Abstract
Septins are able to polymerize into long apolar filaments and have long been considered to be a component of the cytoskeleton alongside intermediate filaments (which are also apolar in nature), microtubules and actin filaments (which are not). Their central guanosine triphosphate (GTP)-binding domain, which is essential for stabilizing the filament itself, is flanked by N- and C-terminal domains for which no direct structural information is yet available. In most cases, physiological filaments are built from a number of different septin monomers, and in the case of mammalian septins this is most commonly either three or four. Comprehending the structural basis for the spontaneous assembly of such filaments requires a deeper understanding of the interfaces between individual GTP-binding domains than is currently available. Nevertheless, in this review we will summarize the considerable progress which has been made over the course of the last 10 years. We will provide a brief description of each structure determined to date and comment on how it has added to the body of knowledge which is rapidly growing. Rather than simply repeat data which have already been described in the literature, as far as is possible we will try to take advantage of the full set of information now available (mostly derived from human septins) and draw the reader’s attention to some of the details of the structures themselves and the filaments they form which have not be commented on previously. An additional aim is to clarify some misconceptions.
Keywords: Septins, Filament assembly, GTP-binding domain, N- and C-terminal domains, Interfaces, Crystal structures
A brief summary
Septins are cytoskeletal proteins capable of self-association, polymerization and binding to cell membranes (Kinoshita 2003; Bridges et al. 2014; Garcia et al. 2016). They were originally identified in yeast mutants deficient in the completion of the cell cycle (Hartwell 1971) and were subsequently localized to the septum during cell division, hence the name. They are present in a variety of eukaryotic cells, playing a fundamental role in cytokinesis as well as in the formation of diffusion barriers. The latter serve to restrict membrane components to a particular cellular component, for example the primary cilium, by limiting the rate of lateral diffusion (Hu et al. 2010). They also participate in the regulation of a series of important cellular processes, including the release of neurotransmitters, and in microtubule dynamics (Surka et al. 2002; Kinoshita et al. 2004; Hu et al. 2010; Bai et al. 2013). More recently they have also been implicated in host–bacterium interactions (Mostowy et al. 2010). Consistent with the localization where they were first discovered, at the bud-neck of budding yeast, septins have been shown to recognize regions of positive membrane curvature and to be enriched at these sites (Byers and Goetsch 1976; Bridges et al. 2016).
Humans present 13 septin genes, SEPT1 to SEPT12 and SEPT14, which are capable of generating a wide variety of protein products due to post-translational modification and alternative splicing. Here we will refer to the canonical protein products as SEPT1 to SEPT12 and SEPT14 (without the italics), according to the nomenclature established by Macara et al. (2002). Based on sequence similarity, they are classified into four different groups (Kinoshita 2003). Although several different nomenclatures have been proposed, it is common to refer to each group by a well-studied member; therefore SEPT1, SEPT2, SEPT4 and SEPT5 are part of the SEPT2 group; SEPT6, SEPT8, SEPT10, SEPT11 and SEPT14 form the SEPT6 group; the SEPT3 group is formed by SEPT3, SEPT9 and SEPT12; the SEPT7 group contains only SEPT7. This is how we will refer to them throughout the remainder of this review. Most remarkable is that, upon polymerization, septins form filaments which are typically comprised of more than one component, i.e. they are hetero-filaments involving a number of different septin monomers. The exact number involved depends on the species. The filament is apolar and non-helical (see below) and involves a linear arrangement of septin monomers strung together as a “string of beads” in which each septin of the hetero-complex occupies a specific position.
The division into groups forms the foundation for the understanding of the “Kinoshita rule,” a guideline implied by the studies of Kinoshita (2003) which rationalizes and predicts septin composition within a filament. This can best be understood with reference to the most well-known mammalian septin filament, which is composed of SEPT2, SEPT6 and SEPT7. The "rule" anticipates that each particular septin may be substituted by another from the same group while still retaining a viable filament. Thus, SEPT2, for example, could be substituted by SEPT1, SEPT4 or SEPT5 while SEPT6 could be replaced by SEPT8, SEPT10, SEPT11 or SEPT14. It was later shown that mammalian septin filaments could be based on combinations of four different septins (rather than three) and that these included also a member of the SEPT3 group (Kim et al. 2011; Sellin et al. 2011). Finally, experimental evidence indicates that the division into groups might be used to predict the nucleotide hydrolytic capability of septins (Huang et al. 2006; Sirajuddin et al. 2007; Macedo et al. 2013; Zeraik et al. 2014).
Based on these observations, a simple combinatorial argument suggests the possible existence of 60 different combinations of four different septins (one representative from each of the four groups). Nevertheless, the generality of Kinoshita’s (2003) hypothesis is far from totally established. Tissue-specific expression (Mostowy and Cossart 2012) is one reason for believing that the number of physiologically relevant combinations is, in fact, far lower than that which is predicted based on the above reasoning. Furthermore, it seems likely that some degree of selectivity may exist between specific septins from different groups. While this does not violate Kinoshita’s rule it may well lead to preferred combinations limiting the number which are physiologically relevant. Finally, some non-canonical arrangements have even been reported in the literature (Nagata et al. 2004).
Septins are guanine nucleotide binding proteins. Although many present GTPase activity, others have been shown not to hydrolyze guanosine triphosphate (GTP), and the role of nucleotide hydrolysis is largely unknown (Huang et al. 2006; Macedo et al. 2013; Zeraik et al. 2014). However, it is almost certainly involved in controlling assembly (Zent and Wittinghofer 2014; Weems and McMurray 2017) and possibly membrane association (Zeraik et al. 2014). The nucleotide binds to the GTP-binding domain (also called the G-domain or GTPase domain) which presents the highest degree of conservation between septins and includes three common GTP-binding motifs (G1, G3 and G4). G1 is also known as the P-loop or Walker A box, while G3 resides in a region known as switch II. G4 is important for the selective binding of guanine nucleotides (Fig. 1). The G-domain ends in the septin unique element (SUE), a stretch of approximately 50 residues which is characteristic of septins and distinguishes them from other small GTP-binding proteins. It is intimately related to the fact that septins form filaments (Fig. 1).
Fig. 1.
Septins and their filaments. a A schematic representation of a mammalian septin showing the three domains (N, G and C). The polybasic region (P) is found at the interface between the N- and G-domains while the nucleotide-binding sequences (G1, G3 and G4) are all located within the G-domain. The final 50 residues of the G-domain corresponds to the septin unique element (SUE) which distinguishes septins from other small GTPases. Little is known about the structure of the N- and C-domains although the latter normally includes heptad pseudo-repeats characteristic of coiled-coil structures. b The structure of a septin monomer (center) is shown within the context of its two neighbors along the filament. This gives rise to the NC- and G-interfaces. The nucleotide is bound at the latter, and its full binding site includes residues coming from both of the adjacent subunits. The SUE (purple) can be seen to contribute to both interfaces but most notably to the NC-interface which is unique to septins. It is this interface which allows septins to polymerize. The G1, G3 and G4 motifs are shown according to the same color scheme as used in a. They are essential for the selective recognition of guanosine triphosphate (GTP)
In addition to this central domain, all septins present N-terminal and C-terminal regions of variable length (the N- and C-domains), and little is known about their structure and function (Fig. 1a). While there is experimental evidence, at least for some septins, that the N-domains are intrinsically disordered (Garcia et al. 2006), the C-domains contain the heptad repeats characteristic of coiled coils and are anticipated to be important for filament assembly. All of the structures which will be described in the following section are of the G-domain, either in the form of a hetero-complex or from an individual septin.
In addition to contributing to contacts made between adjacent septin subunits along the filament (Sirajuddin et al. 2007), several studies indicate that the N-terminal domain may function as a platform for the interaction with other elements of the cytoskeleton and membranes (Bai et al. 2013). It may also play a role in controlling filament assembly, at least in yeast (Weems and McMurray 2017). In most septins the N-terminal domain presents a sequence adjacent to the G-domain that is rich in basic residues, commonly referred to as the polybasic region or α0 helix. In mouse SEPT4, this basic region has been shown to bind membranes via interaction with phosphatidylinositol 4,5-bisphosphate (PIP2) (Zhang et al. 1999a). The N-terminal domain of Saccharomyces cerevisiae Cdc10 has also been shown to participate in the interaction with lipid monolayers containing the same phospholipid. Finally, the N-terminal domain of SEPT9 has recently been shown to bind microtubules and F-actin (Bai et al. 2013; Smith et al. 2015).
Similar to the N-terminal domains, the C-domains of septins also vary considerably in terms of sequence and length, but in this case the variation appears to follow a trend in each septin group. For example, human septins of the SEPT6 and SEPT7 groups present a long C-terminal domain with more than 100 residues, while this region presents only an intermediate length in members of the SEPT2 group and is smallest of all in the SEPT3-like septins. As mentioned above, with the exception of the SEPT3 group, the sequences of the C-terminal domains are predicted to form coiled coils, and biophysical studies have shown that the C-termini of septins of the SEPT6 group associate with high affinity with those of SEPT7, forming stable hetero-dimeric coiled coils (Marques et al. 2012; Sala et al. 2016). The pairing of the coiled coil regions of SEPT6-like septins with SEPT7 is consistent with the predicted number of heptad repeats, which is greater than that expected for SEPT2 and its group members.
Besides their importance for interfacial interactions, the C-terminal domains may play other important roles, such as in the formation of inter-filament cross-bridges (Bertin et al. 2008) and, in the case of a SEPT10 homolog from Schistosoma mansoni, in binding to liposomes (Zeraik et al. 2016).
Having introduced the three septin domains, it is now worth making a brief comment on construct nomenclature. Throughout this text we will use the letters N, G and C after the septin name to refer to the domain content of the crystallized construct. For example, the SEPT3-GC construct includes both the GTPase domain and the C-terminal domain, while SEPT2-NG indicates a construct containing only the N-terminal and the GTPase domains. Similarly, the numbers in parentheses following the construct, where present, refer to the corresponding range of amino acid residues present therein. In an attempt to facilitate comprehension we will try, wherever possible, to be coherent in the use of color for the representation of the four different groups. The SEPT2, SEPT6, SEPT7 and SEPT3 groups will be represented in red, blue, yellow and green, respectively, unless otherwise stated.
