Abstract
Polypeptides can fold into tertiary structures while they are synthesized by the ribosome. In addition to the amino acid sequence, protein folding is determined by several factors within the cell. Among others, the folding pathway of a nascent polypeptide can be affected by transient interactions with other proteins, ligands, or the ribosome, as well as by the translocation through membrane pores. Particularly, the translation machinery and the population of tRNA under different physiological or adaptive responses can dramatically affect protein folding. This review summarizes the scientific evidence describing the role of translation kinetics and tRNA populations on protein folding and addresses current efforts to better understand tRNA biology. It is organized into three main parts, which are focused on: (i) protein folding in the cellular context; (ii) tRNA biology and the complexity of the tRNA population; and (iii) available methods and technical challenges in the characterization of tRNA pools. In this manner, this work illustrates the ways by which functional properties of proteins may be modulated by cellular tRNA populations.
Keywords: In vivo protein folding, Translation kinetics, Codon usage, Codon usage bias, tRNA, tRNA population, tRNA population adaptation, Translation machinery, tRNA quantification, tRNA sequencing, tRNA modified bases
Introduction
The research on native and denatured states of proteins has almost a century of history. It seems interesting to begin this review by quoting Hsien Wu’s work on protein denaturation (Wu 1931; Edsall 1995), just prior to the fundamental communication of Mirsky and Pauling (1936). Wu defined denaturation as “a change in the natural protein molecule whereby it becomes insoluble in solvents in which it was previously soluble.” Later, when the structure of proteins was yet unknown, he noted: “whatever may be the constitution of the protein molecule, its configuration (today we would say conformation) is not completely defined by its structural formula even if this be known.” And Wu concluded: “Evidence is adduced in support of the hypothesis that the molecule of natural, soluble protein is not a flexible open chain of polypeptide but has a compact structure. The force of attraction between the polar groups in a single molecule of proteins holds them together in an orderly way, just as the force of attraction between different molecules holds many molecules together in a crystal. In denaturation or coagulation the compact and orderly structure is disorganized.” The way was open.
For a long time, protein folding was mainly studied in vitro. Anfinsen (1973) showed that several in vitro denatured proteins refold spontaneously from their random disordered state into a well-defined unique structure, recovering their original biological activity. From this denatured state, successive conformational changes culminate when the protein reaches a thermodynamically stable conformation—its native state. Their experimental results have led to the paradigm that the amino acid sequence of a protein contains all the information needed to acquire its tridimensional structure.
What is the path traveled by a denatured polypeptide to get to the final conformation? The number of all possible conformations of a polypeptide chain is too large, incompatible with cellular protein synthesis times, in which protein sequences fold into unique native states in seconds. The Levinthal paradox illustrated that there is no time to randomly search among all conformational possibilities for an unfolded polypeptide (Levinthal 1968). Levinthal proposed that proteins fold through some directed processes of an unknown nature.
“Folding funnel” models illustrate that the lowest energy structure generally corresponds to the native structure of the protein (Wolynes et al. 1995; Karplus 1997) (Fig. 1). The model of protein folding as a funnel-shaped energy landscape has simplified the problem of protein folding and allowed the development of algorithms for protein structure prediction (Wolynes et al. 1995). One of the most basic models used to understand protein folding is the hydrophobic–hydrophilic (HP) residue model, which accounts for hydrophobicity as the major driving force leading to compact, desolvated structures, also allowing local secondary structure formation (Gruebele et al. 2016). Recent reviews have updated knowledge and advances regarding folding energy landscape, kinetics, and thermodynamics, through theory, models, and experimental approaches (Gruebele et al. 2016).
Fig. 1.
Free energy landscape for protein folding. The diagram illustrates how proteins fold into their native structures traveling through different pathways and by minimizing free energy, from a denatured state until reaching the native structure, or remaining in a metastable misfolded form
Interestingly, although conditions for protein folding in vitro and in vivo are extremely different (temperature, concentration, pH, purity), the folding energy landscape has been extended to understand both in vitro and in vivo protein folding, in the crowded cellular environment (Hartl and Hayer-Hartl 2009; Hartl et al. 2011). In this model, proteins trapped in a misfolded state (Fig. 1) could overcome this state and evolve to the native state by interacting with the cellular chaperone systems (see below). Moreover, de novo protein folding can be more efficient than in vitro refolding, which means that many proteins fold productively in the cell but can aggregate under in vitro refolding conditions (Evans et al. 2008). In this paper, we focus on protein folding within the cell, with emphasis on the contribution of the translation machinery and the population of tRNA to this process.
Ribosomal protein synthesis and co-translational folding in a cellular context
The maturation of newly synthesized polypeptides into correctly processed, translocated, and natively folded proteins is intimately linked to protein synthesis (Gloge et al. 2014). During biosynthesis on the ribosome, elongating nascent polypeptides begin to fold following the so-called co-translational folding. Post-translational folding also occurs in vivo, for example in proteins that adopt a stable conformation inside the cavity of hsp60 chaperone (see below), or a combination of both co- and post-translational folding, as it occurs in the folding of multidomain proteins in Escherichia coli (Ciryam et al. 2013). Moreover, co-translationally, proteins can be covalently modified, assembled into complex structures, translocated to different compartments, aggregated, ubiquitinated, and degraded (Turner and Varshavsky 2000; Comyn et al. 2014).
A range of biochemical and biophysical strategies evidence the folding of nascent polypeptides still bound to ribosomes. Among the earlier strategies, it is worth mentioning the detection of rhodanese enzymatic activity in polypeptides bound to bacterial ribosomes in a transcription/translation cell-free E. coli system (Kudlicki et al. 1995). In addition, in the same expression system, the use of antibodies (anti-coumarin) allowed the study of amino terminal nascent peptides labeled with coumarin, from three different proteins (Tsalkova et al. 1998). Furthermore, in semipermeabilized cells, it was shown using limited proteolysis, that CFTR (cystic fibrosis transmembrane conductance regulator) folds mostly co-translationally, domain by domain (Kleizen et al. 2005). Recently, the use of FRET (fluorescence resonance energy transfer) opened the way to study structural transitions of ribosome-bound folding intermediates, generated through in vitro translation of truncated RNA transcripts (Kim et al. 2015). Finally, co-translational protein folding has been studied by NMR spectroscopy providing atomic-resolution information on ribosome-nascent chain complexes isolated from E. coli (Cassaignau et al. 2016).
In prokaryotes, the nascent chain synthesis occurs at a rate of 10–20 amino acids per second. It emerges in a vectorial manner from the ribosomal exit tunnel and enters the crowded cellular environment where total protein concentrations can exceed 300 mg/mL (Gershenson and Gierasch 2011). To ensure efficient folding, different classes of molecular chaperones and proteins mediating processing and translocation interact with the nascent polypeptide. An integrated network of chaperones and degradation machineries are required to maintain protein homeostasis (Balchin et al. 2016). Several evolutionary conserved families of molecular chaperones guide proteins along productive folding pathways, avoiding and sometimes reversing misfolding and aggregation. The major chaperone families (Hsp40, Hsp60, Hsp70, Hsp90, Hsp100, and the small Hsp) prevent misfolding in the cytosol by binding to hydrophobic motives and by promoting productive folding (Balchin et al. 2016). In addition, the enzymes prolyl isomerase and protein disulfide-isomerase accelerate slow steps in protein folding and can be relevant in vivo (Balchin et al. 2016).
During translation, the ribosome acts as a platform for the binding of different factors that interact with the nascent chain (Nyathi and Pool 2015). Those factors that participate in peptide folding, processing, and subcellular targeting include, among others, the chaperones Trigger factor, Hsp70, and NAC (nascent-polypeptide associated complex), and the enzyme MetAP that removes the initial methionine of some proteins or enzymes involved in further modifications as N-myristoyl or N-acetyl transferases. In E. coli, SecA is an essential component of the Sec machinery, which participates in transporting proteins across the cytoplasmic membrane. SecA binds both ribosomes and nascent polypeptides that are Sec substrates (Huber et al. 2016). In eukaryotes, secretory proteins posses a hydrophobic signal sequence that is recognized co-translationally by SRP (signal recognition particle), a ribonucleoprotein composed of six proteins and RNA. SRP bound to the ribosome at the exit site binds the emerging signal sequence and then targets the complex (ribosome-nascent chain-SRP) to the endoplasmic reticulum membrane via the interaction with an SRP receptor (Nyathi and Pool 2015).
Diverse stress conditions promote protein misfolding. Misfolded proteins expose hydrophobic motives and can be directed towards the refolding and degradation machineries or nucleate as intermolecular aggregations. Protein aggregates can be cytotoxic and are frequently associated with degenerative diseases (Lim and Yue 2015). Aggregation however, also sequesters potentially toxic protein species and, therefore, provides protective functions (Ungelenk et al. 2016). Particularly, sHsps promote sequestration of misfolded proteins for storage in native-like conformation (Ungelenk et al. 2016).
Besides chaperones systems, the ribosomal surface itself influences protein folding through transient electrostatic interactions with the emerging nascent chain (Cabrita et al. 2016). Furthermore, ribosomes from prokaryotes, eukaryotes, and mitochondria also assist protein folding (PFAR, protein folding activity of ribosomes) in vitro and in vivo. This activity of ribosomes has been mapped to the domain V of the longer rRNA in the large subunit of the ribosome. PFAR is inhibited by 6-aminophenanthridine (6AP) and guanabenz (GA), the first two identified drugs that specifically inhibit the folding activity of the ribosomes without affecting protein translation. Interestingly, 6AP and GA inhibit prion propagation in yeast (Voisset et al. 2011, 2017; Blondel et al. 2016).
Translation kinetics
Early investigations on translation showed that the kinetics of this process is not uniform. During the synthesis of colicins in E. coli, the presence of discrete bands in denaturant electrophoresis gels showed a variable rate of translation (Varenne et al. 1982). The authors proposed that such variations could result from secondary structures of mRNA, but in the case of the studied colicins, they showed that it was related to differences in tRNA availability (Varenne et al. 1984). Furthermore, Varenne et al. (1984) proposed that, for a given codon, the stochastic search of the cognate ternary complex (aminoacyl-tRNA-EF-Tu-GTP) is the rate-limiting step in the elongation cycle, whereas the transpeptidation and translocation steps are much faster.
