Abstract
Circular dichroism (CD) spectroscopy is a fast, powerful, well-established, and widely used analytical technique in the biophysical and structural biology community to study protein secondary structure and to track changes in protein conformation in different environments. The use of the intense light of a synchrotron beam as the light source for collecting CD measurements has emerged as an enhanced method, known as synchrotron radiation circular dichroism (SRCD) spectroscopy, that has several advantages over the conventional CD method, including a significant spectral range extension for data collection, deeper access to the lower limit (cut-off) of conventional CD spectroscopy, an improved signal-to-noise ratio to increase accuracy in the measurements, and the possibility to collect measurements in highly absorbing solutions. In this review, we discuss different applications of the SRCD technique by researchers from Latin America. In this context, we specifically look at the use of this method for examining the secondary structure and conformational behavior of proteins belonging to the four main classes of the hierarchical protein domain classification CATH (Class, Architecture, Topology, Homology) database, focusing on the advantages and improvements associated with SRCD spectroscopy in terms of characterizing proteins composed of different structural elements.
Keywords: Circular dichroism spectroscopy, Conformational changes, Protein conformation, Protein secondary structure, Synchrotron radiation circular dichroism spectroscopy
Introduction
The phenomenon of circular dichroism (CD) is based on the difference of absorption between a left- and a right-handed circularly polarized light, which arises after such radiation passes through an optically active (chiral) molecule (Wallace and Janes 2009; Ranijbar et al. 2009). In the CD spectrum of a substance, the slight differences in absorbance between these two components of light are measured as a function of the wavelength. Ultraviolet (UV) CD spectroscopy is widely used as an analytical technique to investigate the native fold of soluble (Kelly et al. 2005) and membrane (Miles et al. 2016) proteins, and to follow the conformational changes in their secondary structure (Greenfield 2006), because the CD spectra of proteins are extremely sensitive to the molecular conformations they adopt. At the far-UV region (190– 250 nm), one of the main chromophores to be monitored in the CD spectrum of a protein is the peptide bond (Kelly et al. 2005). Consequently, the electronic transitions in amide groups of the peptide backbone can be described as specific differential absorption bands whose positions are affected by the particular regular structure adopted by the polypeptide chain.
The technique of CD is a fast and powerful spectroscopic method and highly complimentary to other biophysical methods used to characterize proteins and their interactions with partners at molecular level (Poklar 2017), such as the small angle X-ray scattering, different approaches of fluorescence spectroscopy, spin paramagnetic resonance, nuclear magnetic resonance, surface plasmon resonance, and calorimetry. The broad dissemination and the large use of UV-CD spectroscopy among the biochemistry/biophysics community to study proteins is supported by a list of practical advantages of the technique (Wallace and Janes 2009), such as easy sample preparation (proteins are diluted in an appropriated buffer or deposited in dehydrated films), small amount of material required (usual protein concentration is about 0.1–0.15 mg/ml), nondestructive procedure (after measurement, sample recovery is possible), fast measurements (collecting one spectrum may take ~10 min), easy interpretation of spectral bands in terms of protein conformation (all canonical types of secondary structure have bands at well-defined positions within the far-UV region), and accessibility to online softwares/webservers (Sreerama et al. 2000; Whitmore et al. 2004; Micsonai et al. 2015) that have been developed to estimate protein secondary structure from the characteristic CD spectrum profile.
However, although CD spectrometry is a widely applied method, under specific conditions CD measurements may occasionally present severe limitations that hamper the collection of a good quality spectrum (Kelly et al. 2005; Wallace and Janes 2009). Aggregated/turbid samples or solutions containing large particles (including lipid vesicles) may cause light scattering that can interfere with the quality of the CD measurement. Moreover, some of the commonly used buffers (such as HEPES, borate/acetate, citrate) and/or components used for exposing protein structure to different environments (such as sodium chloride, glycerol, urea, guanidine chloride, β-mercaptoethanol, dithiothreitol) may interact with far-UV light to increase the buffer’s absorption of light. As a consequence, the quality of the spectrum will be affected, especially below the 200 nm region, where an abrupt decrease in the intensity of the light source emitting from CD equipment (Xenon arc lamp) is observed (Wallace 2009). Therefore, estimations of the structural content from a CD spectrum collected under any of these conditions may be poor and very limited.
A recent alternative to overcome the limitations found in the conventional CD technique uses the radiation from a synchrotron beam as the light source to collect CD measurements (Sutherland et al. 1992; Wallace 2000, 2009). The higher flux of photons of a synchrotron beam within the low wavelength UV region results in a significant spectral range extension for data collection. The additional wavelength range added to the CD spectrum is further converted into extra structural information on the protein studied. This method is called synchrotron radiation circular dichroism (SRCD) spectroscopy and represents not only a more accurate alternative, but also a more precise, and more sensible approach to the investigation of changes (even subtle) in the secondary structure of proteins and to the monitoring of their dynamic conformational behavior.
Many research groups have used SRCD spectroscopy to examine and estimate the secondary structure content of proteins (Lees et al. 2006; Matsuo and Gekko 2013; Garcia et al. 2013; Hussain et al. 2016; Lopes et al. 2016; Miler et al. 2016). The technique has been successfully employed to characterize the insertion and orientation of helical peptides in lipid bilayers (Bürck et al. 2016) or compare the behavior of globular and natively unfolded proteins (Ruskamo et al. 2012; Yoneda et al. 2017), investigate protein/peptide structure in model membranes (Miles et al. 2008; Dreschler et al. 2009), assess the dynamic and flexible conformation of unordered proteins (Lopes et al. 2014a, b; Bremer et al. 2017), evaluate protein:protein complex formation (Cowieson et al. 2008; Lopes et al. 2013), detect conformational changes induced by the binding to ligands (Lima et al. 2013), perform fold recognition and obtain an accurate distinction between parallel and antiparallel β-sheets (Micsonai et al. 2015) and identify the conformational changes associated with a mutant protein (Wallace et al. 2004). Likewise, a remarkable application of SRCD spectroscopy is defining of the structure at the residue level of the C-terminal domain in a prokaryotic voltage-gated sodium channel (Powl et al. 2010). The high precision, reproducibility, and accuracy of the SRCD method (not commonly achieved with conventional CD spectroscopy) are required to produce the level of details described in such applications.