Our principal objective in the remainder of this short review is to provide a broad overview of progress that has been made in the structural biology of septins over the last 10 years. Although we have aimed to be as complete as possible with respect to the description of the crystal structures themselves, we make no pretense towards absolute completeness concerning the functional implications, which are far reaching. Rather, we will call attention to the points which we consider to be the most interesting from the point of view of a structural biologist. Some excellent recent reviews already exist on the more general aspects of septins, their interactions and their filament assembly (Mostowy and Cossart 2012, Fung et al. 2014; Neubauer and Zeiger 2017a, b).
Recollection on septin structural studies conducted during the last 10 years
The SEPT2–SEPT6–SEPT7 hetero-complex and the SEPT2 G-domain (SEPT2-G)
The first study to present crystallographic data on septins was the milestone work of Sirajuddin at al. (2007), which presented both the structure of the human SEPT2 GTPase domain (SEPT2-G, 1-315) at 3.4 Å resolution [Protein Data Bank (PDB) code 2QA5] and of the human wild-type SEPT2–SEPT6–SEPT7 hetero-filament at 4 Å resolution (PDB code 2QAG). Released in 2007, and despite considerable subsequent effort, this is the only structure of a septin hetero-complex published to date. It is also the only crystal structure obtained from crystals containing full-length septins. The publication of this seminal work stimulated the community to rethink old ideas about septin assembly and led to considerable speculation in the literature. Some, but not all of this, was borne out by a series of crystal structures of individual septins which would follow shortly afterwards. Weirich and colleagues, for example, provided a particularly thought-provoking review in 2008 (Weirich et al. 2008), shortly after the publication of the hetero-complex structure and clearly heavily influenced by it.
Despite the limitations imposed by the low resolution of the original structure, including the lack of electron density for many loops and side chains, and the ambiguous interpretation of some regions, the filament structure unequivocally shows the SEPT2–SEPT6–SEPT7 order along the filament; in Fig. 2a they are shown in red, blue and yellow respectively.
Fig. 2.
Structure of the septin filament. a The hetero-complex formed by septin protein products SEPT2 (red), SEPT6 (blue) and SEPT7 (yellow). Throughout this review these colors will be systematically used to represent these respective septin groups. The hexameric core particle is indicated, and this can be used to reproduce an entire filament by systematic translations (thick arrow). True and pseudo-twofold axes are represented by solid and dashed vertical arrows, respectively. b Schematic representation of a filament based on an octameric core particle. In this case a member of the SEPT3 group (SEPT9) has been included at the terminal positions (shown in green). Two types of generic interface, G and NC, alternate along the filament. These can be further subdivided into NC, NChet and NChom for the NC-interfaces and GDD and GDT for the G-interfaces (further explanation is given in the text)
The use of crystallographic symmetry operations allows the observation of a continuous filament inside the crystal, formed by translational repeats of a linear hexamer composed of septins 7–6–2–2–6–7, in that order. Figure 2a shows that the hexameric core particle has internal symmetry possessing a twofold rotation axis perpendicular to the filament and bisecting the two copies of SEPT2 which occupy the central positions. This has occasionally been described incorrectly as “mirror“ symmetry. Upon polymerization, two copies of SEPT7 from different hexamers come together, leading to the appearance of additional twofold axes as well as to the translational symmetry described above, which relates hexamers along the filament. Furthermore, since all septin monomers are homologs and therefore present similar three-dimensional structures, the filament as a whole also presents pseudo-twofold rotation axes at the SEPT6–SEPT7 and SEPT2–SEPT6 interfaces. Of course, all symmetry elements which result from polymerization are only strictly valid for an infinitely long filament. It is the presence of the twofold symmetry that leads to the formation of an apolar filament (a filament with indistinguishable termini) different to that observed for actin filaments or microtubules (which have plus and minus ends). True twofolds are shown as solid arrows and pseudo-twofolds as dotted arrows in Fig. 2.
In the filament, each septin uses two different interfaces to interact with its neighbors. These have been baptized the G- and NC-interfaces (Sirajuddin et al. 2007) and alternate along the filament. The former is formed, in part, by the nucleotide binding site and the nucleotide itself interacts with residues coming from both monomers (described in section "The G- and NC-interfaces"). The NC-interface is formed largely by the N- and C-terminal regions of the G-domain. However, the SEPT2–SEPT6–SEPT7 complex also reveals that the N-domain (including the polybasic region) also contributes to this interface via domain-swapping (in reality this is probably more appropriately described as ″secondary-structure swapping" since the N-terminal region which interacts intimately with the neighboring monomer does not form a compact domain-like structure). Nevertheless, some caution should be exercised in interpreting some of the details of this structure due to its limited resolution. Figure 1b shows the importance of the SUE in contributing to both interfaces. However, this is clearly more marked in the case of the NC-interface where the C-terminal helix (α6) is a prominent feature. This is consistent with the fact that many small GTPases dimerize via the G-interface but do not form polymers. Clearly, an intact NC-interface (and thus a SUE) is essential for filament formation, a unique feature of septins.
The SEPT2–SEPT2 and SEPT6–SEPT7 interactions are mediated by NC-interfaces while the SEPT2–SEPT6 and SEPT7–SEPT7 interactions are generated by G-interfaces. Even at 4 Å resolution the electron density for the nucleotide in the different septins within the filament is observable, and indicates that SEPT2 and SEPT7 are bound to guanosine diphosphate (GDP), while a GTP molecule is observed bound to SEPT6. This is consistent with a lack of catalytic activity associated with the SEPT6 group (Sirajuddin et al. 2007).
Figure 2b shows a schematic representation of a filament based on an octameric core particle in which a member of the SEPT3 group (SEPT9), shown in green, has been included in a position consistent with observations reported recently (Nakahira et al. 2010; Kim et al. 2011; Sandrock et al. 2011; Sellin et al. 2011). Octamer-based filaments may well be the most relevant in physiological terms. In this case, the G- and NC-interfaces which alternate along the filament can be further subdivided into five distinct classes, two of which are G and three of which are NC. The GDD-interface is found between SEPT7 and SEPT9 and is expected to involve GDP molecules bound to both monomers. The GDT-interface is observed between SEPT2 and SEPT6 and has a GDP molecule bound to the former and a GTP molecule bound to the latter. The NC-interfaces include NC- (which lacks a coiled coil) between two SEPT9 monomers, NChet (involving a heterotypic coiled coil between SEPT7 and SEPT6) and NChom (involving a homotypic coiled coil between monomers of SEPT2.
Another feature of the pioneering structural study of the SEPT2–SEPT6–SEPT7 complex by Sirajuddin at al. (2007) is that no density was observed for the C-terminal domains, although the crystal was shown to contain full-length septins. Remarkably, the electron microscopy and X-ray crystallography data show that the C-terminal domains, predicted to form coiled coils, are not required for filament assembly. Although the study did not demonstrate that the filaments formed by constructs lacking the C-termini present the same septin content as the wild-type filament, this finding strengthened the notion of the importance of the G- and NC-interfaces of the G-domain in filament assembly.
The native SEPT2–SEPT2 interaction in the hetero-complex is mediated by an NC-interface. However, the content of the asymmetric unit in the human SEPT2-G structure is a dimer employing the G-interface, and it is this interface which is observed to stabilize the dimer in solution. This finding provided the first structural evidence of promiscuity in septin interactions, a feature that would later be observed in several other studies. This concept establishes that, when lacking the physiologically relevant binding partners, septins are capable of associating with each other through non-native interactions. Since in vivo human septin filaments are generally formed by four (or possibly three) different septins, the crystal structures in which individual septins are observed to form a filament all show examples of promiscuous septin interactions. However, to date, no crystal structure of a single full-length septin is available; therefore, the promiscuity might originate at least in part from the lack of the N- and C-terminal regions in the septin constructs used for crystallization. It is also probably induced by the high concentrations used for protein crystallization and the obligatory requirement for protein–protein contacts to arise during the formation of the crystal itself.
Taken together, the available structural data indicate that the GTPase domain alone may not contain all of the structural determinants of correct and selective filament assembly. In this regard a series of biophysical studies show that the C-terminal domains of septins from different groups interact with nanomolar affinity and, at least to some extent, may be responsible for the assembly of native NC-interfaces where the C-domain may play a dominant role in selective assembly (Marques et al. 2012; Sala et al. 2016).
Although available data on electron density are not sufficiently definitive to allow the unequivocal assignment of the sequence register, data on the three septins in the filament structure reveal a density for the α0 helix that indicates it contains the polybasic region. The structure of the human SEPT2 GTPase domain (PDB code 2QA5) also presents the N-terminal α0 helix, although with essentially the same limitations imposed by the poor resolution as for the hetero-complex. The structural interpretation of the electron density provided by the authors implies that the α0 helix is tucked into the NC-interface in a “domain swapped” arrangement. This can most clearly be seen at the interface between SEPT6 (in blue) and SEPT7 (in yellow) in Fig. 2a. However, it is far from clear if this orientation of the helix represents that which is relevant for membrane binding. It is tempting to speculate that a polyacidic region found in the NC-interface may play a role in providing electrostatic neutralization for α0 while stored within the interface.
In addition to the structural data, the authors performed a series of mutations and analytical gel filtration experiments, the results of which established that nucleotide-bound SEPT2 forms G-interface dimers in solution and that dimerization is induced by the presence of excess nucleotide. The study also established the notion that some septin interfaces might be disturbed by high salt concentration and that a high salt concentration and glycerol might be necessary to avoid precipitation in the purification steps before the crystallization.
Shortly after the release of the filament structure, the Structural Genomics Consortium (SGC) released the structure of SEPT2-G (residues 22–320) bound to GDP at 2.6 Å resolution (PDB code 2QNR). The construct used to obtain this crystal lacked the first 21 N-terminal residues (including the polybasic region) and the C-terminal domain predicted to form a coiled coil. This strategy was designed—and subsequently proven—to yield better diffracting crystals, and it was readily adopted by other research groups (Sirajuddin et al. 2009). Over the years, this structure was used as template for molecular replacement to solve several septin structures (Serrão et al. 2011; Macedo et al. 2013; Zeraik et al. 2014; Brausemann et al. 2016).