The sequencing of the first structural genes revealed that, for a given amino acid, different codons were used with dissimilar frequency (Fiers et al. 1976). Genes coding for ribosomal proteins (and other abundant proteins) use almost exclusively a reduced number of codons, whereas genes coding for less abundant proteins employ a broader ensemble of codons. Similarly, at the same time, Pedersen (1984) observed differences in the translation rate during the biosynthesis of six proteins encoded by different codon usage; that is, mRNAs contained different proportions of abundant/rare codons.
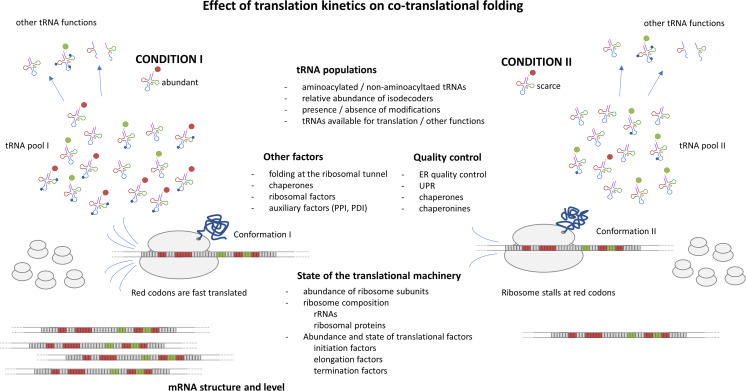
These observations led to the postulation made almost 30 years ago, which claims that the rates at which regions of polypeptides are translated affect protein folding, and that gene sequences have evolved to temporally separate the synthesis of defined domains of proteins (Purvis et al. 1987). In recent years, this subject has gained great attention from the scientific community; however, a deep understanding of the role of the translation kinetics on protein folding remains unclear (Kirchner et al. 2017) (Fig. 2).
Fig. 2.
Schematic representation of translation kinetics effects on co-translational folding of proteins. Two conditions are illustrated, representing ribosomes reading the same codon: (I) ribosomes proceed at high speed in the presence of a relative abundance of the corresponding cognate aminoacylated tRNA (aa-tRNA); (II) ribosomes are slowed down or stalled when the cognate aa-tRNA is scarce. Different conformations of the polypeptide emerging from the translating ribosome are produced. Aminoacylated tRNAs are represented by different colors in their 3′ end. Red and green populations of aa-tRNAs are the cognate partners of the red and green codons, respectively. Modified bases are indicated with blue dots. Note that a small fraction of non-aminoacylated tRNA is also included. The figure schematizes different mechanisms involved in protein folding and homeostasis. PPI: peptidyl-prolyl isomerase; PDI: protein disulfide-isomerase; ER: endoplasmic reticulum; UPR: unfolded protein response
Although tRNA abundance has been proposed as a major determinant in translation kinetics, in fact, it has not been determined precisely in most organisms (see below). On the other hand, thermodynamic parameters of anticodon–codon recognition, which depend on the specific codon, the wobble, and the presence of modified bases in the anticodon loop are also relevant factors in the local translation rate (Novoa and Ribas de Pouplana 2012; Endres et al. 2015).
Based on the observation of a positive correlation between codon usage and tRNA content in E. coli and the yeast Saccharomyces cerevisiae (Ikemura 1985), tRNA abundance was either estimated according to the gene copy number or to the frequency of codon usage in highly expressed proteins. Hence, frequent codons, associated with abundant decoding tRNA, were called “rapid” codons, in reference to the local speed of translation, even though the translation speed was not experimentally determined. Furthermore, it has been described that the usage of frequent codons ensures translation accuracy by reducing frameshifts and errors in the incorporation of amino acids. On the other hand, less frequently used codons are called “rare” or “slow” codons.
tRNAs, codon usage, and protein folding
As mentioned before, a good correlation between tRNA abundance and codon usage was described in prokaryotes, and their effects on protein folding have been demonstrated by different approaches (Fig. 2). Indeed, rare codons (read by non-abundant isoacceptor tRNA) are preferentially located in particular regions: encoding the N-terminal end or unstructured regions of proteins, encoding beta-sheets, turns or links between secondary structured regions, links between consecutive domains, or signal peptides of secreted proteins (Thanaraj and Argos 1996; Zalucki et al. 2011; Hess et al. 2015). To gain insight into the role of rare codons located in specific locations, relevant works are summarized below. (i) During the expression in E. coli of EgFABP, a small fatty acid binding protein from E. granulosus, rare codons encoding a turn between two alpha helices were substituted by frequent ones. The expression of one frequent synonymous variant showed reduced solubility and triggered the activity of a heat shock promoter driving the expression of a reporter gene, indicating the presence of unfolded or misfolded proteins (Cortazzo et al. 2002). Therefore, in this case, the use of frequent codons, instead of being an advantage, led to protein misfolding and in vivo aggregation. (ii) During the expression of recombinant proteins in E. coli, the overproduction of less abundant tRNA or the substitution of rare codons by frequent ones led to a significant yield increase but with lower solubility with accumulation in inclusion bodies (Rosano and Ceccarelli 2009). (iii) More recently, the presence of rare codons encoding links between domains allowed a significant increase of the solubility of the epoxide hydrolases expressed in E. coli (Hess et al. 2015). (iv) In a very elegant way, Zhang et al. (2009) mapped the folding status of translation intermediates and explored whether the local discontinuous translation at certain regions in the mRNA sequence was needed to efficiently coordinate the rate of elongation of the peptide chain and its co-translational folding. Focusing on the translation of the multidomain Suf1 in E. coli and based on codon usage frequency, the calculation of translation kinetics allowed the identification of four slow translating regions in Suf1 mRNA. Both the addition of low-abundant tRNA and the substitution of rare codons by frequent ones have shown changes in the translation kinetics, in the proteolysis profile, and in the type of folding intermediates (Zhang and Ignatova 2009). In sum, evidence strongly indicates that the modulation of translation kinetics in prokaryotes related to tRNA abundance and the choice of synonymous codons affect different processes, including ribosomal traffic, translation accuracy, and protein abundance, as well as topogenesis, protein solubility, and folding (Aguirre et al. 2011; Fernández-Calero et al. 2016).
In eukaryotes, several studies carried out in Saccharomyces and Neurospora have shown a relation between codon usage, RNA structures, and protein activity. Taking a recent example in Neurospora, a genome-wide study on codon usage showed that non-optimal codons preferentially encode intrinsically disordered regions of proteins, whereas in structured domains, more optimal codons were used (Zhou et al. 2015). This observation was experimentally verified by the expression of the circadian clock gene frequency, in which the change of synonymous codons affected its function in vivo (Zhou et al. 2015).
The relevance of synonymous mutations in higher eukaryotes is mainly recognized for their association with diseases. More than 50 diseases have been linked to silent mutations in different proteins (Sauna and Kimchi-Sarfaty 2011). The first identified silent mutations were shown to affect the normal splicing pattern but, more recently, other important effects have been described (Fåhraeus et al. 2016; Fernández-Calero et al. 2016; Gartner et al. 2013; Sauna and Kimchi-Sarfaty 2011; Lamolle et al. 2006; Hunt et al. 2014).
It is worth mentioning the study of synonymous polymorphisms in the MDR1 gene, one of the major drug transporters in humans. This gene encodes P-gp (P-glycoprotein), which actively effluxes a wide range of compounds from cells and is involved in multidrug-resistant cancers. The role of P-gp synonymous single-nucleotide polymorphism from a common haplotype was examined in polarized epithelial cells stably expressing recombinant P-gp (Fung et al. 2014). The synonymous variants in MDR1 did not influence mRNA expression, protein level, or translocation to the apical membrane. However, one synonymous MDR1 single-nucleotide polymorphism showed a significant impact on the stability and the overall folding of P-gp, without affecting ATPase activity. P-gp conformational alterations have subtly changed drug efflux function and the interaction with P-gp inhibitors, leading to altered drug cellular cytotoxicity (Fung et al. 2014). The effect of synonymous polymorphisms in MDR1 provided evidence for the first time that silent mutations can affect structural and functional properties of proteins in mammalian cells and produce major impacts on pharmacology (Kimchi-Sarfaty et al. 2007; Komar 2016; Sauna and Kimchi-Sarfaty 2011; Fung et al. 2014
Within the same research field, our work focuses on the folding of the ERα (estrogen receptor alpha), aiming to understand how the translation machinery and the cellular context of biosynthesis affect its functional properties. ERα is a multidomain nuclear receptor that activates or represses the transcription of specific genes. Regulation is achieved through recruitment of the receptor to DNA response elements either directly through interaction with DNA elements or through protein–protein interactions with other transcriptional factors (Paech et al. 1997; Yi et al. 2002). Upon binding to its natural ligand estradiol, the receptor interacts with specific cofactors for binding to DNA. This protein–DNA complex regulates the expression of different sets of genes, depending on both the cell and the promoter context (Mérot et al. 2004; Wijayaratne and McDonnell 2001; McDonnell and Norris 2002). Interestingly, the activity of ERα is regulated by SERMs (selective estrogen receptor modulators), compounds whose relative agonist/antagonist effect is tissue-specific (Wijayaratne and McDonnell 2001; McDonnell and Norris 2002) and that are crucial for the treatment of breast cancer and osteoporosis.
Initially, we studied the ERα synthetized in two eukaryotic derived in vitro translation systems: rabbit reticulocytes and wheat germ extracts. By limited proteolysis, we showed that the ERα adopts different soluble conformations in those cellular extracts, and exhibits different affinity for estradiol (Horjales et al. 2007). The results revealed that certain components of the cellular extracts affected differentially the translation of the mRNA, leading to different conformations and ligand affinity of the receptor.
Afterwards, we studied the expression of ERα variants in HepG2 and HeLa transfected cells. ER-Ala87 is a synonymous polymorphism, poorly characterized, present in 5–10% of the population, depending on ethnic groups. In transfected HepG2 and HeLa cells, our results showed a significant alteration of the functionality of ERAla87, mainly in transactivation activity and subcellular localization, depending on the cell type (Fernández-Calero et al. 2014). We proposed that a conformational variant might be originated upon translation of ERAla87, as a consequence of a modification of the translational kinetics, probably due to differences in the availability of tRNA species that recognize either the GCG or the GCC codons (Fernández-Calero et al. 2016).