In this paper, we review the use of the SRCD method in studies conducted by researchers in Latin America, describing the different applications of the technique to investigate the secondary structure of proteins that belong to the four classes of CATH (Class, Architecture, Topology, Homology) database (Orengo et al. 1997) and their conformational behavior when exposed to changes in physical chemical properties and/or in the presence of molecular partners. We focus on the advantages of the SRCD method and the improvements gained with its use.
SRCD to estimate protein secondary structure
Class 1: The all-α proteins
Determining the presence of the regular secondary structure α-helix in a protein structure is a relatively straightforward identification process by conventional CD and SRCD spectroscopy due to the large electric macrodipole of the α-helix interacting with the polarized light. The helix dipole arises due to the orientation of the peptide bond, resulting in the alignment of the individual peptide dipole moments parallel to the α-helix axis (~97% of the peptide dipole moments point in the direction of the helix axis; Wada 1976). Proteins with a secondary structure composed entirely of α-helices or those containing a major core of α-helices with small number of isolated β-elements on its periphery belong to the class of all-α proteins. All-α proteins are quite good systems to be investigated by CD spectroscopy because of the high magnitudes of the α-helix spectral bands, which present negative bands at 222 and 208 nm and a positive maximum band at 192 nm (Fig. 1).
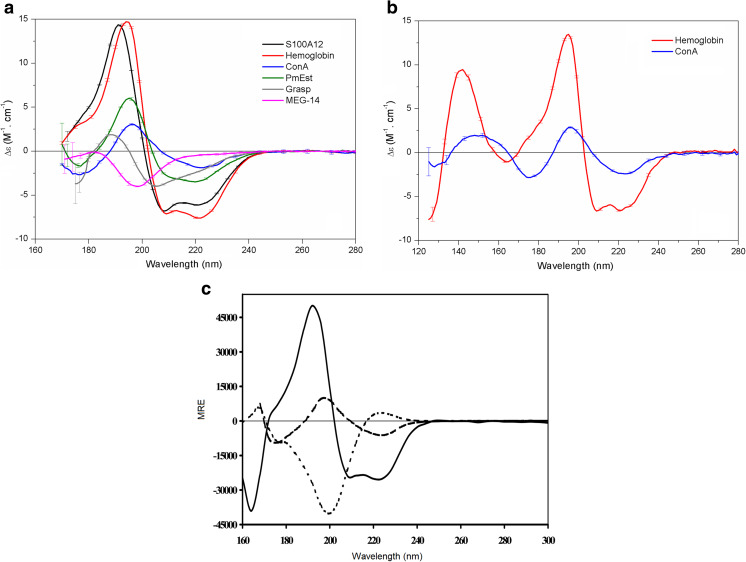
Fig. 1.
Synchrotron radiation circular dichroism (SRCD) spectra of proteins representative of the four different CATH (Class, Architecture, Topology, Homology) classes. a SRCD spectra of the proteins S100A12 (black), hemoglobin (red), concanavalin A (ConA; blue), PmEst (green), CnGRASP (gray), and MEG-14 (magenta) in aqueous solution. Adapted from Garcia et al. (2013), Yoneda et al. (2017), Lopes et al. (2013, 2016), and Mendes et al. (2016), with permission. b SRCD spectra of the dehydrated film of hemoglobin (red) and ConA (blue). Adapted from Yoneda et al. (2017). c SRCD spectra of proteins containing mostly α-helical (solid line), mostly β-strand (dashed line), and mostly polyproline (dotted line) structures. Adapted from Wallace and Janes 2009. Error bars express the standard deviation (SD) of each measurement
The S100 proteins are members of this class and share a highly conserved helical folding across different species. The recombinant S100A12 (previously described in Garcia et al. 2008) is a typical all-α protein that had its secondary structure first investigated with conventional CD and then with SRCD spectroscopy (Garcia et al. (2013). Porcine S100A12 is a relatively small (11 kDa) calcium binding protein with two EF-hand motifs and an additional zinc binding site at the C-terminus. The major helix content estimated for S100A12 with CD spectroscopy (~75%) is in agreement with its classification in the CATH class. SRCD spectroscopy, however, was employed to monitor more accurately the secondary structure content of the recombinant protein (Garcia et al. 2013) and to investigate the intermolecular interactions relevant to its role as a peripheral membrane protein. Such investigations require the use of biomembrane models (phospholipids vesicles) in high concentrations, which may promote light scattering that in turn hampers the quality of the CD spectrum in the CD method.
The SRCD spectrum of S100A12 (Fig. 1a) presented spectral bands in the far-UV wavelength region that are typical of a mainly α-helical protein, such as the negative peaks at 222 nm and 208 nm and the positive maximum peak at 192 nm. In addition, the SRCD spectrum of this calcium binding protein in aqueous solution showed an additional shoulder in the low-wavelength region at 180–185 nm that was not accessed in previous studies. Estimations of the structural content from the S100A12 SRCD spectra revealed that the protein is mainly composed of α-helices (63%), but with a significant content of either turns or unordered structure (~22%). Therefore, a mismatching of 12% in the helix content was evidenced when estimations performed with conventional CD (Garcia et al. 2008) were compared with those performed by SRCD (Garcia et al. 2013). The authors of these studies attributed the mismatching to the limitations of the conventional CD method, especially in terms of monitoring the positive peak at 192 nm (which was completely observed with SRCD spectroscopy). Moreover, deconvolution of the S100A12 SRCD spectrum into structural content (Table 1) was much closer to that seen in the crystal structure of human S100A12 [63% helix in Protein Data Bank (PDB) ID 2WCE] or to the bioinformatics predictions for the homology model to the porcine S100A12 (65% helix), revealing that the SRCD method may allow for an increased accuracy for estimating the secondary structure of proteins.
Table 1.