The Septin fold
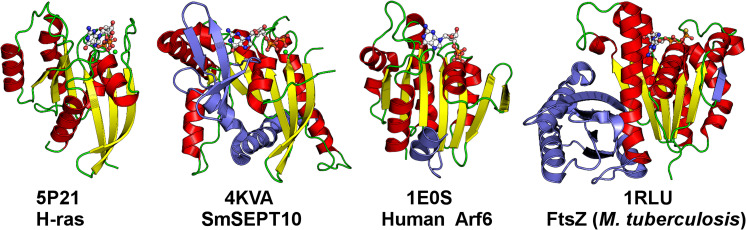
The fold of the G-domain is similar to that observed in all small GTP-binding proteins, typified by RAS p21. Many variants of the fold have been observed over the years, but all are based on a six-stranded β-sheet surrounded by α-helices in which the β2 strand generally runs antiparallel to the remainder (Fig. 1b). Figure 3 shows some variations on the theme in which different embellishments are observed. In the case of septins, the most notable additional feature is the SUE, which is important for filament formation (Fig. 1b). This SUE forms an additional small three-stranded β-sheet composed of β8, β9 and β10+β7 (which are really a single continuous strand but have been defined, for historical reasons, as two) as well as two α-helices (α5 and α6). The small sheet is often incompletely ordered in crystal structures of isolated G-domains. An additional helix (α5′) is observed between α4 and β6. This additional helix is close to the polyacidic region found at the NC-interface. The nomenclature of the principal elements of secondary structure are shown in Fig. 1b. These principal elements are also indicated, together with other important structural features, on an alignment of the G-domains for representative sequences in Fig. 4.
Fig. 3.
Variations in the RAS GTP-binding fold. The topology of H-RAS p21 is based on a central six-stranded β-sheet (yellow) surrounded by five α-helices (red). Additional elements in septins [represented by SmSEPT10 (SEPT10 from Schistosoma mansoni)] include the SUE (blue) and the alpha helix (α5′), representing embellishments. Arf6 (ADP ribosylation factor 6) is involved in endosomal trafficking, and its membrane association is controlled by the orientation of an N-terminal helix (blue), analogous to the α0 helix observed in septins. Re-orientation of the helix depends on the hydrolysis of GTP. FtsZ is a bacterial tubulin homolog which polymerizes into a filamentous ring during cell division. The β-sheet in FtsZ is fully parallel due to the reorientation of the β2 strand and there is a large C-terminal extension (blue) which is important for polymerization. Although FtsZ forms filaments, the architecture of these filaments is completely different from that of septins and they are polar in nature, like tubulin
Fig. 4.
Sequence alignment of representative septin G-domains. SEPT2, SEPT7, SEPT6 and SEPT3 respectively represent each of the four septin groups, and Sm10 is included for its high-resolution structure which permits an accurate assignment of the secondary structure elements. These are indicated in gray and olive shading for the β-strands and α-helices, respectively, together with their standard nomenclature. Structural features, such as the switches, P-loop and the polybasic and polyacidic regions (blue and red boxes, respectively) are explicitly indicated. The G-motifs common to most small GTP-binding proteins are shown in green boxes, while septin-specific motifs (Pan et al. 2007) are shown in black boxes with an orange background. The latter are characteristic of septins and serve to distinguish them, together with the SUE (dashed box), from other small GTPases. Selected residues of structural or functional importance are indicated with colored triangles, the significance of which is given below the alignment. The numbers are sequential and do not correspond to any particular septin
SEPT2-G bound to a GTP analog
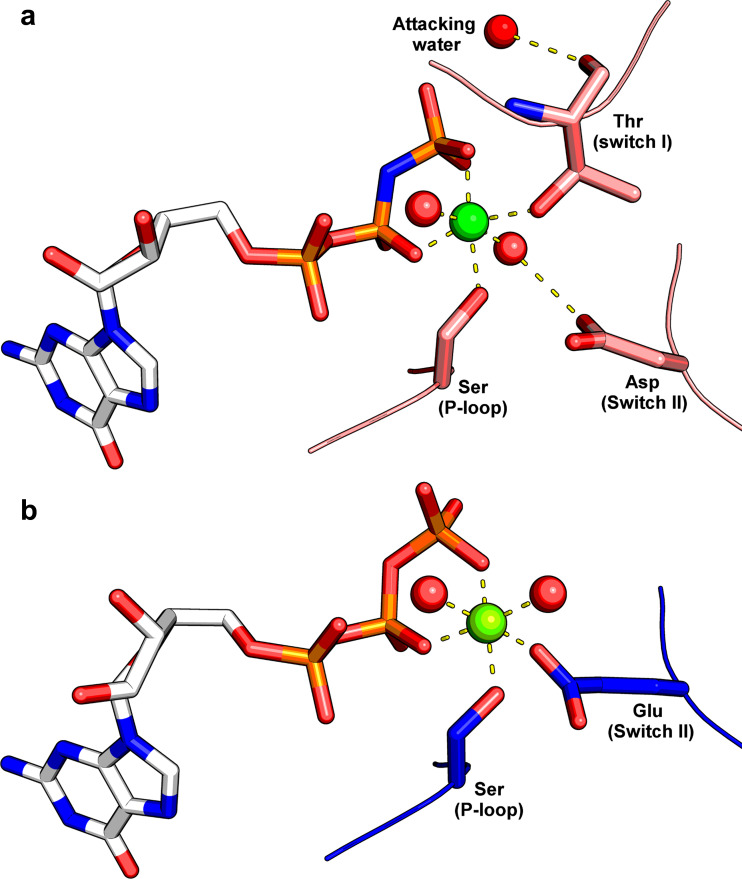
In 2009 the 2.9-Å resolution structure of mouse SEPT2-G (residues 33–306), lacking both the N- and C-termini and bound to the slowly hydrolyzable GTP analog GppNHp (5′-guanylyl imidodiphosphate), shed light on the structural role of the γ-phosphate (PDB code 3FTQ) (Sirajuddin et al. 2009). Unlike the previously released GDP-bound structures, in the presence of GppNHp both the switch I and switch II regions are ordered (Fig. 5), and a magnesium ion (Mg2+) is observed coordinated by Ser51 from the P-loop, amino acid residue Thr78 from the switch I, the β- and γ-phosphates from GppNHp and two water molecules (Fig. 6a). The ordering of the switch regions follows the universal mechanism for small GTPases (Wittinghofer and Pai 1991) and involves the formation of hydrogen bonds to the γ-phosphate by the main chain amides of Thr78 (homolog of Thr35 in RAS) and Gly104 from the G3 motif of switch II. This structural study was complemented with a series of biochemical assays in which the authors evaluated the effect of a series of mutations focusing on the capacity to bind GDP and to hydrolyze GTP. As anticipated, the deletion of the N-terminal domain had no influence on nucleotide affinity. On the other hand, mutating Thr78 to alanine led to a 21-fold reduction in the affinity for GDP, and its mutation to glycine promoted a twofold reduction in the hydrolytic rate for GTP. In SEPT2, Thr78 also coordinates the Mg2+ counterion and the water molecule in position for in-line attack (Fig. 6b). However, Thr78 (the homolog of Thr 35 in RAS) is not conserved in the SEPT6 group (Fig. 4). These observations are consistent with the mutation studies described above and with the observation that GTP binds to SEPT6 in the hetero-complex, indicating that SEPT6 presents poor or no hydrolytic activity.
Fig. 5.
Conformational changes to the switch regions. An overlay of the Cα traces for SEPT2 bound to GDP (2QNR) and to 5′-guanylyl imidodiphosphate (GppNHp; 3FTQ) are shown. The switch I region is incomplete in the GDP-bound structure (red) but becomes ordered (green) in the presence of GTP due to specific interactions involving amino acid residue Thr78, whose amide forms a hydrogen bond with the γ-phosphate and whose side chain coordinates the magnesium ion (Mg2+) which binds concomitantly with the GTP. A significant part of switch II, which is also disordered in the GDP complex (purple), only assumes a well-defined structure upon binding with GTP (pale blue). Amino acid residue Gly104, from the G3 motif, plays a central role in this process as it donates a hydrogen bond to the γ-phosphate. Thr78 and Gly104 represent the essential elements of the universal switch mechanism (Wittinghofer and Pai 1991)
Fig. 6.
Magnesium binding site. Coordination of Mg2+ in a catalytically active septin (SEPT2) (a) and in a catalytically inactive septin (SmSEPT10, a member of the SEPT6 group) (b). The coordinates are taken from Protein Data Bank (PDB) files 3FTQ and 4KVA respectively. The most notable difference is the absence of Thr78 from switch I in the case of the inactive enzyme; Thr78 is absent from all of the SEPT6 group septins. This threonine is taken to be essential for catalysis as it secures a water molecule in a position for in-line attack
The authors describe a distortion to the β-sheet in this complex in which the β2 and β3 strands are tilted by approximately 20o. The authors suggest that this is the result of the presence of the γ-phosphate and therefore a genuine conformational rearrangement which depends on the nature of the bound nucleotide. However, as discussed in section "Non-filamentous crystal structures and crystallization artifacts", it seems more likely that this is, in fact, an artifact induced by crystal packing. None of the subsequent structures bound to GTP (or one of its analogs) which were solved present a similar distortion.
SEPT7-G
In 2011 two groups almost simultaneously published the crystal structures of the GTPase domain of SEPT7 bound to GDP. Zent et al. (2011) obtained the SEPT7 structure (residues 29–297) at 3.3 Å resolution (PDB code 3T5D), and Serrão et al. ( 2011) resolved the slightly longer construct (residues 29–299) at 3.35 Å (PDB code 3TW4).
