Abstract
The tarantula skeletal muscle X-ray diffraction pattern suggested that the myosin heads were helically arranged on the thick filaments. Electron microscopy (EM) of negatively stained relaxed tarantula thick filaments revealed four helices of heads allowing a helical 3D reconstruction. Due to its low resolution (5.0 nm), the unambiguous interpretation of densities of both heads was not possible. A resolution increase up to 2.5 nm, achieved by cryo-EM of frozen-hydrated relaxed thick filaments and an iterative helical real space reconstruction, allowed the resolving of both heads. The two heads, “free” and “blocked”, formed an asymmetric structure named the “interacting-heads motif” (IHM) which explained relaxation by self-inhibition of both heads ATPases. This finding made tarantula an exemplar system for thick filament structure and function studies. Heads were shown to be released and disordered by Ca2+-activation through myosin regulatory light chain phosphorylation, leading to EM, small angle X-ray diffraction and scattering, and spectroscopic and biochemical studies of the IHM structure and function. The results from these studies have consequent implications for understanding and explaining myosin super-relaxed state and thick filament activation and regulation. A cooperative phosphorylation mechanism for activation in tarantula skeletal muscle, involving swaying constitutively Ser35 mono-phosphorylated free heads, explains super-relaxation, force potentiation and post-tetanic potentiation through Ser45 mono-phosphorylated blocked heads. Based on this mechanism, we propose a swaying-swinging, tilting crossbridge-sliding filament for tarantula muscle contraction.
Keywords: Myosin filaments, Myosin heads, Muscle, Myosin interacting-heads motif, Tarantula
Introduction
Our understanding of how muscle contraction occurs, the sliding filament model, is based on two pioneering works (Huxley and Niedergerke 1954; Huxley and Hanson 1954) showing that on contraction the sarcomere shortens by the sliding of two protein filament sets, the thin and thick filaments (see Hitchcock-DeGregori and Irving 2014). A need to understand this model boosted the studies of thin actin-containing and thick myosin-containing filaments. These studies showed that sliding of both filaments was due to heads protruding from thick filaments interacting with thin filaments (see Huxley 2004a, b).
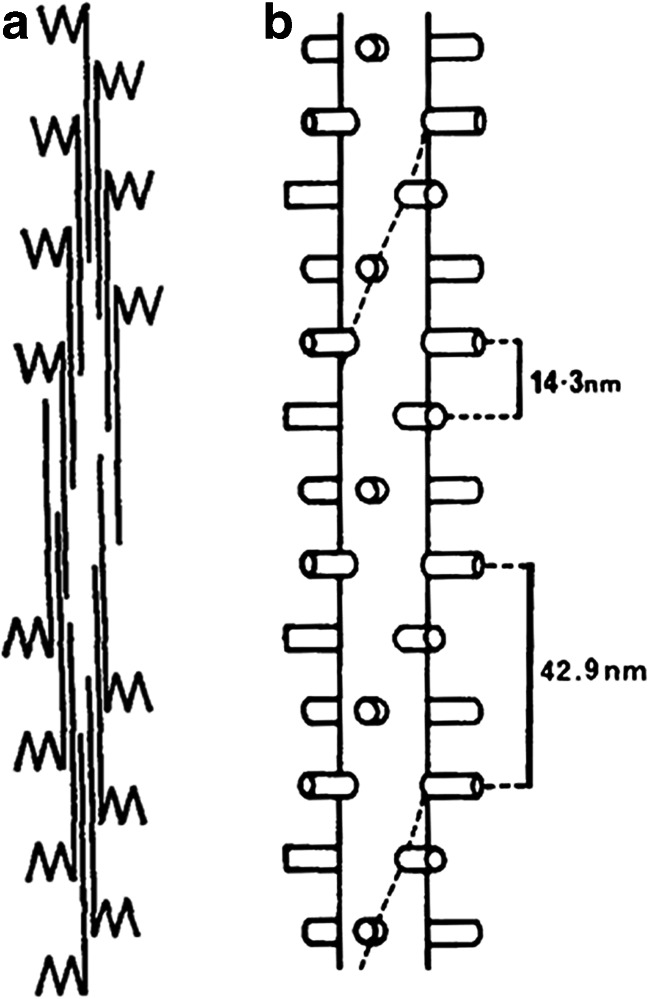
The work of Hugh Huxley in 1963 on isolated thick filaments from rabbit and chicken skeletal muscle by electron microscopy (EM) (Huxley 1963), using an improved negative staining method (Huxley and Zubay 1960), led to his bipolar thick filament model in which the myosin molecules were arranged with their tails packed antiparallel on both sides of a central bare zone forming a straight backbone with heads protruding on its surface (Fig. 1a). Four years later, Huxley reported low-angle X-ray diffraction patterns of frog skeletal muscle showing that heads formed helices with a 14.3-nm separation between “crowns” of two or more heads and a 42.9-nm helical repeat (Fig. 1b) (Huxley and Brown 1967). This model inspired Huxley’s seminal ideas on the mechanism of muscular contraction, which he called the swinging, tilting crossbridge-sliding filament mechanism (Huxley 1969, 2004b) and started the analysis of thick filament structure and function (reviewed by Squire 1975, 1981, 1986, 2009; Barral and Epstein 1999; Craig and Padrón 2004; Squire et al. 2005; Hooper and Thuma 2005; Craig and Woodhead 2006; Hooper et al. 2008; AL-Khayat 2013; Vandenboom 2017).
Fig. 1.
Models of thick filaments proposed by Hugh E. Huxley based on EM of negatively stained thick filaments (a) and low angle X-ray diffraction patterns (b) of skeletal vertebrate muscle. (a) and (b) reproduced with permission from Huxley (1963) and Huxley and Brown (1967) and the Journal of Molecular Biology
Seminal work on the structure of thick filaments was achieved by John Squire who proposed a general model for the arrangement of heads on the thick filament (Squire 1971, 1972, 1975) and models for the packing of myosin tails in the filament backbone as the curved molecular crystal model and the subfilament model (Squire 1973; Chew and Squire 1995). Pioneering work carried out by John Wray provided X-ray diffraction evidences on the helical structure of thick filaments from invertebrate muscles (Wray et al. 1975) and myosin tails packing on subfilaments (Wray 1979), suggesting these subfilaments should be tilted along the filaments axis, except in the horseshoe crab Limulus polyphemus (Arthropoda, Merostomata) and the tarantula Brachypelma sp. (Arthropoda, Arachnida) in which they should run parallel as shown by a characteristic on-equator reflection at 1/4 nm−1 seen in the tarantula X-ray diffraction pattern (Wray 1979, 1982).
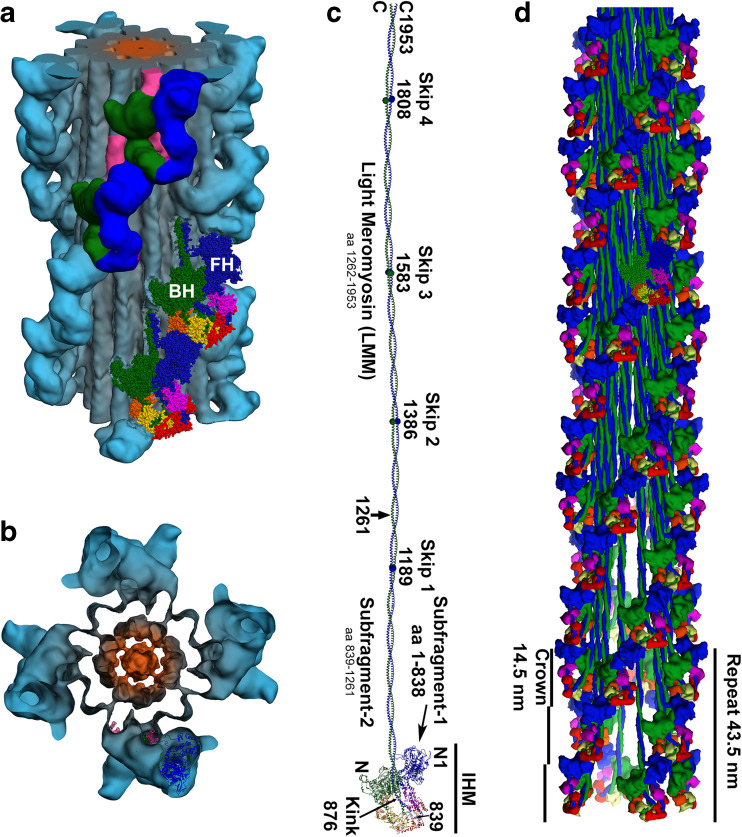
Further advances in thick filament structure came about by the use of a higher resolution EM technique, cryo-EM, which allows observation of rapidly frozen specimens in their native intact state (Fernández-Morán 1965, 1966; Taylor and Glaeser 1974, 1976) (reviewed by Padrón 1999, 2001). Kenneth Taylor and coworkers observed by cryo-EM that the two heads of the myosin heavy meromyosin (HMM) in 2D crystals from chicken smooth muscle make an asymmetric arrangement and suggested how the relaxed (switched OFF) state was achieved (Wendt et al. 1999, 2001; Liu et al. 2003). Taylor and coworkers called the two heads blocked and free heads (Wendt et al. 2001). Soon after, cryo-EM allowed observation of both heads together with its sub-fragment 2 (S2)—missing in the 2D crystals—in tarantula Aphonopelma sp. relaxed striated muscle native thick filament, suggesting that this “interacting-head” structure may underlie the relaxed state of thick filaments in both smooth and striated muscles over a wide range of species (Woodhead et al. 2005). This single molecular entity found in tarantula thick filaments was called the myosin interacting-heads motif (IHM) (Alamo et al. 2008) and its consequences on structure and function are the focus of this review. We review the IHM evolution and its implications on disease in Alamo et al. (2017a).
The myosin II molecule is one of the several members of the myosin superfamily (Coluccio 2008), and is composed by two myosin II heavy chains (MHCIIs), two essential light chains (ELCs) and two regulatory light chains (RLCs) (Sellers 1999; Coluccio 2008). The C-terminus end of the two MHCII coiled-coils forms the myosin tail with the N-terminus extremes arranged on two globular heads, to which a pair of light chains (ELC and RLC) are bound.
Why tarantula muscle?
In spite of diffraction patterns of relaxed muscle that showed heads helically ordered in thick filaments in the sarcomere (Fig. 1b) (Huxley and Brown 1967), electron micrographs of negatively stained thick filaments from relaxed muscle showed that the heads instead were fully disordered (Huxley 1963), probably due to the EM preparative steps. Further diffraction studies by Wray reported a helical order on several invertebrate muscles (Wray et al. 1975; Wray 1979). In 1981, Wray suggested, based on his unpublished patterns (Fig. 2a), that Brachypelma sp. tarantula muscle thick filaments showed evidence of a very well ordered array of heads helices, suggesting tarantula as an ideal specimen for EM. Compared to the rabbit and frog patterns (Huxley and Padrón 1984; Egelman and Padrón 1984), tarantula muscles gave a very detailed pattern, indicating a high degree of helical heads order (Wray 1982). It was 19 years after the first skeletal muscle thick filament electron micrographs that the pioneer EM work by Robert Kensler and Rhea Levine finally succeeded preserving the heads helical order on negatively stained thick filaments from Limulus muscles (Levine et al. 1982), leading to the first published 3D reconstruction of a thick filament (Stewart et al. 1981).
Fig. 2.