In a more global way, whether codon usage fine tunes mRNA translation in mammals is still an open and controversial matter, being the subject of important efforts nowadays. In order to understand whether synonymous variants can be related to translation kinetics and protein folding, and, in turn, be associated with diseases, global genomic and transcriptomic approaches are being performed. However, contradictory results are reported. For instance, the tRNA pool in mammals appears to be equally efficient at translating any transcriptome, regardless of cell type or condition (Rudolph et al. 2016). While some works have suggested that preferentially used codons are not translated faster than unpreferred ones or that rare codons do not correlate with ribosome pausing (Qian et al. 2012; Guo et al. 2010; Pop et al. 2014), other reports have suggested that differences in tRNA population are associated with diverse effects on translation (Lampson et al. 2013; Goodarzi et al. 2016). This is the case of a mutation of a tissue-specific tRNA expressed in the mouse nervous system, which led to ribosome stalling and that was implicated as the cause of neurodegeneration (Ishimura et al. 2014). Another example is related to the activity of CFTR (cystic fibrosis transmembrane conductance regulator). Global analyses of tRNA concentration were integrated with studies on CFTR conformation (by thermal stability and proteolytic susceptibility) and protein activity (Kirchner et al. 2017). In this way, it was shown that a single synonymous CFTR variant modified the normal local speed of mRNA translation, in a tissue-specific tRNA-dependent manner, and altered both the conformational dynamics and the functionality of CFTR (Kirchner et al. 2017). The increase of the cellular tRNAThr(CGU) concentration rescued both the expression and single-channel conduction defects of T2562G-CFTR (Kirchner et al. 2017). Therefore, this work showed a direct link between the abundance of a specific tRNA and the functionality of CFTR.
Taken together, the above examples illustrate how, during the synthesis of proteins in the crowded cellular context, the population of tRNA might modulate the translation kinetics and the folding of proteins. In the next section, we focus on tRNA biology, the possible variations, and the dynamics of the tRNA population as adaptive responses to pathophysiological changes.
tRNA biology
As already mentioned, the journey of a translating ribosome is strongly associated with the available tRNA population, properly aminoacylated. This raises several questions regarding quantitative and qualitative characterization of the tRNA population and it requires accurate methodologies for determining the precise state of the population—structural, functional, and location—of each of its components.
The state of the tRNA population in the cell
Beyond the progressive elucidation of the mechanisms that regulate protein biosynthesis, the translation machinery as a whole was assumed as constitutive or, at least, as highly constitutive. This was particularly the case of the ribosome structure and tRNA population. However, evidence has progressively accumulated showing a broad variation of the composition of the ribosomes (Sauert et al. 2015; Guimaraes and Zavolan 2016; Sloan et al. 2016) and of the tRNA populations in response to environmental signals, during differentiation and diseases.
Early reports on specialized cells, with an extremely biased protein expression profile, revealed high frequencies in the use of selected codons and high concentrations of the specific corresponding decoder tRNA. This was the case for isoacceptor tRNA-Ala and tRNA-Ser species present in the posterior gland of the silkworm Bombyx mori and the aminoacyl-tRNA population of human reticulocytes (Garel et al. 1976; Sprague et al. 1977; Hentzen et al. 1981; Hatfield et al. 1982). Other observations, using cell-free protein synthesis systems, stressed that the rate of translation of a given mRNA was optimal in the presence of tRNA from the homologous tissue (Le Meur et al. 1976; Sharma and Beezley 1976). It is worth mentioning early works emphasizing the biological importance of post-transcriptional modifications of tRNA (Björk and Neidhardt 1975; Ny and Björk 1977; Labuda et al. 1982; Vacher et al. 1984; Meier et al. 1985; Grosjean et al. 1995).
In recent years, together with the description of new functions for tRNA molecules and the development of new analytical technologies, several questions have emerged. Are tRNA genes differentially expressed in different cell states? Do the modified base patterns change depending on the cell state or fate? Is the cellular tRNA pool homogeneous or is it heterogeneously distributed in different cell compartments? Clearly, the answers to these questions are crucial to determine their role in relation to translational kinetics and protein folding.
tRNA functions: expanding the concept of an adaptor molecule
tRNA are a major component of the protein synthesis machinery, ensuring the fidelity of codon recognition in the decoding site of ribosomes and presenting the cognate amino acid in the peptidyl-transferase center. tRNA also play other roles and participate in: regulatory mechanisms during protein synthesis; regulation of gene expression; non-ribosomal peptide bond formation; post-translational protein modification; phospholipid modifications in cell membranes; targeting protein degradation; quality control surveillance pathways; stress response; regulation of metabolic processes; secondary metabolism (Huang and Hopper 2016); priming reverse transcription; inhibition of apoptosis via complexation with cytochrome C; antimicrobial and protein folding (for reviews, see Giegé 2008; Kirchner and Ignatova 2015; Duechler et al. 2016). Recently, the production of fragments of some particular tRNA species (3′ or 5′ halves, and 3′ or 5′ quarters of a tRNA molecule, for instance) was associated with different cell conditions (responses to different stress conditions, hormonal effectors, or cancer processes) and could play regulatory functions (Keam and Hutvagner 2015). Interestingly, tRNA fragments appeared as secreted to the extracellular medium in microvesicles; thus, a possible role in cellular communication has been proposed (Garcia-Silva et al. 2010; Tosar et al. 2015). This variety of functions suggests a complex management of the cellular tRNA population and the existence of fine regulatory mechanisms involving a multiplicity of specific interactions with a great diversity of molecules.
tRNA genes are differentially expressed in different cell states
Over the last few years, investigations based on holistic approaches have progressively converged to shed more light on the biological roles of tRNA. The characterization of the tRNA population by deep sequencing (Pang et al. 2014), microarrays (Dittmar et al. 2006; Gingold et al. 2014), and chromatin analysis at tRNA loci (Pang et al. 2014) indicate that tRNA genes are actually differentially regulated. For instance, in S. cerevisiae, specific changes in the tRNA copy number were associated with stress responses (Pang et al. 2014). Moreover, after a semi-quantification of tRNA in human cells, the existence of two distinct translation programs during proliferation and differentiation was proposed (Gingold et al. 2014). Furthermore, it was shown that differences in the tRNA repertoire of proliferating and differentiated cells correspond to codon usage preferences of proliferation- or differentiation-regulated genes (Gingold et al. 2014). This indicates that tRNA levels are concerted with changes in the transcriptome, in order to optimize codon usage of expressed genes. Among other works supporting this view, it is worth mentioning that breast cancer cells show differences in tRNA population compared to normal tissue, suggesting a fine tuning of tRNA pools to translate mRNAs involved in tumor progression (Pavon-Eternod et al. 2009). The changes in tRNA population strongly suggest a precise coordination between transcription and translation involving regulatory mechanisms to ensure the adaptation of the translation machinery to different cell states (Topisirovic and Sonenberg 2014).
How is the differential expression of tRNA genes achieved? In eukaryotes, RNA polymerase III transcribes tRNA genes, and this activity is regulated by different signaling pathways in response to growth factors, nutrient levels, mitogens, or stress (Dang 2012; Grewal 2015; Khanna et al. 2015). Through protein kinase mTORC1, Maf 1 is a major repressor (Kantidakis et al. 2010; Grewal 2015). mTORC1 integrates mitogenic signals with the nutritional status of the cell, contributing to maintaining the metabolic balance and cellular homeostasis (reviewed in Saxton and Sabatini 2017). However, it remains unclear how the differential expression of tRNA genes is regulated in different cell types and states.
Epigenetics also contributes to the differential expression of tRNA genes. As shown by ChIP-seq experiments, the expression of tRNA genes by RNA polymerase III depends on the cell type (including cancer and stem cells) (Bhargava 2013; Park et al. 2017) and on the chromatin status. DNA CpG methylation, histone modification, as well as the opening of heterochromatin are associated with the activation or repression of the RNA polymerase III. Beyond the relevant studies on the epigenetic regulation of tRNA genes, questions related to the differential states of chromatin remain open.
tRNA base modifications: expanding the complexity of the tRNA population
There are roughly a hundred different post-transcriptional modifications described for tRNA (reviewed in: Grosjean et al. 2010; El Yacoubi et al. 2012; Machnicka et al. 2014), presenting each individual tRNA an average of ten modified bases. Many of them are conserved in all the kingdoms, while some are unique to each branch of life (Fig. 3).
Fig. 3.

Modified nucleosides in the tRNA anticodon loop of eukaryotes. Sites of modified nucleosides in tRNAs and a selection of modified bases present in the anticodon are indicated. Filled black circles positions that can carry base modification; light gray circles: anticodon positions (usually bases 34–36); dark gray circles: base immediately 3′ of the anticodon (usually 37). Modifications of the first base of the anticodon (wobble position) and position 37 that play a critical role in reading frame maintenance and fidelity are shown. Derivatives of adenosine: ms2t6A (2-methylthio-N6-threonylcarbamoyladenosine), ms2i6A (2-methylthio-N6-isopentenyladenosine), I (inosine), m1I (1-methylinsoine). Derivatives of cytidine: Cm (2′-O-methylcytidine), m5C (5-methylcytidine), m3C (3-methylcytidine). Derivatives of guanosine: Gm (2′-O-methylguanosine), m1G (1-methylguanosine), Q (queuosine), yW (wybutosine). Derivatives of uridine: cm5U (5-carboxymethyluridine), ncm5U (5-carbamoylmethyluridine), mcm5U (5-methoxycarbonylmethyluridine), mcm5s2U (5-methoxycarbonylmethyl-2-thiouridine), mcm5Um (5-methoxycarbonylmethyluridine), Y (pseudouridine). Data and chemical structures of modified nucleosides from: Machnicka et al. (2013); El Yacoubi et al. (2012); Tuorto and Lyko (2016)
Since the discovery of pseudouridine in bulk yeast tRNA and for a long time, RNA modified bases were considered a rare exception to the canonical world of nucleic acids, but, nowadays, such modifications have gained a great relevance (Grosjean 2015). The processes of modification–demodification are becoming associated with a number of human pathologies, thus fostering therapeutic research with emphasis on cancer and degenerative diseases (Sarin and Leidel 2014; Torres et al. 2014).