While these structures were obtained under different crystallization conditions, both crystallized in space group P61 and present very similar cell dimensions, being effectively isomorphous. Both contain a G-interface dimer in the asymmetric unit, and unlike SEPT2 bound to GDP, both present continuous electron density for the complete switch II region, indicating that this region contributes to G-interface dimer stability. Although GDP-bound SEPT7 clearly behaves as a dimer in solution under a variety of conditions, it forms filaments in the crystal in the same fashion as SEPT2 (Fig. 7). Additionally, performing a series of mutations and analytical gel filtration experiments, Zent et al. found that the SEPT7 G-interface is more stable to increased salt concentrations than SEPT2 and that SEPT7 dimerization via its G-interface is nucleotide dependent.
Fig. 7.
Filaments observed in crystal structures of different septins. In each structure a small segment of a filament is shown which has been generated using crystallographic symmetry. An example of each group is given, with SmSEPT10 used as a representative of the SEPT6 group for which an example of a human septin structure has yet to be reported. The NC- and G-interfaces are evident in all cases. Many of these may be “promiscuous” and not relevant to physiological filaments. The SEPT3 filament (green) is somewhat foreshortened with respect to the others
With the determination of the structure of SEPT7, a more complete picture became available regarding the promiscuous interactions mentioned previously. A comparison of the filaments observed in the crystal structures of several different G-domains is shown in Fig. 7. Within the hetero-filament, SEPT2 forms a homotypic NC-interface and a heterotypic G-interface with SEPT6. SEPT7, on the other hand, forms a heterotypic NC-interface with SEPT6 and a homotypic G-interface, the result of the polymerization of the hexameric core particles. However, in the crystal structures of the individual G-domains both interfaces are, obviously, homotypic. The non-physiological interfaces which arise in the crystal structures (the SEPT2 G-interface and the SEPT7 NC-interface) are considered to be “promiscuous” interactions as they are not expected to exist in a physiological combination of septins competent for forming hetero-filaments. They have been observed in almost all crystal structures subsequently described, and their appearance is the result of crystal packing which generates favorable non-covalent interactions that stabilize the lattice. Presumably, this is possible due to the conservation across septin groups of many of the residues important for stabilizing both the G- and NC-interfaces. However, whether so-called promiscuous interfaces are always non-physiological or not is still debatable as current knowledge of filament architecture is insufficient. Filaments may exist in which such interfaces become physiological, as for example, in the homo-filament of SEPT2 (Huang et al. 2006).
SEPT3-GC
In 2013 the crystal structure of SEPT3, which lacks the N-terminal domain, corresponding to residues 59–350, was published at 2.88 Å resolution (PDB code 3SOP) together with a series of biophysical and biochemical data (Macedo et al. 2013). This SEPT3-GC structure was obtained in the GDP bound state, and once again a linear filament was observed in the crystal (Fig. 7), with the asymmetric unit containing a G-interface dimer. A Mg2+ ion was observed, coordinated by the β-phosphate of the GDP, Ser75 from the P-loop, Thr102 from switch I (the homolog of Thr78 in SEPT2 and Thr35 in Ras) and three water molecules (Fig. 6b). The arrangement resembles that of the SEPT2 structure bound to GppNHp, but with one of the water molecules substituting for the γ-phosphate. Once more, electron density for the C-terminal domain (the final 21 residues) was not observed.
Analytical gel filtration and small-angle X-ray scattering (SAXS) data showed that unlike all other previously studied septins, SEPT3-GC is a monomer in solution under high salt conditions. Moreover, SEPT3-GC was obtained without a bound nucleotide, a characteristic that has readily been exploited in the design of isothermal titration calorimetry experiments and nucleotide hydrolysis assays. This construct has been shown to hydrolyze GTP more efficiently than SEPT2-GC, and analytical size exclusion chromatography data indicate that SEPT3-GC molecules associate as dimers in the presence of GTPγS [guanosine 5′-O-(gamma-thio)triphosphate, a non-hydrolyzable or slowly hydrolyzable G-protein-activating analog of GTP)] in a salt-dependent fashion. In addition, these authors (Macedo et al. 2013) used mutations to show that the dimers formed in the presence of GTPγS associate through the G-interface.
Following the observation that the monomeric SEPT3-GC presents a threonine in the G-interface that replaces a tyrosine observed in all other septin structures, analytical size exclusion chromatography employing the mutant T282Y was used to show that this mutation is sufficient to change the oligomeric state of SEPT3-GC from a monomer to a dimers, implicating the tyrosine as an important determinant of G-interface stability.
One of the most notable features of the SEPT3-GC structure is that the filament generated by crystallographic symmetry shows significant foreshortening when compared with others (Fig. 7). This is due to squeezing of the NC-interface where the two monomers come closer by approximately 8Å (see section "The G- and NC-interfaces").
Non-mammalian septins
SEPT10-G from Schistosoma mansoni
The first structural study that focused on a non-mammalian septin was published in 2014 (Zeraik et al. 2014) and presented structures of the G-domain of septin 10 from Schistosoma mansoni (SmSEPT10-G, 39-306) bound to GDP at 1.93 Å resolution (PDB code 4KV9) and to GTP at 2.14 Å resolution (PDB code 4KVA). These structures provided the first “high-resolution” glance of a septin structure, thereby allowing for a more detailed description of several features, including an unambiguously interpretable electron density for strand β2. This part of the structure, which is adjacent to the switch I region, is often poorly ordered, particularly in GDP-bound structures. Comparison of structures solved prior to SmSEPT10-G show that it had been interpreted in several different ways due to the difficulty in establishing the correct register of the sequence with respect to the map in the absence of clearly defined sidechain density. As observed in all other septin structures, with the exception of SEPT2 bound to GppNHp, the crystals of SmSEPT10-G, lacking both N- and C-terminal domains, formed filaments inside the crystal lattice, another example of septin promiscuity (Fig. 7).
SmSEPT10 is a homolog of human SEPT10 and therefore belongs to the SEPT6 group, whose members are expected to present very low GTPase activity. In accordance with this expectation, the authors have shown that SmSEPT10 is unable to hydrolyze GTP and that it binds nucleotides with lower affinity than SEPT2 and SEPT3 (Huang et al. 2006; Macedo et al. 2013). Additionally, it was observed that Mg2+ ions are necessary for GTP binding (Fig. 6b), but not for GDP binding.
Perhaps the most remarkable novelty discussed in the SmSEPT10 study is the finding that the β3-strand presents different registers in relation to its neighbors (the β1 and β2 strands) in the presence of GDP and GTP. The unprecedented high resolution of the S. mansoni structures enabled observation of this β3-strand slippage, which allows nucleotide-dependent communication between the G- and NC-interfaces. The details of the strand slippage, including the re-arrangement of the hydrogen bonding, is shown in Fig. 8a. The result leads to an increase in the size of the hairpin loop between the β2 and β3 strands which projects further into the NC-interface in the GTP-bound complex. The authors suggest that the projection of the β2–β3 hairpin into the NC-interface in the GTP-bound form is incompatible with retaining the α0 helix buried, as observed in the original study of Sirajuddin et al. (2007). However, this could not be observed directly as α0 had been eliminated from the construct used in this study. Nevertheless, the observation lead the authors to speculate that GTP hydrolysis at the G-interface may control the orientation of the α0 helix at the NC-interface and therefore modulate membrane association.
Fig. 8.
Strand slippage in septins. a, c Diagrams of hydrogen bonding for the first three β-strands of the central sheet in SmSEPT10 (a) and SEPT2 (c). The slippage of the strands upon progression from the GTP-bound complex to the GDP-bound complex is different in both cases. In SmSEPt10 the central strand (β3) shifts with respect to its neighbors on both sides (β1 and β3 strands), whereas in SEPT2 the β2 and β3 strands (on the left) move together with respect to β1. This can be readily seen by following the alignment of the residues highlighted in blue, which are indicated merely as points of reference. b, d Diagram showing the structural consequences of the slippage for SmSEPT10 (b) and SEPT2 (d). In SEPT2 the GTP-bound form (pink) shows an ordering of the β2–β3 hairpin in the GTP-bound complex compared to the GDP complex (red), leading to an extension of the β-sheet into the NC-interface. Ordering of the hairpin also occurs in SmSEPT10 . The GTP complex is shown in dark blue, with β3 (the slipping strand) in orange and the GDP complex in light blue and the corresponding strand in yellow
These authors and others (McMurray 2014) have not been slow to point out the questionable relevance of this observation given that SmSEPT10 belongs to the catalytically deficient SEPT6 group and, therefore, would always be expected to be bound to GTP in a physiological context. However, re-examination of the SEPT2-G structures for which both GDP and GTP complexes are also available sheds some additional light on this question. Figure 8b shows that strand slippage also occurs in this case despite the fact that it was never mentioned by the authors in their original report (Sirajuddin et al. 2009). There is, however, a subtle difference between the two cases. Whereas in the original SmSEPT10 structure the β3 strand is observed to slip with respect to its neighbors on both sides (β1 and β2 strands), in the case of SEPT2, the β2 and β3 strands move as a rigid unit with respect to the β1 strand (Fig. 8a, c). Consequently, in the case of SEPT2 only approximately half the number of hydrogen bonds need to be broken and reformed during slippage.
However, some caution should be exercised in analyzing these results due to the medium resolution of the SEPT2 structures and the difficulty in interpreting the density for the β2 strand, as mentioned. It therefore remains unclear whether the differences described for the two cases are genuine or not. What is most important is that SEPT2 is a catalytically active septin and, whatever the details, clearly presents strand slippage at the very least between strands β3 and β1. This represents a mechanism for transmitting information from the G-interface to the NC-interface along the filament which may well be of functional relevance. This mechanism is expected to involve the reorientation of the polybasic helix α0. Whether this is a general phenomenon applicable to all catalytically active septins remains to be seen and clearly requires further study.