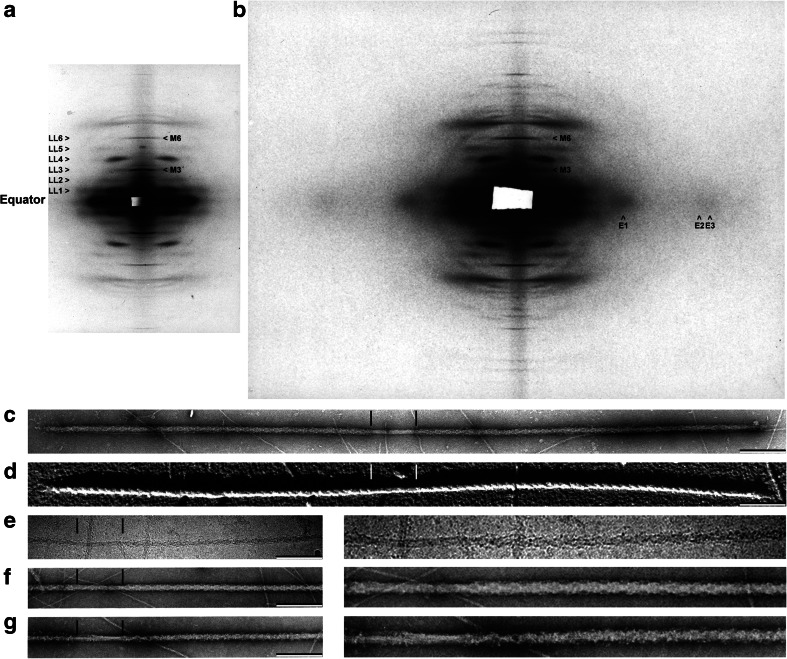
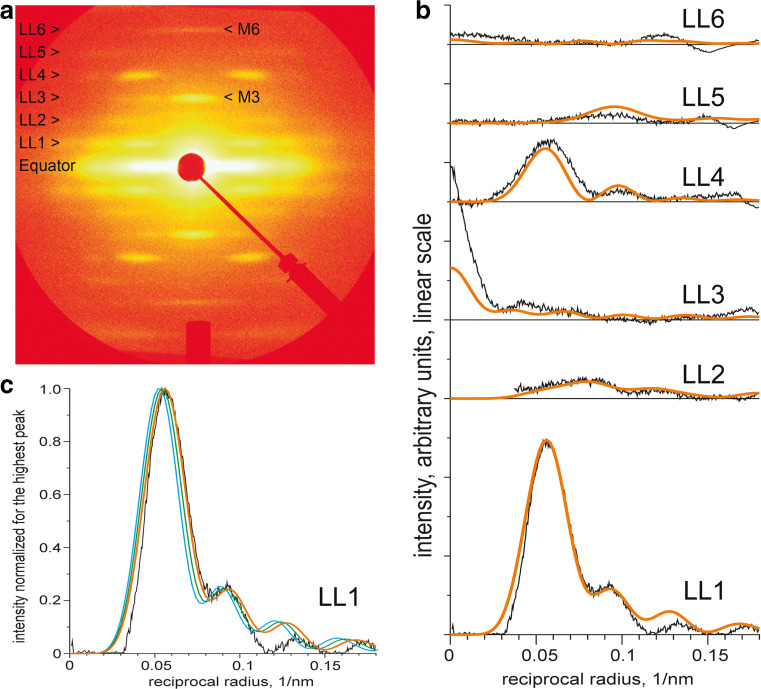
Structural evidence for the thick filament of tarantula striated muscle. a Low-angle X-ray diffraction pattern of demembranated relaxed Brachypelma leg muscle showing layer lines LL1–6 spaced 1/43.5 nm−1 and meridian spots M3 (1/14.5 nm−1) and M6 (1/7.25 nm−1). b Middle-angle pattern showing equatorial reflections E1 (1/4.3 nm−1), E2 (1/2.4 nm-1) and E3 (1/2.0 nm−1). Electron micrographs of isolated relaxed thick filaments from Brachypelma negatively stained (c), metal shadowed (d) or Aphonophelma frozen-hydrated (e) showing oblique helical tracks spaced at 43.5 nm. These helices are established by heads in the relaxed state (f) that become disrupted under Ca2+-activation by Ca2+-activated myosin light chain kinase (MLCK) (g). Insets in (e–g) are zoomed 2 times. The 210-nm bare zone in (c–g) is delimited by vertical lines. Bars 200 nm. (a) and (b) provided by Dr. John Wray, (c) and (d) reproduced with permission from Crowther et al. (1985) and the Journal of Molecular Biology, (e) reproduced with permission from Woodhead et al. (2005) and Nature, (f) and (g) reproduced with permission from Craig et al. (1987) and the Journal of Cell Biology
The pattern of demembranated tarantula relaxed muscle showed six layer lines spaced 1/43.5 nm−1 with two meridian reflections at 1/14.5 nm−1 and 1/7.25 nm−1 (Fig. 2a), indicating the presence of heads in a perfect helical arrangement, organized with a helical repeat of 43.5 nm, exactly three times the axial spacing of 14.5 nm (Wray 1982). These patterns supported the idea that tarantula has an exceptional helically ordered thick filament. In addition, the tarantula middle-angle pattern showed prominent equatorial reflections at 1/4.3 nm−1 (Fig. 2b), suggesting a more clearly visible subfilament structure than in Limulus. All these hints corroborated that the tarantula thick filaments were an excellent specimen to be studied by negative staining and EM as we soon confirmed (Craig and Padrón 1982). Brachypelma sp. muscle thick filaments are very long (4–5 μm) with a centrally located bare zone 20 nm in diameter and 210 nm long, with heads protruding radially up to 32 nm (Fig. 2c) (Crowther et al. 1985). Unidirectionally platinum shadowing showed prominent right-handed helices on these thick filaments (Fig. 2d) (Crowther et al. 1985). Negatively stained thick filaments appeared helically ordered (Fig. 2c), showing oblique tracks at 43.5 nm and transverse banding with a spacing of 14.5 nm corresponding to the repeating levels of heads from four helices (Fig. 2c) (Crowther et al. 1985), (Fig. 2f) (Craig et al. 1987). Thick filaments, analyzed by optical diffraction, showed six layer lines that mimicked closely the X-ray pattern layer lines. Thick filaments exhibited longitudinal striations with ~4 nm side-to-side spacing (Fig. 3a) (Craig and Padrón 1982) consistent with Wray’s tarantula subfilament model (Fig. 3f) (Wray 1979, 1982). A helical 3D reconstruction of these filaments calculated from low electron dose electron micrographs exhibited four helical densities (Fig. 4a1) (Crowther et al. 1985). The thick filaments from tarantulas Eurypelma sp. (Levine et al. 1982, 1983) and Aphonopelma sp. (Hidalgo et al. 2001b, d) were found to be composed mostly by myosin and paramyosin (PM) molecules. This protein composition has simplified the understanding of their structure by X-ray diffraction and EM. In contrast, vertebrate skeletal and cardiac muscle thick filaments lack PM but have other accessory proteins as myosin binding protein C (MyBP-C) and titin.
Fig. 3.
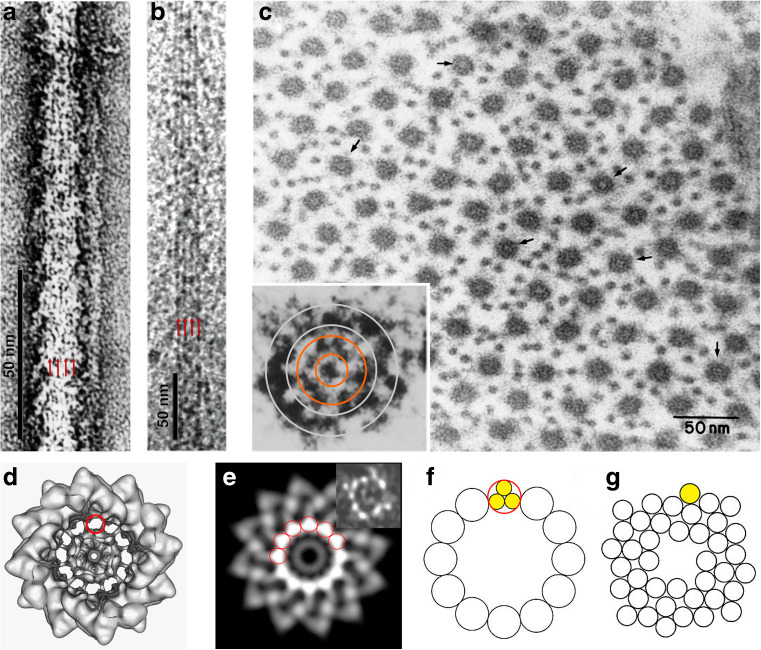
Structural evidence for the tarantula thick filament backbone. Longitudinal views of negatively stained (a) and frozen-hydrated (b) tarantula muscle thick filaments showing longitudinal striations parallel to the filament axis with ~4.0-nm spacing (red arrows) and (c) transverse ultrathin (~30 nm) section of relaxed tarantula muscle, showing sections of filament backbones (black arrows) with an external ring of 12–16 features ~3.9-nm-thick (inset, between gray circles) surrounding a middle ring of 8–11 features ~3.3 nm thick (inset, between orange circles). d Surface rendition of one 43.5-nm repeat of the 3D–recontruction of frozen-hydrated thick filament (Fig. 4a5) showing 12 density tubes interpreted as ~4-nm subfilaments (red circle). e Density projection (contrast reversed: protein white) of this repeat showing an annulus of 12 white features interpreted as “subfilaments” (red circles). The inset shows a projection of an unsymmetrized reconstruction with features similar to subfilaments at a similar radius, in spite of being much noisier. Twelve subfilaments model (red circle) containing three tails (yellow) (f) based on Wray (1979) and (g) curved molecular crystal model based on Squire (1973) for the tarantula thick filament, both with 36 tails. (b), (d) and (e) reproduced with permission from Woodhead et al. (2005) and Nature, inset in (e) personal communication from Dr. Roger Craig
Fig. 4.
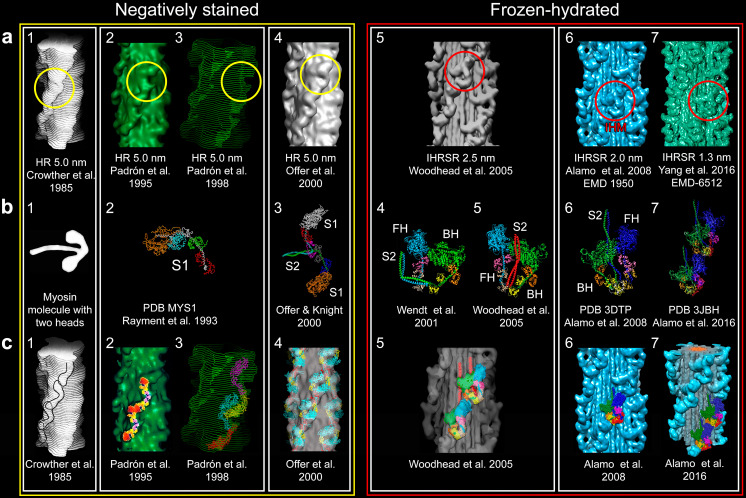
For the elucidation of the tarantula thick filament structure three requisites were needed to be fulfilled: a a better resolution of 3D reconstructions from negatively stained isolated filaments (a1–4 resolution 5.0 nm) or frozen-hydrated (a5 2.5 nm, a6 2.0 nm and a7 1.3 nm) with enhanced 3D reconstruction and visualization techniques (HR helical reconstruction, IHRSR iterative helical real space reconstruction), b the myosin head structure (b2) (PDB MYS1) and improved two-heads (b3–7) models and (c) fitting approaches ranging from eye-fitting (c1), 2D fitting (c2), envelope 3D fitting (c3), density rigid 3D fitting (c4, 5) and flexible 3D fitting (c6, 7). The unambiguous interpretation that lead to the myosin interacting-heads motif (IHM) (Woodhead et al. 2005) (a5, red circle) in the relaxed tarantula thick filament, belonging to the Class I proposed by Squire et al. (2005) and Squire (2009), required a resolution higher that 2.5 nm achieved by the IHRSR technique, frozen-hydrated specimens (a5) and an asymmetric head-pair model (b4) (Wendt et al. 2001), but with the S2 properly positioned (b5). In retrospect, the motif (red circles) was present and could be picked out on the 5-nm resolution 3D maps using the right density cutoffs (yellow circles). Bare zone is at the top here as well as in Figs. 6, 8, 9, and 10. Myosin sub-fragment 1 (S1), sub-fragment 2 (S2), free head (FH), blocked head (BH). The following images were reproduced with permission as follows: a1, c1 Crowther et al. (1985) and the Journal of Molecular Biology, a2, c2 Padrón et al. (1995) and the Journal of Structural Biology, c4 Offer et al. (2000) and the Journal of Molecular Biology, a5, b5, c5 Woodhead et al. (2005) and Nature, and c7 Alamo et al. (2016) and the Journal of Molecular Biology
Lesson I: the IHM arrived unexpectedly
The search for the arrangement of heads in the tarantula thick filament
In 2005, Squire conceptualized four Classes of configurations for myosin heads organization on the relaxed thick filament surface according to how the head-to-head interactions stabilized the helical structures in resting muscle: in Class I, the two heads of one myosin molecule might interact with each other in a parallel fashion; in Class II the two heads might separate laterally and interact with a head from an adjacent myosin molecule in the same crown; in Class III the two heads might separate in an antiparallel or splayed way with heads of successive crowns making interactions between them; and in Class IV the heads do not interact with each other at all (Squire et al. 2005; Squire 2009).
Negative staining EM did not resolve both heads in 3D reconstructions of tarantula thick filament
The first helical 3D–reconstruction of tarantula thick filaments (Fig. 4a1) reached a 5.0-nm resolution (Crowther et al. 1985). Following the proposal of John Haselgrove, based on X-ray diffraction analysis, the 3D map densities were interpreted as repeated features containing two equally sized domains springing from a common region on the backbone which looks very much like the individual myosin molecules images seen in shadowed preparations (Fig. 4b1) (Elliott and Offer 1978) or in negatively stained filaments with extended heads (Knight and Trinick 1984). This suggested that the heads were pointing in opposite directions along the thick filament axis (Haselgrove 1980; Levine et al. 1982). Using improved techniques, the 5.0-nm resolution thick filament 3D map was enhanced (Padrón et al. 1995, 1998; Offer et al. 2000) (Fig. 4a2–4). Once the atomic structure of the myosin head sub-fragment 1 (S1) PDB MYS1 was reported (Rayment et al. 1993) (Fig. 4b2), the 2D shape and 3D envolvent fittings using heads pointing to opposite directions was attempted (Padrón et al. 1995, 1998) (Fig. 4c2, 3). The availability of a two-head model including the head-tail junction for the myosin molecule (Fig. 4b3) (Offer and Knight 1996) allowed fitting it to the 3D reconstruction densities (Offer et al. 2000) (Fig. 4c4). These four initial fitting attempts (Fig. 4c1–4) were ambiguous since at a 5-nm resolution the fitting did not allowed resolving the densities of both heads which were assumed to be identical. The suggested arrangement, in which the heads between adjacent crowns interacts between them along the long-pitched helices, is called the “Class III” model (Squire et al. 2005; Squire 2009). Therefore, by the year 2000, the tarantula thick filaments were thought to be of Class III although a higher resolution than 5.0 nm was required for an unambiguous fitting.
Negative staining EM did not resolve both heads in thick filament 3D reconstructions of other species
Similar interpretations were suggested from other arthropods negatively stained thick filament 3D reconstructions such as Limulus (Stewart et al. 1981, 1985; Levine et al. 1982) and scorpion Vejovis spinigera (Arachnida) (Kensler et al. 1985; Stewart et al. 1985), and from the thick filament 3D reconstruction of the abductor muscle in the scallop Placopecten magellanicus (Mollusca) (Vibert and Craig 1983). The number of helices in the thick filaments was found to be either 3 in vertebrates (frog, Kensler and Stewart 1983; rabbit, Kensler and Stewart 1993), 4 in arthropods (Limulus, (Stewart et al. 1981; tarantula, Crowther et al. 1985; Padrón et al. 1993a; and scorpion, Stewart et al. 1985) or 7 in mollusks (scallop, Vibert and Craig 1983; Craig et al. 1991). Other EM studies on negatively stained thick filaments have been reported for invertebrates such as the indirect flight muscle (IFM) of the giant water bug Lethocerus indicus (Insecta) (Clarke et al. 1986; Morris et al. 1991), as well as for skeletal muscle in several vertebrates such as goldfish Carassius auratus (Kensler and Stewart 1989; Eakins et al. 2002), frog Rana pipiens (Kensler and Stewart 1986; Stewart and Kensler 1986), chicken (Kensler and Woodhead 1995) and rabbit (Kensler and Stewart 1993). However, none of these studies achieved enough resolution to resolve both heads in the thick filaments 3D reconstruction. Therefore, by the year 2000, all the invertebrate and vertebrate thick filaments studied were thought to be of Class III (Squire et al. 2005).