The modified bases play important roles in the structure and function of all RNA molecules. In regard to tRNA modifications, their functions include: stability and flexibility of the tRNA structure (Motorin and Helm 2010), translational fidelity via codon–anticodon interaction (Saint-Léger and Ribas de Pouplana 2015), reading frame maintenance (Waas et al. 2007; Delaunay et al. 2016; Klassen et al. 2016a), tRNA discrimination (Pang et al. 2014), nonsense suppression (Benko et al. 2000), tRNA stability (Alexandrov et al. 2006; Kotelawala et al. 2008; Dewe et al. 2012), proteome integrity (Nedialkova and Leidel 2015), sensitivity to aminoglycoside antibiotics acting at the decoding site of ribosomes (Kalhor and Clarke 2003), response to stress (Kamenski et al. 2007; Chan et al. 2012; Gu et al. 2014), and recognition of tRNA as either self or nonself by TLR7 receptor (Kaiser et al. 2014; Rimbach et al. 2015). For general reviews, see Motorin and Helm (2011), Hopper (2013) and Phizicky and Hopper (2015).
Modification of tRNA bases involves a large set of enzymes exhibiting high specificity for the target tRNA, for the base or nucleoside, and for its location in the tRNA structure. They include: methyl-transferases, adenosine-deaminases, pseudouridine synthetases, thiouridylases, and transglycosylases, among others (for a complete list of enzymes, see http://modomics.genesilico.pl; Machnicka et al. 2013). Most of the modifications take place directly on the nucleosides included in the tRNA (Helm and Alfonzo 2014). Some specific modifications are introduced by transglycosylation, replacing a base of the target tRNA by a modified base, which, in turn, could be further modified in situ once incorporated into the tRNA. Notably, this is the case of the nucleoside queuosine (Q, Fig. 3) present in all kingdoms, whose corresponding base (queuine) is exclusively synthesized in prokaryotes. In eukaryotes, except for yeast, after cellular uptake of the precursor (queuine) and the synthesis of QMP, a tRNA–guanine transferase exchanges specifically a G in first position of the anticodon by Q, into tRNA charging Asn, His, Asp, and Tyr. In the latter two, an additional in situ glycosylation takes place (addition of mannose or galactose, respectively) (Fergus et al. 2015).
Here, we describe with more detail three complex modifications, located in the anticodon loop (Fig. 3). The first two are widely present in prokaryotes, archaea, and eukaryotes:
Modifications of U (usually at position 34) in the first position of UNN anticodons, decoding NNA codons;
The N6-threonyl-carbamoyladenosine (t6A) and its derivatives in position 37, adjacent to anticodons decoding almost all ANN codons, including the initiator tRNA-Met;
Wybutosine (yW) and its derivatives, a heavily modified guanine in position 37, present in eukaryotic and archaeal tRNA-Phe.
These modifications—as any modification in the anticodon loop—play key roles in codon–anticodon recognition in the decoding site of the ribosome (Demeshkina et al. 2010). Their alterations result in severe consequences during translation, such as misreading, frame shifts, translation shifted to another distant site, as well as diverse pleiotropic cellular effects, including pathological states (Wei and Tomizawa 2011; Delaunay et al. 2016).
The three cases are clear examples of the links between tRNA modification and the cellular metabolic state. Indeed, most of the modifications require ubiquitous metabolites and co-enzymes for the transfer of methyl groups, acetyl groups, amino acids, isoprenoids, sugars, etc., that are crucial at the crossroads of basic metabolic pathways: S-adenosyl methionine (a major methyl donor), thiamine pyrophosphate, riboflavin, pyridoxal phosphate, biotin, folic acid, cyanocobalamin, among others (Helm and Alfonzo 2014). The cellular concentration of some of these compounds varies by several folds, depending on the metabolic state and the cellular fate (Fernández-Arroyo et al. 2015).
For instance, in yeast, the modifications of U34 (Fig. 3) require 15 proteins for methylation and 11 proteins for the supplementary thiolations. “Elongator”, a multimeric protein complex with methyl-transferase activity, participates in the first steps. “Elongator” is also related to other cellular processes, such as elongation by RNA polymerase II, histone acetylation, telomeric gene silencing, and DNA repair (Chen et al. 2011; Karlsborn et al. 2014). Noteworthy in yeast, the phenotype produced by a defective U34 modification (morphological and growth alterations) was reverted by the overexpression of the involved tRNA (Klassen et al. 2016b). These observations strongly suggest that the defective U34 modification affected the thermodynamics of the interactions at the ribosome.
The t6A modification requires several enzymatic steps (Perrochia et al. 2013; Thiaville et al. 2014). In Archaea and Eukarya, this process involves a universal protein (Sua5) and the complex KEOPS/EKC (Srinivasan et al. 2011). KEOPS/EKC was early associated with different cellular processes: transcription regulation, telomere homeostasis, genome instability, chromosome segregation, and metabolic regulation (Srinivasan et al. 2011). The t6A modification has been associated with cell growth regulation (Rojas-Benitez et al. 2015) and the levels of t6A-modified tRNA strongly influence TORC1 activity (Rojas-Benitez et al. 2015).
Concerning the yW base and its derivatives, it is interesting to note its complex biosynthesis in which up to six molecules of S-adenosyl-methionine are involved in successive methylations catalyzed by different enzymes (Young and Bandarian 2013). Taken together, these observations suggest a strong link between the modification of tRNA and the cellular metabolic state.
tRNA population: diversity and adaptation
The variations in tRNA gene expression and the diversity of post-transcriptional modifications described above lead us to consider that each particular cellular state has a characteristic tRNA population.
Cellular tRNA populations could then be described in the following manner: tRNA isoacceptors, recognized by an aminoacyl-tRNA synthetase and charged with specific amino acid; isodecoders, those isoacceptor tRNAs that recognize a given codon. In many cases, particularly in higher eukaryotes, the high number of tRNA genes lead to different species of isodecoders. Finally, an isoacceptor–isodecoder species may, in turn, exhibit diverse states of modification. Therefore, a particular tRNA species could be partitioned in a number of variants or subspecies (Hopper 2013).
Then, considering the links between the state of tRNA modifications with metabolism and cell fate described above, and the complexity of the tRNA population, it should be conceived that particular cell states have a defined distribution of tRNA variants, adapted to the ongoing cellular program and to the metabolic conditions. Finally, it is worth mentioning that Ribas de Pouplana and colleagues recently proposed that translation upgrades, such as codon usage adaptations or modulation of the tRNA pool or changes in tRNA modifications, may lead to the synthesis of novel protein structures and functions and, in this way, drive speciation (Ribas de Pouplana et al. 2017).
The study of the cellular tRNA population: scope of current methodologies and present challenges
How can we determine the tRNA population involved in translation in a particular cell context? If we do not properly discriminate between tRNA subspecies, how do we know which tRNA are really available for translation? tRNA quantification presents both biological and technical challenges. First, the diversity of tRNA subspecies within a cell needs to be identified and quantified simultaneously, although not all of them are necessarily involved in translation. Secondly, some characteristics of tRNAs, such as similar sequence length and homology, the very stable secondary structures, and frequent base modifications, hamper the development of techniques able to precisely characterize tRNA populations (Ferro and Ignatova 2015).
Attempts to quantify tRNA have been made in a variety of species. The first purification and sequencing of a tRNA by Holley and co-workers (Holley 1963; Holley et al. 1965; Apgar et al. 1962) opened the way for the development of a large number of methods for specific tRNA purification, generally based on chromatography (Dirheimer and Ebel 1967; Nishimura et al. 1987) or electrophoresis procedures (Martin et al. 1977; Ikemura 1989) and which allowed the quantification of isoacceptors (Bagshaw et al. 1970; White and Tener 1973; Kanduc 1997).
At present, a wide palette of techniques based on RNA hybridization is available. Most of them rely on the design of specific probes, which recognize individually each tRNA sequence. Probe designing is quite challenging because of their small size and the high tRNA sequence homology. Moreover, direct hybridization of DNA probes to modified bases can be tricky, considering that one of the main functions of some modifications (such as m1A, m22G, and m2G) is precisely to prevent Watson–Crick base pairing and participate in the tRNA loops maintenance. Northern blotting was employed for the identification of tRNA (Alwine et al. 1977) and the relative quantification of tRNA (Fu et al. 2009). This method involves a size separation of RNAs by denaturant gel electrophoresis, followed by its transfer to a membrane and a posterior hybridization with labeled probes. This is a laborious technique since each northern blot allows the detection of only one tRNA. Nevertheless, RNA hybridization approaches allowed great progress in the knowledge of tRNA biology and the characterization of tRNA populations. For instance, Ishimura and colleagues employed northern blotting to identify and quantify a specific tRNA isoacceptor specifically expressed in the central nervous system (Ishimura et al. 2014). Variations of northern blotting-based techniques have been developed to answer specific questions. The PHA6 northern blot assay is based on a DNA probe complementary to the anticodon loop which hybridizes better in the absence of i6A37 and a probe complementary to a different region of the same tRNA as a quantitative internal control. The procedure can be used to calibrate and measure differences in i6A37 modification (Lamichhane et al. 2011, 2013a, b, 2016; Yarham et al. 2014).
Immuno-northern blotting combines northern blotting with a subsequent immunoblotting using antibodies against modified nucleosides, thus detecting specific modifications (Mishima et al. 2015). Differences in electrophoretic patterns allows to distinguish aminoacylated from non-aminoacylated tRNA, due to their different mobility.
Microarray technology, frequently used for tRNA quantification, is also based on probe design and RNA hybridization. Pan and co-workers were the first to apply this technology for tRNA quantification. They designed microarray chips to quantify tRNA from different species, such as B. subtilis, E. coli, and H. sapiens (Dittmar et al. 2004, 2005, 2006). In this technique, tRNA are labeled taking advantage of their common 3′ feature. Fluorescent hairpin oligonucleotides with their 3′ end complementary to the 3′-CCA tRNA end are linked to tRNA, and then hybridized to a platform which contains all the designed probes. These probes are distributed along the platform in such a way that several tRNA can be simultaneously quantitated. Even though tRNA microarrays allow the quantification of a high number of isodecoders, only tRNA that differ on at least eight nucleotides are distinguished. More recently, an improved method also developed by Pan and co-workers allowed the detection of tRNA isoacceptors with a single nucleotide resolution (Pavon-Eternod et al. 2009). This methodology, however, only detects differences of one nucleotide in the anticodon loop, while it does not allow the discrimination of isodecoders that differ by one nucleotide elsewhere on the tRNA.