Yeast septins
In 2016 a study discussing the crystal structure of apo Cdc11 from Saccharomyces cerevisiae (residues 20–298) at 2.85 Å resolution (PDB code 5AR1) was published (Brausemann et al. 2016). This study represents the first time a septin structure was obtained without a bound nucleotide, and attempts to co-crystalize or soak the crystals with GDP apparently yielded poorly diffracting crystals. Only a monomer was observed in the asymmetric unit, and although a G-interface dimer was generated by crystallographic symmetry, a linear filament was not observed in the crystal lattice. Interestingly, the authors were able to use site-directed mutagenesis and size exclusion chromatography to shown that, in solution, pure full-length Cdc11 forms an NC-interface dimer, which is expected to be formed due to the C-terminal coiled coil domain. This observation is consistent with the known order of yeast septins within the octameric core particle (Bertin et al. 2008).
The structure, which was refined with anisotropic temperature factors at 2.85Å resolution, presents a number of unexpected features, particularly in the region corresponding to the SUE. In particular, helix α5 is completely missing from the structure. This absence appears to be at odds with the sequence for cdc11 which shows considerable conservation in this region when compared with human septins and which is expected to represent a conserved structural element. It will be of interest to see whether future structural studies of the yeast septins reveal these unusual structural features to be conserved—or not.
Chlamydomonas septin
In 2017 the crystal structure of the GTPase domain of the Chlamydomonas reinhardtii septin (CrSEPT 86–393) bound to GTPγS was solved at 2.04 Å resolution by single wavelength anomalous dispersion phasing (Pinto et al. 2017). It is important to note that the C. reinhardtii genome codes for only this single septin. Size exclusion chromatography data show that this construct is purified as a monomer in a nucleotide-free state and that a dimer is observed after incubation with GTPγS. Consistent with this finding, a G-interface dimer is observed in the asymmetric unit of the crystal. Conversely, crystallographic symmetry operations do not generate NC-interfaces; therefore, a linear filament is not observed in the crystal lattice. The authors employed transmission electron microscopy to show that this construct (residue 86–393) forms a homo-filament at low salt concentrations, and immunofluorescence experiments allied to confocal microscopy were used to observe that, in vivo, septins localize preferentially at the base of the flagella.
In terms of nucleotide binding and hydrolysis, isothermal titration calorimetry data was used to show that the binding affinity of CrSEPT to GTPγS in the presence of Mg2+ is typical, but surprisingly GTP hydrolysis assays revealed that this septin presents the highest GTPase activities observed for all septins. In search of the structural determinants of such a high hydrolytic rate, the authors found that Arg239 from the adjacent subunit interacts across the G-interface with the γ-phosphate of the bound GTPγS. Mutating this arginine to alanine had little impact on the GTPγS binding affinity, but it did eliminate the GTPase activity almost completely, indicating that the two CrSEPT molecules in a dimer might behave as GTPase-activating proteins for each other. This arginine therefore acts as a classical “arginine finger” (Bourne 1997; Zhang et al. 1999b).
Some of the characteristics of all available septin structures are summarized in Table 1. The structure of an individual monomer in each case is given in Fig. 9 together with the corresponding PDB code and the nature of the bound ligand. Although this information is publically available, not all of these structures have been fully described in the literature.
Table 1.
Summary of the available septin structures.
| Septin | Organism | Amino acid residues | Domains | Resolution ( Å) | nucleotide | Mg2+ | Protein Data Bank ID | Reference |
|---|---|---|---|---|---|---|---|---|
| SEPT2–SEPT6–SEPT7 | Homo sapiens | Full length | NGC | 4.0 | GDP/GTP | - | 2QAG | Sirajuddin et al. 2007 |
| SEPT2 | H. sapiens | 1–315 | NG | 3.4 | GDP | No | 2QA5 | Sirajuddin et al. 2007 |
| SEPT2 | H. sapiens | 22–320 | G | 2.6 | GDP | No | 2QNR | Sirajuddin et al. 2007 |
| SEPT2 | Mus musculus | 33–306 | G | 2.9 | GppNHp | Yes | 3FTQ | Sirajuddin et al. 2009 |
| SEPT7 | H. sapiens | 29–297 | G | 3.3 | GDP | No | 3T5D | Zent et al. 2011 |
| SEPT7 | H. sapiens | 29–299 | G | 3.35 | GDP | No | 3TW4 | Serrão et al. 2011 |
| SEPT3 | H. sapiens | 59–350 | GC | 2.88 | GDP | Yes | 3SOP | Macedo et al. 2013 |
| SmSEPT10 | Schistosoma mansoni | 39–306 | G | 1.93 | GDP | No | 4KV9 | Zeraik et al. 2014 |
| SmSEPT10 | S. mansoni | 39–306 | G | 2.14 | GTP | Yes | 4KVA | Zeraik et al. 2014 |
| Cdc11 | Scerevisiae cerevisiae | 20–298 | G | 2.85 | None | No | 5AR1 | Brausemann et al. 2016 |
| CrSEPT | Chlamydomonas reinhardtii | 86–393 | G | 2.04 | GTPγS | Yes | 5IRR | Pinto et al. 2017 |
| SEPT9 | H. sapiens | 27–-567 | GC | 2.73 | GDP | Yes | 4YQF | To be published |
| SEPT9 | H. sapiens | 278–567 | GC | 2.89 | GTPγS | Yes | 5CYP | To be published |
| SEPT9 | H. sapiens | 278–567 | GC | 2.04 | GDP | Yes | 5CYO | To be published |
| SEPT3 | H. sapiens | 43–329 | G | 1.83 | GDP | Yes | 4Z54 | To be published |
| SEPT3 | H. sapiens | 60–330 | G | 1.86 | GppNHp | Yes | 4Z51 | To be published |
SEPT Septin,Sm Schistosoma mansoni, Cr Chlamydomonas reinhardtii, GDP guanosine diphosphate, GTP guanosine triphosphate, GppNHp 5′-guanylyl imidodiphosphate (a GTP analog), GTPγS guanosine 5′-O-(gamma-thio)triphosphate (a G-protein-activating analog of GTP)
Fig. 9.
A table of septin monomers from each of the crystal structures deposited in the PDB. Although publically available some of these structures have yet to be described in detail in the literature. The colors red, blue, yellow and green identify the SEPT2, SEPT6, SEPT7 and SEPT3 group members, respectively. Cdc11 from Saccharomyces cerevisiae can not be readily classified by homology with human septins and is shown in purple. The single Chlamydomonas septin is normally classified as SEPT7-like and is shown in orange
Insights derived from septin structures
One of the first lessons crystallographers learn when studying septins is that these filament-forming proteins are not easy to work with. Individually, full-length septins usually display low solubility and are prone to precipitation even when kept on ice in a buffer containing high concentrations of salt and glycerol. This low solubility is aggravated when it is necessary to prepare pure septin samples at high concentration for crystallization assays (Valadares and Garratt 2016). The authors of several studies explicitly report trying to work with full-length septins (Sirajuddin et al. 2009; Zent et al. 2011; Macedo et al. 2013; Brausemann et al. 2016), but later having to resort to constructs lacking either the N-terminal or C-terminal domains, or both. In fact, the only crystal structure of full-length septins is the human hetero-complex at 4 Å resolution (Sirajuddin et al. 2007), and available data indicate that removal of the N- and/or C-terminal domains is almost a pre-requisite for obtaining crystals that diffract to high resolution. The reason stems from the filament-forming activity of septins, as well as from the properties of these domains. Native septins present a tendency to auto assemble into hetero-oligomers and filaments, and, in vitro, this might lead to precipitation. Some septins have been described to readily form amyloid-like aggregates (Garcia et al. 2007; Damalio et al. 2012). Furthermore, at least part the N-termini of some septins, for example SEPT4 and SEPT9, is unstructured (Garcia et al. 2006), which may hinder crystallization or contribute to the formation of poorly diffracting crystals. With respect to the C-termini, only the structures of the human septin hetero-complex and of SEPT3-GC present this domain, but in both cases no electron density is observed for this region, indicating that these domains are either unstructured or present flexibility with respect to the filament-forming G-domains and are therefore present in several different orientations in the crystal. Finally, the C-termini of members of the SEPT6 and SEPT7 families are expected to form hetero coiled coils with each other (Marques et al. 2012; Sala et al. 2016), which implies that, when pure, these septins might present this domain at least partially unstructured due to the absence of their ideal interaction partner and the consequent inability to form the native heterotypic coiled coil.
Particularly in the case of the early structures, the low to medium resolutions obtained clearly limit the interpretation of the electron density. Discontinuities in the electron density map associated with the lack of unambiguous side chain electron density for some residues results in a challenge in assigning the correct register of the polypeptide chain. Perhaps the best examples of these difficulties are observed in the case of the β2 strand and the α0 helix in all structures which contain them. Figure 9 shows that, where present, the α0 helix is often disconnected from the remainder of the structure in terms of continuous electron density.
The available septin structures vary regarding nucleotide content. The first structures indicated that septins were necessarily bound to a nucleotide. However SEPT3-GC was obtained in a nucleotide-free state and afterwards co-crystallized with GDP and a Mg2+ counterion. In contrast, the SEPT2, SEPT7 and SmSEPT10 structures bound to GDP present no magnesium ion. Both SEPT2 in complex with GppNHp and SmSEPT10 in complex with GTP present Mg2+ counterions. Later, the Saccharomyces cerevisiae Cdc11 structure was obtained in the absence of a nucleotide. These results indicate that while GTP binding is probably accompanied by Mg2+ ion recruitment, the interaction of GDP is more malleable. The lack of catalytic activity associated with the SEPT6 group of septins is a curiosity which suggests a functional role for the γ-phosphate of the GTP that remains bound to them. However, this role has yet to be elucidated. Since the GTP resides at the G-interface, it may have a role in establishing some degree of selectivity in the interaction involving SEPT6 and SEPT2. The lack of activity per se is more completely understood. The important role played by the threonine residue from switch I (Thr78 in SEPT2) was initially established by mutational studies (Sirajuddin et al. 2009), but it was only with the determination of the structure of the catalytically inactive SmSEPT10 bound to GTP that the details became apparent. In the structure of the GTP complex the switch I region remains disordered, different to SEPT2, and the absence of the switch I threonine residue results in both a different Mg2+ coordination and, critically, the absence of the catalytic water, which in the structure of SEPT2 lies poised for in-line attack on the γ-phosphate.
