cannot be readily observed in living rodent models. For example, using a tagged marker for F-actin, Nawaz et al. (2015) could define actin dynamics during myelin growth in vivo. In this study, immediately prior to axonal contact, F-actin was spatially restricted to the furthest edges of the expanding OL processes and following contact localized spirally along the axons as the OL plasma membrane iteratively wrapped the selected axonal segment (Nawaz et al., 2015). Further, these studies were recapitulated in purified mouse OLs in culture (Zuchero et al., 2015), highlighting zebrafish as a complementary approach to studying myelination.
Although the vast majority of OL and SC molecular markers are well conserved from zebrafish to mammals, some differences have been reported. Genomics studies reveal that zebrafish myelin genes such as myelin basic protein (mbp), myelin protein zero (mpz), and proteolipid protein (plp) share significant homology to their mammalian counterparts (Bai, Sun, Stolz, & Burton, 2011; Nawaz, Schweitzer, Jahn, & Werner, 2013; Schweitzer, Becker, Schachner, Nave, & Werner, 2006). However, during early stages of PNS myelination, zebrafish SCs do not encode key myelin compaction genes mpz and plp, suggesting that myelin compaction may be delayed in zebrafish compared to mammals (Brösamle & Halpern, 2002). Moreover, in zebrafish, mpz expression persists in both the CNS and PNS myelinated axons, in contrast to the mammalian homolog P0, which is found exclusively in the PNS (Bai et al., 2011). In addition, several proteins such as Zwilling-A, Zwilling-B, 36k, and Claudin K are distinctively present in zebrafish myelin; their homologs are either absent or not involved in myelin compaction in mammals (reviewed in Ackerman & Monk, 2015).
A deeper understanding of the cell biological functions of myelin proteins can only be achieved using fine structural imaging methods, including electron microscopy. While many aspects of myelin structure are indeed conserved between multiple fish species and mammals, we direct the reader to excellent reviews on important disparities between myelin structure in fish and rodent systems (Avila, Tevlin, Lees, Inouye, & Kirschner, 2007; Möbius, Nave, & Werner, 2016). Differences in myelin protein composition certainly lead to some variations in observed morphology, as electron microscopic analysis in zebrafish is generally performed at time points when myelin is not yet compact. Moreover, conventional chemical fixations are not adequate to preserve the myelin ultrastructure in zebrafish, and so many labs employ microwave-assisted tissue processing techniques (Czopka & Lyons, 2011). Additionally, exciting advances in high-pressure freezing techniques can provide improved resolution in the analysis of myelin ultrastructure in zebrafish as more laboratories acquire expertise in this technology (Möbius et al., 2016).
The myelin ultrastructure of the larval zebrafish can be visualized via transmission electron microscopy (TEM) to study OL and SC development in the spinal cord and the lateral line, respectively (Fig. 2A). A cross section of the larval zebrafish shows the spinal cord situated in the center of the animal and the two PLLns situated on either side (Fig. 2B). A closer magnification of the spinal cord shows several myelinated axons including the Mauthner axon, a critical neural circuit for zebrafish fast escape response (Korn & Faber, 2005; Fig. 2C). Similarly, myelinated axons are also observed in the PLLn (Fig. 2D).
FIGURE 2. Transmission electron microscopy (TEM) for zebrafish myelinated axons.
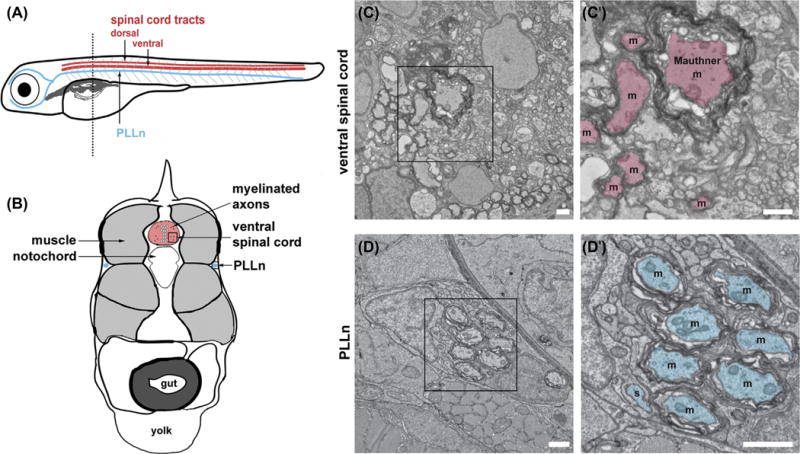
(A) Schematized cartoon depicting the myelinated axons commonly studied via TEM in larval zebrafish. Dotted line indicates the anteriore–posterior position of cross section depicted in panels B–D. (B) Diagram represents the cross section of a larval zebrafish and indicates positions of the spinal cord [red] and the PLLn on either side of the body [blue]. (C–D) Transmission electron micrographs between body segments five to seven at 5 days postfertilization. Boxes in left panels denote regions of interest shown in right panels. All panels, scale bar 1 = μmm. (C–C′) Ventral spinal cord with midline to the left. Myelinated axons in right panel are pseudocolored in red and marked with m. Note large caliber Mauthner axon. (D–D′) PLLn with midline to the left. Axons in right panel are pseudocolored in blue to mark myelinated (m) or sorted (s).
Myelinated axons can also be dissected from adult zebrafish and visualized via TEM. A protocol for TEM preparation of adult lateral line is described in the following section.
1.1 SOLUTIONS REQUIRED
Modified Karnovsky’s fixative (4% paraformaldehyde + 2% glutaraldehyde in
0.1-M sodium cacodylate, pH 7.4)
0.1-M sodium cacodylate
2% osmium tetraoxide in 0.1-M sodium cacodylate
25%, 50%, 75%, 95% ethanol in sterile filtered (i.e., MilliQ, MQ) water; 100% ethanol; propylene oxide; EMBED solution (see Section “Recipe” in Czopka & Lyons, 2011).
1.2 FIXATION AND EMBEDDING OF ADULT NERVE
Euthanize individual zebrafish by immediate and prolonged immersion in ice water or by prolonged overdose with tricaine (pH 7.4), until gill movements have ceased. With a clean razor blade, quickly sever the spinal cord.
- Fix the whole adult fish in Karnovsky’s solution in 2-mL Eppendorf tubes. For optimal fixation, the nerves need to be fixed uniformly and as fast as possible. To accelerate tissue processing, the immersed fish is microwaved in a water bath in either a Pelco microwave processor (model 3451; Ted Pella Mountain Lakes, CA) or a countertop microwave with inverter technology. It is crucial to keep the tissue at about 15–18 °C to prevent overheating the samples. While the temperature can be regulated via water recirculation in the Pelco system, a water/ice bath may be used as an alternative and the temperature can be monitored with a thermometer and adjusted after each cycle.
- Step 1: 250 W for 1 min, off for 1 min, 250 W for 1 min
- Step 2: 450 W for 1 min, off for 1 min, 450 W for 1 min
Upon fixation, the adult PLLn is located and removed using forceps. The ganglia of the PLLn are situated posterior to the ear, and the nerves run along the body on the surface of the fish. The dissected nerve is postfixed in Karnovsky’s for 1–2 h at room temperature or 4 °C overnight in a 12-well culture plate.
Remove Karnovsky’s and wash 3 × 15′ with 0.1-M sodium cacodylate at room temperature (RT). Samples can be stored at 4 °C, although we have found that prolonged storage (more than a month) at this stage can result in poorer quality images.
Postfix in 2% osmium tetraoxide in 0.1-M sodium cacodylate. The optimal incubation time and dilution vary between samples, but we regularly fix for 1 h.
At this point, gently move the osmium-treated nerves from the culture dishes to a solvent-proof Eppendorf tube using toothpicks or a paintbrush. Care must be taken to ensure that the samples do not dry.
MQ water washes—3 × 15′ at RT.
- Sequential ethanol dehydration steps:
- 25%—20′ at RT
- 50%—20′ at RT
- 75%—20′ at RT (if necessary, tissue can be stored at 4 °C overnight after this step)
- 95%—20′ at RT
- 100%—20′ at RT × 2
- Propylene oxide steps:
- propylene oxide—15′ at RT
- 1:1 propylene oxide: EMBED overnight
- 100% EMBED 4 h at RT
Embed and polymerize overnight in oven at 65 °C or until blocks are solid. Samples are trimmed, sectioned, and stained as previously described (Czopka & Lyons, 2011).
2. GENETIC ANALYSIS OF MYELIN DEVELOPMENT IN ZEBRAFISH
2.1 FORWARD GENETIC SCREENS
A major advantage of the zebrafish model system is the ability to perform large-scale forward genetic screens (Driever et al., 1996; Haffter & Nüsslein-Volhard, 1996). Zebrafish produce large clutches (hundreds of embryos per pair of mating adults), have a reasonable generation time, and can be housed as adults in a relatively small space. To randomly introduce mutations, male zebrafish are exposed to N-ethyl-N-nitrosourea (ENU), outcrossed to wild-type females, mutations are driven to homozygosity, and larvae are screened for phenotypes of interest in the F3 generation (Fig. 3). In this scheme, transgenes to screen for phenotypes in the F3 progeny may be introduced by outcrossing in the F1 generation. Historically, causative mutations were identified via a mapcross to an outbred strain, followed by polymerase chain reaction (PCR)-based linkage analysis (Postlethwait & Talbot, 1997). More recently, whole-genome sequencing (WGS) and RNA sequencing have become more affordable and thus more efficient options to rapidly identify a lesion of interest, as they can be used without a map-crossing step or parental information (Henke, Bowen, & Harris, 2013; Hill et al., 2013; Miller, Obholzer, Shah, Megason, & Moens, 2013).
FIGURE 3. Generation of mutants via forward genetic screen.