Another set of methods developed for tRNA identification and quantification employ reverse transcription (RT). Synthesis of DNA from an RNA template including modified nucleotides has its drawbacks where nucleotide misincorporation or interruptions in cDNA synthesis are frequent. Even in the absence of modifications, the highly stable secondary structure can make synthesis stop. RT-based techniques for the identification of modified residues were considerably improved by employing chemical reagents that specifically react with a given modified base (reviewed in Motorin et al. 2007), thus allowing their identification and localization in the RNA sequence. Despite these advances, it is not yet possible to simultaneously detect different modifications in tRNA.
Among RT strategies, four-leaf clover quantitative RT-PCR (FL-PCR) was designed to specifically quantify individual mature tRNA (Honda et al. 2015). In FL-PCR, T4 RNA ligase ligates a stem-loop adapter to mature tRNA. Subsequent TaqMan qRT-PCR amplifies only unmodified regions of the tRNA-adapter ligation products. This procedure, by avoiding the retrotranscription of the anticodon, limits the identification of different isodecoders. Also, by RFLP (restriction fragment length polymorphism) studies, different post-transcriptional modifications at specific sites of different tRNA types have been detected, taking advantage of the fact that base changes on reverse transcription PCR amplicons generated as a consequence of post-transcriptional modifications might create or abolish endonuclease restriction sites (Wulff et al. 2017).
High-throughput RNA sequencing opened new possibilities for tRNA quantification. The general small RNA sequencing procedure usually applied for tRNA sequencing consists of: (i) ligation of 3′ and 5′ adaptors to RNA molecules, (ii) RT-PCR, (iii) amplification of cDNA, (iv) size selection, (v) sequencing by synthesis. This method presents several biases: ligation of adaptors is not equally efficient on all molecules, modifications cannot be detected, the fact of being a method based on RT-PCR means that just a few molecules can be fully sequenced. Nevertheless, several groups have used RNA sequencing datasets to detect modified tRNA residues, exploiting the fact that not all sequencing errors are technical artifacts, but, conversely, they often conceal biological marks of post-transcriptional RNA modifications sites (Ebhardt et al. 2009; Iida et al. 2009; Findeiss et al. 2011; Ryvkin et al. 2013; Torres et al. 2015). By analyzing mismatches between sequencing reads and the genomic region where those reads mapped, as well as by analyzing special read patterns, they have been able to distinguish known and potentially novel post-transcriptional base modifications on tRNAs and, in some cases, also allowing a “relative” quantification of tRNA species (Torres et al. 2015).
Variations of this technique overcome some of these biases. In 2009, a direct RNA sequencing method was developed (Ozsolak et al. 2009), where poly(A)(+) tails are added to RNA and hybridized to a surface coated with poly(dT) oligonucleotides before sequencing-by-synthesis. Still, the authors report a 4% error rate. This fact limits its use, considering the high tRNA sequence homology. Lowe and co-workers developed the AlkB-facilitated RNA methylation sequencing (ARM-seq), which demethylates m1A, m3C, and m1G, commonly found in tRNA (Cozen et al. 2015). Besides, comparative methylation analysis using ARM-seq provides a transcriptome-scale map of these modifications.
Pan and co-workers also developed the DM-tRNA-seq method (Zheng et al. 2015). They used AlkB to remove m1A, m3C, and m1G methylations, and a thermostable group II intron reverse transcriptase (TGIRT) to synthesis cDNA from highly structured tRNA. The TGIRT reaction does not require adapter ligation and produces longer and full-length cDNAs. Another variation called tRNA-seq (Pang et al. 2014) proposes a first step ligation to a linker to the 3′ end of purified tRNA, followed by cDNA synthesis. cDNAs are then subjected to another round of linker ligation at the new 3′ end, followed by PCR amplification and standard next-generation sequencing. The authors argue that the first ∼30 nt starting from the 3′ tRNA end provide unique identification of all tRNA and contain the fewest modified bases along most tRNA. Thus, the use of tRNA-seq minimizes modification-induced polymerase fall-off during reverse transcription, and captures truncated fragments when reverse transcription terminates at modified ribonucleosides. Even though several procedure variations have been designed for tRNA sequencing, none can actually identify and quantify all tRNA subspecies.
Methods based on mass spectrometry (MS) are also employed to identify and quantify tRNA subspecies. Among them, matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) (Hossain and Limbach 2007, 2009), liquid chromatography-mass spectrometry (LC-MS) (Castleberry and Limbach 2010; Wetzel and Limbach 2012), and liquid chromatography tandem MS (LC-MS/MS) (Wetzel and Limbach 2012) have been developed. These methods consist of: (i) enzymatic digestion of tRNA by ribonucleases (e.g., RNase T1), which generates unique or “signature” digestion products; (ii) detection by MS or by a combination of chromatography and MS. These methods allow the identification and quantification of small tRNA pools like those from E. coli or M. capricolum. One of the main drawbacks of MS-based methods is that the identification of tRNA requires the prior knowledge of all tRNA sequences and modifications present in the sample.
At the moment, there are no methods allowing the precise quantification of tRNA isodecoders and subspecies in a high-throughput fashion. Nevertheless, the approximations developed so far have allowed great progress on the characterization of tRNA populations. Yet, there is more to come. Advances on MS-based methods seem encouraging. In addition, expected results from tRNA sequencing look promising through the Oxford Nanopore third-generation sequencing technology. This sequencing by degradation consists of nanoscale holes through which a RNA molecule can pass, nucleotide by nucleotide. An ionic current through the nanopores allows the identification of successive nucleotides. Recently, this technology employed for the identification of modified RNA bases has successfully detected m7G on 16S rRNA (Smith et al. 2017).
Conclusions and perspectives
At present, important efforts are being focused on the elucidation of the mechanisms related to in vivo protein folding, fostered by the increasing number of pathologies associated with protein misfolding. Although it has come a long way, some complex questions still remain open. Our review focuses on the contribution of translation kinetics on protein folding and describes recent evidence regarding the role of the translational machinery and the dynamics of the tRNA population in this process.
Although several molecular mechanisms have been elucidated, the mechanisms by which codon usage and tRNA availability affect translation kinetics and protein folding remain unclear. A major difficulty has been the precise determination of tRNA population and its variation in response to cellular states. Central options defining states and cellular fates involve homeostatic mechanisms to reach new equilibria and include metabolic changes associated with anaplerosis–cataplerosis and energy production. Beyond transcription regulation, these changes involve an adaptation of the translation machinery at the post-transcriptional level.
The development of more accurate methods for the characterization of tRNA population are expected, as they will certainly contribute to the elucidation of the questions regarding the role of tRNA and the translation machinery on the folding of proteins.
Acknowledgements
The authors would like to thank Paula Tucci for the helpful discussions. This work was partially supported by Fondo Clemente Estable (Agencia Nacional de Investigación e Innovación, ANII), PEDECIBA, Uruguay. T. Fernández-Calero was the recipient of fellowships from ANII and CAP-Universidad de la República.
Compliance with ethical standards
Conflict of interest
Mónica Marín declares that she has no conflict of interest. Tamara Fernández-Calero declares that she has no conflict of interest. Ricardo Ehrlich declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Latin America’ edited by Pietro Ciancaglini and Rosangela Itri.
Contributor Information
Mónica Marín, Phone: 5982 5252095, Email: marin@fcien.edu.uy.
Tamara Fernández-Calero, Email: tamfer@pasteur.edu.uy.
Ricardo Ehrlich, Email: ehrlich@pasteur.edu.uy.