Promiscuity
A charming characteristic of septin crystal structures is that due to the filament-forming nature of these proteins, several of the crystal contacts observed in these structures provide a great deal of meaningful native-like information. The use of crystallographic symmetry operations allows the inspection of interfaces not observed in the asymmetric unit but that in some cases are physiologically relevant. For example, the native SEPT2 NC-interface can only be observed in the SEPT2 structures and in the filament structure after the use of crystallographic symmetry operations. In fact, with the exception of the hetero-complex structure, all observed interfaces in the asymmetric unit of septin crystal structures are G-interfaces. It is important to keep in mind that, since all the structures of individual septins represent constructs lacking the N- and/or the C-terminal domains, the NC-interfaces observed are expected to be incomplete; in particular, the contacts of the α0 helix and the C-terminal coiled coils are expected to play a role in these interfaces. Accordingly, the SEPT2–SEPT2 native interface is of the NC-type, but SEPT2-NG has been shown to form a G-interface dimer in solution (Sirajuddin et al. 2007). Likewise, pure Saccharomyces cerevisiae Cdc11 has been shown to exist as NC-interface dimers in solution (Brausemann et al. 2016), which is not entirely an unexpected result, since Cdc11 occupies the terminal position in the S. cerevisiae octameric linear filament and associates with another Cdc11 molecule from a neighboring octamer through its NC-interface (Bertin et al. 2008). However, only G-interface dimers are observed after the application of symmetry operations to the crystal structure of S. cerevisiae Cdc11 (residues 20–298), which is curious at the very least. Further studies would be beneficial in this case.
The above discussion invariably leads to the captivating issue of septin plasticity or promiscuity. Assuming that the native/physiological interfaces are those present in the human septin hexameric filament and in the octameric filament 9–7–6–2–2–6–7–9 (Sirajuddin et al. 2007; Kim et al. 2011; Sellin et al. 2011), several non-physiological interfaces are observed in the crystal structures in which a linear filament is observed in the expanded crystal lattice. Following that reasoning, the term promiscuous might be used when referring to the G-interfaces observed in all SEPT2 structures and in the SEPT3 structure, as well as in the NC-interfaces observed in the SEPT7 structures. In the case of promiscuity due to the NC-interfaces of septins, since all structures of individual septins lack the N- and/or the C-terminal domains, it might be possible that the promiscuity arises, at least partially, from the absence of these domains. Conversely, in all structures of individual septins, the G-interface is intact and this plasticity is still observed.
It is thought that promiscuity occurs in the absence of a physiologically relevant binding partner and that constructs lacking the N- and/or the C-terminal domain are more likely to display this behavior. However, this plasticity in the recruitment of an interaction partner might present a role in vivo. At least for the G-interface, this hypothesis is supported by the results of analytical gel filtration experiments showing that SEPT2 forms G-interface dimers in solution (Sirajuddin et al. 2007) and by the analysis of the conservation of the residues that contact the adjacent subunit in the G-interface of the available structures. The analysis of the human SEPT2, SEPT3 and SEPT7 structures indicate that of the 22 residues observed close to the G-interface of the neighboring subunit, eight are conserved in all 13 human septins, and seven others are conserved in all members of at least two of the four septin groups. Although this proposal is not in accordance with the Kinoshita rule, it might significantly increase the number and complexity of in vivo septin filaments.
The G- and NC-interfaces
A full understanding of both physiological and promiscuous interfaces will only be reached after a complete analysis of both interface types and of their subdivision into the five categories described previously (NC, NChom, NChet, GDD and GDT). However, once again, it is necessary to issue a note of caution regarding the structural data currently available because the clearest views of the interfaces to have been described to date have been derived from isolated G-domains. These may be promiscuous in nature and therefore not fully reflect the true situation in a functional filament.
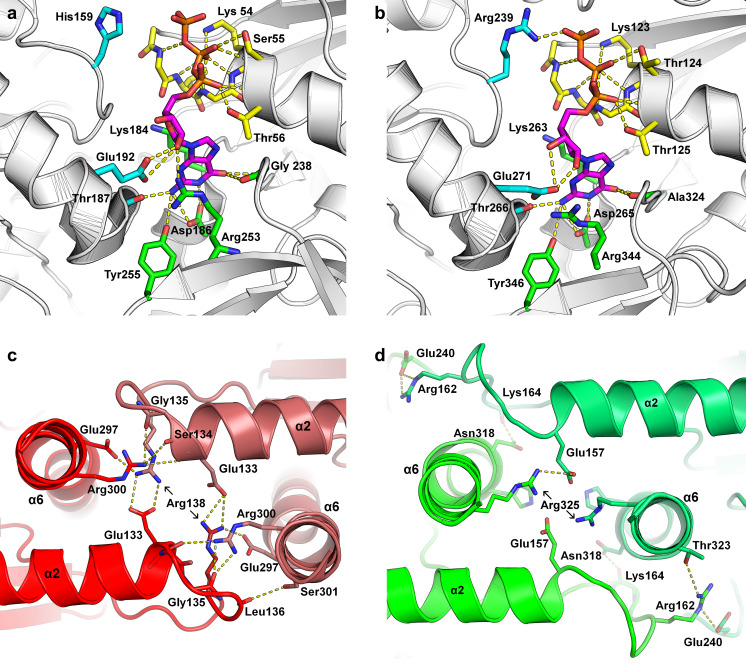
The more recently published higher resolution structures have allowed for a fuller description of the G-interface and its limited degree of variation. The canonical G-interface with GTP bound is shown in Fig. 10a. The P-loop (yellow) is principally responsible for binding the phosphate moieties, including the penultimate residue Thr56 (Fig. 4), not always observed in other small GTP-binding proteins, which forms a hydrogen bond to the α-phosphate. Further interactions from the same subunit include Asp186, which is responsible for GTP selectivity, Lys184 and Arg253, which form classical stacking interactions on either side of the base, and Tyr255, which is important for G-dimer interface stability. Interactions involving residues from the neighboring monomer across the G-interface are shown in light blue in Fig. 10; these include the conserved Glu192, which also forms a cross-interface salt bridge with R253, thereby integrating nucleotide binding with G-dimer formation. Thr187 is not completely conserved, being an alanine in SEPT6, as it uses its mainchain to interact with the base. His159 shows considerable variation across structures, occasionally interacting directly with the phosphates but frequently presenting no electron density at all. The histidine is striking for its conservation and resides within one of the septin-specific sequential motifs identified by Pan et al. (2007). The significance of these conserved regions for septin function (SEPT1 to SEPT2 in Fig. 4) has yet to be fully elucidated.
Fig. 10.
Conservation and variability at the G- and NC-interfaces. a The canonical G-interface as seen in SmSEPT10 bound to GTP. The P-loop (yellow) is principally responsible for binding the phosphate moieties (orange) and the Mg2+ (not shown for clarity). Further interactions with GTP occur with residues from the same subunit (green) or the neighboring subunit (light blue). b The G-interface in Chlamydomonas septin, which is very similar to the G-interface of all other septins, with the exception of the arginine finger (Arg239). c The canonical NC-interface as seen in SEPT2. d The squeezed NC-interface of SEPT3 bound
The situation is rather different in Chlamydomonas septin (Fig. 10b) where the homolog of His159 (His238), although present, is displaced and replaced by the neighboring Arg239 (Chlamydomonas numbering) which forms the arginine finger that is responsible for its elevated catalytic activity. In some septins (such as SEPT3 and SEPT7) the C-terminal portions of the switch II regions of two monomers pair up across the G-interface. However, such interactions may be of little importance since these interfaces are not expected to be physiological, at least not in the case of an octamer-based filament.
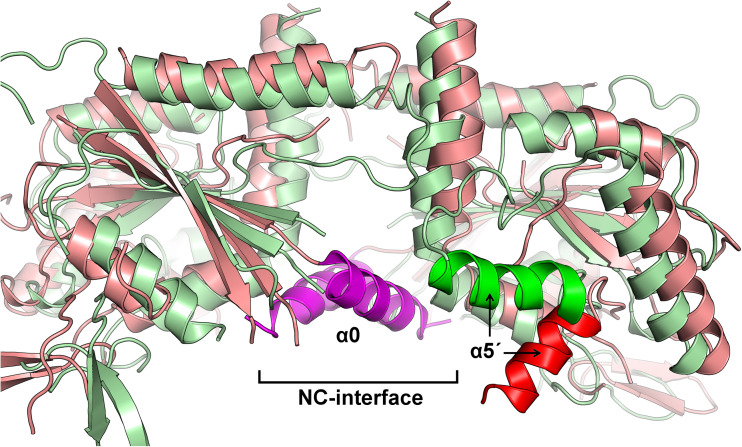
The details of the NC-interface are somewhat less clear due to the fact that the highest resolution structures available systematically lack the α0 helix which is known, from the structure of the heterocomplex (SEPT2–SEPT6–SEPT7), to be an integral part of the interface. However, a comparison of the two structures available for SEPT2-G (one with and one without α0) show that the absence of the helix does not significantly perturb the remainder of the interface, suggesting that it is probably safe to draw structural conclusions without running too much risk of describing artifacts. Figure 10c shows the canonical NC-interface observed in all structures thus far reported, with the exception of SEPT3-GC. The most prominent feature of the interface is a network of salt bridges involving charged residues coming from helix α6 and a region involving the C-terminus of α2 together with the following loop (Figs.4 and 10c). Both provide a Glu and an Arg to the network, and these are highly conserved across septins including, curiously, both Chlamydomonas and cdc11, despite the fact that these septins do not present the NC contact in the crystal structures reported. The network also includes a classical interaction involving the helix dipole of α2 and Arg300. In SEPT3-GC, the interface is squeezed together by about 8 Å, leading to a foreshortening of the filament (Fig. 7). This leads to a complete rearrangement of the salt-bridge network (Fig. 10d). New contacts also emerge as a result, which include the participation of Glu240 from the polyacidic region (Fig. 4) which precedes the α5′ helix. This helix is a structural feature characteristic of septins, and it is of note that in SEPT3 it presents a completely different orientation to that seen in all other septins and is aligned approximately parallel to the main filament axis (Fig. 11). Given the terminal position of SEPT3 in the octameric core particle (where it would occupy an equivalent position to SEPT9 in Fig. 2), an NC-interface between two copies of SEPT3 would be expected to arise upon filament polymerization. The unusual nature of this interface is therefore probably physiological and not a crystallization artifact. However, the relevance of the difference between SEPT3 and other septins remains to be clarified although it may represent a means to favor a physiological interface over a promiscuous one.