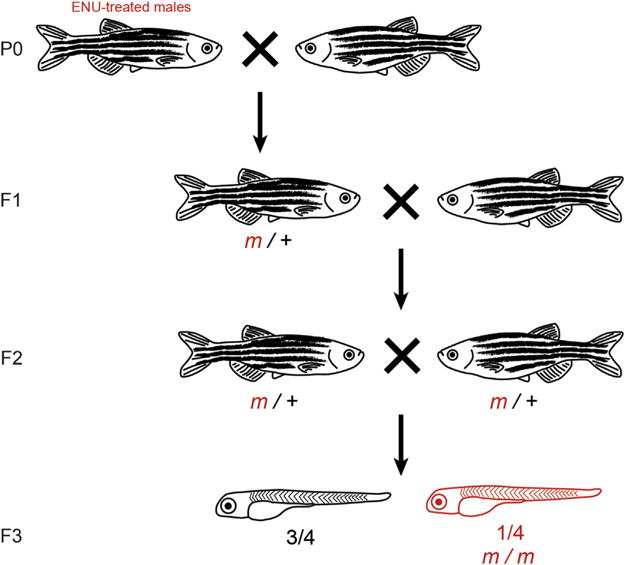
Schematic for a traditional three-generation screen. Generations (P0, F1, F2, F3) are labeled to the left. P0 males are treated with N-ethyl-N-nitrosourea (ENU) to induce random mutations, which are driven to homozygosity in the F3 generation using the crossing scheme shown.
Because of the relatively rapid development of myelinated axons in zebrafish and advent of transgenic tools for in vivo observation, large-scale genetic screens are a tool uniquely suited to the zebrafish to discover novel pathways that drive myelination. In the Appel laboratory, larvae were screened for aberrant olig2:EGFP expression in the CNS (Snyder, Kearns, & Appel, 2012), whereas the Talbot group screened for both CNS and PNS mutant phenotypes using ISH for mbp (Pogoda et al., 2006), and a complementary screen was performed to analyze sodium channel clustering at the nodes of Ranvier between myelin sheaths along an axon (Monk, Voas, Franzini-Armstrong, Hakkinen, & Talbot, 2013; Voas et al., 2007). The key myelin regulators uncovered in these screens are summarized in Table 3 and in the following sections. Most recently, the Monk laboratory screened for phenotypes using both mbp:mCherry and mbp ISH in combination with lhx1:GFP, which marks axons in the CNS and PNS (Ackerman et al., in preparation; Harty et al., in preparation; Herbert et al., in preparation). Although these screens have been quite expansive (>600 genomes in each of the mbp-based screens) and have identified many genes required for myelin development (Table 3), screens for myelination mutants in zebrafish have not yet reached saturation.
Table 3.
Myelination Genes Revealed by Forward Genetic Screening in Zebrafish
| Allele | Gene (If Known) | Myelinating Glia Phenotype | Function | Screen/Citation |
|---|---|---|---|---|
|
| ||||
| st14, st48 | erbb3b | Reduced mbp in LLn | Nrg/Erb signaling | Lyons et al. (2005) |
| st50, st61 | Erbb2 | Reduced mbp in LLn | Nrg/Erb signaling | Lyons et al. (2005) |
| st20 | Foxa2 | Reduced mbp in hindbrain | Forkhead transcription factor | Norton et al. (2005) |
| st47 | Reduced mbp in CNS | Pogoda et al. (2006) | ||
| st64 | Reduced mbp in ALLn | Pogoda et al. (2006) | ||
| i26 | aldh1a2 | PLLn amyelination | Aldehyde dehydrogenase | Kazakova et al. (2006) |
| M807 | Med12 | PLLn/CNS amyelination | Transcription mediator complex subunit | Kazakova et al. (2006) |
| tm79a | dzip1 | Loss of mbp in posterior PLLn | DAZ interacting zinc finger protein, regulator of Hedgehog signaling | Kazakova et al. (2006) |
| tt258 | Loss of mbp in posterior PLLn | Kazakova et al. (2006) | ||
| st25, st53 | Nsfa | Reduced mbp in LLn and CNS | N-ethylmaleimide-sensitive factor, synaptic vesicle fusion | Woods, Lyons, Voas, Pogoda, and Talbot (2006) |
| st60 | aII-spectrin | Reduced mbp in PNS/CNS | Cortical cytoskeleton component | Voas et al. (2007) |
| st23 | Kbp | Reduced mbp in LLn, CNS amyelination | Intracellular transport | Lyons, Naylor, Mercurio, Dominguez and Talbot (2008) |
| st49, st63 | adgrg6/gpr126 | PNS amyelination | GPCR Gas signaling | Monk et al. (2009) |
| st43 | kif1b | Reduced mbp in LLn, punctate mbp in CNS | mRNA transport | Lyons, Naylor, Scholze, and Talbot (2009) |
| vu56 | fbxw7 | Excess CNS myelination | F-box subunit of E3 ubiquitin ligase | Kearns et al. (2015) and Snyder et al. (2012) |
| vu76 | dync1h1 | Reduced PNS myelination; reduced OPCs | Dynein heavy chain subunit | Langworthy and Appel (2012) and Yang et al. (2015) |
| vu166 | Pescadillo | Reduced OPCs | Ribosome biogenesis, cell proliferation | Simmons and Appel (2012) |
| st67 | sec63 | Reduced PLLn and CNS myelination | ER translocon machinery | Monk et al. (2013) |
| st51 | notch3 | Reduced mbp in CNS | Notch signaling | Zaucker et al. (2013) |
| vu57 | hmgcs1 | OPC migration defects, reduced CNS mbp | Isoprenoid and cholesterol synthesis | Mathews et al. (2014) |
| st78 | znf16l | Reduced mbp in CNS | Zinc finger transcription factor | Sidik and Talbot (2015) |
As a complementary approach to ENU-based forward genetic screens, Kazakova et al. reasoned that genes required for the establishment of neuronal architecture may also be required in myelin development. Therefore, they assayed mbp expression among a subset known early neurodevelopment mutants (Kazakova et al., 2006). One of the mutants, motionless, was later shown to be mediator complex subunit 12 (med12) (Wang et al., 2006), which cooperates with sox10 to promote myelinating glia differentiation in mammals (Vogl et al., 2013).
2.2 REVERSE GENETICS APPROACHES
Classical forward genetic screens for myelin mutants have provided a powerful approach to unraveling new genes that regulate formation of myelinated axons. While forward genetic screens allow multiple entry points to interrogate mechanisms involved in myelination via cloning and sequencing of the mutated genes, reverse genetics is an important complement. The last decade has seen important additions to the gene editing toolbox in zebrafish, particularly with the recent discovery and application of transcription activator-like effector nucleases (TALENs) and the type II prokaryotic clustered regularly interspaced short palindromic repeat (CRISPR/Cas9) system (Gaj, Gersbach, & Barbas, 2013).
Briefly, TALENs consist of bipartite fusion proteins that recognize specific DNA sequences each fused to the FokI restriction endonuclease. We frequently use the TALEN targeter tool (https://tale-nt.cac.cornell.edu/) to find candidate binding sites and assemble TALEN from TALE repeats using the Golden Gate approach. Once assembled, the TALEN is cloned into an appropriate TALEN backbone to generate mRNA expression plasmids. Following transcription, the TALEN mRNA is injected at the one-cell stage. While TALEN assembly is generally more straightforward for genome editing compared to previous technologies, TALENs are labor intensive and maintaining a library of TALE repeats can be expensive. On the other hand, CRISPR technology entails the generation of a guide RNA (gRNA) that is injected along with Cas9 nuclease. There are several online tools to design gRNAs for the target gene including ZiFiT, CRISPR design tool, and CHOPCHOP to name a few (Montague, Cruz, Gagnon, Church, & Valen, 2014; Ran et al., 2013; Sander et al., 2010). The advantages and disadvantages of these techniques are reviewed elsewhere (Gaj et al., 2013); importantly, both approaches allow for targeted, rapid genome editing. Using these facile techniques, it is possible to manipulate the zebrafish genome and recover mutants with small inframe indels that are ideal for structure/function analyses during myelination (Petersen et al., 2015). Excitingly, recent studies in different developmental processes in zebrafish have utilized the CRISPR/Cas9 system to target multiple genomic loci simultaneously via multiplexed pools of gRNAs (Shah, Davey, Whitebirch, Miller, & Moens, 2016). For further reading, we suggest several excellent reviews and seminal research articles (Auer & Del Bene, 2014; Boch et al., 2009; Gagnon et al., 2014; Gaj et al., 2013; Jao, Wente, & Chen, 2013; Talbot & Amacher, 2014).
While TALENs and CRISPR/Cas9 technology allow for precise, targeted mutations in the genome, these techniques are less effective for studying genes that are either important for early embryonic development or have pleiotropic functions at later stages. The constitutive and global loss of these factors may be lethal and preclude further studies of the cellular autonomy of these factors beyond early development. To circumvent this problem, several alternate approaches have been proposed. For instance, cell-specific gene mutations have been carried out by expressing a CRISPR/Cas9 vector under the control of tissue-specific promoters of interest (Ablain, Durand, Yang, Zhou, & Zon, 2015), and knockdowns can be performed in a spatiotemporal manner via photoactivatable caged antisense morpholinos to either block mRNA splicing or protein translation (Ruble, Yeldell, & Dmochowski, 2015). These approaches are technically demanding and are restricted to certain developmental stages. Moreover, recent studies have demonstrated toxic side effects of using morpholinos that have been erroneously attributed to knocking down a specific gene (reviewed in Schulte-Merker & Stainier, 2014). Taken together, morpholinos can be informative in transiently disrupting protein function when used with stringent controls and coupled with corresponding mutant data for the target gene.
In addition to caged morpholinos, expression can be spatially and temporally controlled in zebrafish through site-specific recombination using Cre or Flp recombinases (Hans, Kaslin, Freudenreich, & Brand, 2009), tamoxifen-inducible Cre/Lox technology (Hans et al., 2009), tetracycline-inducible systems (Huang et al., 2005), and the Gal4/UAS system (Halpern et al., 2008). These techniques are conceptually elegant but have minor limitations in zebrafish, as new myelin-specific drivers need to be generated. Nonetheless, the fast generation time and relatively large clutch sizes allow one to screen for mutants within a short period of time. As tools and inducible constructs are developed and become more widely available, in the future, it will be interesting to perform cell-specific gene inactivation to determine autonomy and transient tissue-specific functions during myelination.