Estimations of the secondary structure content of proteins by conventional circular dichroism and synchrotron radiation circular dichroism methods
| Protein | Method | α-Helix (%) | β-Strand (%) | Disordered (%) | Other (%) | NMRSD |
|---|---|---|---|---|---|---|
| S100A12 | CD | 75 | - | 9 | 16 | 0.028 |
| SRCD | 63 | 3 | 22 | 12 | 0.011 | |
| Homology model | 65 | 4 | 26 | 5 | - | |
| PmEst | CD | 39 | 21 | 30 | 10 | 0.059 |
| SRCD | 31 | 26 | 33 | 10 | 0.064 | |
| Homology model | 33 | 18 | 37 | 12 | - | |
| MEG-14 | CD | 14 | 15 | 46 | 25 | 0.068 |
| SRCD | 2 | 10 | 78 | 10 | 0.034 | |
| Disorder prediction | - | - | 80–100 | - | - |
Another canonical α-helical protein studied with SRCD was hemoglobin. The major helix content of hemoglobin was seen both in aqueous solution (Fig. 1a) and when the protein was measured in a partially dehydrated film (Fig. 1b) deposited on the surface of quartz or calcium fluoride plates. The three-dimensional (3D) structure of globular proteins is stabilized by electrostatic forces, hydrogen bonds, hydrophobic interactions, van der Walls forces, and a substantial amount of water molecules. The removal of water may affect the structure and stability of proteins depending on how the water is removed: by fast removal, as in the lyophilization process, or slow removal, as in dry films.
Using SRCD, Yoneda et al. (2017) investigated the removal of water from proteins in a soft manner (dry films) and described the use of partially dehydrated films as a potential and useful approach to examine the structure of proteins with SRCD, since the secondary structure of globular proteins were shown to exhibit only minor changes upon removal of bulk water during film formation. However, these authors also pointed out that the group of proteins possessing significant irregular structure under dehydration conditions demonstrated a very distinct structural behavior . As bulk water was removed (during film formation), the disordered content of the protein changed, and there was a large gain of regular structure, with a larger number of folded conformations. This increase in regular structure was evidenced by an increase in the band around 224 nm in their CD spectra. The approach of collecting SRCD measurements in dehydrated films allowed accessing the region of vacuum-UV wavelengths below 170 nm, which is not possible in aqueous solution because of the strong water absorption peak near this wavelength. Therefore, the authors could collect an extended SRCD spectrum up to ~125 nm and monitor additional spectral bands within this region. For a major α-helix protein like hemoglobin, the additional spectral bands were described as a broad minimum at 165 nm and an intense maximum at 142 nm (Fig. 1b).
Class 2: The β-rich proteins
The β-strand is another regular secondary structure of a polypeptide chain that puts the backbone in a more extended conformation than it assumes in an α-helix. The β-strands can be arranged together in β-sheets with optimal dihedral angles and close hydrogen bonding distances, in which chains are nearly completely extended. Using data collected from a laboratory-based machine, the correct assignment and the estimations of the structural content of a β-rich protein are not as precise as that performed for an all-α protein due to the almost insignificant net dipole moment of a β-strand structure, which results in spectral bands of small magnitudes presenting with a negative band at 216 nm and a positive band at 198 nm (Fig. 1c). Moreover, in the region accessed by the conventional CD method, both α-helices and β-strands present minima and maxima at very similar positions, making it difficult to make a clear distinction between these individual spectral bands when both regular structures are present. Therefore, the use of the SRCD method for studying β-rich proteins has the important advantage of extending the spectral range to the low wavelength UV-region where the magnitudes and the peak positions for α-helices and β-strands are better distinguished.
The structural content of the all-β protein concanavalin A (ConA) from Canavalia ensiformis was investigated with SRCD by Yoneda et al. (2017). The structural content of ConA is predominantly organized in β-strands (~48%) with an almost negligible content of helices (~3%). The approach using dehydrated film to obtain the SRCD spectrum of ConA (Fig. 1b) enabled the authors to extend the spectral range significantly to up to ~125 nm. Accessing the low wavelength UV-region of this β-rich protein helped to overcome the limitations found with the CD method, enabling the β-content to be distinguished. The additional peaks for characterizing the β-strand structure in ConA were a minimum at 175 nm (while at this wavelength, the spectral band for α-helix is a positive maximum) and a broad positive maximum at 160–140 nm (while negative values are found for the α-helix structure).
When working with proteins in aqueous solutions, going deep to wavelengths of below 170 nm is not possible. In any event, the information provided with conventional CD method is limited at an early stage, at ~190 nm (Wallace and Janes 2009). Therefore, the CD spectrum is less accurate in terms of estimating the structural content in all-β proteins than the spectrum collected the SRCD method, which could range to 170 nm, thereby accessing some of the additional bands for this regular secondary structure between 190 and 170 nm.
Class 3: The mixed α-β proteins
Proteins with a secondary structure containing a mix of α-helix and β-strands are found in two different arrangements: (1) the α/β-proteins, which in addition to the α-helix content present high amounts of β-strands, with both elements alternating along the backbone; (2) the (α + β)-proteins, which present two separate domains, one composed of α-helices and other major composed of β-strands. The ability to access the low wavelength region with SRCD, which is where the two canonical structures, the α-helix and β-strands, present different peak positions and spectral bands of opposite magnitudes, truly improves the quantitative analyses of both mixed α/β- and (α + β)-proteins.
Similar to the failure observed in the class of all-β proteins, the estimations of secondary structure in this mixed class tend to be equally problematic and less reliable using conventional CD spectroscopy. These limitations of CD spectroscopy are due to the high diversity of possible β-structures, which is manifested not only in antiparallel or parallel orientation of the neighboring β-strands, but also in variations in length, extent, direction, degree of twist, and distortion of the β-sheets (Micsonai et al. 2015), and also to the lower spectral amplitudes of the β-content, which can cause a super estimation of the helical content over the β-structure.
An example of a protein containing this mixed α-β folding was studied by Lopes et al. (2016) with SRCD spectroscopy. The protein (named PmEst) was originally produced by the extremophile organism Petrotoga mobilis and exhibits hydrolytic activity on short acyl chains esters. While no crystal structure for PmEst is available, the homology model predicted for its 3D structure was used in comparison with the structural content estimated with the SRCD method and bioinformatics analyses (Lopes et al. 2016). Estimations of structural content for PmEst based on the SRCD method were more closely correlated with the homology model than those performed with laboratory-bench CD (see values in Table 1). Moreover, it is possible to observe that the SRCD spectrum of PmEst (Fig. 1a), classified as an α/β-protein, represents an almost a linear combination between the spectra observed for the all-α and all-β proteins, presenting spectral bands at the same positions expected for an all-α protein, but with reduced spectral amplitudes (similar to the all-β proteins).