References
- Aguirre B, Costas M, Cabrera N, Mendoza-Hernández G, Helseth DL, Jr, Fernández P, de Gómez-Puyou MT, Pérez-Montfort R, Torres-Larios A, Gómez Puyou A. A ribosomal misincorporation of Lys for Arg in human triosephosphate isomerase expressed in Escherichia coli gives rise to two protein populations. PLoS One. 2011;6:e21035. doi: 10.1371/journal.pone.0021035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Alwine JC, Kemp DJ, Stark GR. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Apgar J, Holley RW, Merrill SH. Purification of the alanine-, valine-, histidine-, and tyrosine-acceptor ribonucleic acids from yeast. J Biol Chem. 1962;237:796–802. [PubMed] [Google Scholar]
- Bagshaw JC, Finamore FJ, Novelli GD. Changes in transfer RNA in developing brine shrimp. Developmental Biology. 1970;23(1):23–35. doi: 10.1016/s0012-1606(70)80005-7. [DOI] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353(6294):aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- Benko AL, Vaduva G, Martin NC, Hopper AK. Competition between a sterol biosynthetic enzyme and tRNA modification in addition to changes in the protein synthesis machinery causes altered nonsense suppression. Proc Natl Acad Sci U S A. 2000;97(1):61–66. doi: 10.1073/pnas.97.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava P. Epigenetic regulation of transcription by RNA polymerase III. Biochim Biophys Acta. 2013;1829(10):1015–1025. doi: 10.1016/j.bbagrm.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Björk GR, Neidhardt FC. Physiological and biochemical studies on the function of 5-methyluridine in the transfer ribonucleic acid of Escherichia coli. J Bacteriol. 1975;124(1):99–111. doi: 10.1128/jb.124.1.99-111.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M, Soubigou F, Evrard J, Nguyen PH, Hasin N, Chédin S, Gillet R, Contesse MA, Friocourt G, Stahl G, Jones GW, Voisset C. Protein folding activity of the ribosome is involved in yeast prion propagation. Sci Rep. 2016;6:32117. doi: 10.1038/srep32117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita LD, Cassaignau AM, Launay HM, Waudby CA, Wlodarski T, Camilloni C, Karyadi ME, Robertson AL, Wang X, Wentink AS, Goodsell LS, Woolhead CA, Vendruscolo M, Dobson CM, Christodoulou J. A structural ensemble of a ribosome-nascent chain complex during co-translational protein folding. Nat Struct Mol Biol. 2016;23(4):278–285. doi: 10.1038/nsmb.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassaignau AM, Launay HM, Karyadi ME, Wang X, Waudby CA, Deckert A, Robertson AL, Christodoulou J, Cabrita LD. A strategy for co-translational folding studies of ribosome-bound nascent chain complexes using NMR spectroscopy. Nat Protoc. 2016;11(8):1492–1507. doi: 10.1038/nprot.2016.101. [DOI] [PubMed] [Google Scholar]
- Castleberry CM, Limbach PA. Relative quantitation of transfer RNAs using liquid chromatography mass spectrometry and signature digestion products. Nucleic Acids Res. 2010;38(16):e162. doi: 10.1093/nar/gkq578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Huang B, Eliasson M, Rydén P, Byström AS. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 2011;7(9):e1002258. doi: 10.1371/journal.pgen.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P, Morimoto RI, Vendruscolo M, Dobson CM, O’Brien EP. In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome. Proc Natl Acad Sci U S A. 2013;110(2):E132–E140. doi: 10.1073/pnas.1213624110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comyn SA, Chan GT, Mayor T. False start: cotranslational protein ubiquitination and cytosolic protein quality control. J Proteome. 2014;100:92–101. doi: 10.1016/j.jprot.2013.08.005. [DOI] [PubMed] [Google Scholar]
- Cortazzo P, Cerveñansky C, Marín M, Reiss C, Ehrlich R, Deana A. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem Biophys Res Commun. 2002;293(1):537–541. doi: 10.1016/S0006-291X(02)00226-7. [DOI] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12(9):879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay S, Rapino F, Tharun L, Zhou Z, Heukamp L, Termathe M, Shostak K, Klevernic I, Florin A, Desmecht H, Desmet CJ, Nguyen L, Leidel SA, Willis AE, Büttner R, Chariot A, Close P. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J Exp Med. 2016;213(11):2503–2523. doi: 10.1084/jem.20160397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Yusupova G, Yusupov M. Interactions of the ribosome with mRNA and tRNA. Curr Opin Struct Biol. 2010;20(3):325–332. doi: 10.1016/j.sbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Dewe JM, Whipple JM, Chernyakov I, Jaramillo LN, Phizicky EM. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA. 2012;18:1886–1896. doi: 10.1261/rna.033654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirheimer G, Ebel JP. Fractionation of ribonucleosides by chromatography on Sephadex G l0. Bull Soc Chim Biol (Paris) 1967;49(4):447–448. [PubMed] [Google Scholar]
- Dittmar KA, Mobley EM, Radek AJ, Pan T. Exploring the regulation of tRNA distribution on the genomic scale. J Mol Biol. 2004;337(1):31–47. doi: 10.1016/j.jmb.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Sørensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6(2):151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duechler M, Leszczyńska G, Sochacka E, Nawrot B. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell Mol Life Sci. 2016;73(16):3075–3095. doi: 10.1007/s00018-016-2217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009;37:2461–2470. doi: 10.1093/nar/gkp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsall JT. Hsien Wu and the first theory of protein denaturation (1931) Adv Protein Chem. 1995;46:1–5. [PubMed] [Google Scholar]
- El Yacoubi B, Bailly M, de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- Endres L, Dedon PC, Begley TJ. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 2015;12(6):603–614. doi: 10.1080/15476286.2015.1031947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MS, Sander IM, Clark PL. Cotranslational folding promotes beta-helix formation and avoids aggregation in vivo. J Mol Biol. 2008;383(3):683–692. doi: 10.1016/j.jmb.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fåhraeus R, Marin M, Olivares-Illana V. Whisper mutations: cryptic messages within the genetic code. Oncogene. 2016;35(29):3753–3760. doi: 10.1038/onc.2015.454. [DOI] [PubMed] [Google Scholar]
- Fergus C, Barnes D, Alqasem MA, Kelly VP. The queuine micronutrient: charting a course from microbe to man. Nutrients. 2015;7(4):2897–2929. doi: 10.3390/nu7042897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Arroyo S, Cuyàs E, Bosch-Barrera J, Alarcón T, Joven J, Menendez JA. Activation of the methylation cycle in cells reprogrammed into a stem cell-like state. Oncoscience. 2015;2(12):958–967. doi: 10.18632/oncoscience.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calero T, Astrada S, Alberti A, Horjales S, Arnal JF, Rovira C, Bollati-Fogolín M, Flouriot G, Marin M. The transcriptional activities and cellular localization of the human estrogen receptor alpha are affected by the synonymous Ala87 mutation. J Steroid Biochem Mol Biol. 2014;143:99–104. doi: 10.1016/j.jsbmb.2014.02.016. [DOI] [PubMed] [Google Scholar]
- Fernández-Calero T, Cabrera-Cabrera F, Ehrlich R, Marín M. Silent polymorphisms: can the tRNA population explain changes in protein properties? Life (Basel) 2016;6(1):E9. doi: 10.3390/life6010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro I, Ignatova Z. Quantifying the ‘escapers’ among RNA species. Biochem Soc Trans. 2015;43(6):1215–1220. doi: 10.1042/BST20150158. [DOI] [PubMed] [Google Scholar]
- Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, Min Jou W, Molemans F, Raeymaekers A, Van den Berghe A, Volckaert G, Ysebaert M. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260(5551):500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Findeiss S, Langenberger D, Stadler PF, Hoffmann S. Traces of post-transcriptional RNA modifications in deep sequencing data. Biol Chem. 2011;392:305–313. doi: 10.1515/BC.2011.043. [DOI] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583(2):437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- Fung KL, Pan J, Ohnuma S, Lund PE, Pixley JN, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. MDR1 synonymous polymorphisms alter transporter specificity and protein stability in a stable epithelial monolayer. Cancer Res. 2014;74(2):598–608. doi: 10.1158/0008-5472.CAN-13-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, Rovira C, Robello C, Goldenberg S, Cayota A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol. 2010;171(2):64–73. doi: 10.1016/j.molbiopara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Garel JP, Hentzen D, Schlegel M, Dirheimer G. Structural studies on RNA from Bombyx mori L.: I.—Nucleoside composition of enriched tRNA species from the posterior silkgland purified by countercurrent distribution. Biochimie. 1976;58:1089–1100. doi: 10.1016/s0300-9084(76)80087-9. [DOI] [PubMed] [Google Scholar]
- Gartner JJ, Parker SC, Prickett TD, Dutton-Regester K, Stitzel ML, Lin JC, Davis S, Simhadri VL, Jha S, Katagiri N, Gotea V, Teer JK, Wei X, Morken MA, Bhanot UK, NISC Comparative Sequencing Program. Chen G, Elnitski LL, Davies MA, Gershenwald JE, Carter H, Karchin R, Robinson W, Robinson S, Rosenberg SA, Collins FS, Parmigiani G, Komar AA, Kimchi-Sarfaty C, Hayward NK, Margulies EH, Samuels Y. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci U S A. 2013;110(33):13481–13486. doi: 10.1073/pnas.1304227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenson A, Gierasch LM. Protein folding in the cell: challenges and progress. Curr Opin Struct Biol. 2011;21(1):32–41. doi: 10.1016/j.sbi.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R. Toward a more complete view of tRNA biology. Nat Struct Mol Biol. 2008;15(10):1007–1014. doi: 10.1038/nsmb.1498. [DOI] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sørensen KD, Andersen LD, Andersen CL, Hulleman E, Wurdinger T, Ralfkiær E, Helin H, Grønbæk K, Ørntoft T, Waszak SM, Dahan O, Pedersen JS, Lund AH, Pilpel Y. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Gloge F, Becker AH, Kramer G, Bukau B. Co-translational mechanisms of protein maturation. Curr Opin Struct Biol. 2014;24:24–33. doi: 10.1016/j.sbi.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165(6):1416–1427. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SS. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim Biophys Acta. 2015;1849(7):898–907. doi: 10.1016/j.bbagrm.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Grosjean H, Sprinzl M, Steinberg S. Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie. 1995;77(1–2):139–141. doi: 10.1016/0300-9084(96)88117-x. [DOI] [PubMed] [Google Scholar]
- Grosjean H, de Crécy-Lagard V, Marck C. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010;584(2):252–264. doi: 10.1016/j.febslet.2009.11.052. [DOI] [PubMed] [Google Scholar]
- Grosjean H. RNA modification: the Golden Period 1995–2015. RNA. 2015;21(4):625–626. doi: 10.1261/rna.049866.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruebele M, Dave K, Sukenik S. Globular protein folding in vitro and in vivo. Annu Rev Biophys. 2016;45:233–251. doi: 10.1146/annurev-biophys-062215-011236. [DOI] [PubMed] [Google Scholar]