2.3 GENETIC REGULATORS OF MYELINATION: LESSONS FROM ZEBRAFISH
The utility of the zebrafish model depends upon faithful recapitulation of the myelination program between teleosts and mammals. To date, microscopic, biochemical, and genetic analyses demonstrate strong similarity of zebrafish myelin to that of rodents, and most of the markers and mutations used to study myelin in mammals have been found to be important in zebrafish myelin development as well (most recently reviewed in Ackerman & Monk, 2015). As an example, Neuregulin-ErbB2/3 signaling is an established, necessary pathway for SC development (Newbern & Birchmeier, 2010), and the conservation of the myelination program in zebrafish is perhaps most profoundly illustrated by the unbiased recovery of mutations in ErbB2/3 signaling in one of the first large-scale forward genetic screens for myelin mutants (Lyons et al., 2005). Subsequent in vivo analyses in zebrafish were able to parse roles for Nrg-ErbB signaling in directed migration, proliferation, and radial sorting of SCs (Lyons et al., 2005; Perlin, Lush, Stephens, Piotrowski, & Talbot, 2011; Raphael, Lyons, & Talbot, 2011), thus illustrating the unique importance of zebrafish in myelination studies.
As mentioned, zebrafish genetic screens have revealed established regulators of myelinating glial cell development. Screens can also uncover genes previously implicated in myelinopathies, which establish new models for disease research. For instance, patients with CADASIL syndrome (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy, OMIM #125310) present with neurodegeneration accompanied by white matter deficits as a result of an autosomal dominant mutation in NOTCH3 (Joutel et al., 1996). Recently, a mutant from the Talbot lab screen for mbp abnormalities was revealed to a lesion in notch3, and the zebrafish adults present with a hemorrhage phenotype similar to the cardiovascular dysfunction in CADASIL patients (Zaucker, Mercurio, Sternheim, Talbot, & Marlow, 2013). In addition to establishing a new zebrafish disease model, this mutant may lend more nuanced insight to how OPC proliferation and specification is regulated by Notch signaling (Park & Appel, 2003).
The true power of zebrafish genetic screens is exemplified by novel, crucial myelination genes discovered therein, which frequently complement the cell biology of myelination known from other systems. For instance, excess OPCs are produced in a mutant for fbxw7, which encodes an F-box substrate recognition subunit of E3 ubiquitin ligase. In fbxw7 mutants, Notch signaling is elevated, and inhibition of Notch signaling is sufficient to suppress the fbxw7 phenotype (Snyder et al., 2012). Further studies in zebrafish demonstrated that fbxw7 mutants also have elevated mammalian protein Target Of Rapamycin (mTOR) signaling (Kearns, Ravanelli, Cooper, & Appel, 2015), which results in hypermyelination in mammals (Narayanan, Flores, Wang, & Macklin, 2009). Thus, zebrafish screens have identified a functionally relevant component of the mTOR pathway in myelinating glia. Another example is the discovery of sec63 as a regulator of both CNS and PNS myelination via its function in the endoplasmic reticulum (ER) translocon machinery. Mutants in sec63 have fragmented and swollen ER in glia and other tissues, as well as upregulated markers of unfolded protein response and ER stress (Monk et al., 2013). Because myelinating glia must synthesize large amounts of protein and membrane during differentiation and wrapping, deficits in secretory pathways are implicated in myelinopathies, and these effects can be reduced in cultured OLs by treatment with the ER stress blocker guanabenz (reviewed in Clayton & Popko, 2016). Further investigation of sec63 in zebrafish can examine how cellular stress pathways influence myelination potential.
One of the most critically important myelination genes initially discovered in zebrafish was the adhesion G protein-coupled receptor (aGPCR) gpr126/adgrg6, which is essential for SC myelination in the PNS. Though it had been known for decades that elevation of cyclic adenosine monophosphate (cAMP) was essential for SC wrapping and terminal differentiation, the transducer that increased cAMP levels was not known. Starting with a forward genetic screen, the Talbot lab identified two alleles of gpr126 in which CNS myelination is normal but SCs fail to wrap. This phenotype could be rescued with addition of forskolin, a pharmacological adenylyl cyclase activator (Monk et al., 2009), which suggested that Gpr126 was the sought-after GPCR that elevated cAMP to promote SC wrapping. Subsequent structure-function analysis in zebrafish demonstrated two critically important facets to Gpr126: first, that Gpr126 has bimodal, domain-dependent functions in SC development and second, that Gpr126 contains a cryptic tethered agonist sufficient to activate signaling and promote myelination (Liebscher et al., 2014; Petersen et al., 2015). Furthermore, these studies implicated Laminin-211, a component of the SC basal lamina, as a novel binding partner for Gpr126, which may mechanically activate this a GPCR at a particular phase of SC development. This finding, as well as biochemical experiments demonstrating collagen IV binds zebrafish Gpr126 (Paavola, Sidik, Zuchero, Eckart, & Talbot, 2014), highlights the need to pursue how the ECM cooperates with cell surface receptors in glial cell development in zebrafish.
The discovery of Gpr126 as a critical regulator of SC myelination has spurred reverse genetic analyses to test the role of other adhesion family GPCRs in myelination. Recently, new zebrafish alleles of a related adhesion GPCR, gpr56, were created to analyze function in myelinating glia. In complementary papers using both zebrafish and mouse models, Gpr56 was demonstrated to be an evolutionarily conserved regulator of OL development. In Gpr56 mutants, loss of RhoA signaling results in precocious OPC differentiation at the expense of proliferation such that hypomyelination is observed at later stages (Ackerman, Garcia, Piao, Gutmann, & Monk, 2015; Giera et al., 2015). Together, the critical roles of Gpr126 and Gpr56 in myelination suggest that additional adhesion GPCRs may be required for glial cell development and differentiation, and zebrafish represent as an excellent model to probe the biological functions of adhesion GPCRs in myelinating glia.
Recently, a new zinc finger transcription factor, Znf16l, was identified in a forward genetic screen due to loss of mbp expression in the CNS. A combination of imaging studies demonstrated that Znf16l cell autonomously promotes proliferation, migration, and wrapping in the OL lineage. Though Znf16l does not appear conserved in mammals, the znf16l phenotype could be rescued by expression of mouse Zfp488, which itself appears absent from the zebrafish genome (Sidik & Talbot, 2015). Zfp488 is also a zinc finger transcription factor expressed in OLs and is necessary for CNS myelination and remyelination in mammals (Soundarapandian et al., 2011; Wang et al., 2006). Thus, these experiments highlight the conserved function of Zfp16l and Zfp488 in OL development and place these zinc finger proteins within the transcriptional hierarchy of OL development.
Together, these recent studies and previous work demonstrate the strong advantages of the zebrafish myelination model for gene discovery and for dissecting the conserved function of novel molecular regulators. Given recent advances in reverse genetic techniques in zebrafish, future investigations into known regulators of myelination may prove fruitful for uncovering molecular and cellular mechanisms using this optically and genetically tractable model.
3. PHARMACOLOGICAL MANIPULATION OF MYELINATED AXONS IN ZEBRAFISH
3.1 SMALL MOLECULES AND PEPTIDES ALTER MYELINATION
Pharmacological manipulation of zebrafish development is robust and well established (Bruni, Lakhani, & Kokel, 2014; Rennekamp & Peterson, 2015; Tan & Zon, 2011; Taylor, Grant, Temperley, & Patton, 2010). The ability to treat externally developing, free-swimming zebrafish larvae with small molecules and peptides is a substantial advantage over mammalian models. Although it is necessary to consider that compounds can have off-target effects, drug treatments are rapid compared to generating double or triple mutants and thus can be a powerful approach to identify or validate candidate pathways of interest. For instance, aldh1a2 was identified in a shelf screen as having CNS mbp defects (Kazakova et al., 2006); aldh1a2 encodes an enzyme involved in synthesis of retinoic acid (RA) (Begemann, Schilling, Rauch, Geisler, & Ingham, 2001), which is required for OPC differentiation and CNS myelination (Huang et al., 2011). Treatment of aldh1a2 mutant larvae with exogenous RA at 10–16 hpf was sufficient to rescue mbp expression in the CNS, while the effect was less potent at a later stage (Kazakova et al., 2006). Together, these data highlight a critical developmental window at which RA signaling is necessary for OL development.
As described previously, the elevation of cAMP via Gpr126 is a critically important step in SC myelination (Jessen & Mirsky, 1991; Mogha et al., 2013; Monk et al., 2009). To demonstrate that cAMP elevation can rescue myelination defects in gpr126 mutants, larvae were treated with forskolin at 45–52 hpf, when SCs are sorting but typically have not yet myelinated PLLn axons (Monk et al., 2009). Similar to RA signaling, cAMP elevation may also be required at a critical window of SC development as forskolin treatment at earlier or later stages is not sufficient for rescue (Glenn & Talbot, 2013; Monk et al., 2009), and longer treatments (e.g., 24 h) cause lethality, even at lower doses. Importantly, forskolin was used to demonstrate that the genetic differentiation program may be uncoupled from physical wrapping in SCs. In gpr126 mutants with radial sorting defects, forskolin is unable to rescue sorting or wrapping, but SCs in forskolin-treated gpr126 mutants nonetheless begin to express mbp (Petersen et al., 2015). Interestingly, gpr126 mutants at 30 days postfertilization (dpf) following a pulsed exposure of forskolin from 50 to 55 hpf had a small number of compact, myelinated axons (Glenn & Talbot, 2013). These data suggest that the lack of myelin seen in the gpr126 mutants is suppressed by transient elevation of forskolin to initiate myelination, but gpr126 is not required for myelin maintenance up to 1 month.