Class 4: The unordered proteins
Many of the biophysical methods routinely used to investigate the structure of folded proteins fail when applied to investigate the natively unfolded proteins. These proteins, also referred to as intrinsically disordered proteins (IDPs), are a large and relevant group of functional proteins that lack a persistent structure in native conditions, presenting high conformational flexibility and a striking ability to undergo conformational changes upon interactions with molecular partners. Some IDPs can be fully disordered and do not contain any rigid/fixed 3D structure, thereby lacking any regular content, such as α-helix or β-strands. CD spectroscopy is a very useful tool to study these quite dynamic molecules since it is suitable for identifying the disordered content (due to the presence of the negative band at ~198 nm in disordered proteins), following their dynamic behavior, and tracking their disordered-to-order transitions as a response to interactions with a partner.
One of the classic members of the IDPs studied with SRCD by Lopes et al. (2013) was found in the genome of the flatworm parasite Schistosoma mansoni. The genome of this parasite possesses a group of genes with a remarkable and unusual structure (very small size and symmetrical exons), called micro-exon genes (MEGs). Due to the alternatively spliced transcripts to be translated from each gene, the primary structures of proteins coded by MEGs exhibit high variability. The soluble domain of a protein belonging to this group, named MEG-14 (previously described in Lopes et al. 2013), was found to have no homology with any other known families of proteins in any organism. The primary structure of MEG-14 is in fact intriguing due to the remarkable presence of charged and proline residues spread throughout its sequence and the absence of aromatic residues; these two properties provide the protein with a high propensity to adopt more extended (not folded) conformations. Moreover, experimental evidence reported by Lopes et al. (2013) confirms the complete disordered state of MEG-14 in aqueous solution, such as the unusual migration of MEG-14 during sodium dodecyl sulfate-polyacrylamide electrophoresis, its elution profile in size exclusion chromatography, and the results of bioinformatics analysis with disordered predictions. Nonetheless, the use of SRCD spectroscopy revealed MEG-14 to be a complete disordered protein that is able to assume a more partially folded conformation in response to its binding to partners or changes in the environmental factors.
The major peak observed in the SRCD spectrum of MEG-14 in aqueous solution (Fig. 1a) is a strong negative band at 198 nm, the presence of which is in agreement with its quite disordered state. This typical SRCD spectrum of MEG-14 confirmed both its high disordered content and the absence of any of the canonical secondary structures of a globular protein; since no peaks above 205 nm were observed, this protein lacked any α-helix or β-strands elements. Moreover, as synchrotron radiation was used, an additional spectral band to study IDPs was revealed, which is a small positive peak between 180 and 185 nm. Estimations of secondary structure in MEG-14 were much better after the low wavelength region was considered in the analysis, suggesting ~80% disorder in the protein, which is consistent with the sequence-based predictions (Table 1).
Kumagai et al. (2017) also reported the importance of accurately reaching the low UV-wavelength region on CD data when working with IDPs, because the major CD peaks for analyzing the disordered structure occur at wavelengths of <200 nm. The wavelength extension was shown to clearly improve the observation of the full peaks at ~198 nm and ~185 nm in the SRCD spectrum of an IDP, but also to improve the signal-to-noise level, reducing the errors bars (improving the accuracy of measurement) below 200 nm. Therefore, the SRCD method has many advantages over the conventional CD method when studying IDPS because it allows accessing additional electronic transitions in this group of disordered proteins and provides higher information content in the spectrum which can significantly improve the structural analysis.
In contrast to MEG-14, which is a full IDP, a well-structured protein can also present one or more regions where a local intrinsic disorder is found; these regions are referred to as intrinsically disordered regions (IDRs). Alternatively, a certain degree of disorder within a protein may be found at a specific domain or linker, as in a protein composed of a combination of structured and disordered regions. Such different types of IDRs are equally important to the set of functions that the proteins can have to mediate/participate in various cell mechanisms (van der Lee et al. 2014). A protein containing IDRs studied with SRCD was investigated by Mendes et al. (2016) when characterizing a member of the Golgi reassembly and stacking proteins (GRASP) family. GRASP are proteins required for the stacking of Golgi cisternae, allowing lateral connections, and for the tethering of vesicles targeted for fusion with the Golgi apparatus (Kinseth et al. 2007, Feng et al. 2013). These proteins have two PDZ-like domains in the N-terminal region and a variable and non-conserved serine- and proline-rich domain at the C-terminus. Many functions related to Golgi functionality are attributed to GRASP due to the high number of partners with which they can interact. Since no 3D structure for full-length GRASP is available, except to only PDZ domains structures (Truschel et al. 2011; Feng et al. 2013), other low resolution techniques, such as spectroscopies, are suitable. The specfic SRCD spectrum of the full-length GRASP from the fungus Cryptococcus neoformans (CnGRASP) (Fig. 1a) was due to a mixed contribution of α/β structures and a large amount of unordered protein; however, the level of disorder differed from that observed for MEG-14. The SRCD spectrum of CnGRASP showed small shifts in the position of the bands assigned to the disordered structure towards the peaks of regular secondary structures (like an α-helix), moving the peaks from 185 and 198 nm to 189 and 205 nm, respectively. Additionally, the small magnitude of the negative peak at 222 nm and a contribution of the signal at 215 nm (feature of β-strands) provide evidence of partially folded regions in CnGRASP.
As a general rule, proteins presenting high contents of α-helix have a stronger CD signal than β-rich and/or unordered/disordered proteins. Therefore, when other types of regular secondary structure or disordered content present together with a helical structure, they can appear as small shifts in the peak positions of the SRCD spectrum of the α-helix and/or a reduction in the amplitude of the peaks. From the deconvolution of SRCD spectrum and structure prediction, CnGRASP was estimated to have, indeed, multiple disordered regions coexisting with structured regions (Mendes et al. 2016).
SRCD to track conformational changes in protein structure
Changes in protein conformation can occur as a response to modifications of the physical chemical properties of the environment (pH, polarity, temperature, ionic strength, concentration, etc.) as these changes may affect the maintenance of the non-covalent bonds that preserve the specific protein structure. Many different biophysical techniques can be employed to track such changes; however, the most common tools are CD/SRCD spectroscopy.