Cryo-EM studies finally allowed resolving both heads
To increase 3D reconstruction resolution to resolve both heads, attempts were carried out to capture thick filament structure by low-temperature methods as rapid freezing using liquid helium and nitrogen cryogens and freeze substitution (Fernández-Morán 1960), modified for muscle studies (Padrón et al. 1988). Rapidly frozen, freeze-substituted muscle of tarantula Avicularia avicularia allowed the observing of helices of heads directly in thick filaments in the sarcomere (Padrón et al. 1992) and the calculating of helical 3D reconstruction in situ (Padrón et al. 1995). Rapidly frozen frog sartorius muscle slammed against a copper block cooled by liquid helium (Padrón et al. 1988) allowed the observation of head helices (Craig et al. 1992) and MyBP-C (Luther et al. 2008, 2011). It was concluded that with low-temperature techniques it was possible to preserve the thick filament helically ordered structure as seen on diffraction patterns (Fig. 2a). The next step to increase the resolution was to avoid freeze-substitution by direct rapid plunge–freeze copper grids containing a thin layer of filament homogenate to obtain a very thin layer of frozen-hydrated relaxed helically ordered filaments. Grids, kept in liquid nitrogen, were transferred to a liquid nitrogen cryo-holder and inserted in a liquid nitrogen cryo-electron microscope for recording electron micrographs at low electron dose. The first electron micrographs of frozen-hydrated helically ordered thick filaments were achieved in Lethocerus IFM by Menetret et al. (1988) and later in scallop (Vibert 1992). Nevertheless, achieving helically ordered tarantula thick filaments was very difficult as the concentration of thin filaments in the filament homogenate was very high. Therefore, devising an appropriate purification procedure, using gelsolin to disrupt the thin filaments, was needed to produce a purified preparation of native tarantula Aphonopelma thick filaments (Hidalgo et al. 2001c). This allowed the recording of the first electron micrographs of frozen-hydrated tarantula helically ordered thick filaments (Fig. 2e) and to calculate its helical 3D reconstruction by 2003. Regrettably, it was not possible to separate the overlapping Bessel functions on the layer lines as required to calculate a helical 3D recontruction which was possible for the more highly contrasted negatively stained specimens (Crowther et al. 1985). Apart from successful cryo-EM of intact near in vivo isolated frozen-hydrated helically ordered tarantula thick filaments (Fig. 2e), the essential remaining step was to calculate the 3D recontruction by using the robust iterative helical real space reconstruction (IHRSR) algorithm based on single-particle methods developed 3 years earlier (Egelman 2000) instead of helical 3D reconstruction. This last crucial step finally allowed the calculation of three similar IHRSR 3D maps by mid-2004, at last achieving enough resolution (2.5 nm) to finally resolve both heads unambiguously (Fig. 4a5) (Woodhead et al. 2005).
The myosin interacting-heads motif (IHM)
The arrangement of both heads on the tarantula striated muscle thick filament (Fig. 4a5) (Woodhead et al. 2005) unexpectedly showed the asymmetric densities found in vertebrate smooth muscle by Taylor and coworkers (Fig. 4b4) (Wendt et al. 1999, 2001; Liu et al. 2003). Resolving the tube density joining the two heads densities was the critical step to interpret the two heads densities (Fig. 4a5) (Woodhead et al. 2005). Roger Craig interpreted the asymmetric densities of the frozen-hydrated 3D reconstruction (Fig. 4a5) as shown in Fig. 4c5, by properly repositioning the S2 in the asymmetric model for chicken smooth muscle HMM 2D crystals proposed by Taylor and coworkers (Fig. 4b4 versus b5) (Wendt et al. 1999, 2001; Liu et al. 2003). The motif unambiguously fitted the 2.5-nm tarantula thick filament 3D reconstruction densities, including the S2 density tube (Fig. 4c5) (Woodhead et al. 2005) which was missing in the 2D crystal densities. This entity was later named the myosin interacting-heads motif (IHM) (Alamo et al. 2008). For an account of how the IHM structure was achieved see Padrón (2013).
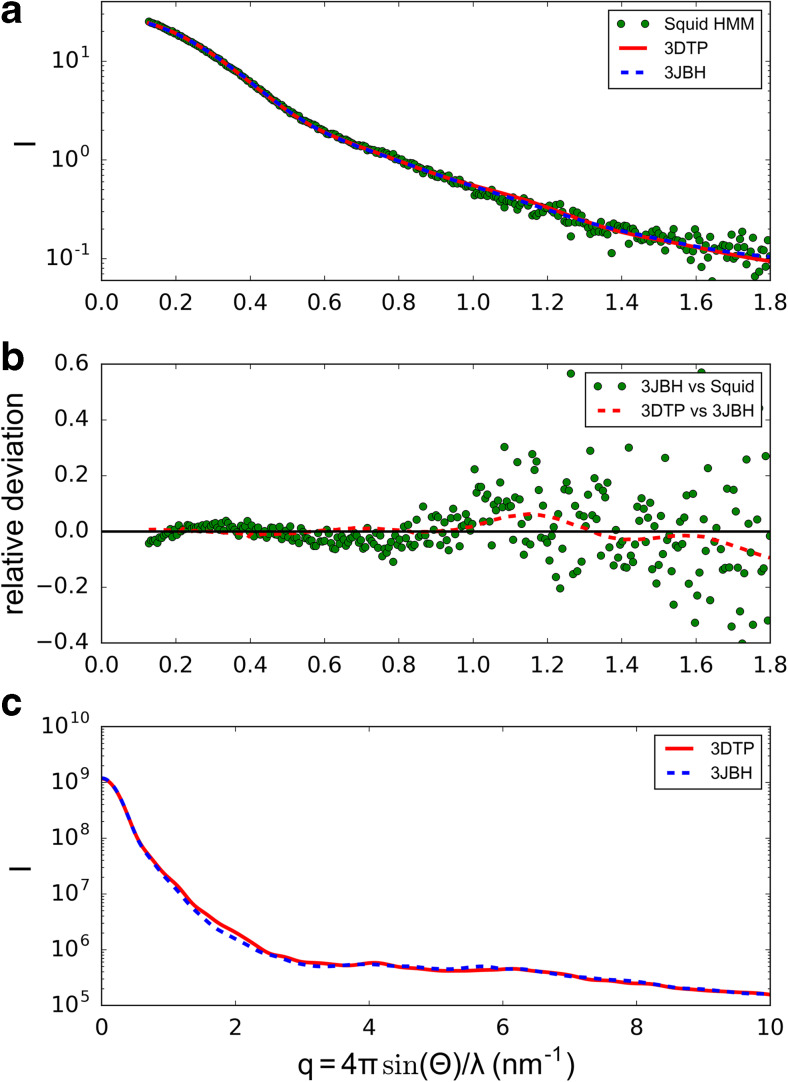
In contrast to all previous EM studies, the two heads arrangement was of the Class I proposed by Squire (Squire et al. 2005; Squire 2009) and not of Class III as previously thought for all species. A further increase on resolution up to 2.0 nm was quickly achieved by improving the IHRSR 3D reconstruction method (Fig. 4a6) (Alamo et al. 2008). This 2.0-nm 3D reconstruction showed an additional density in the myosin RLC region due to the much larger tarantula RLC N-terminal extension (NTE) mass (Aphonopelma sp. 5671.4 Da). Then, a hybrid IHM model was built (PDB 3DTP) (Fig. 4b6) that fitted better this 3D–reconstruction (Alamo et al. 2008). The model included the smooth muscle chicken S1 PDB 1I84 model (Liu et al. 2003), the human cardiac S2 crystal structure PDB 2XFM (Blankenfeldt et al. 2006) and a homologous RLC structure using the tarantula A. avicularia RLC sequence (GenBank EU090070) and its predicted NTE secondary structure. A single species homologous tarantula myosin IHM (PDB 3JBH) (Fig. 4b7) built using the tarantula Aphonopelma sp. MHCII (KT619079) (Alamo et al. 2016), ELC and RLC (KT390185 and KT390186) (Zhu et al. 2009) sequences fitted to the 2.0-nm 3D reconstruction was recently reported (Fig. 4c7) (Alamo et al. 2016). A higher resolution (1.3 nm) has been achieved for the frozen-hydrated tarantula thick filament by improvements on the preparation of the filaments and the use of electron detectors (Fig. 4a7) (Yang et al. 2015a, b). Small-angle X-ray solution scattering (SAXS) studies of isolated squid HMM (Loligo peaelii), conducted in both the absence and presence of the regulatory cation Ca2+, showed a clear distinction between a compact “off” state and an extended “on” state. The pair distance distribution function (PDDF) derived from the SAXS data for squid HMM goes from being unimodal in the Ca2+-free “off” state to bimodal in the Ca2+-bound “on” state. This result implies a Ca2+-mediated transformation from a compact state to an extended state in which some domains have become spatially separated (Gillilan et al. 2013). In addition, the computed scattering profiles for 3DTP and 3JBH models (Fig. 5a) closely agree with the measured scattering profile of squid HMM in Ca2+-free (EGTA) conditions (Alamo et al. 2016). These results strongly inferred the IHM presence on squid and strengthened the evidence for an evolutionary ancient compact off state (Gillilan et al. 2013).
Fig. 5.
a Calculated small-angle X-ray solution scattering (SAXS) profile of PDB 3DTP (red line) and 3JBH (blue dashed line) matches the experimental squid heavy meromyosin SAXS profile (green dots) (Gillilan et al. 2013). Integrated scattering intensity (I in arbitrary units) is given as a function of momentum transfer, q = 4π sin(θ)/λ, with a scattering angle of 2θ and a wavelength of λ. b Relative deviation between squid HMM experimental data and PDB 3JBH (green dots) is calculated as (Imodel-Iexp)/Iexp. The corresponding difference in scattering between PDB 3DTP and PDB 3JBH models is also shown on the same scale (red dashed line). Comparison shows that the models cannot be distinguished based on currently available scattering data. c Further calculations of PDB 3DTP and PDB 3JBH show that the scattering of the two models is nearly identical at wide angles. SAXS and error calculation described in Alamo et al. (2017b)
The search for the arrangement of myosin tails in the tarantula thick filament backbone
The frozen-hydrated 3D map showed 12 ~4-nm tubular densities parallel to the filament axis (Fig. 3d) (Woodhead et al. 2005), interpreted as if each tube contained more than one 2-nm tails forming 12 subfilaments (Woodhead et al. 2005). The projected density of a 43.5-nm repeat confirmed this interpretation (Fig. 3e) (Woodhead et al. 2005). It was also consistent with parallel striations to the axis observed in Brachypelma sp. negatively stained thick filaments (Fig. 3a) (Craig and Padrón 1982; Levine et al. 1983; Crowther et al. 1985) and Aphonopelma sp. frozen-hydrated thick filaments (Fig. 3b) (Woodhead et al. 2005), as well as with backbone features observed in ultrathin sections of muscle from other tarantulas such as Eurypelma sp. (Levine et al. 1983) and A. avicularia (Guerrero and Padrón 1992) (Fig. 3c) and with A. avicularia thick filament fraying in four features only near the filament tips (Padrón et al. 1993b) (cf. 3 subfilaments all along the vertebrate thick filaments (Maw and Rowe 1980). This evidence and the ~1/4-nm−1 equatorial reflection (Fig. 2b) suggested that, in tarantula thick filament, possibly three tails pack together on a subfilament, and that 12 subfilaments form the backbone outer part (Figs. 3b, 6a, gray). Twelve tubes are seen in Limulus, Tityus and Schistosoma four-stranded filament 3D reconstructions, while 21 or 9 subfilaments are expected in the seven-stranded or three-stranded scallop and cardiac muscle as proposed by Wray’s subfilament model (Fig. 3f) (Wray 1979). Nevertheless, the recent near-atomic (0.55 nm) resolution achieved for the Lethocerus IFM frozen-hydrated thick filament of Hu et al. (2016) with sufficient resolution in the region of the backbone where the myosin tails pack, enabled the tracking of the myosin tail α-helices, showing unambiguously for the first time that the tails pack in a layer molecular crystal arrangement proposed 44 years ago by Squire (1973) (Fig. 3g). Additional near-atomic cryo-EM studies should reveal if tails in the backbone of tarantula thick filaments are packed on 12 subfilaments as proposed by Wray (1979) and as supported by evidence in Fig. 3a–e or if it is as in Lethocerus.
Fig. 6.
Longitudinal (a) and transverse (b) views of frozen-hydrated tarantula thick 3D reconstruction, filtered to 2-nm resolution (EMD-1950) showing four IHMs helices (blue), packed tails (gray), and PM core (orange). The 3D reconstruction shows four 14.5-nm crowns, each with four IHMs. Densities of two IHMs (top) are shown, with blocked (BH, green) and free (FH, blue) heads and its subfragment 2 (S2, magenta). The model 3JBH (bottom), made from two adjacent IHMs models, shown as spheres, was flexibly fitted to the map. Blocked and free heads MHCIIs in green and blue, ELCs in magenta (FH) and orange (BH), and RLCs in red (FH) and yellow (BH). c Tarantula myosin II molecule based on 3JBH plus tail built with tarantula MHCII sequence (GenBank KT619079). d Tarantula pseudo-filament model built with tarantula myosin II molecules in (c)
The search for the arrangement of PM rods in the tarantula thick filament backbone
The tarantula thick filament contains PM (Aphonopelma GenBank KT692662), an accessory protein present only on invertebrate muscles. The PM to myosin (M) heavy chain ratio (PM/M) in tarantula muscle is 0.31–0.37 (Levine et al. 1983; Hidalgo et al. 2001b). PM molecules are thought to occupy the core of most invertebrate filaments serving as scaffolding for supporting the myosin tails above it on very long filaments (Fig. 6a, orange). The tarantula myosin and PM heavy chain sequences (Aphonopelma GenBank KT619079 and KT692662, respectively), the PM/M ratio and the measurement by quantitative scanning transmission electron microscopy (STEM) of the mass/length M/L ratio of isolated tarantula thick filaments in the bare zone (M/L ~99 kDa/nm) and the heads region (M/L ~174 kDa/nm) (Hidalgo et al. 2001b, d) are important constrains that will help solving the tarantula and Lethocerus PM core substructure by further near-atomic or atomic resolution cryo-EM studies (cf. Hu et al. 2016).