Treatment with small peptides is also sufficient to modify myelination via the Gpr126-regulated program. A small tethered agonist termed the Stachel (German for “stinger”) is encoded within all tested adhesion GPCRs, and in vitro studies suggest that the Stachel is necessary and sufficient for activation of signaling (Demberg, Rothemund, Schöneberg, & Liebscher, 2015; Liebscher et al., 2014; Stoveken, Hajduczok, Xu, & Tall, 2015). Given that structure-function analyses demonstrated the Stachel is necessary in vivo for myelination in zebrafish, we also wanted to test if the Stachel was sufficient for Gpr126 signaling in myelination. Exogenous treatment of hypomorphic gpr126 larvae with a 16-amino acid Stachel peptide fragment suppressed mbp expression defects, demonstrating that the Stachel is sufficient to activate the Gpr126-regulated myelination program (Liebscher et al., 2014). More recently, Gpr126 was shown to interact with the flexible tail of neuronal prion protein Prp. By treating gpr126 hypomorphic mutant zebrafish with soluble Prp peptide, we also demonstrated that Prp can partially suppress Mbp defects in these mutants, further exploiting the utility of peptide rescue for myelination and demonstrating the conservation the Prp-Gpr126 interaction in zebrafish (Küffer et al., 2016).
Temporal requirements of Nrg-ErbB signaling in SC development were also revealed via pharmacological manipulation in zebrafish. Mutations in ErbB receptors caused a reduction in myelinated axons, but it was not clear whether this was due to defects in fate specification, migration, proliferation, myelination, or a combination thereof. By pulsing the pan-ErbB inhibitor AG1478 at different stages of larval development, it was revealed that ErbB signaling is continuously required for directed migration of SCPs but that proliferation is not necessary for migration. However, ErbB signaling is also required in postmigratory SCs for proliferation and radial sorting (Lyons et al., 2005). Subsequent experiments with AG1478 demonstrated that both ErbB signaling and proliferation are required for radial sorting but that proliferation itself is not dependent on ErbB (Raphael et al., 2011). In summary, these experiments highlight how the zebrafish myelination model is uniquely suited for temporal analysis of myelin development.
3.2 DRUG SCREENS FOR NOVEL MYELIN REGULATORS
In addition to pathways of interest, zebrafish are an exquisitely powerful system for in vivo drug discovery for therapeutics that can protect or restore the myelin sheath. Myelin is evident in zebrafish by 72 hpf, though cell specification and morphogenetic behaviors to initiate myelination begin earlier. At these time points, zebrafish larvae can be arrayed in 96-well plates using a P1000 pipette with the tip cutoff to create a larger bore; with this technique, several hundred larvae can be precisely arrayed in minutes. We have found that one to three larvae can survive for 24 h in a single well containing 250-mL liquid, allowing multiple larvae to be screened for each compound. Daily water changes are accomplished using a multichannel pipette and permit the larvae to survive in 96-well format up to 5 dpf.
In the first large-scale drug screen for remyelination therapeutics, Buckley et al. screened for enhanced olig2:GFP proliferation or migration in larvae treated with 1170 commercially available compounds. Hits from the primary screen, approximately 2% of drugs tested, were subsequently assayed for their effect on mbp levels. From this screen, PP2 was identified as a Src kinase inhibitor that decreased mbp transcript (Buckley et al., 2010). This effect was likely mediated through Fyn kinase, which has been shown to be an important regulator of OL myelination in mammals and zebrafish (Colognato, Ramachandrappa, Olsen, & Ffrench-Constant, 2004; Czopka, Ffrench-Constant, & Lyons, 2013; Laursen, Chan, & Ffrench-Constant, 2009; Osterhout, Wolven, Wolf, Resh, & Chao, 1999).
New screens for small molecules that promote myelination may instead focus on zebrafish models in which myelination are defective. For instance, hypomorphic gpr126 mutants have reduced, but not absent, mbp expression and myelination (Monk et al., 2009; Petersen et al., 2015), and gpr56 mutants have a similar hypomorphic phenotype in the CNS (Ackerman et al., 2015). Given that GPCRs are highly druggable and represent significant targets within existing libraries, these proteins represent promising targets for development of future therapies. In this situation, homozygous mutant larvae may be assayed for an increase in mbp compared to control via available transgenes. In addition to identifying small molecule targets of aGPCRs, which are known regulators of myelin, this approach may identify compounds that alter the activity of other proteins, lending insight to distinct pathways that drive myelination.
4. PLASTICITY, MAINTENANCE, AND REGENERATION OF MYELINATED AXONS IN ZEBRAFISH
4.1 NEURONAL ACTIVITY AND MYELINATION
Plasticity of the nervous system is evolutionary conserved and necessary for development, learning, and memory across phyla. More recently, it has been shown that not only are synapses highly plastic, but myelin also is altered by changes in neuronal activity during development and in response to the environment (Fields, 2015; Gibson et al., 2014; Makinodan, Rosen, Ito, & Corfas, 2012). Studies in zebrafish have demonstrated how OLs can respond to neuronal activity. Transgenic expression of tetanus toxin (TTX), which prevents synaptic vesicle exocytosis, was sufficient to decrease OPC specification and myelination when expressed in neurons (Hines, Ravanelli, Schwindt, Scott, & Appel, 2015; Mensch et al., 2015). A complementary experiment demonstrated that treatment with pentylenetetrazole, which enhances neuronal signaling, promoted myelination in zebrafish larvae (Mensch et al., 2015). Time-lapse in vivo analyses demonstrated that neuronal activity did not affect OL process extension or “sampling” of axon segments, but rather influenced whether nascent sheaths were stabilized over time. Importantly, stabilized myelin sheaths appeared at sites of synaptic vesicle exocytosis labeled with Syp-EGFP, suggesting that synaptic vesicles are local cues that drive OL myelin maturation (Hines et al., 2015). However, not all neuron subtypes mediate myelination via activity; a recent study demonstrated that TTX expression modifies the myelination pattern of reticulospinal neurons, but not of commissural primary ascending neurons (Koudelka et al., 2016). Together, these results suggest a mechanism for the differences in myelination patterns and activity-dependent myelin changes observed in mammals.
A disadvantage of the zebrafish model system for assaying myelin changes in response to activity compared to mammalian models is the lack of well-established electrophysiological assays for neuronal conduction velocity. Furthermore, there are currently no behavioral tests for zebrafish that model neurological disorders or assess myelin function. Nonetheless, the genetic techniques available in zebrafish, including optogenetics and the recently developed magnetically controlled “Magneto” protein (Wheeler et al., 2016) are uniquely rapid and could complement electrophysiological and behavioral studies in mammals. Future studies in neuronal activity and myelination in the zebrafish might explore these avenues and couple them to in vivo imaging throughout development, rendering the zebrafish model an extremely powerful system to study activity-dependent myelination.
4.2 ANALYSIS OF REMYELINATION IN ZEBRAFISH
Defects in myelin in demyelinating diseases or following injury can lead to devastating symptoms and ultimately paralysis. However, no effective therapeutics exists to prevent demyelination or promote remyelination. Here, we highlight studies investigating the molecular control of remyelination in zebrafish; these studies provide the basis for novel therapies for patients with nerve injury and dysmyelinating diseases. Unlike mammals, zebrafish have the remarkable ability to regenerate their damaged tissues, and this phenomenon has been studied in multiple organs including heart, fin, and the CNS (Gemberling, Bailey, Hyde, & Poss, 2013). As mentioned in Section 3, zebrafish also serve as a suitable chassis for drug screens. Indeed, with respect to CNS remyelination, Buckley et al. (2010) have carried out a pharmacological screen for promyelinating compounds with the goal of opening up an avenue for potential therapeutics to stimulate remyelination in multiple sclerosis (MS).
Spontaneous remyelination occurs in patients with early stages of MS and in animal models of demyelination; however, this regenerative potential declines with disease chronicity and age. Using a focal demyelination paradigm whereby foam soaked in detergent lysophosphatidylcholine was placed next to the zebrafish optic nerve, Munzel et al. observed demyelination followed by remyelination within 4 weeks. Importantly, there was no axonal damage, suggesting an autonomous function in the OLs (Münzel, Becker, Becker, & Williams, 2014). Interestingly, myelin is fully regenerated in young adults (4–7 months) but inefficacious in aged fish (15–18 months), suggesting that the cellular ability to regenerate is limited with increasing age similar to mammals. Moreover, Munzel et al. observed reduced phagocytotic microglia/macrophage recruitment to the site of lesion in old zebrafish suggesting myelin debris clearance could be impaired with age. It remains unknown whether inefficient remyelination in aged fish is due to impaired myelin debris clearance or instead due to reduced intrinsic potential to myelinate.
A major strength of zebrafish in regeneration studies is the ability to perform live-cell imaging to observe cellular behavior following injury. To test the regenerative potential of OLs and track remyelination in vivo, OLs can be genetically ablated using the nitroreductase (NTR)-mediated cell ablation techniques (Chung et al., 2013; Fang et al., 2014; Kim et al., 2015). Briefly, transgenic fish express NTR, a bacterial enzyme, in myelinating glia. When the fish are exposed to a prodrug metronidazole (MTZ), NTR binds and converts MTZ to a cytotoxin leading to targeted cell ablation (Curado, Stainier, & Anderson, 2008). Moreover, the drug can be washed away to allow for regeneration. Using the NTR/MTZ system in OLs, Kim et al. (2015) observed that treating the fish with sulfasalazine, an antiinflammatory compound, promoted remyelination. In addition to uncovering therapeutic targets for coaxing regeneration, the NTR/MTZ system can also be used to elucidate cell lineage relationships during regeneration. During regeneration in the postamputated fin, cell lineage tracing experiments suggest that regenerated tissues arise from preexisting cells and retain their developmental identity (reviewed in Pfefferli & Jazwinska, 2015). Recent evidence from juvenile and adult mice suggests that the OLs in the CNS are heterogeneous with differential propensity to myelinate (Marques et al., 2016). If the zebrafish CNS also contains distinct subpopulations of OLs, this would open up exciting avenues for cell lineage studies that will examine if the subpopulations of OLs recapitulate their cell fate following ablation and if there are distinct OLs that are more adept at remyelination in the CNS.