- Gu C, Begley TJ, Dedon PC. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014;588(23):4287–4296. doi: 10.1016/j.febslet.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes JC, Zavolan M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016;17(1):236. doi: 10.1186/s13059-016-1104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16(6):574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hatfield D, Varricchio F, Rice M, Forget BG. The aminoacyl-tRNA population of human reticulocytes. J Biol Chem. 1982;257(6):3183–3188. [PubMed] [Google Scholar]
- Helm M, Alfonzo JD. Posttranscriptional RNA modifications: playing metabolic games in a cell’s chemical Legoland. Chem Biol. 2014;21(2):174–185. doi: 10.1016/j.chembiol.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzen D, Chevallier A, Garel JP. Differential usage of iso-accepting tRNASer species in silk glands of Bombyx mori. Nature. 1981;290:267–269. doi: 10.1038/290267a0. [DOI] [PubMed] [Google Scholar]
- Hess AK, Saffert P, Liebeton K, Ignatova Z. Optimization of translation profiles enhances protein expression and solubility. PLoS One. 2015;10(5):e0127039. doi: 10.1371/journal.pone.0127039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley RW. Large-scale preparation of yeast "soluble" ribonucleic acid. Biochem Biophys Res Commun. 1963;10:186–188. doi: 10.1016/0006-291x(63)90048-2. [DOI] [PubMed] [Google Scholar]
- Holley RW, Everett GA, Madison JT, Zamir A. Nucleotide sequences in the yeast Alanine transfer ribonucleic acid. J Biol Chem. 1965;240:2122–2128. [PubMed] [Google Scholar]
- Honda S, Shigematsu M, Morichika K, Telonis AG, Kirino Y. Four-leaf clover qRT-PCR: a convenient method for selective quantification of mature tRNA. RNA Biol. 2015;12(5):501–508. doi: 10.1080/15476286.2015.1031951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics. 2013;194(1):43–67. doi: 10.1534/genetics.112.147470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horjales S, Cota G, Señorale-Pose M, Rovira C, Román E, Artagaveytia N, Ehrlich R, Marín M. Translational machinery and protein folding: evidence of conformational variants of the estrogen receptor alpha. Arch Biochem Biophys. 2007;467(2):139–143. doi: 10.1016/j.abb.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Hossain M, Limbach PA. Mass spectrometry-based detection of transfer RNAs by their signature endonuclease digestion products. RNA. 2007;13(2):295–303. doi: 10.1261/rna.272507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, Limbach PA. Multiple endonucleases improve MALDI-MS signature digestion product detection of bacterial transfer RNAs. Anal Bioanal Chem. 2009;394(4):1125–1135. doi: 10.1007/s00216-008-2562-2. [DOI] [PubMed] [Google Scholar]
- Huang HY, Hopper AK. Multiple layers of stress-induced regulation in tRNA biology. Life (Basel) 2016;6(2):E16. doi: 10.3390/life6020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D, Jamshad M, Hanmer R, Schibich D, Döring K, Marcomini I, Kramer G, Bukau B. SecA cotranslationally interacts with nascent substrate proteins in vivo. J Bacteriol. 2016;199(2):e00622-16. doi: 10.1128/JB.00622-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt RC, Simhadri VL, Iandoli M, Sauna ZE, Kimchi-Sarfaty C. Exposing synonymous mutations. Trends Genet. 2014;30(7):308–321. doi: 10.1016/j.tig.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Iida K, Jin H, Zhu JK. Bioinformatics analysis suggests base modifications of tRNAs and miRNAs in Arabidopsis thaliana. BMC Genomics. 2009;10:155. doi: 10.1186/1471-2164-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Purification of RNA molecules by gel techniques. Methods Enzymol. 1989;180:14–25. doi: 10.1016/0076-6879(89)80088-6. [DOI] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, Senju S, Nishimura Y, Chuang JH, Ackerman SL. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345(6195):455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S, Rimbach K, Eigenbrod T, Dalpke AH, Helm M. A modified dinucleotide motif specifies tRNA recognition by TLR7. RNA. 2014;20(9):1351–1355. doi: 10.1261/rna.044024.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor HR, Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol. 2003;23:9283–9292. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenski P, Kolesnikova O, Jubenot V, Entelis N, Krasheninnikov IA, Martin RP, Tarassov I. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol Cell. 2007;26:625–637. doi: 10.1016/j.molcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Kanduc D. Changes of tRNA population during compensatory cell proliferation: differential expression of methionine-tRNA species. Arch Biochem Biophys. 1997;342(1):1–5. doi: 10.1006/abbi.1996.9869. [DOI] [PubMed] [Google Scholar]
- Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci U S A. 2010;107(26):11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsborn T, Tükenmez H, Mahmud AKMF, Xu F, Xu H, Byström AS. Elongator, a conserved complex required for wobble uridine modifications in eukaryotes. RNA Biol. 2014;11(12):1519–1528. doi: 10.4161/15476286.2014.992276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M. The Levinthal paradox: yesterday and today. Fold Des. 1997;2(Suppl 1):S69–S75. doi: 10.1016/s1359-0278(97)00067-9. [DOI] [PubMed] [Google Scholar]
- Keam SP, Hutvagner G (2015) tRNA-derived fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life (Basel) 5(4):1638–1651 [DOI] [PMC free article] [PubMed]
- Khanna A, Pradhan A, Curran SP. Emerging roles for Maf1 beyond the regulation of RNA polymerase III activity. J Mol Biol. 2015;427(16):2577–2585. doi: 10.1016/j.jmb.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Yoon JS, Shishido H, Yang Z, Rooney LA, Barral JM, Skach WR. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348(6233):444–448. doi: 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kirchner S, Cai Z, Rauscher R, Kastelic N, Anding M, Czech A, Kleizen B, Ostedgaard LS, Braakman I, Sheppard DN, Ignatova Z. Alteration of protein function by a silent polymorphism linked to tRNA abundance. PLoS Biol. 2017;15(5):e2000779. doi: 10.1371/journal.pbio.2000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet. 2015;16(2):98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- Klassen R, Bruch A, Schaffrath R. Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA Biol. 2016;12:1–8. doi: 10.1080/15476286.2016.1267098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R, Ciftci A, Funk J, Bruch A, Butter F, Schaffrath R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016;44(22):10946–10959. doi: 10.1093/nar/gkw705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B, van Vlijmen T, de Jonge HR, Braakman I. Folding of CFTR is predominantly cotranslational. Mol Cell. 2005;20(2):277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Komar AA. The Yin and Yang of codon usage. Hum Mol Genet. 2016;25(R2):R77–R85. doi: 10.1093/hmg/ddw207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelawala L, Grayhack EJ, Phizicky EM. Identification of yeast tRNA Um(44) 2′-O-methyltransferase (Trm44) and demonstration of a Trm(44) role in sustaining levels of specific tRNA(Ser) species. RNA. 2008;14:158–169. doi: 10.1261/rna.811008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicki W, Chirgwin J, Kramer G, Hardesty B. Folding of an enzyme into an active conformation while bound as peptidyl-tRNA to the ribosome. Biochemistry. 1995;34(44):14284–14287. doi: 10.1021/bi00044a003. [DOI] [PubMed] [Google Scholar]
- Labuda D, Grosjean H, Striker G, Pörschke D. Codon:anticodon and anticodon:anticodon interaction. Evaluation of equilibrium and kinetic parameters of complexes involving a G:U wobble. Biochim Biophys Acta. 1982;698(3):230–236. doi: 10.1016/0167-4781(82)90152-x. [DOI] [PubMed] [Google Scholar]
- Lamolle G, Marin M, Alvarez-Valin F. Silent mutations in the gene encoding the p53 protein are preferentially located in conserved amino acid positions and splicing enhancers. Mutat Res. 2006;600(1–2):102–112. doi: 10.1016/j.mrfmmm.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Lamichhane TN, Blewett NH, Maraia RJ. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA. 2011;17:1846–1857. doi: 10.1261/rna.2628611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Blewett NH, Crawford AK, Cherkasova VA, Iben JR, Begley TJ, Farabaugh PJ, Maraia RJ. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol Cell Biol. 2013;33:2918–2929. doi: 10.1128/MCB.00278-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Mattijssen S, Maraia RJ. Human cells have a limited set of tRNA anticodon loop substrates of the tRNA isopentenyltransferase TRIT1 tumor suppressor. Mol Cell Biol. 2013;33:4900–4908. doi: 10.1128/MCB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Arimbasseri AG, Rijal K, Iben JR, Wei FY, Tomizawa K, Maraia RJ. Lack of tRNA-i6A modification causes mitochondrial-like metabolic deficiency in S. pombe by limiting activity of cytosolic tRNATyr, not mito-tRNA. RNA. 2016;22(4):583–596. doi: 10.1261/rna.054064.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson BL, Pershing NL, Prinz JA, Lacsina JR, Marzluff WF, Nicchitta CV, MacAlpine DM, Counter CM. Rare codons regulate KRas oncogenesis. Curr Biol. 2013;23(1):70–75. doi: 10.1016/j.cub.2012.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Meur MA, Gerlinger P, Ebel JP. Messenger RNA translation in the presence of homologous and heterologous tRNA. Eur J Biochem. 1976;67(2):519–526. doi: 10.1111/j.1432-1033.1976.tb10718.x. [DOI] [PubMed] [Google Scholar]
- Levinthal C. Are there pathways for protein folding? J Chim Phys. 1968;65:44–45. [Google Scholar]
- Lim J, Yue Z. Neuronal aggregates: formation, clearance, and spreading. Dev Cell. 2015;32(4):491–501. doi: 10.1016/j.devcel.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, Helm M, Bujnicki JM, Grosjean H. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41(Database issue):D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Olchowik A, Grosjean H, Bujnicki JM. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014;11(12):1619–1629. doi: 10.4161/15476286.2014.992273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RP, Schneller JM, Stahl AJ, Dirheimer G. Study of yeast mitochondrial tRNAs by two-dimensional polyacrylamide gel electrophoresis: characterization of isoaccepting species and search for imported cytoplasmic tRNAs. Nucleic Acids Res. 1977;4(10):3497–3510. doi: 10.1093/nar/4.10.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296(5573):1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- Meier F, Suter B, Grosjean H, Keith G, Kubli E. Queuosine modification of the wobble base in tRNAHis influences ‘in vivo’ decoding properties. EMBO J. 1985;4(3):823–827. doi: 10.1002/j.1460-2075.1985.tb03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérot Y, Métivier R, Penot G, Manu D, Saligaut C, Gannon F, Pakdel F, Kah O, Flouriot G. The relative contribution exerted by AF-1 and AF-2 transactivation functions in estrogen receptor alpha transcriptional activity depends upon the differentiation stage of the cell. J Biol Chem. 2004;279(25):26184–26191. doi: 10.1074/jbc.M402148200. [DOI] [PubMed] [Google Scholar]