Future search for IHM on thick filament and HMM by EM and SAXS studies
Further EM studies using methods to isolate and visualize relaxed native thick filaments (Craig 2012) should take into account the bound nucleotide state (Clarke et al. 1986; Craig and Padrón 1989), the pre-power stroke state head conformation (Zoghbi et al. 2004; Llinas et al. 2015) and the blebbistatin stabilizing effect (Zhao et al. 2008). IHM detection in only one half of a single thick filament in the tarantula Gramostola rosea using electron tomography could be a quick alternative EM approach for IHM detection (Márquez et al. 2014).
Solving the IHM near atomic and the HMM atomic structures
Crystallization of squid HMM suggests that its atomic structure should eventually be solved (O’Neall-Hennessey et al. 2013). However as a HMM crystal includes only one IHM, the atomic structure of the intermolecular IHM–IHM and IHM–backbone interactions cannot be achieved by crystallography unless a two-interacting HMMs crystal could be crystallized, similar to the 3JBH model (cf. Fig. 6a). Achieving a near-atomic resolution HMM structure by near-atomic cryo-EM should be possible by single particle averaging of frozen-hydrated HMMs (Yang et al. 2017). However, frozen-hydrated interacting HMM dimers should be required to achieve near atomic information of the IHM–IHM intermolecular interactions. In consequence, the only way IHM–IHM and IHM–backbone intermolecular near-atomic information could be simultaneously achieved is by obtaining the near-atomic resolution IHM structure in situ actually forming IHM–IHM helices and IHM–backbone interactions on frozen-hydrated thick filaments. However achieving the near-atomic resolution IHM structure on frozen-hydrated thick filaments seems to be a difficult task on view that only ~2-nm resolution has been achieved on the heads region of the 0.55-nm near-atomic resolution Lethocerus 3D map (Hu et al. 2016). This is probably a consequence of the intrinsic flexibility of the IHM paired heads arrangement (Yang et al. 2015a) especially of the free swaying heads as explained below (Brito et al. 2011; Sulbarán et al. 2013). Interestingly, it has been recently reported that a lack of negative charge in S2 may explain why Lethocerus thick filaments display an IHM perpendicular to the filament axis, rather than a parallel one as seen in tarantula (Fee et al. 2017).
In retrospect, both head densities resolved in frozen-hydrated 3D reconstructions (Fig. 4a5–7, red circles) were also present in negatively stained ones (Fig. 4a1–4, yellow circles) as in fact can be seen with a higher density cutoff (cf. Fig. 4a4) (Padrón and Alamo 2004). The lack of a clearly resolved head–tail junction density in 3D reconstructions together with the wrong assumption that both heads were identical and symmetric structures pointing oppositely delayed 20 years the right interpretation of negatively stained 3D reconstructions in tarantula and other species. The unambiguous IHM presence seen in the negatively stained scorpion thick filament (Pinto et al. 2012) clearly demonstrated that resolving both heads was indeed possible without cryo-EM. So the lesson was finally painfully learned: assigning a Class III model to the tarantula thick filament was a mistake caused by interpretations that were beyond the 3D map resolutions, as these filaments were in fact Class I as envisioned by Squire et al. (2005).
It took 42 years from Huxley proposing his thick filament model (Fig. 1a) (Huxley 1963) to start achieving an understandable quasi-atomic-level of it (Woodhead et al. 2005). Tarantula muscle X-ray diffraction and EM studies were crucial for it. Diffraction studies suggested that tarantula should provide the best ordered thick filaments for EM studies (Fig. 2a). Improvements on isolation, EM and 3D reconstruction of invertebrate thick filaments (Craig 2012) and on preservation of the heads helical arrangement by rapid freezing (Padrón and Alamo 2004) allowed tarantula filament native preservation in relaxed conditions. Recording of low electron dose micrographs by cryo-EM together with the IHRSR method (Egelman 2000) led to a 3D reconstruction with enough resolution to resolve both heads and S2 disclosing the tarantula IHM quasi-atomic structure.
Lesson II: the IHM at work: the two myosin II heads work better in cooperation
Soon after the helical arrangement of heads was detected in the relaxed state (i.e. Mg.ATP presence, low [Ca2+]) in tarantula muscle by X-ray diffraction (Fig. 2a) and by EM on negatively stained thick filaments (Fig. 2c ,d), it was shown by EM that RLC phosphorylation induced loss of heads helical order (Fig. 2g vs. f) (Craig et al. 1987) and by equatorial X-ray diffraction patterns that heads moved out by ~6 nm (Pante et al. 1986; Sosa et al. 1986, 1988; Panté et al. 1988; Padrón et al. 1991). The RLC phosphorylation status was assessed by urea/glycerol polyacrylamide gel electrophoresis (PAGE) suggesting that two different RLCs were present and that both could be phosphorylated potentiating contraction (Craig et al. 1987). However, further studies showed that the phosphorylation mechanism included only a single RLC that can be mono- or biphosphorylated (Hidalgo et al. 2001a). This evidence triggered interest on understanding the basis of the IHM relaxed state and how both heads move away and disorder upon RLC phosphorylation on thick filament activation.
The earlier tarantula IHM 3DTP quasi-atomic model
IHM structure imposes different heads environments explaining heads ordered release on activation
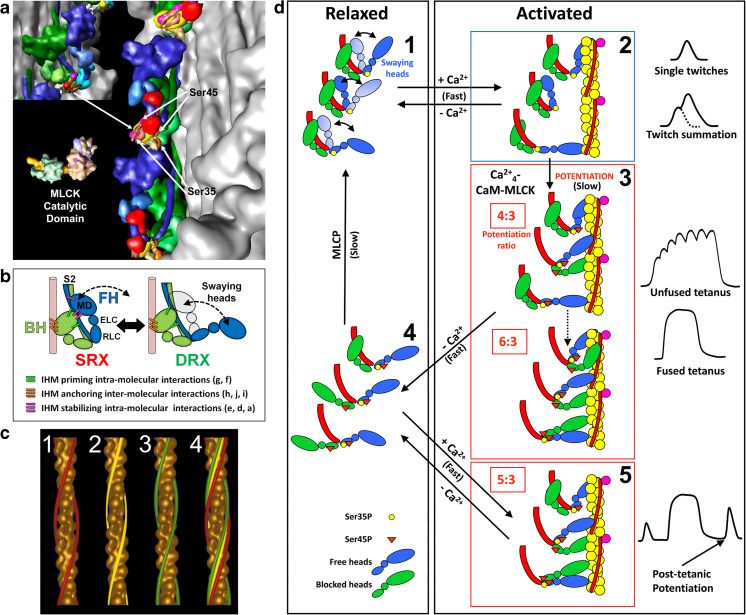
The 3DTP model has both IHM heads asymmetrically arranged; so their RLC phosphorylatable serines, Ser35 and Ser45, were not similarly exposed to its specific kinases, protein kinase C (PKC) and myosin light chain kinase (MLCK) (Alamo et al. 2008; Brito et al. 2011) (Fig. 7a). These differences preset the order in which these heads are released on phosphorylation and activation. The free head, which has the appropriate conformation and is properly located, is released first so that it can interact with actin in the thin filament; followed by the blocked head, if required (Alamo et al. 2008).
Fig. 7.
Functional implications of the tarantula asymmetric IHM 3DTP model on the relaxation and activation of thick filaments. a The IHM asymmetric structure 3DTP (inset) produces different environments for the myosin regulatory light chain (RLC) NTEs of the free (blue) and blocked (green) heads containing phosphorylatable Ser35 and Ser45. This enables the MLCK to access only the serines of the free head. b The constitutively mono-phosphorylated Ser35 on the free heads are ready to interact with the Ca2+-switched on thin filament through the free head swaying mechanism involving the formation and disruption of intramolecular stabilizing interactions (magenta bars) between the swaying free head and the blocked head and interconnecting intermolecular interactions (Fig. 9b). The un-phosphorylated blocked head is docked onto its own S2 by priming intramolecular interactions (green bars) and with its neighbor myosin tail with anchoring intermolecular interactions (orange bars) (Alamo et al. 2016). c Tarantula thin filament is Ca2+- activated according to the steric model of muscle contraction (Huxley 1973) shown by tropomyosin (TM) movements (c4) on thin filament 3D–reconstructions in relaxed (c1, TM strands, red), activating (c2, yellow) and rigor (c3, green). d Model for activation (d1–d2), potentiation (d2–d3), post-tetanic potentiation (d3–d5) and relaxation (d2–d1 and d4–d1) for tarantula striated muscle dual thin and thick filaments regulation. (a) and (d) reproduced with permission from Brito et al. (2011) and the Journal of Molecular Biology, (c) reproduced with permission from Craig and Lehman (2001) and the Journal of Molecular Biology
RLC phosphorylation controls heads release explaining Ca2+-controlled activation mechanism
The Ser35 of free heads are constitutively phosphorylated by PKC while Ser45 can be phosphorylated by MLCK and dephosphorylated by myosin light chain phosphatase (MLCP) (Fig. 7a). (Brito et al. 2011; Sulbarán et al. 2013). In the relaxed state, the blocked head is immobilized as it is docked onto its S2 and neighbor myosin tail, whereas the free head is docked to the blocked head and allowed to sway away (Fig. 7b). This makes the free head capable of interacting with Ca2+-activated thin filaments (Fig. 7c2) on single twitches and twitch summation (Fig. 7d1,2). When [Ca2+] remains long enough above the threshold MLCK is activated by Ca2+, bi-phosphorylating the constitutively Ser35 mono-phosphorylated free heads and mono-phosphorylating the blocked heads recruiting them to permit force potentiation on unfused or fuse tetani (Fig. 7d3) explaining post-tetanic potentiation (Fig. 7d3–5) (Ranke 1865). So the working in cooperation of the two heads in myosin II working in cooperation is crucial to allow these specialized physiological responses of skeletal muscle to sustain relaxation and control the force produced during contraction. This model for activation, potentiation and pos-tetanic potentiation (Brito et al. 2011; Sulbarán et al. 2013) explains its cooperativity so it was called the cooperative phosphorylation-activation (CPA) mechanism (Sulbarán et al. 2013).
The latest tarantula IHM 3JBH quasi-atomic model
The improved tarantula 3JBH model (Fig. 6a) was fitted to the 2.0-nm frozen-hydrated Aphonopelma sp. 3D map and is formed by two adjacent IHMs (Alamo et al. 2016) so it includes their interconnecting intermolecular interactions. Tails (Fig. 6c) pack together (Fig. 6a, gray) around a central PM core (Fig. 6a, orange) forming the backbone with IHMs (blue) protruding on four helices (Fig. 6b). The close agreement between the latest 3JBH quasi-atomic model (Alamo et al. 2016) and the experimental squid HMM SAXS data (Fig. 5) strengthens its value as a utilizable IHM model. The layer-lines intensities LL1–6 calculated from the pseudo-filament model (Fig. 6d) match well the intensity profiles of the tarantula muscle X-ray diffraction pattern layer lines (Fig. 8). This suggests that in intact relaxed tarantula muscle, heads are organized as in the pseudo-filament model (Fig. 6d) with IHMs protruding ~2 nm above the backbone.
Fig. 8.
a Low-angle synchrotron radiation X-ray diffraction pattern (60 s exposure) of skinned relaxed tarantula Brachypelma sp. leg muscle showing layer-lines LL1–6 and meridian spots M3 and M6. b Intensity profiles of LL1–6 (black lines) from this pattern. The LL1–6 intensities (orange) calculated from the tarantula thick pseudo-filament model (Fig. 6d) using the 3JBH-based tarantula myosin molecule model (Fig. 6c) matches well the experimental intensities up to ~1/0.125 nm−1. c Effect on the LL1 profile of a small shift (green: +0.5 nm, cyan: +1.0 nm) in the radial position of the IHM. This suggests that in intact relaxed tarantula muscle heads are organized as in the pseudo-filament model (Fig. 6d) with IHMs protruding ~2 nm above the backbone. Fourier transforms F1 on the l-th layer line of the heads were calculated according to the formula F1 (R,ψ) = Σn G nl (R) exp (−inψ) (Vainstein 1963) where the Fourier–Bessel structural factor G nl is G nl (R) = Σj f jJ n(2πRr j) exp(i[n(π/2 – φj) + 2π z jl/c]), f j is the structural factor of j-th amino acid (number of electrons); r j, ψj, z j are polar coordinates of Cα atom of j-th amino acid (cylindrical coordinates r, ψ, z where the z-axis coincides with the axis of the thick filament); J n is the nth order Bessel function of the first kind; R, ψ are the radial and azimuthal coordinates in the reciprocal space; and c = 3 × 145 Å = 435 Å is the axial size of the unit cell. Layer line intensities were calculated as the azimuthally averaged square of the Fourier transform (Vainstein 1963) as Il = Σn | Fnl|2 with –NB < n < NB, NB = 20. X-ray diffraction patterns in (a) kindly provided by Dr. John Wray
Interactions form IHMs and their helical tracks in the relaxed state
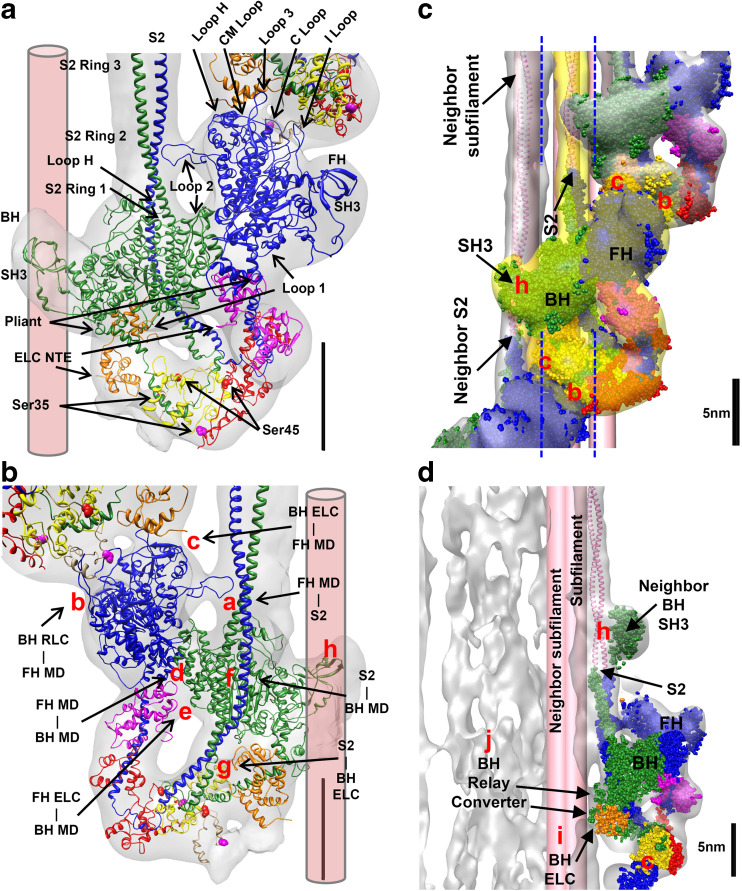
Model 3JBH (Figs. 6a, 9a) improves the earlier 3DTP model since it is built using only Aphonopelma sp. sequences instead of sequences from three species. In addition to three loops (loop 1, loop 2 and loop near ATP pocket V) missing in 3DTP, 3JBH includes all myosin loops (loop H, CM loop, loop 3, C loop and I loop) involved on five intramolecular interactions “a”, “d”, “e”, “f” and “g” (Fig. 9b) and five intermolecular interactions “b”, “c”, “h”, “i” and “j” (Fig. 9c, d) as well as the S2, SH3, catalytic, relay, and converter (Alamo et al. 2016). In the relaxed state, S2 of the IHM emerges from the bare zone side with a slight angle of ~6° (i.e. approximately parallel to filament axis), causing IHMs helices to “float”, separated from the surface by ~2 nm (Fig. 9d). The blocked head is the only part of the IHM that is in contact with the backbone and is covalently connected to it via the S2 and electrostatically connected by the three “anchoring” intermolecular interactions: “h”, “i” and “j” with the neighboring S2 (Fig. 9c).