In the PNS, following nerve injury, SCs become dedicated “repair SCs,” which release neuroprotective factors, elevate cytokines, remove myelin, and axonal debris, and cue macrophage recruitment (reviewed in Jessen & Mirsky, 2016). This repair and regenerative program is orchestrated by bursts of injury-specific genes such as c-Jun, a transcription factor normally downregulated during myelinating SC development (Arthur-Farraj et al., 2012). The role of SCs in peripheral nerve repair has been studied in the PLLn (Graciarena, Dambly-Chaudière, & Ghysen, 2014; Xiao et al., 2015), motor nerves (Rosenberg, Isaacman-Beck, Franzini-Armstrong, & Granato, 2014), and the adult zebrafish maxillary barbels (ZMB) (LeClair & Topczewski, 2010; Moore, Mark, Hogan, Topczewski, & LeClair, 2012). In the larval PLLn, following laser-mediated axonal transection, adjacent SCs extend bridging processes toward the site of injury and begin clearance of axonal debris (Xiao et al., 2015). Similarly, in the transected larval motor nerves, SCs undergo morphological changes and contain fluorescently tagged axonal debris within their membrane, suggesting that SCs are actively involved in debris clearance. However, in mutants lacking SCs, motor growth cone regeneration was unaffected suggesting SCs are dispensible for axonal regrowth (Rosenberg et al., 2014). Moreover, in both the PLLn and motor nerves, in the absence of SCs, regenerated axons are misrouted (Rosenberg et al., 2014; Xiao et al., 2015), indicating that SCs are important for providing directional guidance cues. One of the guidance cues important for directional cue has been suggested to be the collagen modifying enzyme glycosyltransferase lysyl hydroxylase 3 (lh3) (Isaacman-Beck, Schneider, Franzini-Armstrong, & Granato, 2015). While lh3 mutants have impaired axonal guidance following transfection, SC-specific expression of lh3 was sufficient to restore regeneration (Isaacman-Beck et al., 2015).
Peripheral nerve remyelination in zebrafish can be studied across development. In order to examine remyelination in adult axons, Moore et al. performed a time-course analysis in ZMBs following amputation (LeClair & Topczewski, 2010; Moore et al., 2012). ZMBs are optically transparent appendages that are innervated by the facial nerve, chemosensory cells, and epithelial nerves (LeClair & Topczewski, 2010). At 10 days post amputation (dpa), few axons myelinate, and a patchy collagen-rich matrix surrounds the axons. By 28 dpa, the matrix thickens, the ultrastructure of the barbel is restored, and axons are remyelinated, albeit myelination is reduced compared to uninjured controls. Regenerating barbels undergo extensive remyelination and several promyelinating transcription factors are upregulated in the distal regenerating barbel tissues. While Moore et al. focused on a candidate gene expression approach to probe the mechanism of remyelination in the ZMB, future WGS approaches would provide the ability to profile relevant, injury-induced targets and compare these large expression data sets to mammalian remyelination paradigms. Taken together, the zebrafish peripheral nerves provide an unprecedented view of the complex cell–cell and cell–matrix interactions involved in SC repair following injury in vivo.
CONCLUSIONS
Zebrafish is a powerful vertebrate system to study myelination using both forward and reverse genetic approaches, in vivo analysis during development and repair, and for performing drug screens to uncover novel drivers of myelination and remyelination. Genetic screens in zebrafish have uncovered several regulators of myelination that are conserved in mammals. Moreover, genome-wide association studies have identified candidate genes associated with schizophrenia (reviewed in Roussos & Haroutunian, 2014), multiple sclerosis (Andlauer et al., 2016), and peripheral neuropathies (Safka Brožková, Nevšímalová, Mazanec, Rautenstrauss, & Seeman, 2012). The advent of genome-editing techniques will allow researchers tools to analyze how single nucleotide polymorphisms contribute to disease progression. The identification of these variants allow for a better molecular handle on the etiology of the several diseases where myelin is impaired. In addition to finding novel genes, zebrafish are amenable to chemical screens demonstrating the feasibility of zebrafish to discover compounds for future therapeutics. Future studies will leverage the many strengths of zebrafish to build a functional network of genetic pathways involved in myelination, a cost-effective and rapid approach to drug discovery, and continue to solidify the importance of zebrafish research in basic science and clinical medicine.
Acknowledgments
We thank members of the Monk laboratory for helpful discussions and comments, David Lyons for TEM protocol suggestions, and Lisette Mateo and Christina Rozario for assistance with figure generation. We apologize to our colleagues whose primary work we were unable to cite due to space limitations. This work was supported by grants from the National Institutes of Health (NS079445, HD08601 to KRM; NS087786 to SCP), the Muscular Dystrophy Association (MDA293295 to KRM), and KRM is a Harry Weaver Scholar of the National Multiple Sclerosis Society.
References
- Ablain J, Durand EM, Yang S, Zhou Y, Zon LI. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Developmental Cell. 2015;32(6):756–764. doi: 10.1016/j.devcel.2015.01.032. http://doi.org/10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman SD, Garcia C, Piao X, Gutmann DH, Monk KR. The adhesion GPCR Gpr56 regulates oligodendrocyte development via interactions with Ga12/13 and RhoA. Nature Communications. 2015;6:6122. doi: 10.1038/ncomms7122. http://doi.org/10.1038/ncomms7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman SD, Monk KR. The scales and tales of myelination: using zebrafish and mouse to study myelinating glia. Brain Research. 2015 doi: 10.1016/j.brainres.2015.10.011. http://doi.org/10.1016/j.brainres.2015.10.011. [DOI] [PMC free article] [PubMed]
- Almeida RG, Czopka T, Ffrench-Constant C, Lyons DA. Individual axons regulate the myelinating potential of single oligodendrocytes in vivo. Development. 2011;138(20):4443–4450. doi: 10.1242/dev.071001. http://doi.org/10.1242/dev.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida RG, Lyons DA. Intersectional gene expression in zebrafish using the split KalTA4 system. Zebrafish. 2015;12(6):377–386. doi: 10.1089/zeb.2015.1086. http://doi.org/10.1089/zeb.2015.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andlauer TFM, Buck D, Antony G, Bayas A, Bechmann L, Berthele A, et al. Novel multiple sclerosis susceptibility loci implicated in epigenetic regulation. Science Advances. 2016;2(6):e1501678. doi: 10.1126/sciadv.1501678. http://doi.org/10.1126/sciadv.1501678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur-Farraj PJ, Latouche M, Wilton DK, Quintes S, Chabrol E, Banerjee A, et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 2012;75(4):633–647. doi: 10.1016/j.neuron.2012.06.021. http://dx.doi.org/10.1016/j.neuron.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer TO, Del Bene F. CRISPR/Cas9 and TALEN-mediated knock-in approaches in zebrafish. Methods (San Diego, California) 2014;69(2):142–150. doi: 10.1016/j.ymeth.2014.03.027. http://doi.org/10.1016/j.ymeth.2014.03.027. [DOI] [PubMed] [Google Scholar]
- Avila RL, Tevlin BR, Lees JPB, Inouye H, Kirschner DA. Myelin structure and composition in zebrafish. Neurochemical Research. 2007;32(2):197–209. doi: 10.1007/s11064-006-9136-5. http://doi.org/10.1007/s11064-006-9136-5. [DOI] [PubMed] [Google Scholar]
- Bai Q, Parris RS, Burton EA. Different mechanisms regulate expression of zebrafish myelin protein zero (P0) in myelinating oligodendrocytes and its induction following axonal injury. Journal of Biological Chemistry. 2014;289(35):24114–24128. doi: 10.1074/jbc.M113.545426. http://doi.org/10.1074/jbc.M113.545426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Q, Sun M, Stolz DB, Burton EA. Major isoform of zebrafish P0 is a 23.5 kDa myelin glycoprotein expressed in selected white matter tracts of the central nervous system. The Journal of Comparative Neurology. 2011;519(8):1580–1596. doi: 10.1002/cne.22587. http://doi.org/10.1002/cne.22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebra-fish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128(16):3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. doi: 10.1126/science.1178811. http://doi.org/10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Brösamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39(1):47–57. doi: 10.1002/glia.10088. http://doi.org/10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Bruni G, Lakhani P, Kokel D. Discovering novel neuroactive drugs through high-throughput behavior-based chemical screening in the zebrafish. Frontiers in Pharmacology. 2014;5:153. doi: 10.3389/fphar.2014.00153. http://doi.org/10.3389/fphar.2014.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley CE, Marguerie A, Roach AG, Goldsmith P, Fleming A, Alderton WK, Franklin RJM. Drug reprofiling using zebrafish identifies novel compounds with potential pro-myelination effects. Neuropharmacology. 2010;59(3):149–159. doi: 10.1016/j.neuropharm.2010.04.014. http://doi.org/10.1016/j.neuropharm.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Chung AY, Kim PS, Kim S, Kim E, Kim D, Jeong I, et al. Generation of demyelination models by targeted ablation of oligodendrocytes in the zebrafish CNS. Molecules and Cells. 2013;36(1):82–87. doi: 10.1007/s10059-013-0087-9. http://doi.org/10.1007/s10059-013-0087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton BLL, Popko B. Endoplasmic reticulum stress and the unfolded protein response in disorders of myelinating glia. Brain Research. 2016 doi: 10.1016/j.brainres.2016.03.046. http://doi.org/10.1016/j.brainres.2016.03.046. [DOI] [PMC free article] [PubMed]