Fig. 11.
Helix α5′. Superposition of cartoon representations of the G-domains of SEPT2 (red) and SEPT3 (green) around the NC-interface. The α0 helix in the SEPT2 structure is shown buried within the NC interface. Helix α5′ presents a very different orientation in SEPT3 when compared to all other structures (here represented by SEPT2) and lies almost parallel to the main axis of the filament
Non-filamentous crystal structures and crystallization artifacts
Along with the Saccharomyces cerevisiae Cdc11 apo structure and the CrSEPT from Chlamydomonas reinhardtii, the only other structure that does not present a linear filament in the crystal lattice is the structure of SEPT2 G-domain bound to GppNHp (Sirajuddin et al. 2009). In this case, the structure was obtained in the space group P212121, and symmetry expansion of the asymmetric unit does not generate NC-interfaces but results in the extension of the central mixed β-sheet by lateral association with a neighboring subunit. In this outcome, two β2-strands from adjacent molecules associate in an antiparallel way, effectively connecting the β-sheets of the two septin molecules involved (Fig. 12). It is difficult to attribute such a drastic organizational difference in relation to all other structures to the nucleotide content of this structure, and this case probably constitutes a crystal packing artifact (see section "SEPT2-G bound to a GTP analog"). Since the strands at the edges of β-sheets might present unsatisfied hydrogen bonds, and the lateral association of two β-sheets fulfills this hydrogen bonding potential, this phenomena is not completely unexpected.
Fig. 12.
Crystal contacts in SEPT2-G bound to GppNHp. The distortion of the β-sheet in the complex of SEPT2 with a GTP analog appears to be the result of crystal packing and may not represent an intrinsic property of the GTP-bound complex. A G-interface dimer is shown at the top of the figure. A second dimer, related to the first by crystallographic symmetry, is shown at the bottom right. The β-hairpins formed by strands β2 and β3 (blue) from one of the monomers from each of the two dimers are hydrogen bonded together, leading to the formation of a contiguous β-sheet. This particular aspect of the structure should, therefore, be treated with some caution
A question not well studied is that of the direct interaction between regions of the N-terminal domain (other than the α0 helix) with GTPase domains in the filament (Sirajuddin et al. 2007). In the human hetero-hexamer structure the N-terminal region of SEPT6 interacts extensively with the GTPase domain of SEPT7, an interaction that is observed in the region between the NC-interface and the switch I of SEPT7. While the low resolution of this crystal structure makes it impossible to point out exactly which residues are involved in this interaction, the position and shape of the electron density is strongly suggestive that the interaction does indeed exists. This observation indicates that, in addition to the C-terminal domains of SEPT6 and SEPT7, which have been shown to interact to form a high-affinity coiled coil, the N-terminal domain may also contribute affinity and selectivity to filament assembly.
Follow-up studies and future perspectives
Despite their obvious impact on the way in which septin filaments are now perceived and in the design of subsequent functional studies, the known septin structures have been used relatively little in follow-up studies designed to exploit structural aspects per se. However, “fence” (diffusion barrier) formation was studied by Lee et al. (2014) in a theoretical study of PIP2 defusion. The study revealed that septin filaments, when inserted into the membrane to sufficient depth, were able to impact on PIP2 diffusion. On the other hand the studies of Angelis et al. (2014) heavily relied on medium resolution structures to perform in silico molecular docking with forchlorfenuron (FCF), a compound known to affect filament assembly. The authors suggest that FCF binds directly to the nucleotide-binding site (possibly in more than one pose) and takes advantage of residues coming from the G1 and G4 nucleotide-binding motifs for the formation of hydrogen bonds. Souza and Barbosa (2010) described the homology modeling of human SEPT8, and in a recent study Weems and McMurray (2017) generated a series of models for the yeast septins cdc3, cdc10, cdc12 and shs1 based on the crystal structure of cdc11 and used these to aid experimental work, leading to the proposal of a sequential mechanism for filament assembly in Saccharomyces cerevisiae.
The lack of further studies is probably a reflection on the fact that we have only recently begun to scratch the surface of the structural biology of septins and their filaments. Clearly there is a need to invest greater effort in the determination of more crystal structures. In the case of human septins, for the most part, up until now we have been forced to draw conclusions from the structures of a single representative of each group. Variation between group members is a subject which has barely been touched upon but may be extremely relevant for septin physiology. The recent deposition of a second member of the SEPT3 group (SEPT9) is the first step in that direction, and a full description of this septin is expected to be published soon (manuscript in preparation). The lack of structural information on the N- and C-terminal domains is also a significant deficit.
The situation for other species is even worse. The most notable deficiency is the paucity of structural information on yeast septins, with the recent description of the apo cdc11 structure being the only example available to date. It is to be hoped that more effort will be focused in this direction in the future as the wealth of biological information on yeast septins is rich and fertile territory for structure–function correlations (McMurray 2016). It would also be fascinating to see the structures of septins bound to some of their interaction partners (Neubauer and Zeiger 2017a, b).
However, what is most critical is the lack of a high-resolution structure of a hetero-filament, from whatever species. A human septin hetero-complex based on an octameric core particle, including members from all four groups would be highly desirable but has so far proved elusive. Intrinsic flexibility of the structure may be the main reason, and crystallization attempts may be destined to fail. In this respect, the recent spectacular advances in cryo-electron microscopy may represent a possible solution (Merk et al. 2016). Structures at higher and higher resolution of smaller and smaller particles are being reported almost weekly and provide the potential to elucidate not only three-dimensional structures, but variation within them (such as the frequently reported bending at the center of the core particle). Such advances may represent the key to unlocking the mystery of the exquisite molecular recognition which leads to individual septins finding their rightful position along the hetero-filament. Already studies using cryo-electron microscopy tomography of yeast septins have provided considerable insight (Sadian et al. 2013). However, in order to fully understand this process it is necessary to have at hand a high-resolution view of each of the five different interfaces which must form in order to generate an octamer-based filament. This is the challenge for the future.
Acknowledgements
We would like to acknowledge the financial support of the Brazilian funding agencies CNPq, FAPESP and FAPDF. We would also like to specifically mention the dedication of students and post-docs over the last 10 years (too many to mention by name, but they know who they are) who have contributed significantly to the structural biology of septins.
Compliance with ethical standards
Conflicts of interest
Napoleão Fonseca Valadares declares that he has no conflicts of interest. Humberto d´Muniz Pereira declares that he has no conflicts of interest. Ana Paula Ulian de Araujo declares that she has no conflicts of interest. Richard Charles Garratt declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Footnotes
This article is part of a Special Issue on ‘Latin America’ edited by Pietro Ciancaglini and Rosangela Itri.