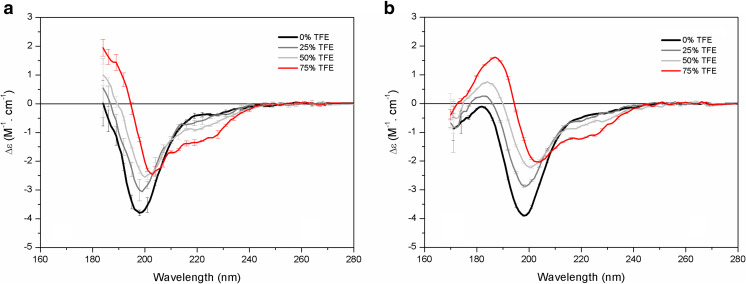
Trifluoroethanol (TFE) is a solvent known to promote conformational changes in the structure of proteins, inducing them to fold into more ordered conformations (Jasanoff et al. 1997). In order to demonstrate the advantages of using SRCD over conventional CD method, Kumagai and coworkers (Kumagai et al. 2017) conducted a series of comparative experiments with MEG-14 and soybean trypsin inhibitor (STI), a protein containing ~50% disordered content. The results showed that the improvements gained with SRCD are important in terms of increasing the accuracy of detecting changes, improving the estimations of disordered content, and discriminating between the partially folded states of a natively unfolded protein. Both CD (Fig. 2a) and SRCD (Fig. 2b) approaches were able to monitor the significant α-helix induction in the disordered proteins in the presence of TFE, which were observed by the gradual increase of the two minima at 208 and 222 nm and the positive maximum at 192 nm. Although all changes were observed in the conventional far-UV region, large error bars of <200 nm affected the analysis of the peak at ~198 nm when the conventional method was used, while the SRCD approach enabled a more accurate analysis of the changes on the entire negative peak at 198 nm. The SRCD method also showed additional transitions at the 184 nm region. In addition, the signal-to-noise ratio was improved and data were of better quality with the SRCD method, with a lower number of repeated scans than required in the conventional CD method, thereby reducing the time for data collection. Therefore, the improved signal-to-noise level of SRCD might also be considered as a significant advantage of the method.
Fig. 2.
Tracking conformational changes on a protein with SRCD spectroscopy. Comparison of conventional CD (a) and SRCD (b) spectra of MEG-14 in 10 mM sodium phosphate buffer, pH 7.4 (black), and in the presence 25% (gray), 50% (light gray), and 75% (red) trifluoroethanol (TFE). Adapted from Kumagai et al. (2017) with permission
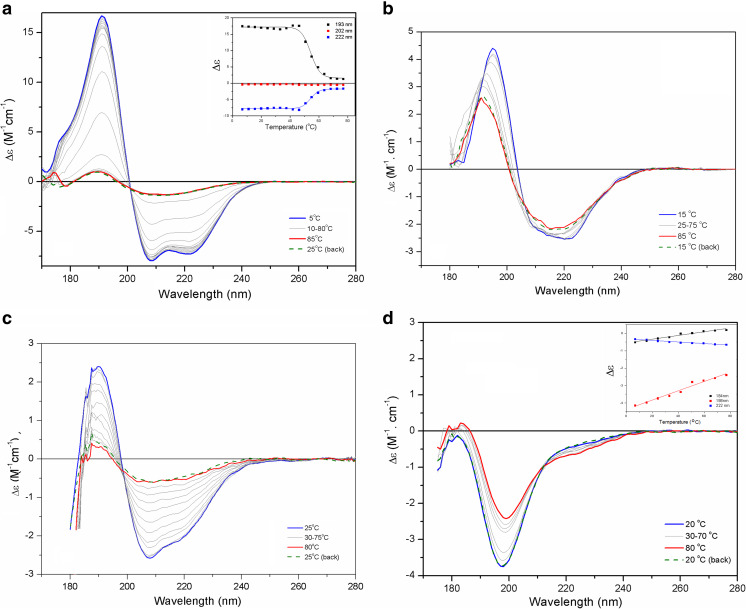
The unfolding or the thermal denaturation of a protein is a process widely investigated using CD spectroscopy, following the loss of a protein’s native conformation via the gradual decrease of the typical spectral bands in the native protein. The occurrence of conformational changes in a protein as an effect of temperature changes has been performed with proteins belonging to each of the CATH classes (Fig. 3a–d). Garcia et al. (2008) conducted a thermal melting assay of S100A12 over a range of temperatures from 15 °C to 80 °C with SRCD; the process was marked by a huge decrease in the spectral bands for the helical content with increasing temperature. Monitoring the spectral changes at fixed ellipticity values (at 222, 202, and 193 nm) resulted in quite good sigmoidal curves, suggesting that S100A12 denaturation occurred in a two-state process, based on the existence of an isodichroic (isosbestic) point in the SRCD curves, which is a good indicator of a true equilibrium process.
Fig. 3.
Evaluating thermal stability of proteins with SRCD spectroscopy. SRCD spectra of the thermal melting of S100A12 (a), PmEst (b), CnGRASP (c), and MEG-14 (d). Inset: Ellipticity values taken at 193, 202, and 222 nm during the thermal melting of S100A12 and at the values of 184, 198, and 222 nm for MEG-14. Adapted from Garcia et al. (2013), Lopes et al. (2013, 2016), and Mendes et al. (2016), with permission
Lopes et al. (2016) studied the thermal behavior of the enzyme PmEst and found that this enzyme was able to retain its native structure quite stably until 55 °C, which is in agreement with the preservation of its hydrolytic activity. When significant changes on the SRCD spectrum of the enzyme were noted (above 55 °C), abolishment of the hydrolytic activity of the enzyme was also observed, suggesting that although the enzyme was not complete unfolded, it may assume an inactive conformation.
The effect of temperature in the class of IDPs can also go in the opposite direction of that for globular proteins. The full IDPs usually undergo conformational changes, tending to increase their amount of regular structure with increases in temperature (Recveur-Brechot et al. 2006; Uversky 2009). Such disorder-to-order conformational changes were shown in MEG-14 by Lopes et al. (2013) as an almost linear dependence with temperature increase. Albeit subtle, the amplitude changes observed in the SRCD spectra of MEG-14 (Fig. 3c) in the peaks at 198 and 184 nm followed by an increase in the ellipticity values at 222 nm are in agreement with the reduction of its disordered content due to the increase in the strength of hydrophobic interactions at higher temperatures, taking the protein into a more partially folded conformation. However, the transitions caused by the temperature increase were quite reversible, with the disordered protein resuming its complete flexible and unfolded state after exposure to cooling temperature of 25 °C. The thermal melting of proteins containing IDRs may show a more predictable behavior than that for globular proteins, but with lower cooperativity between the native/denatured transition. This typical behavior was observed for CnGRASP, which gradually lost its regular secondary structure content with a low cooperativity as temperature increased; the thermal unfolding occurred in an irreversible process.