- Colognato H, Ramachandrappa S, Olsen IM, Ffrench-Constant C. Integrins direct Src family kinases to regulate distinct phases of oligodendrocyte development. The Journal of Cell Biology. 2004;167(2):365–375. doi: 10.1083/jcb.200404076. http://doi.org/10.1083/jcb.200404076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court FA, Wrabetz L, Feltri ML. Basal lamina: Schwann cells wrap to the rhythm of space-time. Current Opinion in Neurobiology. 2006;16(5):501–507. doi: 10.1016/j.conb.2006.08.005. http://doi.org/10.1016/j.conb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Curado S, Stainier DYR, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nature Protocols. 2008;3(6):948–954. doi: 10.1038/nprot.2008.58. http://doi.org/10.1038/nprot.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czopka T, Ffrench-Constant C, Lyons DA. Individual oligodendrocytes have only a few hours in which to generate new myelin sheaths in vivo. Developmental Cell. 2013;25(6):599–609. doi: 10.1016/j.devcel.2013.05.013. http://doi.org/10.1016/j.devcel.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czopka T, Lyons DA. Dissecting mechanisms of myelinated axon formation using zebrafish. Methods in Cell Biology. 2011;105:25–62. doi: 10.1016/B978-0-12-381320-6.00002-3. http://doi.org/10.1016/B978-0-12-381320-6.00002-3. [DOI] [PubMed] [Google Scholar]
- Das A, Crump JG. Bmps and id2a act upstream of Twist1 to restrict ectomesen-chyme potential of the cranial neural crest. PLoS Genetics. 2012;8(5):e1002710. doi: 10.1371/journal.pgen.1002710. http://doi.org/10.1371/journal.pgen.1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demberg LM, Rothemund S, Schöneberg T, Liebscher I. Identification of the tethered peptide agonist of the adhesion G protein-coupled receptor GPR64/ADGRG2. Biochemical and Biophysical Research Communications. 2015;464(3):743–747. doi: 10.1016/j.bbrc.2015.07.020. http://doi.org/10.1016/j.bbrc.2015.07.020. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, et al. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128(21):4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010;330(6005):779–782. doi: 10.1126/science.1190927. http://doi.org/10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- Fang Y, Lei X, Li X, Chen Y, Xu F, Feng X, et al. A novel model of demye-lination and remyelination in a GFP-transgenic zebrafish. Biology Open. 2014;4(1):62–68. doi: 10.1242/bio.201410736. http://doi.org/10.1242/bio.201410736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields RD. A new mechanism of nervous system plasticity: activity-dependent myelination. Nature Reviews Neuroscience. 2015;16(12):756–767. doi: 10.1038/nrn4023. http://doi.org/10.1038/nrn4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Ahkmetova L, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9(5):e98186. doi: 10.1371/journal.pone.0098186. http://doi.org/10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. http://doi.org/10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends in Genetics. 2013;29(11):611–620. doi: 10.1016/j.tig.2013.07.003. http://doi.org/10.1016/j.tig.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson EM, Purger D, Mount CW, Goldstein AK, Lin GL, Wood LS, et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science. 2014;344(6183):1252304. doi: 10.1126/science.1252304. http://doi.org/10.1126/science.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giera S, Deng Y, Luo R, Ackerman SD, Mogha A, Monk KR, et al. The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nature Communications. 2015;6:6121. doi: 10.1038/ncomms7121. http://doi.org/10.1038/ncomms7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DT, Maischein HM, Nüsslein-Volhard C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron. 2002;34(4):577–588. doi: 10.1016/s0896-6273(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Glenn TD, Talbot WS. Analysis of Gpr126 function defines distinct mechanisms controlling the initiation and maturation of myelin. Development. 2013;140(15):3167–3175. doi: 10.1242/dev.093401. http://doi.org/10.1242/dev.093401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graciarena M, Dambly-Chaudière C, Ghysen A. Dynamics of axonal regeneration in adult and aging zebrafish reveal the promoting effect of a first lesion. Proceedings of the National Academy of Sciences. 2014;111(4):1610–1615. doi: 10.1073/pnas.1319405111. http://doi.org/10.1073/pnas.1319405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Nüsslein-Volhard C. Large scale genetics in a small vertebrate, the zebrafish. The International Journal of Developmental Biology. 1996;40(1):221–227. [PubMed] [Google Scholar]
- Halpern ME, Rhee J, Goll MG, Akitake CM, Parsons M, Leach SD. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish. 2008;5(2):97–110. doi: 10.1089/zeb.2008.0530. http://doi.org/10.1089/zeb.2008.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PLoS One. 2009;4(2):e4640. doi: 10.1371/journal.pone.0004640. http://doi.org/10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke K, Bowen ME, Harris MP. Identification of mutations in zebrafish using next-generation sequencing. Ausubel FM, et al., editors. Current protocols in molecular biology. 2013:104. doi: 10.1002/0471142727.mb0713s104. [Unit 7.13] http://doi.org/10.1002/0471142727.mb0713s104. [DOI] [PubMed]
- Hill JT, Demarest BL, Bisgrove BW, Gorsi B, Su YC, Yost HJ. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Research. 2013;23(4):687–697. doi: 10.1101/gr.146936.112. http://doi.org/10.1101/gr.146936.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines JH, Ravanelli AM, Schwindt R, Scott EK, Appel B. Neuronal activity biases axon selection for myelination in vivo. Nature Neuroscience. 2015;18(5):683–689. doi: 10.1038/nn.3992. http://doi.org/10.1038/nn.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JK, Jarjour AA, Nait-Oumesmar B, Kerninon C, Williams A, Krezel W, et al. Retinoid X receptor gamma signaling accelerates CNS remyelination. Nature Neuroscience. 2011;14(1):45–53. doi: 10.1038/nn.2702. http://doi.org/10.1038/nn.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Jou TS, Ho YL, Lee WH, Jeng YT, Hsieh FJ, Tsai HJ. Conditional expression of a myocardium-specific transgene in zebrafish transgenic lines. Developmental Dynamics. 2005;233(4):1294–1303. doi: 10.1002/dvdy.20485. http://doi.org/10.1002/dvdy.20485. [DOI] [PubMed] [Google Scholar]
- Isaacman-Beck J, Schneider V, Franzini-Armstrong C, Granato M. The lh3 gly-cosyltransferase directs target-selective peripheral nerve regeneration. Neuron. 2015;88(4):691–703. doi: 10.1016/j.neuron.2015.10.004. http://doi.org/10.1016/j.neuron.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences. 2013;110(34):13904–13909. doi: 10.1073/pnas.1308335110. http://doi.org/10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. Schwann cell precursors and their development. Glia. 1991;4(2):185–194. doi: 10.1002/glia.440040210. http://doi.org/10.1002/glia.440040210. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nature Reviews Neuroscience. 2005;6(9):671–682. doi: 10.1038/nrn1746. http://doi.org/10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. The repair Schwann cell and its function in regenerating nerves. The Journal of Physiology. 2016 doi: 10.1113/JP270874. http://doi.org/10.1113/JP270874. [DOI] [PMC free article] [PubMed]
- Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383(6602):707–710. doi: 10.1038/383707a0. http://doi.org/10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- Jung SH, Kim S, Chung AY, Kim HT, So JH, Ryu J, et al. Visualization of myelination in GFP-transgenic zebrafish. Developmental Dynamics. 2010;239(2):592–597. doi: 10.1002/dvdy.22166. http://doi.org/10.1002/dvdy.22166. [DOI] [PubMed] [Google Scholar]
- Kazakova N, Li H, Mora A, Jessen KR, Mirsky R, Richardson WD, Smith HK. A screen for mutations in zebrafish that affect myelin gene expression in Schwann cells and oligodendrocytes. Developmental Biology. 2006;297(1):1–13. doi: 10.1016/j.ydbio.2006.03.020. http://doi.org/10.1016/j.ydbio.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Kearns CA, Ravanelli AM, Cooper K, Appel B. Fbxw7 limits myelination by inhibiting mTOR signaling. Journal of Neuroscience. 2015;35(44):14861–14871. doi: 10.1523/JNEUROSCI.4968-14.2015. http://doi.org/10.1523/JNEUROSCI.4968-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee YI, Chang KY, Lee DW, Cho SC, Ha YW, et al. Promotion of remyelination by sulfasalazine in a transgenic zebrafish model of demyelination. Molecules and Cells. 2015;38(11):1013–1021. doi: 10.14348/molcells.2015.0246. http://doi.org/10.14348/molcells.2015.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nature Neuroscience. 2006;9(12):1506–1511. doi: 10.1038/nn1803. http://doi.org/10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- Korn H, Faber DS. The Mauthner cell half a century later: a neurobiological model for decision-making? Neuron. 2005;47(1):13–28. doi: 10.1016/j.neuron.2005.05.019. http://doi.org/10.1016/j.neuron.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Koudelka S, Voas MG, Almeida RG, Baraban M, Soetaert J, Meyer MP, et al. Individual neuronal subtypes exhibit diversity in CNS myelination mediated by synaptic vesicle release. Current Biology. 2016;26(11):1447–1455. doi: 10.1016/j.cub.2016.03.070. http://doi.org/10.1016/jcub.2016.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucenas S, Takada N, Park HC, Woodruff E, Broadie K, Appel B. CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nature Neuroscience. 2008;11(2):143–151. doi: 10.1038/nn2025. http://doi.org/10.1038/nn2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küffer A, Lakkaraju AK, Mogha A, Petersen SC, Airich K, Doucerain C, Aguzzi A. The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature. 2016;536(7617):464–468. doi: 10.1038/nature19312. http://www.nature.com/nature/journal/v536/n7617/full/nature19312.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon trans-genesis constructs. Developmental Dynamics. 2007;236(11):3088–3099. doi: 10.1002/dvdy.21343. http://doi.org/10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Langworthy MM, Appel B. Schwann cell myelination requires Dynein function. Neural Development. 2012;7:37. doi: 10.1186/1749-8104-7-37. http://doi.org/10.1186/1749-8104-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen LS, Chan CW, Ffrench-Constant C. An integrin-contactin complex regulates CNS myelination by differential Fyn phosphorylation. Journal of Neuroscience. 2009;29(29):9174–9185. doi: 10.1523/JNEUROSCI.5942-08.2009. http://doi.org/10.1523/JNEUROSCI.5942-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClair EE, Topczewski J. Development and regeneration of the zebrafish maxillary barbel: a novel study system for vertebrate tissue growth and repair. PLoS One. 2010;5(1) doi: 10.1371/journal.pone.0008737. http://dx.doi.org/10.1371/journal.pone.0008737C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levavasseur F, Mandemakers W, Visser P, Broos L, Grosveld F, Zivkovic D, Meijer D. Comparison of sequence and function of the Oct-6 genes in zebrafish, chicken and mouse. Mechanisms of Development. 1998;74(1–2):89–98. doi: 10.1016/s0925-4773(98)00067-7. [DOI] [PubMed] [Google Scholar]