Fig. 9.
The tarantula IHM 3JBH quasi-atomic model shown flexibly fitted to 3D reconstruction (in gray) of frozen-hydrated relaxed tarantula thick filament (EMD-1950) (Alamo et al. 2008). The model is made by a free (FH, blue) and blocked (BH, green) heads, seen from its back in (b) and is established by five intramolecular interactions: two priming interactions “f” and “g” between the blocked head and the S2 (BH–S2 precursor) and three stabilizing interactions “a”, “d” and “e” between the free head and the blocked head and S2, that keep the IHM stable. The four helices of IHMs above the backbone surface (Figs. 6a, 7a, 9d) are established by five inter-molecular interactions: the heads along each helix are connected by two inter-chaining intermolecular interactions “b” and “c” established between the free head motor domain (MD) and the RLC and ELC of the neighboring blocked head regulatory domain shown in (c) and maintained above the backbone surface by three anchoring intermolecular interactions: “h” with the neighboring S2 shown in (b–d), and “i” and “j” with a neighbor “subfilament” shown on the 90° rotated view in (d). For clarity, in (c), the surface of the 3D map corresponding to the IHM in the center is highlighted in yellow, and the S2 of the model of the two IHMs on the left has been extended as coiled-coil α-helices (pink). Since the backbone structure of tarantula thick filament is not known, the two neighboring “subfilaments” tails are depicted in (d) as cylinders with ~2.2 nm diameters. In (d), it is shown that the model of the IHMs is present only in the slice between the two blue dotted lines in (a), causing the neighboring “h” interaction to be far and the “i” and “j” to be closer to the reader. In relaxed state, the S2 of the IHM emerges from the bare zone (top) with a slight angle of 6° (d), causing the helix of IHMs to “float”, separated from the backbone surface by ~2 nm. The blocked head is the only part of the IHM that is in contact with the backbone and is covalently connected to it via the S2 and electrostatically connected by the three “anchoring” intermolecular interactions: “h” (blocked head SH3 domain) with the extended S2 of an adjacent tail and “i” (blocked head relay/converter) and “j” (blocked head ELC) with the neighboring S2. The MHCII in 3JBH shows six surface loops (2, H, CM, 3, C, and I) that are involved in the interactions. Also, the ELC in the 3JBH model shows the extra two amino acids that are missing in the chicken ELC sequence. Bar 5 nm. (a–d) Reproduced with permission from Alamo et al. (2016) and the Journal of Molecular Biology
The IHM is a dynamic structure exhibiting four key functions not displayed by single myosin heads
As a consequence of the heads asymmetry, the IHM exhibits four distinctive functions not observed in isolated S1 myosin fragments:
Self-inhibition of both heads ATPases
The intrinsic asymmetric structure of the IHM suggested a simple mechanism to explain relaxation by switching off the two heads in different ways as proposed by Taylor and coworkers (Wendt et al. 1999; Woodhead et al. 2005). In addition, 3JBH model explains the reduced ATPase activity in blocked heads and intermittently in free heads (Fig. 7b, d1, 2) (Alamo et al. 2016).
Swaying heads capability
The mobility of the free head, that sways back and forth by Brownian motion through breaking and restoring stabilizing IHM interactions make it a dynamic structure (Fig. 7b, d1, 2): the free heads Ser35 readily accessible to PKC (Fig. 7a) are the only heads constitutively phosphorylated. Constitutively, Ser35 mono-phosphorylated free heads—which amount up to a half of the thick filament heads—are less strongly bound to the thick filament backbone and may oscillate occasionally between attached and detached states (“swaying” heads; Fig. 7b, d1, 2). Several lines of evidence support the existence of swaying heads. First, the presence of a central diffuse scattering disk circularly symmetrical in the low-angle X-ray diffraction patterns from relaxed and non-overlap rigor skeletal muscles behaving as solution scattering and that comes predominantly from myosin heads, in contrast to full-overlap rigor in which this disk is compressed diagonally, indicating that heads have a bent shape and preferred orientation consistent with a 45° angle of attachment to actin (Poulsen and Lowy 1983; Huxley and Padrón 1984). This suggests that in relaxed skeletal muscle two head populations are simultaneously present: one helically ordered and one disordered (i.e. swaying heads). Second, on activation, the heads are released by RLC phosphorylation and disordered around the backbone surface (Fig. 2g) (Craig et al. 1987; Padrón et al. 1991), a necessary step towards the interactions of these released heads with the thin filament on contraction. Third, negatively stained turkey smooth muscle myosin shows flexible free heads, detaching from their partner blocked heads and S2 adopting different motor domain orientations (Burgess et al. 2007). Fourth, the greater mobility for the free head than for the blocked head is also revealed by the lower density of the free head than for the blocked head in the 3D reconstructions (Pinto et al. 2012; Yang et al. 2015a). Free heads are the only movable parts of the relaxed tarantula thick filament (Brito et al. 2011; Alamo et al. 2016) (Fig. 9d) as they have fewer intra- and inter-molecular interactions than the blocked heads (Alamo et al. 2008, 2016). This is supported by the result that the free heads are the ones that collapse onto the backbone while the blocked head already rests on it (Pinto et al. 2012). A comparison of negative stain and frozen-hydrated tarantula thick filament 3D reconstructions (Fig. 4a4 vs. a5) shows that free heads protrude above the backbone surface in relaxing native conditions (Fig. 4a5) collapsing by moving ~3 nm closer to the backbone following negative staining and drying (Fig. 4a4). This shows that free heads are movable by this EM procedure (Craig 2012), whilst the blocked heads are much less movable (Pinto et al. 2012). The center of mass radius (~14.7 nm) of the IHMs determined from the 3rd meridian layer line in the low-angle X-ray diffraction pattern of relaxed tarantula striated muscle (LL3 in Fig. 8b) is similar to the one calculated for the Fourier transform of the frozen-hydrated thick filament (~15.2 nm) but larger than for the negatively stained one (~12.6 nm). The importance of this intrinsic flexibility has been discussed by concluding it limits the 3D reconstruction resolution (Yang et al. 2015a). Fifth, in vitro motility assays have shown that rabbit F-actin filaments slides at 4 μm/s along relaxed isolated thick filaments of tarantula A. avicularia striated muscle (Brito et al. 2011; Sulbarán et al. 2013); direct Ca2+-binding on ELC (i.e. scallop muscle) promotes heads release from the filament surface in milliseconds (Zhao and Craig 2003a, b). Sixth, it has been found that the small fraction of constitutively ON motors (i.e. swaying heads) allows the muscle to respond immediately to Ca2+-activation when the external load is low (cf. Figs. 7d1, 2, 10d, 11b) (Linari et al. 2015). Seventh, X-ray diffraction of rabbit skeletal muscle suggests that RLC phosphorylation move heads away from thick towards thin filaments (Yamaguchi et al. 2016) confirming tarantula equatorial X-ray diffraction results (Padrón et al. 1991). Eighth, spectroscopic studies of the super-relaxed state of skeletal muscle supports the putative RLC–RLC interface identified by cryo-EM showing that both the divalent cation bound to RLC and the RLC NTE play a role in the stability of the super-relaxed state (Nogara et al. 2016). And ninth, other experimental evidence supports the presence of swaying heads (Alamo et al. 2008, 2015, 2016; Brito et al. 2011; Sulbarán et al. 2013; Espinoza-Fonseca et al. 2015; Yamaguchi et al. 2016; reviewed by (Vandenboom 2017).
Fig. 10.
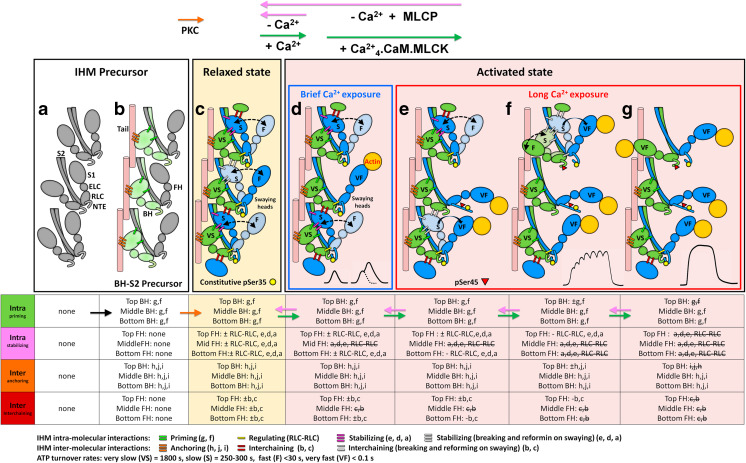
Cooperative phosphorylation activation (CPA) mechanism for recruiting active heads in tarantula thick filament activation. a Thick filament model with heads disordered around S2 with RLC NTEs un-phosphorylated. Heads do not make any inter- or intramolecular interactions (see interaction table at the bottom). b On myogenesis only one head of each myosin, in pre-powerstroke state (Zoghbi et al. 2004; Llinas et al. 2015) can establish the two required priming intramolecular interactions docking them onto their own S2 as “blocked” heads (“BH–S2 precursor”, light green) and to a neighbor myosin tail (pink cylinder) on the backbone by anchoring intermolecular interactions (Alamo et al. 2016). The other partner (“free”) head, also in pre-powerstroke state, remains disordered without making interactions with the docked blocked head as needed for establishing an IHM. c Precursor IHMs become fully functional in relaxed state (yellow box) after Ser35 of disordered free head (blue) becomes mono-phosphorylated (yellow circle) by a temporarily activated PKC which can only phosphorylate the fully exposed Ser35 of free heads, since blocked heads Ser35 (green) are not accessible (Alamo et al. 2008; Brito et al. 2011; Sulbarán et al. 2013). To fully assemble a functional IHM, each partner free head should establish three intramolecular stabilizing interactions and one regulating RLC-RLC intramolecular interaction with its partner docked blocked head (Alamo et al. 2016). The constitutive Ser35 mono-phosphorylation (Brito et al. 2011; Sulbarán et al. 2013) cannot be dephosphorylated (Brito et al. 2011; Sulbarán et al. 2013). Free heads Ser35 mono-phosphorylation promotes their swaying away and back by Brownian motion (“swaying” heads; Fig. 7b) by breaking and reforming the intramolecular stabilizing interactions (dotted curved arrows, “±” in table) (Brito et al. 2011). d–g The CPA mechanism supports the recruiting of heads for controlled force production during brief (red box) and long Ca2+ exposures on unfused or fused tetani (Brito et al. 2011; Sulbarán et al. 2013; Espinoza-Fonseca et al. 2015; Alamo et al. 2015, 2016) as explained in the text (cf. Fig. 11c, d). The very slow, slow, fast and very fast tarantula ATP turnover life times are labeled with VS, S, F and VF on each head. The mechanism explains the structural origin of these life times according to each head type. The table shows how priming intramolecular and stabilizing intermolecular interactions are sequentially established in relaxation (c), being progressively removed (crossed out) on activation (c–f), ending in a disordered array (f) with fewer interactions. Conversely, it also shows how interactions are reformed from (f) to a newly achieved ordered array (c) after lowering [Ca2+] and MLCP dephophorylate Ser45 of bi-phosphorylated and mono-phosphorylated RLCs. Modified from (Alamo et al. 2016)
Fig. 11.