References
- Angelis D, Karasmanis EP, Bai X, Spiliotis ET. In silico docking of forchlorfenuron (FCF) to septins suggests that FCF interferes with GTP binding. PLoS One. 2014;9(5):e96390. doi: 10.1371/journal.pone.0096390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Bowen JR, Knox TK, Zhou K, Pendziwiat M, Kuhlenbäumer G, Sindelar CV, Spiliotis ET. Novel septin 9 repeat motifs altered in neuralgic amyotrophy bind and bundle microtubules. J Cell Biol. 2013;203:895–905. doi: 10.1083/jcb.201308068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Grob P, Park SS, Garcia G, 3rd, Patanwala I, Ng HL, Alber T, Thorner J, Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR. The arginine finger strikes again. Nature. 1997;389:673–674. doi: 10.1038/39470. [DOI] [PubMed] [Google Scholar]
- Brausemann A, Gerhardt S, Schott AK, Einsle O, Große-Berkenbusch A, Johnsson N, Gronemeyer T (2016) Crystal structure of Cdc11, a septin subunit from Saccharomyces cerevisiae. J Struct Biol 193:157–161. 10.1016/j.jsb.2016.01.004 [DOI] [PubMed]
- Bridges AA, Zhang H, Mehta SB, Occhipinti P, Tani T, Gladfelter AS. Septin assemblies form by diffusion-driven annealing on membranes. Proc Natl Acad Sci USA. 2014;111(6):2146–2151. doi: 10.1073/pnas.1314138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges AA, Jentzsch MS, Oakes PW, Occhipinti P, Gladfelter AS. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J Cell Biol. 2016;213(1):23–32. doi: 10.1083/jcb.201512029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69(3):717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damalio JCP, Garcia W, Alves Macêdo JN, de Almeida MI, Andreu JM, Giraldo R, Garratt RC, Ulian Araújo AP. Self assembly of human septin 2 into amyloid filaments. Biochimie. 2012;94:628–636. doi: 10.1016/j.biochi.2011.09.014. [DOI] [PubMed] [Google Scholar]
- Fung KYY, Dai L, Trimble WS (2014) Cell and molecular biology of septins. Inl Rev Cell Mol Biol 310:289–339 [DOI] [PubMed]
- Garcia W, de Araújo APU, Oliveira Neto M, Ballestero MRM, Polikarpov I, Tanaka M, Tanaka T, Garratt RC. Dissection of a human septin: definition and characterization of distinct domains within human SEPT4. Biochemistry. 2006;45(46):13918–13931. doi: 10.1021/bi061549z. [DOI] [PubMed] [Google Scholar]
- Garcia W, Ulian de Araújo AP, Lara F, Foguel D, Tanaka M, Tanaka T, Garratt RC. An intermediate in the thermal unforlding of the GTPase domain of human septin 4 (SEPT4/Bradeion-β) forms amyloid filaments in vitro. Biochemistry. 2007;46:11101–11109. doi: 10.1021/bi700702w. [DOI] [PubMed] [Google Scholar]
- Garcia G, 3rd, Finnigan GC, Heasley LR, Sterling SM, Aggarwal A, Pearson CG, Nogales E, McMurray MA, Thorner J. Assembly, molecular organization, and membrane-binding properties of development-specific septins. J Cell Biol. 2016;212:515–529. doi: 10.1083/jcb.201511029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329:436–439. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YW, Surka MC, Reynaud D, Pace-Asciak C, Trimble WS. GTP binding and hydrolysis kinetics of human septin 2. FEBS J. 2006;273:3248–3260. doi: 10.1111/j.1742-4658.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- Kim MS, Froese CD, Estey MP, Trimble WS. SEPT9 occupies the terminal positions in septin octamers and mediates polymerization-dependent functions in abscission. J Cell Biol. 2011;195:815–826. doi: 10.1083/jcb.201106131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M. Assembly of mammalian septins. J Biochem. 2003;134:491–496. doi: 10.1093/jb/mvg182. [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Kimura K, Matsumoto N, Watanabe M, Fukaya M, Ide C. Mammalian septin Sept2 modulates the activity of GLAST, a glutamate transporter in astrocytes. Genes Cells. 2004;9:1–14. doi: 10.1111/j.1356-9597.2004.00696.x. [DOI] [PubMed] [Google Scholar]
- Lee KI, Im W, Pastor RW. Langevin dynamics simulations of charged model phosphatidylinositol lipids in the presence of diffusion barriers: toward an atomic level understanding of corralling of PIP2 by protein fences in biological membranes. BMC Biophys. 2014;7:13. doi: 10.1186/s13628-014-0013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara IG, Baldarelli R, Field CM, Glotzer M, Hayashi Y, Hsu SC, Kennedy MB, Kinoshita M, Longtine M, Low C, Maltais LJ, McKenzie L, Mitchison TJ, Nishikawa T, Noda M, Petty EM, Peifer M, Pringle JR, Robinson PJ, Roth D, Russell SE, Stuhlmann H, Tanaka M, Tanaka T, Trimble WS, Ware J, Zeleznik-Le NJ, Zieger B. Mammalian septins nomenclature. Mol Biol Cell. 2002;13(12):4111–4113. doi: 10.1091/mbc.e02-07-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo JN, Valadares NF, Marques IA, Ferreira FM, Damalio JC, Pereira HM, Garratt RC, Araujo AP. The structure and properties of septin 3: a possible missing link in septin filament formation. Biochem J. 2013;450:95–105. doi: 10.1042/BJ20120851. [DOI] [PubMed] [Google Scholar]
- Marques IA, Valadares NF, Garcia W, Damalio JC, Macedo JN, de Araújo AP, Botello CA, Andreu JM, Garratt RC. Septin C-terminal domain interactions: implications for filament stability and assembly. Cell Biochem Biophys. 2012;62:317–328. doi: 10.1007/s12013-011-9307-0. [DOI] [PubMed] [Google Scholar]
- McMurray M. Lean forward: Genetic analysis of temperature-sensitive mutants unfolds the secrets of oligomeric protein complex assembly. Bioessays. 2014;36:836–846. doi: 10.1002/bies.201400062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA. Assays for genetic dissection of septin filament assembly in yeast, from de novo folding through polymerization. Methods Cell Biol. 2016;136:99–116. doi: 10.1016/bs.mcb.2016.03.012. [DOI] [PubMed] [Google Scholar]
- Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JL, Subramaniam S. Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell. 2016;165(7):1698–1707. doi: 10.1016/j.cell.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nature Rev. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- Mostowy S, Bonazzi M, Hamon MA, Tham TN, Mallet A, Lelek M, Gouin E, Demangel C, Brosch R, Zimmer C, Sartori A, Konoshita M, Lecuit M, Cossart P. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010;8:433–444. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Nagata K, Asano T, Nozawa Y, Inagaki M. Biochemical and c cell biological analyses of a mammalian septin complex, Sept7/9b/11. J Biol Chem. 2004;279:55895–55904. doi: 10.1074/jbc.M406153200. [DOI] [PubMed] [Google Scholar]
- Nakahira M, Macedo JN, Seraphim TV, Cavalcante N, Souza TA, Damalio JC, Reyes LF, Assmann EM, Alboghetti MR, Garratt RC, et al. A draft of the human septin interactome. PLoS One. 2010;5:e13799. doi: 10.1371/journal.pone.0013799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer K, Zeiger B (2017a) The mammalian septin interactome. Front Cell Dev Biol 5:3. 10.3389/fcell.2017.00003 [DOI] [PMC free article] [PubMed]
- Neubauer K, Zeiger B. The mammalian septin interactome. Front Cell Dev Biol. 2017;5:1–9. doi: 10.3389/fcell.2017.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Malmberg RL, Momany M. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol Biol. 2007;7:103. doi: 10.1186/1471-2148-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto APA, Pereira HM, Zeraik AE, Ciol H, Ferreira FM, Brandão-Neto J, DeMarco R, Navarro MVAS, Risi C, Galkin VE, Garratt RC, Araujo APU (2017) Filaments and fingers: Novel structural aspects of the single septin from Chlamydomonas reinhardtii. J Biol Chem 292(26):10899–10911. 10.1074/jbc.M116.762229 [DOI] [PMC free article] [PubMed]
- Sadian Y, Gastogiannis C, Patasi C, Hofnagel O, Goody RS, Farkasovsky M, Rausner S. The role of Cdc42 and Gic1 in the regulation of septin filament formation and dissociation. eLife. 2013;2:e01085. doi: 10.7554/eLife.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala FA, Valadares NF, Macedo JN, Borges JC, Garratt RC. Heterotypic coiled-coil formation is essential for the correct assembly of the septin heterofilament. Biophys J. 2016;111(12):2608–2619. doi: 10.1016/j.bpj.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrock K, Bartsch I, Blaser S, Busse A, Busse E, Zieger B. Characterization of human septin interactions. Biol Chem. 2011;392:751–761. doi: 10.1515/BC.2011.081. [DOI] [PubMed] [Google Scholar]
- Sellin ME, Sandblad L, Stenmark S, Gullberg M. Deciphering the rules governing assembly order of mammalian septin complexes. Mol Biol Cell. 2011;22:3152–3164. doi: 10.1091/mbc.E11-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrão VH, Alessandro F, Caldas VE, Marçal RL, Pereira HD, Thiemann OH, Garratt RC. Promiscuous interactions of human septins: the GTP binding domain of SEPT7 forms filaments within the crystal. FEBS Lett. 2011;585:3868–3873. doi: 10.1016/j.febslet.2011.10.043. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Hauer F, Kühlmann D, Macara IG, Weyand M, Stark H, Wittinghofer A. Structural insight into filament formation by mammalian septins. Nature. 2007;449:311–315. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Zent E, Wittinghofer A. GTP-induced conformational changes in septins and implications for function. Proc Natl Acad Sci USA. 2009;106:16592–16597. doi: 10.1073/pnas.0902858106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Dolat L, Angelis D, Forgacs E, Spiliotis ET, Galkin VE. Septin 9 exhibits polymorphic binding to F-actin and inhibits myosin and cofilin activity. J Mol Biol. 2015;427:3273–3284. doi: 10.1016/j.jmb.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza TA, Barbosa JA. Cloning, overexpression, purification and preliminary characterization of human septin 8. Protein J. 2010;29:328–335. doi: 10.1007/s10930-010-9256-2. [DOI] [PubMed] [Google Scholar]
- Surka MC, Tsang CW, Trimble WS. The mammalian septin MSF localizes with microtubules and is required for completion of cytokinesis. Mol Biol Cell. 2002;13:3532–3545. doi: 10.1091/mbc.e02-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadares NF, Garratt RC. Septin crystallization for structural analysis. Methods Cell Biol. 2016;136:321–338. doi: 10.1016/bs.mcb.2016.03.017. [DOI] [PubMed] [Google Scholar]
- Weems A, McMurray M. The step-wise pathway of septin hetero-octamer assembly in budding yeast. Elife. 2017;25:6. doi: 10.7554/eLife.23689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Barral Y. The septin family of GTPases: achitecture and dynamics. Nat Rev. 2008;9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A, Pai EF. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem Sci. 1991;16:382–387. doi: 10.1016/0968-0004(91)90156-P. [DOI] [PubMed] [Google Scholar]
- Zent E, Wittinghofer A. Human septin isoforms and the GDP-GTP cycle. Biol Chem. 2014;395(2):169–180. doi: 10.1515/hsz-2013-0268. [DOI] [PubMed] [Google Scholar]
- Zent E, Vetter I, Wittinghofer A. Structural and biochemical properties of Sept7, a unique septin required for filament formation. Biol Chem. 2011;392:791–797. doi: 10.1515/BC.2011.082. [DOI] [PubMed] [Google Scholar]
- Zeraik AE, Pereira HM, Santos YV, Brandão-Neto J, Spoerner M, Santos MS, Colnago LA, Garratt RC, Araújo AP, DeMarco R. Crystal structure of a Schistosoma mansoni septin reveals the phenomenon of strand slippage in septins dependent on the nature of the bound nucleotide. J Biol Chem. 2014;289:7799–7811. doi: 10.1074/jbc.M113.525352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeraik AE, Staykova M, Fontes MG, Nemuraitė I, Quinlan R, Araújo AP, DeMarco R. Biophysical dissection of schistosome septins: Insights into oligomerization and membrane binding. Biochimie. 2016;131:96–105. doi: 10.1016/j.biochi.2016.09.014. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kong C, Xie H, McPherson PS, Grinstein S, Trimble WS. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr Biol. 1999;9:1458–1467. doi: 10.1016/S0960-9822(00)80115-3. [DOI] [PubMed] [Google Scholar]
- Zhang B, Zhang Y, Collins CC, Johnson DI, Zheng Y. A built-in arginine finger triggers the self-stimulatory GTPase-activating activity of rho family GTPases. J. Biol Chem. 1999;274:2609–2612. doi: 10.1074/jbc.274.5.2609. [DOI] [PubMed] [Google Scholar]