- Liebscher I, Schön J, Petersen SC, Fischer L, Auerbach N, Demberg LM, et al. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Reports. 2014;9(6):2018–2026. doi: 10.1016/j.celrep.2014.11.036. http://doi.org/10.1016/j.celrep.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DA, Naylor SG, Mercurio S, Dominguez C, Talbot WS. KBP is essential for axonal structure, outgrowth and maintenance in zebrafish, providing insight into the cellular basis of Goldberg-Shprintzen syndrome. Development. 2008;135(3):599–608. doi: 10.1242/dev.012377. http://doi.org/10.1242/dev.012377. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Naylor SG, Scholze A, Talbot WS. Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nature Genetics. 2009;41(7):854–858. doi: 10.1038/ng.376. http://doi.org/10.1038/ng.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DA, Pogoda HM, Voas MG, Woods IG, Diamond B, Nix R, et al. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Current Biology. 2005;15(6):513–524. doi: 10.1016/j.cub.2005.02.030. http://doi.org/10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Makinodan M, Rosen KM, Ito S, Corfas G. A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science. 2012;337(6100):1357–1360. doi: 10.1126/science.1220845. http://doi.org/10.1126/science.1220845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcão A, Xiao L, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352(6291):1326–1329. doi: 10.1126/science.aaf6463. http://doi.org/10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews ES, Mawdsley DJ, Walker M, Hines JH, Pozzoli M, Appel B. Mutation of 3-Hydroxy-3-Methylglutaryl CoA synthase I reveals requirements for isopre-noid and cholesterol synthesis in oligodendrocyte migration arrest, axon wrapping, and myelin gene expression. Journal of Neuroscience. 2014;34(9):3402–3412. doi: 10.1523/JNEUROSCI.4587-13.2014. http://doi.org/10.1523/JNEUROSCI.4587-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensch S, Baraban M, Almeida R, Czopka T, Ausborn J, El Manira A, Lyons DA. Synaptic vesicle release regulates myelin sheath number of individual oligodendrocytes in vivo. Nature Neuroscience. 2015;18(5):628–630. doi: 10.1038/nn.3991. http://doi.org/10.1038/nn.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AC, Obholzer ND, Shah AN, Megason SG, Moens CB. RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome Research. 2013;23(4):679–686. doi: 10.1101/gr.147322.112. http://doi.org/10.1101/gr.147322.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitew S, Hay CM, Peckham H, Xiao J, Koenning M, Emery B. Mechanisms regulating the development of oligodendrocytes and central nervous system myelin. Neuroscience. 2014;276:29–47. doi: 10.1016/j.neuroscience.2013.11.029. http://doi.org/10.1016/j.neuroscience.2013.11.029. [DOI] [PubMed] [Google Scholar]
- Möbius W, Nave KA, Werner HB. Electron microscopy of myelin: structure preservation by high-pressure freezing. Brain Research. 2016;1641(Pt A):92–100. doi: 10.1016/j.brainres.2016.02.027. http://doi.org/10.1016/j.brainres.2016.02.027. [DOI] [PubMed] [Google Scholar]
- Mogha A, Benesh AE, Patra C, Engel FB, Schöneberg T, Liebscher I, Monk KR. Gpr126 functions in Schwann cells to control differentiation and myelination via G-protein activation. Journal of Neuroscience. 2013;33(46):17976–17985. doi: 10.1523/JNEUROSCI.1809-13.2013. http://doi.org/10.1523/JNEUROSCI.1809-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk KR, Naylor SG, Glenn TD, Mercurio S, Perlin JR, Dominguez C, et al. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science. 2009;325(5946):1402–1405. doi: 10.1126/science.1173474. http://doi.org/10.1126/science.1173474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk KR, Talbot WS. Genetic dissection of myelinated axons in zebrafish. Current Opinion in Neurobiology. 2009;19(5):486–490. doi: 10.1016/j.conb.2009.08.006. http://doi.org/10.1016/j.conb.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk KR, Voas MG, Franzini-Armstrong C, Hakkinen IS, Talbot WS. Mutation of sec63 in zebrafish causes defects in myelinated axons and liver pathology. Disease Models and Mechanisms. 2013;6(1):135–145. doi: 10.1242/dmm.009217. http://doi.org/10.1242/dmm.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Research. 2014;42:W401–W407. doi: 10.1093/nar/gku410. Web Server Issue. http://doi.org/10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AC, Mark TE, Hogan AK, Topczewski J, LeClair EE. Peripheral axons of the adult zebrafish maxillary barbel extensively remyelinate during sensory appendage regeneration. The Journal of Comparative Neurology. 2012;520(18):4184–4203. doi: 10.1002/cne.23147. http://doi.org/10.1002/cne.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JK, Willard BB, Yin X, Jeserich G, Kinter M, Trapp BD. The 36K protein of zebrafish CNS myelin is a short-chain dehydrogenase. Glia. 2004;45(4):378–391. doi: 10.1002/glia.10338. http://doi.org/10.1002/glia.10338. [DOI] [PubMed] [Google Scholar]
- Münzel EJ, Becker CG, Becker T, Williams A. Zebrafish regenerate full thickness optic nerve myelin after demyelination, but this fails with increasing age. Acta Neuropathologica Communications. 2014;2:77. doi: 10.1186/s40478-014-0077-y. http://doi.org/10.1186/s40478-014-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzel EJ, Schaefer K, Obirei B, Kremmer E, Burton EA, Kuscha V, et al. Claudin k is specifically expressed in cells that form myelin during development of the nervous system and regeneration of the optic nerve in adult zebrafish. Glia. 2012;60(2):253–270. doi: 10.1002/glia.21260. http://doi.org/10.1002/glia.21260. [DOI] [PubMed] [Google Scholar]
- Narayanan SP, Flores AI, Wang F, Macklin WB. Akt signals through the mammalian target of rapamycin pathway to regulate CNS myelination. Journal of Neuroscience. 2009;29(21):6860–6870. doi: 10.1523/JNEUROSCI.0232-09.2009. http://doi.org/10.1523/JNEUROSCI.0232-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz S, Sánchez P, Schmitt S, Snaidero N, Mitkovski M, Velte C, et al. Actin filament turnover drives leading edge growth during myelin sheath formation in the central nervous system. Developmental Cell. 2015;34(2):139–151. doi: 10.1016/j.devcel.2015.05.013. http://doi.org/10.1016/j.devcel.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz S, Schweitzer J, Jahn O, Werner HB. Molecular evolution of myelin basic protein, an abundant structural myelin component. Glia. 2013;61(8):1364–1377. doi: 10.1002/glia.22520. [DOI] [PubMed] [Google Scholar]
- Newbern J, Birchmeier C. Nrg1/ErbB signaling networks in Schwann cell development and myelination. Seminars in Cell and Developmental Biology. 2010;21(9):922–928. doi: 10.1016/j.semcdb.2010.08.008. http://doi.org/10.1016/j.semcdb.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton WH, Mangoli M, Lele Z, Pogoda HM, Diamond B, Mercurio S, et al. Monorail/Foxa2 regulates floorplate differentiation and specification of oligoden-drocytes, serotonergic raphé neurones and cranial motoneurones. Development. 2005;132(4):645–658. doi: 10.1242/dev.01611. http://doi.org/10.1242/dev.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout DJ, Wolven A, Wolf RM, Resh MD, Chao MV. Morphological differentiation of oligodendrocytes requires activation of Fyn tyrosine kinase. The Journal of Cell Biology. 1999;145(6):1209–1218. doi: 10.1083/jcb.145.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavola KJ, Sidik H, Zuchero JB, Eckart M, Talbot WS. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Science Signaling. 2014;7(338):ra76. doi: 10.1126/scisignal.2005347. http://doi.org/10.1126/scisignal.2005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Appel B. Delta-Notch signaling regulates oligodendrocyte specification. Development. 2003;130(16):3747–3755. doi: 10.1242/dev.00576. [DOI] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Developmental Biology. 2002;248(2):356–368. doi: 10.1006/dbio.2002.0738. http://www.ncbi.nlm.nih.gov/pubmed/12167410. [DOI] [PubMed] [Google Scholar]
- Perlin JR, Lush ME, Stephens WZ, Piotrowski T, Talbot WS. Neuronal Neuregulin 1 type III directs Schwann cell migration. Development. 2011;138(21):4639–4648. doi: 10.1242/dev.068072. http://doi.org/10.1242/dev.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen SC, Luo R, Liebscher I, Giera S, Jeong SJ, Mogha A, et al. The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with Laminin-211. Neuron. 2015;85(4):755–769. doi: 10.1016/j.neuron.2014.12.057. http://doi.org/10.1016/j.neuron.2014.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferli C, Jaźwińska A. The art of fin regeneration in zebrafish. Regeneration. 2015;2(2):72–83. doi: 10.1002/reg2.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda HM, Sternheim N, Lyons DA, Diamond B, Hawkins TA, Woods IG, et al. A genetic screen identifies genes essential for development of myelinated axons in zebrafish. Developmental Biology. 2006;298(1):118–131. doi: 10.1016/j.ydbio.2006.06.021. http://doi.org/10.1016/j.ydbio.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Talbot WS. Zebrafish genomics: from mutants to genes. Trends in Genetics. 1997;13(5):183–190. doi: 10.1016/s0168-9525(97)01129-3. [DOI] [PubMed] [Google Scholar]