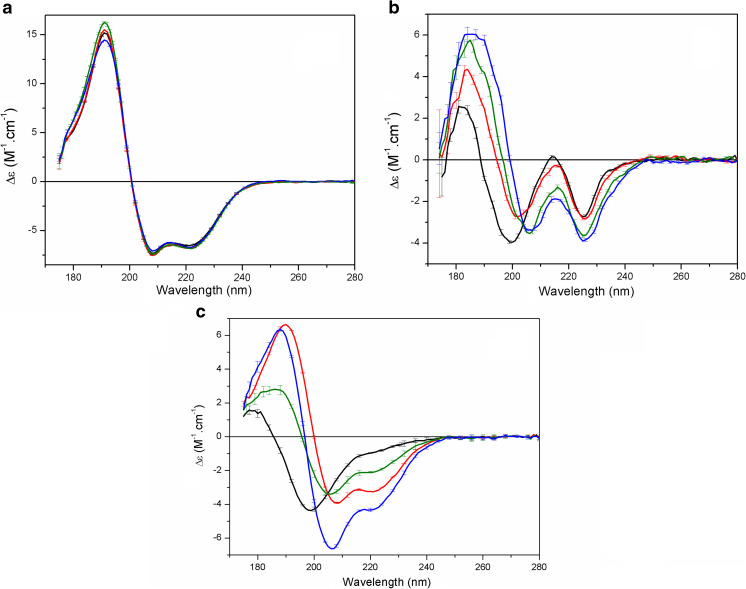
Conformational changes may also take place in a protein upon its interaction with a molecular partner, which could be a small ligand, an ion, a carbohydrate, a lipid or a lipid membrane, another protein, among others. SRCD spectroscopy has been used to study all of those interactions. The binding of S100A12 to large unilamellar vesicles of zwitterionic and negatively charged phospholipids was clearly demonstrated with SRCD. The use of such lipid systems in conventional CD would have promoted light scattering. However, the scattering caused by the presence of vesicles was quite insignificant when using SRCD spectroscopy to study the binding of the S100A12 in 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) vesicles in the presence of calcium and/or zinc ions (Garcia et al. 2013), allowing the use of high lipid to protein molar ratios and the collection of the SRCD spectra down to ~175 nm (Fig. 4a).
Fig. 4.
Studying protein–partner interactions with SRCD spectroscopy. a S100A12 in the presence of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) liposomes (black) without ions and incubated with liposomes and calcium (red), zinc (green), or both ions (blue). b SRCD spectra of the peptide Sit1 in aqueous solution (black) and titrated with 0.3 mM (red), 0.6 mM (green), and 1.2 mM oleoyl-CoA (OCoA; blue). c SRCD spectra of MEG-14 (black) and S100A12 (red), the weighted sum of the MEG-14 and S100A9 spectra (green), and the spectrum of the solution containing the MEG-14/S100A9 complex (blue). Error bars indicate 1 SD in the repeat measurements. Adapted from Garcia et al. (2013), Lopes et al. (2014a, b), and Orcia et al. (2017) with permission
Lopes et al. (2014b) employed SRCD spectroscopy to examine peptide–lipid interactions, using synthetic peptides corresponding to the predicted substrate binding sites of the diacylglycerol acyltransferase 1 (DGAT1), a membrane-bound enzyme on which little structural information is available. DGAT1 was predicted to have a large luminal extramembranous loop which includes the FYxDWWN motif common to acyl-CoA cholesterol acyltransferase, and the putative diacylglycerol binding domain HKWCIRHFYKP found in protein kinase C and diacylglycerol kinases. The authors studied the interactions between synthetic peptides corresponding to these two motifs and substrates associated with the triacylglyceride synthesis (oleoyl-CoA and dioleoylglycerol). One of the DGAT1 peptides, named Sit1, exhibited a SRCD spectrum (Fig. 4a) in aqueous solution typical of disordered structures (a negative band at ~198 nm and small positive band at ~184 nm), with very low per-residue magnitudes due to the high conformational flexibility found in the peptides, and a negative peak at ~228 nm, which was attributed to the excitation interactions of the adjacent Trp aromatic residues in this peptide. However, upon binding to oleoyl-CoA (OCoA) conformational changes suggested an increase in ordering (or decrease in disorder) in Sit1, such as the replacement of the 198 nm band by a peak at ~207 nm, and the red-shift and increase of the 184 nm peak. These SRCD results were put together with other complementary biophysical techniques to predict Sit1 as a potential site of interaction for the acyl-CoA substrate. Also, as the CoA molecules alone caused no changes in the Sit1 SRCD spectrum, the peptide–substrate binding was considered likely to be due primarily to an interaction with the acyl-chain of the substrate (OCoA).
Protein–protein interactions were studied with SRCD by Orcia et al. (2017) who performed the binding of the disordered protein MEG-14 to its validated partner within a human leukocyte cDNA library, the protein S100A9, which has an important role in the signaling of inflammatory processes in humans (definitive host for the tropical parasite S. mansoni). In order to investigate interactions between these two proteins due to complex formation, the SRCD spectrum of each protein alone was obtained and compared to the spectrum of the proteins incubated together (complex MEG-14/S100A9) and the weighted sum spectrum of the two components (taking into account the different number of residues in the two proteins). The SRCD analysis showed that the two proteins physically interact to form a complex and that upon formation of the complex, they undergo a conformational change. The formation of the MEG-14/S100A9 complex showed changes associated with an increase in the α-helix content and a significantly reduction in the disordered content of the individual proteins. These results were in good agreement with binding studies performed using other biophysical techniques.