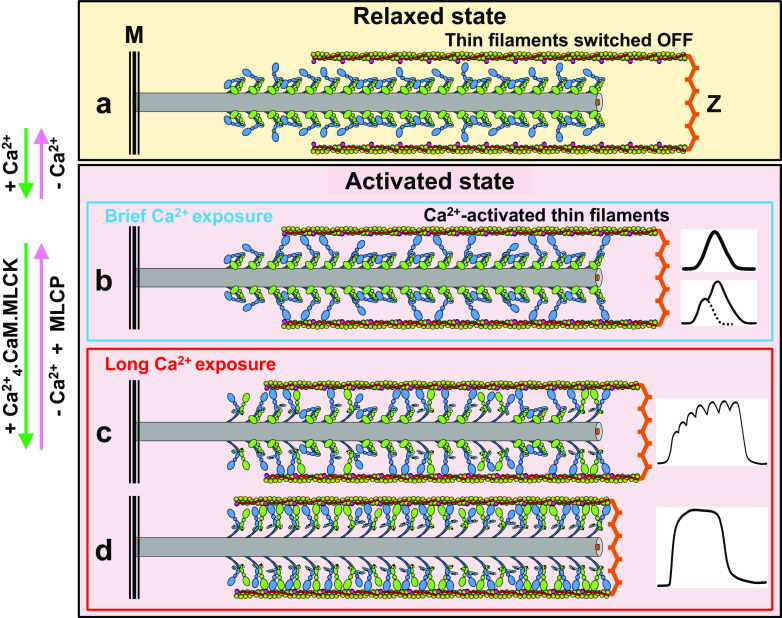
Swaying-swinging, tilting crossbridge-sliding filament mechanism (cf. (Huxley 1969, 2004b)). The number of recruited heads producing force in the tarantula striated muscle half-sarcomere under contraction according to the CPA mechanism (Fig. 10) depends on the time [Ca2+] is below the threshold, as in the relaxed state (a), or above it, as in activated state (b–d). In relaxed state (a) about half of the total number of heads (free heads) could be released swaying heads while about the other half are docked swaying heads and docked blocked heads (Fig. 7d and 10). The released swaying heads move out with a swaying duty cycle Ds = 0.6 (i.e. they are released 60% of cycle time, see text) (Fig. 7b). As thin filaments are not Ca2+-activated (Fig. 7c1) these heads cannot bind actin neither produce force. When [Ca2+] is high thin filaments become activated (b–d, Fig. 7 c2). For brief Ca2+ exposures (blue box), only the detached swaying heads can bind to the thin filament producing force in twitches and twitch summation (b), while for longer Ca2+ exposures CaM.MLCK (red box) become activated (Ca2+ 4.CaM.MLCK). Thus, blocked heads are recruited by phosphorylation swaying away binding to actin, potentiating force in unfused tetani (c). With a much longer Ca2+ exposure, many more of the remaining blocked heads become recruited, producing near maximal force in fused tetani (d). Tarantula thick filaments are ~5 μm long with a ~210-nm central bare zone, so, since there are four free (blue) and blocked (green) heads per 14.5 nm crown, each filament halve has ~1320 free and blocked heads. To simplify, only heads from 20 crowns (1 crown per repeat) are shown on each filament segment halve. From the four IHMs on each crown, only the top and bottom ones are shown. M M-band, Z Z-disk. Not to scale (Padrón 2007)
Cooperative phosphorylation activation (CPA) mechanism
The release of extra free and blocked heads upon activation for force potentiation (Fig. 7d) (Brito et al. 2011) suggested a cooperative phosphorylation mechanism for activation of tarantula thick filaments (Sulbarán et al. 2013) extended by further work (Espinoza-Fonseca et al. 2007, 2008, 2014, 2015; Kast et al. 2010; Alamo et al. 2015, 2016; Yamaguchi et al. 2016). The CPA mechanism is shown in Fig. 10c–g: On activation, a sudden brief increase on [Ca2+] (Fig. 10 blue box) switch-on tarantula thin filaments after Ca2+ binding to troponin C (Craig and Lehman 2001). Released swaying free heads (“swinging” heads) could make crossbridges with actin on switched-on thin filaments generating powerstroke force in single twitches and twitch summation (Brito et al. 2011). If [Ca2+] suddenly decreases, actin-bound swaying heads are released and relaxation is regained (Fig. 10c). When [Ca2+] remains high long enough Fig. 10 (red box), calmodulin (CaM) binds to it creating a complex Ca2+ 4.CaM. This complex activates MLCK by binding to it. The activated Ca2+ 4.CaM.MLCK is capable of phosphorylating only the exposed free head Ser45 (Fig. 10 red triangle). This phosphorylation promotes the RLC NTEs (Fig. 10 blue NTE) extension (Espinoza-Fonseca et al. 2015) and rigidity (Alamo et al. 2015) releasing this free head (Fig. 10e) since it is unable to dock back to its partner blocked head to make the RLC–RLC regulating intramolecular interaction (Espinoza-Fonseca et al. 2015) (Alamo et al. 2015). This free head releasing allows then the access of Ca2+ 4.CaM.MLCK to cooperatively mono-phosphorylate next blocked head Ser45 on the helix of heads, making it to sway away (Fig. 10f, top IHM). This double phosphorylation mechanism enables tarantula striated muscle to respond to activation during contraction explaining force potentiation and post-tetanic potentiation phenomenon (Fig. 7d5) (Brito et al. 2011). O18 mass spectrometry results showed that activation does not produce de novo Ser35 phosphorylation (Biasutto et al., in preparation) suggesting that the number of Ser35 mono-phosphorylated heads remains constant on activation supporting the CPA mechanism and confirming that it is controlled only by Ser45 phosphorylation.
Relationship between the invertebrate CPA mechanism and the vertebrate mechanosensing force generation controlling mechanism
Recent studies describe a thick filament mechanosensing regulatory mechanism in vertebrate skeletal and cardiac muscle where stress in the thick filament recruits additional force-producing heads augmenting the number of myosin motors available to interact with actin-containing thin filaments (Linari et al. 2015; Fusi et al. 2016; Reconditi et al. 2017). These additional recruited heads switch off quickly not producing a time potentiation as does the RLC phosphorylation. Further work will be needed to show if the mechanosensing mechanism is also present in tarantula striated muscle (which lacks MyBP-C and titin) and additional unphosphorylated RLC heads are recruited beside the potentiating RLC Ser45 phosphorylated heads according to the CPA mechanism (Duno et al. in preparation).
Super-relaxation (SRX)
The pioneering work of Cooke and coworkers proposed a new myosin state, that they called super-relaxed state (SRX) which exhibits a slow ATP turnover rate as a mechanism in thermogenesis in resting skeletal (Stewart et al. 2010; Cooke 2011; McNamara et al. 2015). In tarantula muscle, there is an additional very slow ATP turnover rate (Naber et al. 2011; Wilson et al. 2014). The structural origin of the lifetimes detected in tarantula muscle (very slow: >1800 s, slow: 250–300 s, fast: < 30 s and very fast: < 0.1 s) is explained by the CPA mechanism (Fig. 10) as shown by (Alamo et al. 2016) considering the different head types: (1) the very slow life time is associated to the un-phosphorylated blocked heads (exhibiting SRX), whose ATP activity is sterically “blocked” since its actin-binding interface is positioned in the IHM onto the converter domain of the free head; (2) in contrast, in the released swaying free heads, this converter domain stabilization is removed enabling the recovery of its ATPase activity and showing a slow ATP turnover life time. In consequence, the docked and immobile blocked heads as well as the swaying free heads exhibit these two slow lifetimes detected in relaxed state; and (3) the fast and very fast ATP turnover life times are associated to permanently mobile bi-phosphorylated free heads and swaying free and blocked heads which can bind to actin on the activated thin filament.
The CPA mechanism allows quantifying the swaying free head duty cycle and the SRX/DRX energetics ratio:
Swaying free head duty cycle: According to the CPA mechanism for skeletal muscle, the blocked head remains docked while the free head sway away for a time ts being docked only for a time td. Therefore, for a total time t = ts + td, the swaying duty cycle Ds can be defined as Ds = ts/t. Ds and can be estimated in relaxed muscles from measurements of RLC orientation using fluorescence polarization from bifunctional rhodamine probes bound to RLC (Fusi et al. 2015). In relaxed rabbit psoas muscle, ~70% of all heads are parallel to the fiber axis, while ~30% of all heads are perpendicular (Fusi et al. 2015), implying that the head population comprises 50% blocked heads (all parallel), 20% docked free heads (parallel) and 30% swaying free heads (perpendicular). It has been suggested that Ds depends on muscle type and that Ds = 2/(1 + R) where R = Npar/Nper is the ratio of the number of heads detected in parallel (Npar) and perpendicular (Nper) conformations (Alamo et al. 2017b). Therefore, in skeletal muscle, R = 0.70/0.3 = 2.3 so Ds = 0.60, i.e. the heads spend 60% of their time swaying away and 40% docked to the blocked head.
SRX/DRX energetics ratio: It has been suggested that the estimated ratio of heads in the SRX and the disordered relaxed (DRX, Fig. 7b) states is ε = SRX/DRX = (2–Ds)/Ds and that it depends on muscle type (Alamo et al. 2017b). Therefore, as Ds = 0.6 in skeletal muscle is ε = 2.3, it is possible to predict an estimated 2.3-fold reduction of ATP hydrolysis. As skeletal muscle heads rapidly transition out of SRX into DRX (such as occurs with tetanic activation; Hooijman et al. 2011; Brito et al. 2011), the changing population of myosin SRX and DRX heads in skeletal muscle—as predicted by the CPA mechanism—yields energy-saving imposed by the IHM structure retaining the ability to rapidly shift myosin into DRX for force potentiation.
In relaxed state, free heads are ready to interact with thin filaments when activated while blocked heads are not.
Studies on tarantula thick filament activation have shown that, in the relaxed sarcomere, up to half of the heads (the detached swaying free heads) present in the thick filament are conformationally ready and properly located to quickly interact with the thin filament as soon as they are activated by a sudden Ca2+ increase. On the other hand, slightly more than half of the heads (docked swaying heads and docked blocked heads) are not. After being phosphorylated by MLCK, these heads are recruited to interact with the activated thin filament (Fig. 7 d, 10) (Brito et al. 2011). These two different heads populations have functional consequences on the sarcomere relaxation and contraction (Fig. 11) as well as important implications on the effect of mutations associated with HCM due to the involvement of these extra recruited blocked heads on the hypercontractility caused by HCM (Alamo et al. 2017b).
Swaying-swinging, tilting crossbridge-sliding filament mechanism (cf. Huxley 1969, 2004b). As shown in Fig. 11a, according to the CPA mechanism in relaxed state about half of the total number of heads (free heads) could be released swaying heads while about half of the total number heads are transiently docked free swaying heads and docked blocked heads (Fig. 7b, 10). When [Ca2+] briefly increases, the detached swaying heads can interact with the activated thin filament producing force (Fig. 11b). When [Ca2+] remains high for a longer time, the MLCK becomes activated by Ca2+, recruiting additional Ser45 mono-phosphorylated blocked heads resulting in force potentiation (Fig. 11c) until most blocked heads are recruited on tetanus-producing maximal force (Fig. 11d). A sudden decrease of [Ca2+] below the threshold will stop force production and relax the sarcomere such that, if [Ca2+] is suddenly increased, it will produce a force bigger that the initial one, i.e. post-tetanic potentiation. If [Ca2+] remains below the threshold long enough such that the endogenous MLCP dephosphorylate the Ser45, a new induced twitch will produce the same force as originally.
Conclusions
The two myosin heads assemble asymmetrically on an IHM by intramolecular interactions. In filaments, intermolecular interactions with neighbor myosin tails and adjacent IHMs promotes the generation of 3, 4 or 7 head helices on striated muscle, supporting our structural model for its regulation and activation. Future studies on crystallography and cryo-EM would allow solving the HMM near-atomic structure.
The IHM structure endorses additional functions—not seen in single heads—as self-inhibition of both heads ATPases and super-relaxation, free head swaying force on twitches, blocked head force potentiation on tetani and post-tetanic potentiation through a cooperative phosphorylation activation mechanism. Based on this mechanism, we propose a swaying-swinging, tilting crossbridge-sliding filament for tarantula muscle contraction.
Acknowledgements
We thank Dr. John Wray for permission to reproduce unpublished tarantula diffraction patterns, and Drs. Guidenn Sulbarán, Jesús Mavárez and Gustavo Márquez for help with the manuscript. Molecular graphics images were produced using the UCSF Chimera package (Pettersen et al. 2004) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by the National Institutes of Health grant P41 RR-01081). This work was supported in part by Centro de Biología Estructural del Mercosur (www.cebem-lat.org) (to R.P.), Russian Foundation for Basic Research (15-04-02174 to N.K., 16-04-00693 to A.T.), Cornell High Energy Synchrotron Source (CHESS) is supported by the NSF & NIH/NIGMS via NSF award DMR-1332208, and the MacCHESS resources are supported by NIGMS award GM-103485 (to R.E.G.) and the Howard Hughes Medical Institute (to R.P.).
We dedicate this paper to the memory of Dr. Hugh E. Huxley.
Compliance with ethical standards
Conflicts of interest
Lorenzo Alamo declares that he has no conflicts of interest. Natalia Koubassova declares that she has no conflicts of interest. Antonio Pinto declares that he has no conflicts of interest. Richard Gillilan declares that he has no conflicts of interest. Andrey Tsaturyan declares that he has no conflicts of interest. Raúl Padrón declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Latin America’ edited by Pietro Ciancaglini and Rosangela Itri.