- Prendergast A, Linbo TH, Swarts T, Ungos JM, McGraw HF, Krispin S, et al. The metalloproteinase inhibitor Reck is essential for zebrafish DRG development. Development. 2012;139(6):1141–1152. doi: 10.1242/dev.072439. http://doi.org/10.1242/dev.072439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature Protocols. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. http://doi.org/10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raphael AR, Lyons DA, Talbot WS. ErbB signaling has a role in radial sorting independent of Schwann cell number. Glia. 2011;59(7):1047–1055. doi: 10.1002/glia.21175. http://doi.org/10.1002/glia.21175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennekamp AJ, Peterson RT. 15 years of zebrafish chemical screening. Current Opinion in Chemical Biology. 2015;24:58–70. doi: 10.1016/j.cbpa.2014.10.025. http://doi.org/10.1016/j.cbpa.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AF, Isaacman-Beck J, Franzini-Armstrong C, Granato M. Schwann cells and deleted in colorectal carcinoma direct regenerating motor axons towards their original path. Journal of Neuroscience. 2014;34(44):14668–14681. doi: 10.1523/JNEUROSCI.2007-14.2014. http://doi.org/10.1523/JNEUROSCI.2007-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Haroutunian V. Schizophrenia: susceptibility genes and oligodendrog-lial and myelin related abnormalities. Frontiers in Cellular Neuroscience. 2014;8:5. doi: 10.3389/fncel.2014.00005. http://doi.org/10.3389/fncel.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruble BK, Yeldell SB, Dmochowski IJ. Caged oligonucleotides for studying biological systems. Journal of Inorganic Biochemistry. 2015;150:182–188. doi: 10.1016/j.jinorgbio.2015.03.010. http://doi.org/10.1016/j.jinorgbio.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safka Brožková D, Nevsímalová S, Mazanec R, Rautenstrauss B, Seeman P. Charcot-Marie-Tooth neuropathy due to a novel EGR2 gene mutation with mild pheno-typeeusefulness of human mapping chip linkage analysis in a Czech family. Neuromus-cular Disorders. 2012;22(8):742–746. doi: 10.1016/j.nmd.2012.04.002. http://doi.org/10.1016/j.nmd.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Research. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. Web Server Issue. http://doi.org/10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer K, Brösamle C. Zwilling-A and -B, two related myelin proteins of teleosts, which originate from a single bicistronic transcript. Molecular Biology and Evolution. 2009;26(3):495–499. doi: 10.1093/molbev/msn298. http://doi.org/10.1093/molbev/msn298. [DOI] [PubMed] [Google Scholar]
- Schebesta M, Serluca FC. olig1 Expression identifies developing oligodendro-cytes in zebrafish and requires hedgehog and Notch signaling. Developmental Dynamics. 2009;238(4):887–898. doi: 10.1002/dvdy.21909. http://doi.org/10.1002/dvdy.21909. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Stainier DYR. Out with the old, in with the new: reassessing morpholino knockdowns in light of genome editing technology. Development. 2014;141(16):3103–3104. doi: 10.1242/dev.112003. http://doi.org/10.1242/dev.112003. [DOI] [PubMed] [Google Scholar]
- Schweitzer J, Becker T, Schachner M, Nave KA, Werner H. Evolution of myelin proteolipid proteins: gene duplication in teleosts and expression pattern divergence. Molecular and Cellular Neurosciences. 2006;31(1):161–177. doi: 10.1016/j.mcn.2005.10.007. http://doi.org/10.1016/j.mcn.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB. Rapid reverse genetic screening using CRISPR in zebrafish. Zebrafish. 2016;13(2):152–153. doi: 10.1089/zeb.2015.29000.sha. http://doi.org/10.1089/zeb.2015.29000.sha. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park HC, Topczewska JM, Mawdsley DJ, Appel B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods in Cell Science: An Official Journal of the Society for In Vitro Biology. 2003;25(1–2):7–14. doi: 10.1023/B:MICS.0000006847.09037.3a. http://doi.org/10.1023/B:MICS.0000006847.09037.3a. [DOI] [PubMed] [Google Scholar]
- Sidik H, Talbot WS. A zinc finger protein that regulates oligodendrocyte specification, migration and myelination in zebrafish. Development. 2015;142(23):4119–4128. doi: 10.1242/dev.128215. http://doi.org/10.1242/dev.128215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons T, Appel B. Mutation of pescadillo disrupts oligodendrocyte formation in zebrafish. PLoS One. 2012;7(2):e32317. doi: 10.1371/journal.pone.0032317. http://doi.org/10.1371/journal.pone.0032317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaidero N, Möbius W, Czopka T, Hekking LHP, Mathisen C, Verkleij D, et al. Myelin membrane wrapping of CNS axons by PI(3,4,5)P3-dependent polarized growth at the inner tongue. Cell. 2014;156(1–2):277–290. doi: 10.1016/j.cell.2013.11.044. http://doi.org/10.1016/j.cell.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JL, Kearns CA, Appel B. Fbxw7 regulates Notch to control specification of neural precursors for oligodendrocyte fate. Neural Development. 2012;7:15. doi: 10.1186/1749-8104-7-15. http://doi.org/10.1186/1749-8104-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundarapandian MM, Selvaraj V, Lo UG, Golub MS, Feldman DH, Pleasure DE, Deng W. Zfp488 promotes oligodendrocyte differentiation of neural progenitor cells in adult mice after demyelination. Scientific Reports. 2011;1:2. doi: 10.1038/srep00002. http://doi.org/10.1038/srep00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoveken HM, Hajduczok AG, Xu L, Tall GG. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proceedings of the National Academy of Sciences. 2015;112(19):6194–6199. doi: 10.1073/pnas.1421785112. http://doi.org/10.1073/pnas.1421785112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada N, Kucenas S, Appel B. Sox10 is necessary for oligodendrocyte survival following axon wrapping. Glia. 2010;58(8):996–1006. doi: 10.1002/glia.20981. http://doi.org/10.1002/glia.20981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JC, Amacher SL. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish. 2014;11(6):583–585. doi: 10.1089/zeb.2014.1047. http://doi.org/10.1089/zeb.2014.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JL, Zon LI. Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods in Cell Biology. 2011;105:493–516. doi: 10.1016/B978-0-12-381320-6.00021-7. http://doi.org/10.1016/B978-0-12-381320-6.00021-7. [DOI] [PubMed] [Google Scholar]
- Takada N, Appel B. Identification of genes expressed by zebrafish oligodendro-cytes using a differential microarray screen. Developmental Dynamics. 2010;239(7):2041–2047. doi: 10.1002/dvdy.22338. http://www.ncbi.nlm.nih.gov/pubmed/20549738. [DOI] [PubMed] [Google Scholar]
- Taylor KL, Grant NJ, Temperley ND, Patton EE. Small molecule screening in zebrafish: an in vivo approach to identifying new chemical tools and drug leads. Cell Communication and Signaling. 2010;8:11. doi: 10.1186/1478-811X-8-11. http://doi.org/10.1186/1478-811X-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Halpern ME, Postlethwait JH. Goosecoid expression in neurectoderm and mesendoderm is disrupted in zebrafish cyclops gastrulas. Developmental Biology. 1994;164(2):420–429. doi: 10.1006/dbio.1994.1212. http://doi.org/10.1006/dbio.1994.1212. [DOI] [PubMed] [Google Scholar]
- Voas MG, Lyons DA, Naylor SG, Arana N, Rasband MN, Talbot WS. alphaII-spectrin is essential for assembly of the nodes of Ranvier in myelinated axons. Current Biology. 2007;17(6):562–568. doi: 10.1016/j.cub.2007.01.071. http://doi.org/10.1016/j.cub.2007.01.071. [DOI] [PubMed] [Google Scholar]
- Vogl MR, Reiprich S, Küspert M, Kosian T, Schrewe H, Nave KA, Wegner M. Sox10 cooperates with the mediator subunit 12 during terminal differentiation of myelinating glia. Journal of Neuroscience. 2013;33(15):6679–6690. doi: 10.1523/JNEUROSCI.5178-12.2013. http://doi.org/10.1523/JNEUROSCI.5178-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SZ, Dulin J, Wu H, Hurlock E, Lee SE, Jansson K, Lu QR. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133(17):3389–3398. doi: 10.1242/dev.02522. http://doi.org/10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- Wheeler MA, Smith CJ, Ottolini M, Barker BS, Purohit AM, Grippo RM, et al. Genetically targeted magnetic control of the nervous system. Nature Neuroscience. 2016;19(5):756–761. doi: 10.1038/nn.4265. http://doi.org/10.1038/nn.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods IG, Lyons DA, Voas MG, Pogoda HM, Talbot WS. nsf is essential for organization of myelinated axons in zebrafish. Current Biology. 2006;16(7):636–648. doi: 10.1016/j.cub.2006.02.067. http://doi.org/10.1016/j.cub.2006.02.067. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Faucherre A, Pola-Morell L, Heddleston JM, Liu TL, Chew TL, et al. High-resolution live imaging reveals axon-glia interactions during peripheral nerve injury and repair in zebrafish. Disease Models and Mechanisms. 2015;8(6):553–564. doi: 10.1242/dmm.018184. http://doi.org/10.1242/dmm.018184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ML, Shin J, Kearns CA, Langworthy MM, Snell H, Walker MB, Appel B. CNS myelination requires cytoplasmic dynein function. Developmental Dynamics. 2015;244(2):134–145. doi: 10.1002/dvdy.24238. http://doi.org/10.1002/dvdy.24238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Macklin WB. Oligodendrocyte development and myelination in GFP-transgenic zebrafish. Journal of Neuroscience Research. 2005;81(1):1–8. doi: 10.1002/jnr.20516. http://doi.org/10.1002/jnr.20516. [DOI] [PubMed] [Google Scholar]
- Zalc B, Goujet D, Colman D. The origin of the myelination program in vertebrates. Current Biology. 2008;18(12):R511–R512. doi: 10.1016/j.cub.2008.04.010. http://doi.org/10.1016/j.cub.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Zannino DA, Appel B. Olig2+ precursors produce abducens motor neurons and oligodendrocytes in the zebrafish hindbrain. Journal of Neuroscience. 2009;29(8):2322–2333. doi: 10.1523/JNEUROSCI.3755-08.2009. http://doi.org/10.1523/JNEUROSCI.3755-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaucker A, Mercurio S, Sternheim N, Talbot WS, Marlow FL. Notch3 is essential for oligodendrocyte development and vascular integrity in zebrafish. Disease Models and Mechanisms. 2013;6(5):1246–1259. doi: 10.1242/dmm.012005. http://doi.org/10.1242/dmm.012005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Zhao X, Zheng K, Li H, Huang H, Zhang Z, et al. Genetic evidence that Nkx2.2 and Pdgfra are major determinants of the timing of oligodendrocyte differentiation in the developing CNS. Development. 2014;141(3):548–555. doi: 10.1242/dev.095323. http://doi.org/10.1242/dev.095323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Fu MM, Sloan SA, Ibrahim A, Olson A, Zaremba A, et al. CNS myelin wrapping is driven by actin disassembly. Developmental Cell. 2015;34(2):152–167. doi: 10.1016/j.devcel.2015.06.011. http://doi.org/10.1016/j.devcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]