Conclusions and perspectives
Synchrotron radiation circular dichroism spectroscopy is an emerging technique that represents the state-of-the-art of a biophysical method widely used to investigate the secondary structure of proteins and to monitor their conformational behavior in response to changes in the physical–chemical properties of the environment in which they are studied. Researchers in South America have been using this enhanced method in the structural analysis of proteins of different CATH classes The examples of the application of this method reviewed here clearly demonstrate that SRCD spectroscopy possesses a number of practical advantages in terms of monitoring protein structure, protein stability, and protein flexibility/conformational changes. Most of the advantages of SRCD over the conventional CD method are based on the higher flux of photons of the synchrotron radiation, which provides an extended interval for collecting CD data (especially for accessing lower wavelengths in the vacuum-UV region), but also increases the signal-to-noise ratio, providing thereby the potential to collect measurements in high absorbing buffers, as well as a better characterization of protein conformation and better assignment of each individual type of secondary structure. SRCD spectroscopy has been successfully employed to study protein interactions with partners of a distinct nature because of its better signal-to-noise ratio and access to lower wavelengths. Both of these are especially important for examining peptides and proteins upon binding to their partners, especially binding that causes only subtle conformational changes in protein structure, or even those that might include high non-chiral absorption of the partner at low UV-wavelengths.
SRCD measurements have been constantly taken at SRCD beamlines in Europe (UK, France, Denmark, and Germany) and Asia (Japan, Taiwan, and China). Currently, there is no SRCD beamline in operation in any synchrotron facility in the Americas. However, due to the introduction and broad dissemination of the SRCD method within the biophysicist community in Latin America, there is a real urge and discussion for the possible inclusion of this technique at the new Brazilian Synchrotron Light Laboratory (LNLS; http://lnis.cnpem.br) (Sirius Project) in Brazil, which would benefit greatly the use of SRCD on our continent and prompt many other successful examples of applications of the method—hopefully in a near future.
Acknowledgements
JLSL and PSK are grateful for beamtime access at the AU-CD beamline at the ASTRID2 (Aarhus, Denmark) and at the UV-CD12 at t he ANKA (Karlsruhe, Germany) synchrotrons. All authors thank Prof. BA Wallace for supporting the implementation of SRCD in Brazil and for helpful discussions.
Funding information
We thank following institutions for providing financial support in the form of grants: Sao Paulo Research Foundation (FAPESP) grants 15/50347-2 (to APUA), National Council for Scientific and Technological Development (CNPq) grants 150417/2016-0 (to PSK), 407337/2013-0 (to APUA) and 303513/2016-0 (to JLSL)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Latin America’ edited by Pietro Ciancaglini and Rosangela Itri.
References
- Bremer A, Wolff M, Thalhammer A, Hincha DK. Folding of intrinsically disordered plant LEA proteins is driven by glycerol-induced crowding and the presence of membranes. FEBS J. 2017;284(6):919–936. doi: 10.1111/febs.14023. [DOI] [PubMed] [Google Scholar]
- Bürck J, Wadhwani P, Fanghänel S, Ulrich AS. Oriented circular dichroism: A method to characterize membrane-active peptides in oriented lipid bilayers. Acc Chem Res. 2016;49(2):184–192. doi: 10.1021/acs.accounts.5b00346. [DOI] [PubMed] [Google Scholar]
- Cowieson NP, Miles AJ, Robin G, Forwood JK, Kobe B, Martin JL, Wallace BA. Evaluating protein:protein complex formation using synchrotron radiation circular dichroism spectroscopy. Proteins. 2008;70(4):1142–1146. doi: 10.1002/prot.21631. [DOI] [PubMed] [Google Scholar]
- Drechsler A, Miles AJ, Norton RS, Wallace BA, Separovic F. Effect of lipid on the conformation of the N-terminal region of equinatoxin II: A synchrotron radiation circular dichroism spectroscopic study. Eur Biophys J. 2009;39(1):121–127. doi: 10.1007/s00249-009-0445-x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Yu W, Li X, Zhou Y, Hu J, Liu X. Structural insight into Golgi membrane stacking by GRASP65 and GRASP55 proteins. J Biol Chem. 2013;288(39):28418–28427. doi: 10.1074/jbc.M113.478024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AF, Garcia W, Nonato MC, Araújo AP. Structural stability and reversible unfolding of recombinant porcine S100A12. Biophys Chem. 2008;134(3):246–253. doi: 10.1016/j.bpc.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Garcia AF, Lopes JLS, Costa-Filho AJ, Wallace BA, Araujo APU. Membrane interactions of S100A12 (Calgranulin C) PLoS One. 2013;8(12):e82555. doi: 10.1371/journal.pone.0082555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2006;1(6):2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain R, Jávorfi T, Rudd TR, Siligardi G. High-throughput SRCD using multi-well plates and its applications. Sci Rep. 2016;6:38028. doi: 10.1038/srep38028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasanoff A, Fersht AR. Mechanism of helix induction by trifluoroethanol: A framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemist. 1997;36:8413. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta. 2005;1751(2):119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Kinseth MA, Anjard C, Fuller D, Guizzunti G, Loomis WF, Malhotra V. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 2007;130:524–534. doi: 10.1016/j.cell.2007.06.029. [DOI] [PubMed] [Google Scholar]
- Kumagai PS, DeMarco R, Lopes JLS (2017) Advantages of synchrotron radiation circular dichroism spectroscopy to study intrinsically disordered proteins. Eur Biophys J. doi 10.1007/s00249-017-1202-1 [DOI] [PubMed]
- Lees JG, Miles AJ, Wien F, Wallace BA. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006;22(16):1955–1962. doi: 10.1093/bioinformatics/btl327. [DOI] [PubMed] [Google Scholar]
- Lima MA, Hughes AJ, Veraldi N, Rudd TR, Hussain R, Brio AS, Chavante SF, Tersariol II, Siligardi G, Nader HB, Yates EA. Antithrombin stabilisation by sulfated carbohydrates correlates with anticoagulant activity. Med Chem Commun. 2013;4:870–873. doi: 10.1039/C3MD00048F. [DOI] [Google Scholar]