References
- AL-Khayat HA. Three-dimensional structure of the human myosin thick filament: clinical implications. Glob Cardiol Sci Pract. 2013;2013:280–302. doi: 10.5339/gcsp.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo L, Li XE, Espinoza-Fonseca LM, Pinto A, Thomas DD, Lehman W, Padrón R. Tarantula myosin free head regulatory light chain phosphorylation stiffens N-terminal extension, releasing it and blocking its docking back. Mol BioSyst. 2015;11:2180–2189. doi: 10.1039/c5mb00163c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo, L., Pinto, A., Sulbarán, G., Mavárez, J., and Padrón, R. (2017a). Lessons from a tarantula: New insights into myosin interacting-heads motif evolution and its implications on disease. Biophys Rev. doi:10.1007/s12551-017-0292-4 [DOI] [PMC free article] [PubMed]
- Alamo L, Qi D, Wriggers W, Pinto A, Zhu J, Bilbao A, Gillilan RE, Hu S, Padrón R. Conserved Intramolecular interactions maintain myosin interacting-heads motifs explaining tarantula muscle super-relaxed state structural basis. J Mol Biol. 2016;428:1142–1164. doi: 10.1016/j.jmb.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo L, Ware JS, Pinto A, Gillilan RE, Seidman JG, Seidman CE, Padrón R. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife. 2017;6:e24634. doi: 10.7554/eLife.24634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo L, Wriggers W, Pinto A, Bartoli F, Salazar L, Zhao FQ, Craig R, Padrón R. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J Mol Biol. 2008;384:780–797. doi: 10.1016/j.jmb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, Epstein HF. Protein machines and self assembly in muscle organization. BioEssays. 1999;21:813–823. doi: 10.1002/(SICI)1521-1878(199910)21:10<813::AID-BIES3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Blankenfeldt W, Thoma NH, Wray JS, Gautel M, Schlichting I. Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc Natl Acad Sci U S A. 2006;103:17713–17717. doi: 10.1073/pnas.0606741103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito R, Alamo L, Lundberg U, Guerrero JR, Pinto A, Sulbarán G, Gawinowicz MA, Craig R, Padrón R. A molecular model of phosphorylation-based activation and potentiation of tarantula muscle thick filaments. J Mol Biol. 2011;414:44–61. doi: 10.1016/j.jmb.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Chew MW, Squire JM. Packing of alpha-helical coiled-coil myosin rods in vertebrate muscle thick filaments. J Struct Biol. 1995;115:233–249. doi: 10.1006/jsbi.1995.1048. [DOI] [PubMed] [Google Scholar]
- Clarke ML, Hofman W, Wray JS. ATP binding and crossbridge structure in muscle. J Mol Biol. 1986;191:581–585. doi: 10.1016/0022-2836(86)90153-1. [DOI] [PubMed] [Google Scholar]
- Coluccio LM. Myosin: a Superfamily of molecular motors. Dordrecht: Springer; 2008. [Google Scholar]
- Cooke R. The role of the myosin ATPase activity in adaptive thermogenesis by skeletal muscle. Biophys Rev. 2011;3:33–45. doi: 10.1007/s12551-011-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R. Isolation, electron microscopy and 3D reconstruction of invertebrate muscle myofilaments. Methods. 2012;56:33–43. doi: 10.1016/j.ymeth.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Alamo L, Padrón R. Structure of the myosin filaments of relaxed and rigor vertebrate striated muscle studied by rapid freezing electron microscopy. J Mol Biol. 1992;228:474–487. doi: 10.1016/0022-2836(92)90836-9. [DOI] [PubMed] [Google Scholar]
- Craig R, Lehman W. Crossbridge and tropomyosin positions observed in native, interacting thick and thin filaments. J Mol Biol. 2001;311:1027–1036. doi: 10.1006/jmbi.2001.4897. [DOI] [PubMed] [Google Scholar]
- Craig R, Padrón R. Structure of tarantula muscle thick filaments. J Muscle Res Cell Motil. 1982;3:487. [Google Scholar]
- Craig R, Padrón R. Disorder induced in nonoverlapping myosin cross-brdiges by loss of adenosine triphosphate. Biophys J. 1989;56:927–933. doi: 10.1016/S0006-3495(89)82738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, R. Padrón R (2004) Molecular structure of the sarcomere. In: Engel AG, Franzini-Armstrong C (eds) Myology (3rd edn). McGraw-Hill, New York, pp 129–166
- Craig R, Padrón R, Alamo L. Direct determination of myosin filament symmetry in scallop striated adductor muscle by rapid freezing and freeze substitution. J Mol Biol. 1991;220:125–132. doi: 10.1016/0022-2836(91)90386-k. [DOI] [PubMed] [Google Scholar]
- Craig R, Padrón R, Kendrick-Jones J. Structural changes accompanying phosphorylation of tarantula muscle myosin filaments. J Cell Biol. 1987;105:1319–1327. doi: 10.1083/jcb.105.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Woodhead JL. Structure and function of myosin filaments. Curr Opin Struct Biol. 2006;16:204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Padrón R, Craig R. Arrangement of the heads of myosin in relaxed thick filaments from tarantula muscle. J Mol Biol. 1985;184:429–439. doi: 10.1016/0022-2836(85)90292-x. [DOI] [PubMed] [Google Scholar]
- Eakins F, AL-Khayat HA, Kensler RW, Morris EP, Squire JM. 3D structure of fish muscle myosin filaments. J Struct Biol. 2002;137:154–163. doi: 10.1006/jsbi.2002.4453. [DOI] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Egelman EH, Padrón R. X-ray diffraction evidence that actin is a 100 a filament. Nature. 1984;307:56–58. [Google Scholar]
- Elliott A, Offer G. Shape and flexibility of the myosin molecule. J Mol Biol. 1978;123:505–519. doi: 10.1016/0022-2836(78)90204-8. [DOI] [PubMed] [Google Scholar]
- Espinoza-Fonseca LM, Alamo L, Pinto A, Thomas DD, Padrón R. Sequential myosin phosphorylation activates tarantula thick filament via a disorder-order transition. Mol BioSyst. 2015;11:2167–2179. doi: 10.1039/c5mb00162e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Fonseca LM, Colson BA, Thomas DD. Effects of pseudophosphorylation mutants on the structural dynamics of smooth muscle myosin regulatory light chain. Mol BioSyst. 2014;10:2693–2698. doi: 10.1039/c4mb00364k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Fonseca LM, Kast D, Thomas DD. Molecular dynamics simulations reveal a disorder-to-order transition on phosphorylation of smooth muscle myosin. Biophys J. 2007;93:2083–2090. doi: 10.1529/biophysj.106.095802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Fonseca LM, Kast D, Thomas DD. Thermodynamic and structural basis of phosphorylation-induced disorder-to-order transition in the regulatory light chain of smooth muscle myosin. J Am Chem Soc. 2008;130:12208–12209. doi: 10.1021/ja803143g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee, L., Lin, W., Qiu, F., and Edwards, R. J. (2017) Myosin II sequences for Lethocerus indicus. J Musc Res Cell Motil (in press) [DOI] [PMC free article] [PubMed]
- Fernández-Morán H. Low-temperature preparation techniques for electron microscopy of biological specimens based on rapid freezing with liquid helium II. Ann N Y Acad Sci. 1960;85:689–713. doi: 10.1111/j.1749-6632.1960.tb49990.x. [DOI] [PubMed] [Google Scholar]
- Fernández-Morán H. Electron microscopy with high-field superconducting solenoid lenses. Proc Natl Acad Sci U S A. 1965;53:445–451. doi: 10.1073/pnas.53.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Morán H. High-resolution electron microscopy with superconducting lenses at liquid helium temperatures. Proc Natl Acad Sci U S A. 1966;56:801–808. doi: 10.1073/pnas.56.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusi L, Brunello E, Yan Z, Irving M. Thick filament mechano-sensing is a calcium-independent regulatory mechanism in skeletal muscle. Nat Commun. 2016;7:13281. doi: 10.1038/ncomms13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusi L, Huang Z, Irving M. The conformation of myosin heads in relaxed skeletal muscle: implications for myosin-based regulation. Biophys J. 2015;109:783–792. doi: 10.1016/j.bpj.2015.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillilan RE, Kumar VS, O’Neall-Hennessey E, Cohen C, Brown JH. X-ray solution scattering of squid heavy Meromyosin: strengthening the evidence for an ancient compact off state. PLoS ONE. 2013;8:e81994. doi: 10.1371/journal.pone.0081994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero JR, Padrón R. The substructure of the backbone of the thick filaments from tarantula muscle. Acta Microsc. 1992;1:63–83. [Google Scholar]
- Haselgrove JC. A model of myosin crossbridge structure consistent with the low-angle x-ray diffraction pattern of vertebrate muscle. J Muscle Res Cell Motil. 1980;1:177–191. doi: 10.1007/BF00711798. [DOI] [PubMed] [Google Scholar]
- Hidalgo C, Craig R, Ikebe M, Padrón R. Mechanism of phosphorylation of the regulatory light chain of myosin from tarantula striated muscle. J Muscle Res Cell Motil. 2001;22:51–59. doi: 10.1023/a:1010388103354. [DOI] [PubMed] [Google Scholar]
- Hidalgo, C., Medina, R., Padrón, R., Tonino, P. Horowitz R., Fa-Qing Zhao, Alamo, L., Simon, M., and Craig, R. (2001b) Mass measurement and composition of native purified myosin filaments from muscle. 55th Annual Meeting and Symposium Molecular Motors of the Society of the General Physiologists. Woodshole, MA
- Hidalgo C, Padrón R, Horowitz R, Zhao FQ, Craig R. Purification of native myosin filaments from muscle. Biophys J. 2001;81:2817–2826. doi: 10.1016/S0006-3495(01)75923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo C, Padrón R, Medina R, Tonino P, Alamo L, Simon M, Craig R. Mass/length measurement and composition of native purified myosin filaments from muscle. J Gen Physiol. 2001;118:10A. [Google Scholar]
- Hitchcock-DeGregori SE, Irving TC. Hugh E. Huxley: the compleat biophysicist. Biophys J. 2014;107:1493–1501. doi: 10.1016/j.bpj.2014.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooijman P, Stewart MA, Cooke R. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys J. 2011;100:1969–1976. doi: 10.1016/j.bpj.2011.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SL, Hobbs KH, Thuma JB. Invertebrate muscles: thin and thick filament structure; molecular basis of contraction and its regulation, catch and asynchronous muscle. Prog Neurobiol. 2008;86:72–127. doi: 10.1016/j.pneurobio.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SL, Thuma JB. Invertebrate muscles: muscle specific genes and proteins. Physiol Rev. 2005;85:1001–1060. doi: 10.1152/physrev.00019.2004. [DOI] [PubMed] [Google Scholar]
- Hu Z, Taylor DW, Reedy MK, Edwards RJ, Taylor KA. Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 a resolution. Sci Adv. 2016;2:e1600058. doi: 10.1126/sciadv.1600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE. Structural changes in the actin- and myosin containing filaments during contraction. Cold Spring Harb Symp Quant Biol. 1973;37:361–376. [Google Scholar]
- Huxley AF, Niedergerke R. Structural changes in muscle during contraction: interference microscopy of living muscle fibres. Nature. 1954;173:971–972. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- Huxley HE. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- Huxley HE. Fifty years of muscle and the sliding filament hypothesis. Eur J Biochem. 2004;271:1403–1415. doi: 10.1111/j.1432-1033.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- Huxley HE. Recent X-ray diffraction studies of muscle contraction and their implications. Philos Trans R Soc Lond B. 2004;359:1879–1882. doi: 10.1098/rstb.2004.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE, Brown W. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- Huxley HE, Hanson EJ. Changes in cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- Huxley HE, Padrón R. The effect of the ATP analogue AMPPNP on the structure of crossbridges in vertebrate skeletal muscle: X-ray diffraction and mechanical studies. J Muscle Res Cell Motil. 1984;5:613–655. doi: 10.1007/BF00713923. [DOI] [PubMed] [Google Scholar]
- Huxley HE, Zubay G. Electron microscope observations on the structure of microsomal particles from Escherichia Coli. J Mol Biol. 1960;2:10–18. [Google Scholar]
- Kast D, Espinoza-Fonseca LM, Yi C, Thomas DD. Phosphorylation-induced structural changes in smooth muscle myosin regulatory light chain. Proc Natl Acad Sci U S A. 2010;107:8207–8212. doi: 10.1073/pnas.1001941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler RW, Levine RJ, Stewart M. Electron microscopic and optical diffraction analysis of the structure of scorpion muscle thick filaments. J Cell Biol. 1985;101:395–401. doi: 10.1083/jcb.101.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler RW, Stewart M. Frog skeletal muscle thick filaments are three-stranded. J Cell Biol. 1983;96:1797–1802. doi: 10.1083/jcb.96.6.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler RW, Stewart M. An ultrastructural study of cross-bridge arrangement in the frog thigh muscle thick filament. Biophys J. 1986;49:343–351. doi: 10.1016/S0006-3495(86)83647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler RW, Stewart M. An ultrastructural study of crossbridge arrangement in the fish skeletal muscle thick filament. J Cell Sci. 1989;94(Pt 3):391–401. doi: 10.1242/jcs.94.3.391. [DOI] [PubMed] [Google Scholar]
- Kensler RW, Stewart M. The relaxed crossbridge pattern in isolated rabbit psoas muscle thick filaments. J Cell Sci. 1993;105(Pt 3):841–848. doi: 10.1242/jcs.105.3.841. [DOI] [PubMed] [Google Scholar]
- Kensler RW, Woodhead JL. The chicken muscle thick filament: temperature and the relaxed cross-bridge arrangement. J Muscle Res Cell Motil. 1995;16:79–90. doi: 10.1007/BF00125312. [DOI] [PubMed] [Google Scholar]
- Knight P, Trinick J. Structure of the myosin projections on native thick filaments from vertebrate skeletal muscle. J Mol Biol. 1984;177:461–482. doi: 10.1016/0022-2836(84)90295-x. [DOI] [PubMed] [Google Scholar]
- Levine RJ, Kensler RW, Reedy MC, Hofmann W, King HA. Structure and paramyosin content of tarantula thick filaments. J Cell Biol. 1983;97:186–195. doi: 10.1083/jcb.97.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RJC, Kensler RW, Stewart M, Haselgrove JC. Molecular organization of limulus thick filaments. Soc Gen Physiol Ser. 1982;37:37–52. [PubMed] [Google Scholar]
- Linari M, Brunello E, Reconditi M, Fusi L, Caremani M, Narayanan T, Piazzesi G, Lombardi V, Irving M (2015) Force generation by skeletal muscle is controlled by mechanosensing in myosin filaments. Nature 528:276–279 [DOI] [PubMed]
- Liu J, Wendt T, Taylor D, Taylor K. Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J Mol Biol. 2003;329:963–972. doi: 10.1016/s0022-2836(03)00516-3. [DOI] [PubMed] [Google Scholar]
- Llinas P, Isabet T, Song L, Ropars V, Zong B, Benisty H, Sirigu S, Morris C, Kikuti C, Safer D, Sweeney HL, Houdusse A. How actin initiates the motor activity of myosin. Dev Cell. 2015;33:401–412. doi: 10.1016/j.devcel.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther PK, Bennett PM, Knupp C, Craig R, Padrón R, Harris SP, Patel J, Moss RL. Understanding the organisation and role of myosin binding protein C in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. J Mol Biol. 2008;384:60–72. doi: 10.1016/j.jmb.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther PK, Winkler H, Taylor K, Zoghbi ME, Craig R, Padrón R, Squire JM, Liu J. Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc Natl Acad Sci U S A. 2011;108:11423–11428. doi: 10.1073/pnas.1103216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez G, Pinto A, Alamo L, Baumann B, Ye F, Winkler H, Taylor K, Padrón R. A method for 3D-reconstruction of a muscle thick filament using the tilt series images of a single filament electron tomogram. J Struct Biol. 2014;186:265–272. doi: 10.1016/j.jsb.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maw MC, Rowe AJ. Fraying of A-filaments into three subfilaments. Nature. 1980;286:412–414. doi: 10.1038/286412a0. [DOI] [PubMed] [Google Scholar]
- McNamara JW, Li A, dos Remedios CG, Cooke R. The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys Rev. 2015;7:5–14. doi: 10.1007/s12551-014-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetret JF, Hofmann W, Lepault J. Cryo-electron microscopy of insect flight muscle thick filaments. An approach to dynamic electron microscope studies. J Mol Biol. 1988;202:175–178. doi: 10.1016/0022-2836(88)90530-x. [DOI] [PubMed] [Google Scholar]
- Morris EP, Squire JM, Fuller GW. The 4-stranded helical arrangement of myosin heads on insect (Lethocerus) flight muscle thick filaments. J Struct Biol. 1991;107:237–249. [Google Scholar]
- Naber N, Cooke R, Pate E (2011) Slow myosin ATP turnover in the super-relaxed state in tarantula muscle. J Mol Biol 411:943–950 [DOI] [PMC free article] [PubMed]
- Nogara L, Naber N, Pate E, Canton M, Reggiani C, Cooke R. Spectroscopic studies of the super relaxed state of skeletal muscle. PLoS ONE. 2016;11:e0160100. doi: 10.1371/journal.pone.0160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neall-Hennessey E, Reshetnikova L, Senthil Kumar VS, Robinson H, Szent-Gyorgyi AG, Cohen C. Purification, crystallization and preliminary X-ray crystallographic analysis of squid heavy meromyosin. Acta Crystallogr F. 2013;69:248–252. doi: 10.1107/S1744309112049925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offer G, Knight P. The structure of the head-tail junction of the myosin molecule. J Mol Biol. 1996;256:407–416. doi: 10.1006/jmbi.1996.0096. [DOI] [PubMed] [Google Scholar]
- Offer G, Knight PJ, Burgess SA, Alamo L, Padrón R. A new model for the surface arrangement of myosin molecules in tarantula thick filaments. J Mol Biol. 2000;298:239–260. doi: 10.1006/jmbi.2000.3664. [DOI] [PubMed] [Google Scholar]
- Padrón R, Alamo L. The use of negative staining and cryo-electron microscopy to understand the molecular mechanism of myosin-linked regulation of striated muscle contraction. Acta Microscopica. 2004;13:14–29. [Google Scholar]
- Padrón R. Contribución de Humberto Fernández-Morán a la Microscopía Electrónica. Rev Lat Met Mat. 1999;19:5–6. [Google Scholar]
- Padrón R. The contribution of Humberto Fernández-Morán to the electron microscopy. Acta Microsc. 2001;10:54–56. [Google Scholar]
- Padrón R. Filamento de miosina: Modelo atómico. Invest Cienc. 2007;369:28–30. [Google Scholar]
- Padrón R. Two and a half years at the LMB that imprinted my scientific career (1980-1983) In: Huxley HE, editor. Memoirs and Consequences. Cambridge: MRC Laboratory of Molecular Biology; 2013. pp. 315–322. [Google Scholar]
- Padrón R, Alamo L, Craig R, Caputo C. A method for quick-freezing live muscles at known instants during contraction with simultaneous recording of mechanical tension. J Microsc. 1988;151:81–102. doi: 10.1111/j.1365-2818.1988.tb04616.x. [DOI] [PubMed] [Google Scholar]
- Padrón R, Alamo L, Guerrero JR, Granados M, Uman P, Craig R. Three-dimensional reconstruction of thick filaments from rapidly frozen, freeze-substituted tarantula muscle. J Struct Biol. 1995;115:250–257. doi: 10.1006/jsbi.1995.1049. [DOI] [PubMed] [Google Scholar]
- Padrón R, Alamo L, Murgich J, Craig R. Towards an atomic model of the thick filaments of muscle. J Mol Biol. 1998;275:35–41. doi: 10.1006/jmbi.1997.1448. [DOI] [PubMed] [Google Scholar]
- Padrón R, Granados M, Alamo L, Guerrero JR, Craig R. Visualization of myosin helices in sections of rapidly frozen relaxed tarantula muscle. J Struct Biol. 1992;108:269–276. doi: 10.1016/1047-8477(92)90027-8. [DOI] [PubMed] [Google Scholar]
- Padrón R, Guerrero JR, Alamo L, Granados M, Gherbesi N, Craig R. Direct visualization of myosin filament symmetry in tarantula striated muscle by electron microscopy. J Struct Biol. 1993;111:17–21. doi: 10.1006/jsbi.1993.1031. [DOI] [PubMed] [Google Scholar]
- Padrón R, Pante N, Sosa H, Kendrick-Jones J. X-ray diffraction study of the structural changes accompanying phosphorylation of tarantula muscle. J Muscle Res Cell Motil. 1991;12:235–241. doi: 10.1007/BF01745112. [DOI] [PubMed] [Google Scholar]
- Padrón R, Rodríguez J, Guerrero JR, Alamo L. Fraying of thick filaments from tarantula muscle into subfilaments. Acta Microscopica. 1993;2:85–92. [Google Scholar]
- Pante N, Sosa H, Padrón R. Prediction of relative changes in the equatorial x-ray diffraction pattern of striated muscle caused by the activation of muscle contraction. Acta Cient Venez. 1986;37:223–225. [PubMed] [Google Scholar]
- Panté N, Sosa H, Padrón R. X-ray diffraction study of the structural changes accompanying tarantula thick filament phosphorylation. Acta Cient Ven. 1988;39:230–236. [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pinto A, Sanchez F, Alamo L, Padrón R. The myosin interacting-heads motif is present in the relaxed thick filament of the striated muscle of scorpion. J Struct Biol. 2012;180:469–478. doi: 10.1016/j.jsb.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Poulsen FR, Lowy J. Small-angle X-ray scattering from myosin heads in relaxed and rigor frog skeletal muscles. Nature. 1983;303:146–152. doi: 10.1038/303146a0. [DOI] [PubMed] [Google Scholar]
- Ranke J. Tetanus: Eine Physiologische Studie. Leipzig: Wilhelm Engelmann; 1865. [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Reconditi M, Caremani M, Pinzauti F, Powers JD, Narayanan T, Stienen GJ, Linari M, Lombardi V, Piazzesi G. Myosin filament activation in the heart is tuned to the mechanical task. Proc Natl Acad Sci U S A. 2017;114:3240–3245. doi: 10.1073/pnas.1619484114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR. Myosins. Oxford: Oxford University Press; 1999. [Google Scholar]
- Sosa H, Panté N, Padrón R. Determination of the equatorial section of the X-ray diffraction pattern of live tarantula muscle. Acta Cient Ven. 1986;37:587–588. [Google Scholar]
- Sosa H, Panté N, Padrón R. Analysis of the equatorial section of the X-ray diffraction pattern of tarantula striated muscle in different experimental conditions. Acta Cient Ven. 1988;39:51–59. [PubMed] [Google Scholar]
- Squire JM. General model for the structure of all myosin-containing filaments. Nature. 1971;233:457–462. doi: 10.1038/233457a0. [DOI] [PubMed] [Google Scholar]
- Squire JM. General model of myosin filament structure. II. Myosin filaments and cross-bridge interactions in vertebrate striated and insect flight muscles. J Mol Biol. 1972;72:125–138. doi: 10.1016/0022-2836(72)90074-5. [DOI] [PubMed] [Google Scholar]
- Squire JM. General model of myosin filament structure. 3. Molecular packing arrangements in myosin filaments. J Mol Biol. 1973;77:291–323. doi: 10.1016/0022-2836(73)90337-9. [DOI] [PubMed] [Google Scholar]
- Squire JM. Muscle filament structure and muscle contraction. Annu Rev Biophys Bioeng. 1975;4:137–163. doi: 10.1146/annurev.bb.04.060175.001033. [DOI] [PubMed] [Google Scholar]
- Squire JM. The structural basis of muscular contraction. New York: Plenum; 1981. [Google Scholar]
- Squire JM. Muscle: design, diversity, and disease. Menlo Park: Benjamin/Cummings; 1986. [Google Scholar]
- Squire JM. Muscle myosin filaments: cores, crowns and couplings. Biophys Rev. 2009;1:149–160. doi: 10.1007/s12551-009-0017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire JM, Al-Khayat HA, Knupp C, Luther PK. Molecular architecture in muscle contractile assemblies. Adv Protein Chem. 2005;71:17–87. doi: 10.1016/S0065-3233(04)71002-5. [DOI] [PubMed] [Google Scholar]
- Stewart M, Kensler RW. Arrangement of myosin heads in relaxed thick filaments from frog skeletal muscle. J Mol Biol. 1986;192:831–851. doi: 10.1016/0022-2836(86)90032-x. [DOI] [PubMed] [Google Scholar]
- Stewart M, Kensler RW, Levine RJ. Structure of limulus telson muscle thick filaments. J Mol Biol. 1981;153:781–790. doi: 10.1016/0022-2836(81)90418-6. [DOI] [PubMed] [Google Scholar]
- Stewart M, Kensler RW, Levine RJ. Three-dimensional reconstruction of thick filaments from limulus and scorpion muscle. J Cell Biol. 1985;101:402–411. doi: 10.1083/jcb.101.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MA, Franks-Skiba K, Chen S, Cooke R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci U S A. 2010;107:430–435. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulbarán G, Biasutto A, Alamo L, Riggs C, Pinto A, Mendéz F, Craig R, Padrón R. Different head environments in tarantula thick filaments support a cooperative activation process. Biophys J. 2013;105:2114–2122. doi: 10.1016/j.bpj.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. Electron diffraction of frozen, hydrated protein crystals. Science. 1974;186:1036–1037. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. Electron microscopy of frozen hydrated biological specimens. J Ultrastruct Res. 1976;55:448–456. doi: 10.1016/s0022-5320(76)80099-8. [DOI] [PubMed] [Google Scholar]
- Vainstein V. Diffraction of X-ray by chain molecules. New York: American Elsevier Publishing Co.; 1963. [Google Scholar]
- Vandenboom R. Modulation of skeletal muscle contraction by myosin phosphorylation. Compr Physiol. 2017;7:171–212. doi: 10.1002/cphy.c150044. [DOI] [PubMed] [Google Scholar]
- Vibert P. Helical reconstruction of frozen-hydrated scallop myosin filaments. J Mol Biol. 1992;223:661–671. doi: 10.1016/0022-2836(92)90982-p. [DOI] [PubMed] [Google Scholar]
- Vibert P, Craig R. Electron microscopy and image analysis of myosin filaments from scallop striated muscle. J Mol Biol. 1983;165:303–320. doi: 10.1016/s0022-2836(83)80259-9. [DOI] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA. Visualization of head-head interactions in the inhibited state of smooth muscle myosin. J Cell Biol. 1999;147:1385–1390. doi: 10.1083/jcb.147.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci U S A. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Naber N, Pate E, Cooke R. The myosin inhibitor blebbistatin stabilizes the super-relaxed state in skeletal muscle. Biophys J. 2014;107:1637–1646. doi: 10.1016/j.bpj.2014.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padrón R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- Wray JS. Structure of the backbone in myosin filaments of muscle. Nature. 1979;277:37–40. doi: 10.1038/277037a0. [DOI] [PubMed] [Google Scholar]
- Wray JS. Organization of myosin in invertebrate thick filaments. Soc Gen Physiol Ser. 1982;37:29–36. [PubMed] [Google Scholar]
- Wray JS, Vibert PJ, Cohen C. Diversity of cross-bridge configurations in invertebrate muscles. Nature. 1975;257:561–564. doi: 10.1038/257561a0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Kimura M, Li ZB, Ohno T, Takemori S, Hoh JF, Yagi N. X-ray diffraction analysis of the effects of myosin regulatory light chain phosphorylation and butanedione monoxime on skinned skeletal muscle fibers. Am J Physiol Cell Physiol. 2016;310:C692–C700. doi: 10.1152/ajpcell.00318.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Lee K, Dato O, Ikene M, Craig R. 3D Recontruction of the folded, inhibited form of vertebrate smooth muscle myosin II by single particle analysis. Biophys J. 2017;112:266a. [Google Scholar]
- Yang S, Woodhead JL, Zhao FQ, Sulbarán G, Craig R (2015a) An approach to improve the resolution of helical filaments with a large axial rise and flexible subunits. J Struct Biol 193:45–54 [DOI] [PMC free article] [PubMed]
- Yang, S., Zhao, F.Q., Sulbarán, G., Woodhead, J.L., Alamo, L., Pinto, A., Padrón, R., and Craig, R. (2015b) Improved imaging, 3D reconstruction and homology modelling of tarantula thick filaments. Biophys J 589a
- Zhao FQ, Craig R. Ca2+ causes release of myosin heads from the thick filament surface on the milliseconds time scale. J Mol Biol. 2003;327:145–158. doi: 10.1016/s0022-2836(03)00098-6. [DOI] [PubMed] [Google Scholar]
- Zhao FQ, Craig R. Capturing time-resolved changes in molecular structure by negative staining. J Struct Biol. 2003;141:43–52. doi: 10.1016/s1047-8477(02)00546-4. [DOI] [PubMed] [Google Scholar]
- Zhao FQ, Padrón R, Craig R. Blebbistatin stabilizes the helical order of myosin filaments by promoting the switch 2 closed state. Biophys J. 2008;95:3322–3329. doi: 10.1529/biophysj.108.137067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sun Y, Zhao FQ, Yu J, Craig R, Hu S. Analysis of tarantula skeletal muscle protein sequences and identification of transcriptional isoforms. BMC Genomics. 2009;10:117. doi: 10.1186/1471-2164-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi ME, Woodhead JL, Craig R, Padrón R. Helical order in tarantula thick filaments requires the "closed" conformation of the myosin head. J Mol Biol. 2004;342:1223–1236. doi: 10.1016/j.jmb.2004.07.037. [DOI] [PubMed] [Google Scholar]