- Lopes JLS, Miles AJ, Whitmore L, Wallace BA. Distinct circular dichroism spectroscopic signatures of polyproline II and unordered secondary structures: Applications in secondary structure analyses. Protein Sci. 2014;23(12):1765–1772. doi: 10.1002/pro.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JLS, Nobre TM, Cilli EM, Beltramini LM, Araujo APU, Wallace BA. Deconstructing the DGAT1 enzyme: Binding sites and substrate interactions. Biochim Biophys Acta. 2014;1838(12):3145–3152. doi: 10.1016/j.bbamem.2014.08.017. [DOI] [PubMed] [Google Scholar]
- Lopes JLS, Orcia D, Araujo APU, DeMarco R, Wallace BA. Folding factors and partners for the intrinsically disordered protein micro-exon gene 14 (MEG-14) Biophys J. 2013;104(11):2512–2520. doi: 10.1016/j.bpj.2013.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JLS, Yoneda JS, Martins JM, DeMarco R, Jameson DM, Castro AM, Bossolan NR, Wallace BA, Araujo APU. Environmental factors modulating the stability and enzymatic activity of the Petrotoga mobilis esterase (PmEst) PLoS One. 2016;11(6):e0158146. doi: 10.1371/journal.pone.0158146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Gekko K. Circular-dichroism and synchrotron-radiation circular-dichroism spectroscopy as tools to monitor protein structure in a lipid environment. Methods Mol Biol. 2013;974:151–176. doi: 10.1007/978-1-62703-275-9_8. [DOI] [PubMed] [Google Scholar]
- Mendes LFS, Garcia AF, Kumagai PS, Morais FR, Melo FA, Kmetzsch L, Vainstein MH, Rodrigues ML, Costa-Filho AJ. New structural insights into Golgi reassembly and stacking protein (GRASP) in solution. Sci Rep. 2016;6:29976. doi: 10.1038/srep29976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micsonai A, Wien F, Kernya L, Lee YH, Goto Y, Réfrégiers M, Kardos J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci USA. 2015;112(24):E3095–E3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles AJ, Drechsler A, Kristan K, Anderluh G, Norton RS, Wallace BA, Separovic F. The effects of lipids on the structure of the eukaryotic cytolysin equinatoxin II: A synchrotron radiation circular dichroism spectroscopic study. Biochim Biophys Acta. 2008;1778(10):2091–2096. doi: 10.1016/j.bbamem.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Miles AJ, Wallace BA. Circular dichroism spectroscopy of membrane proteins. Chem Soc Rev. 2016;45(18):4859–4872. doi: 10.1039/c5cs00084j. [DOI] [PubMed] [Google Scholar]
- Miller WC, Miles AJ, Wallace BA. Structure of the C-terminal domain of the prokaryotic sodium channel orthologue NsvBa. Eur Biophys J. 2016;45(8):807–814. doi: 10.1007/s00249-016-1125-2. [DOI] [PubMed] [Google Scholar]
- Orcia D, Zeraik AE, Lopes JLS, Macedo JNA, Santos CR, Oliveira KC, Anderson L, Wallace BA, Verjovski-Almeida S, Araujo APU, DeMarco R. Interaction of an esophageal MEG protein from schistosomes with a human S100 protein involved in inflammatory response. Biochim Biophys Acta. 2017;1861(1 Pt A):3490–3497. doi: 10.1016/j.bbagen.2016.09.015. [DOI] [PubMed] [Google Scholar]
- Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH-a hierarchic classification of protein domain structures. Structure. 1997;5(8):1093–1108. doi: 10.1016/S0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Poklar Ulrih N. Analytical techniques for the study of polyphenol-protein interactions. Crit Rev Food Sci Nutr. 2017;57(10):2144–2161. doi: 10.1080/10408398.2015.1052040. [DOI] [PubMed] [Google Scholar]
- Powl AP, O’Reilly AO, Miles AJ, Wallace BA. Synchrotron radiation circular dichroism spectroscopy-defined structure of the C-terminal domain of NaChBac and its role in channel assembly. Proc Natl Acad Sci USA. 2010;107(32):14064–14069. doi: 10.1073/pnas.1001793107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar B, Gill P. Circular dichroism techniques: Biomolecular and nanostructural analyses- a review. Chem Biol Drug Des. 2009;74(2):101–120. doi: 10.1111/j.1747-0285.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- Recveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- Ruskamo S, Chukhlieb M, Vahokoski J, Bhargav SP, Liang F, Kursula I, Kursula P. Juxtanodin is an intrinsically disordered F-actin-binding protein. Sci Rep. 2012;2:899. doi: 10.1038/srep00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287(2):252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- Sutherland JC, Emrick A, France LL, Monteleone DC, Trunk J. Circular dichroism user facility at the National Synchrotron Light Source: Estimation of protein secondary structure. BioTechniques. 1992;13(4):588–590. [PubMed] [Google Scholar]
- Truschel ST, Sengupta D, Foote A, Heroux A, Macbeth MR, Linstedt AD. Structure of the membrane-tethering GRASP domain reveals a unique PDZ ligand interaction that mediates Golgi biogenesis. J Biol Chem. 2011;286(23):20125–20129. doi: 10.1074/jbc.C111.245324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins and their environment: Effects of strong denaturants, temperate pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28:305–325. doi: 10.1007/s10939-009-9201-4. [DOI] [PubMed] [Google Scholar]
- van der Lee R, Buljan M, Lang B, Weatheritt RJ, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114(13):6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A (1976) The alpha-helix as an electric macro-dipole. Adv Biophys:1–63 [PubMed]
- Wallace BA. Conformational changes by synchrotron radiation circular dichroism spectroscopy. Nat Struct Biol. 2000;7(9):708–709. doi: 10.1038/78915. [DOI] [PubMed] [Google Scholar]
- Wallace BA, Wien F, Miles AJ, Lees JG, Hoffmann SV, Evans P, Wistow GJ, Slingsby C. Biomedical applications of synchrotron radiation circular dichroism spectroscopy: Identification of mutant proteins associated with disease and development of a reference database for fold motifs. Faraday Discuss. 2004;126:237–243. doi: 10.1039/b306055c. [DOI] [PubMed] [Google Scholar]
- Wallace BA. Protein characterisation by synchrotron radiation circular dichroism spectroscopy. Q Rev Biophys. 2009;42(4):317–370. doi: 10.1017/S003358351000003X. [DOI] [PubMed] [Google Scholar]
- Wallace BA, Janes RW (eds) (2009) Modern techniques for circular dichroism and synchrotron radiation circular dichroism spectroscopy. Advances in biomedical spectroscopy, vol 1. IOS Press, Amsterdam
- Whitmore L, Wallace BA (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res 32(Web Server issue):W668–73. [DOI] [PMC free article] [PubMed]
- Yoneda JS, Miles AJ, Araujo APU, Wallace BA. Differential dehydration effects on globular proteins and intrinsically disordered proteins during film formation. Protein Sci. 2017;26(4):718726. doi: 10.1002/pro.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]