- Mirsky AE, Pauling L. On the structure of native, denatured, and coagulated proteins. Proc Natl Acad Sci U S A. 1936;22:439–447. doi: 10.1073/pnas.22.7.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima E, Jinno D, Akiyama Y, Itoh K, Nankumo S, Shima H, Kikuchi K, Takeuchi Y, Elkordy A, Suzuki T, Niizuma K, Ito S, Tomioka Y, Abe T. Immuno-northern blotting: detection of RNA modifications by using antibodies against modified nucleosides. PLoS One. 2015;10(11):e0143756. doi: 10.1371/journal.pone.0143756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49(24):4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2(5):611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- Motorin Y, Muller S, Behm-Ansmant I, Branlant C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- Nedialkova DD, Leidel SA. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S, Shindo-Okada N, Crain PF. Isolation of amino acid-specific tRNA by high-performance liquid chromatography. Methods Enzymol. 1987;155:373–379. doi: 10.1016/0076-6879(87)55025-x. [DOI] [PubMed] [Google Scholar]
- Novoa EM, Ribas de Pouplana L. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 2012;28(11):574–581. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Ny T, Björk GR. Stringent regulation of the synthesis of a transfer ribonucleic acid biosynthetic enzyme: transfer ribonucleic acid(m5U)methyltransferase from Escherichia coli. J Bacteriol. 1977;130(2):635–641. doi: 10.1128/jb.130.2.635-641.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyathi Y, Pool MR. Analysis of the interplay of protein biogenesis factors at the ribosome exit site reveals new role for NAC. J Cell Biol. 2015;210(2):287–301. doi: 10.1083/jcb.201410086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. Direct RNA sequencing. Nature. 2009;461(7265):814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson JÅ, Kushner PJ, Scanlan TS. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science. 1997;277(5331):1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- Pang YL, Poruri K, Martinis SA. tRNA synthetase: tRNA aminoacylation and beyond. Wiley Interdisciplinary Reviews: RNA. 2014;5(4):461–480. doi: 10.1002/wrna.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang YL, Abo R, Levine SS, Dedon PC. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res. 2014;42:e170. doi: 10.1093/nar/gku945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JL, Lee YS, Kunkeaw N, Kim SY, Kim IH, Lee YS. Epigenetic regulation of noncoding RNA transcription by mammalian RNA polymerase III. Epigenomics. 2017;9(2):171–187. doi: 10.2217/epi-2016-0108. [DOI] [PubMed] [Google Scholar]
- Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrochia L, Guetta D, Hecker A, Forterre P, Basta T. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t6A tRNA modification. Nucleic Acids Res. 2013;41(20):9484–9499. doi: 10.1093/nar/gkt720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. tRNA processing, modification, and subcellular dynamics: past, present, and future. RNA. 2015;21(4):483–485. doi: 10.1261/rna.049932.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, Koller D. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol Syst Biol. 2014;10:770. doi: 10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis IJ, Bettany AJ, Santiago TC, Coggins JR, Duncan K, Eason R, Brown AJ. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo: a hypothesis. J Mol Biol. 1987;193(2):413–417. doi: 10.1016/0022-2836(87)90230-0. [DOI] [PubMed] [Google Scholar]
- Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8(3):e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas de Pouplana L, Torres AG, Rafels-Ybern À. What froze the genetic code? Life (Basel) 2017;7(2):14. doi: 10.3390/life7020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimbach K, Kaiser S, Helm M, Dalpke AH, Eigenbrod T. 2′-O-Methylation within bacterial RNA acts as suppressor of TLR7/TLR8 activation in human innate immune cells. J Innate Immun. 2015;7(5):482–493. doi: 10.1159/000375460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Benitez D, Thiaville PC, de Crécy-Lagard V, Glavic A. The levels of a universally conserved tRNA modification regulate cell growth. J Biol Chem. 2015;290(30):18699–18707. doi: 10.1074/jbc.M115.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano GL, Ceccarelli EA. Rare codon content affects the solubility of recombinant proteins in a codon bias-adjusted Escherichia coli strain. Microb Cell Factories. 2009;8:41. doi: 10.1186/1475-2859-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph KL, Schmitt BM, Villar D, White RJ, Marioni JC, Kutter C, Odom DT. Codon-driven translational efficiency is stable across diverse mammalian cell states. PLoS Genet. 2016;12(5):e1006024. doi: 10.1371/journal.pgen.1006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvkin P, Leung YY, Silverman IM, Childress M, Valladares O, Dragomir I, Gregory BD, Wang LS. HAMR: high-throughput annotation of modified ribonucleotides. RNA. 2013;19:1684–1692. doi: 10.1261/rna.036806.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Léger A, Ribas de Pouplana L. The importance of codon–anticodon interactions in translation elongation. Biochimie. 2015;114:72–79. doi: 10.1016/j.biochi.2015.04.013. [DOI] [PubMed] [Google Scholar]
- Sarin LP, Leidel SA. Modify or die? RNA modification defects in metazoans. RNA Biol. 2014;11(12):1555–1567. doi: 10.4161/15476286.2014.992279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauert M, Temmel H, Moll I. Heterogeneity of the translational machinery: variations on a common theme. Biochimie. 2015;114:39–47. doi: 10.1016/j.biochi.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12(10):683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma OK, Beezley DN, Roberts WK. Limitation of reticulocyte transfer RNA in the translation of heterologous messenger RNAs. Biochemistry. 1976;15(19):4313–4318. doi: 10.1021/bi00664a027. [DOI] [PubMed] [Google Scholar]
- Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DL, Bohnsack MT. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2016;2:1–16. doi: 10.1080/15476286.2016.1259781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Jain M, Mulroney L, Garalde DR, Akeson M (2017) Reading canonical and modified nucleotides in 16S ribosomal RNA using nanopore direct RNA sequencing. bioRxiv 132274 [DOI] [PMC free article] [PubMed]
- Sprague KU, Hagenbüchie O, Zuniga MC. The nucleotide sequence of two silk gland alanine tRNAs: implications for fibroin synthesis and for initiator tRNA structure. Cell. 1977;11:561–570. doi: 10.1016/0092-8674(77)90074-5. [DOI] [PubMed] [Google Scholar]
- Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 2011;30(5):873–881. doi: 10.1038/emboj.2010.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj TA, Argos P. Protein secondary structural types are differentially coded on messenger RNA. Protein Sci. 1996;5(10):1973–1983. doi: 10.1002/pro.5560051003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville PC, Iwata-Reuyl D, de Crécy-Lagard V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t(6)a), a universal modification of tRNA. RNA Biol. 2014;11(12):1529–1539. doi: 10.4161/15476286.2014.992277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Sonenberg N. Distinctive tRNA repertoires in proliferating versus differentiating cells. Cell. 2014;158:1238–1239. doi: 10.1016/j.cell.2014.08.031. [DOI] [PubMed] [Google Scholar]
- Torres AG, Batlle E, Ribas de Pouplana L (2014) role of tRNA modifications in human diseases. Trends Mol Med 20(6):306–314 [DOI] [PubMed]
- Torres AG, Piñeyro D, Rodríguez-Escribà M, Camacho N, Reina O, Saint-Léger A, Filonava L, Batlle E, Ribas de Pouplana L. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015;43(10):5145–5157. doi: 10.1093/nar/gkv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar JP, Gámbaro F, Sanguinetti J, Bonilla B, Witwer KW, Cayota A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015;43(11):5601–5616. doi: 10.1093/nar/gkv432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsalkova T, Odom OW, Kramer G, Hardesty B. Different conformations of nascent peptides on ribosomes. J Mol Biol. 1998;278(4):713–723. doi: 10.1006/jmbi.1998.1721. [DOI] [PubMed] [Google Scholar]
- Tuorto F, Lyko F (2016 Dec) Genome recoding by tRNA modifications. Open Biol 6(12):160287. Review [DOI] [PMC free article] [PubMed]
- Turner GC, Varshavsky A. Detecting and measuring cotranslational protein degradation in vivo. Science. 2000;289(5487):2117–2120. doi: 10.1126/science.289.5487.2117. [DOI] [PubMed] [Google Scholar]
- Ungelenk S, Moayed F, Ho CT, Grousl T, Scharf A, Mashaghi A, Tans S, Mayer MP, Mogk A, Bukau B. Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding. Nat Commun. 2016;7:13673. doi: 10.1038/ncomms13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher J, Grosjean H, Houssier C, Buckingham RH. The effect of point mutations affecting Escherichia coli tryptophan tRNA on anticodon–anticodon interactions and on UGA suppression. J Mol Biol. 1984;177(2):329–342. doi: 10.1016/0022-2836(84)90460-1. [DOI] [PubMed] [Google Scholar]
- Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a non-uniform process: effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984;180(3):549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Varenne S, Knibikhler M, Cavard D, Morlon J, Lazdunski C. Variable rate of polypeptide chain elongation for colicins a, E2 and E3. J Mol Biol. 1982;159(1):57–70. doi: 10.1016/0022-2836(82)90031-6. [DOI] [PubMed] [Google Scholar]
- Voisset C, Saupe SJ, Blondel M. The various facets of the protein-folding activity of the ribosome. Biotechnol J. 2011;6(6):668–673. doi: 10.1002/biot.201100021. [DOI] [PubMed] [Google Scholar]
- Voisset C, Blondel M, Jones GW, Friocourt G, Stahl G, Chédin S, Béringue V, Gillet R. The double life of the ribosome: when its protein folding activity supports prion propagation. Prion. 2017;11(2):89–97. doi: 10.1080/19336896.2017.1303587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waas WF, Druzina Z, Hanan M, Schimmel P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J Biol Chem. 2007;282:26026–26034. doi: 10.1074/jbc.M703391200. [DOI] [PubMed] [Google Scholar]
- Wei FY, Tomizawa K. Functional loss of Cdkal1, a novel tRNA modification enzyme, causes the development of type 2 diabetes. Endocr J. 2011;58(10):819–825. doi: 10.1507/endocrj.ej11-0099. [DOI] [PubMed] [Google Scholar]
- Wetzel C, Limbach PA. Global identification of transfer RNAs by liquid chromatography-mass spectrometry (LC-MS) J Proteomics. 2012;75(12):3450–3464. doi: 10.1016/j.jprot.2011.09.015. [DOI] [PubMed] [Google Scholar]
- White BN, Tener GM. Chromatography of Drosophila tRNA on BD-cellulose. Can J Biochem. 1973;51(6):896–902. doi: 10.1139/o73-111. [DOI] [PubMed] [Google Scholar]
- Wijayaratne AL, McDonnell DP. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J Biol Chem. 2001;276(38):35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267(5204):1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- Wu H. Studies on denaturation of proteins. XIII. A theory of denaturation. Chin J Physiol. 1931;5:321–344. [PubMed] [Google Scholar]
- Wulff TF, Argüello RJ, Molina Jordàn M, Roura Frigolé H, Hauquier G, Filonava L, Camacho N, Gatti E, Pierre P, Ribas de Pouplana L, Torres AG (2017) Detection of a subset of posttranscriptional transfer RNA modifications in vivo with a restriction fragment length polymorphism-based method. Biochemistry 56(31):4029–4038 [DOI] [PubMed]
- Yarham JW, Lamichhane TN, Pyle A, Mattijssen S, Baruffini E, Bruni F, Donnini C, Vassilev A, He L, Blakely EL, Griffin H, Santibanez-Koref M, Bindoff LA, Ferrero I, Chinnery PF, McFarland R, Maraia RJ, Taylor RW. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet. 2014;10:e1004424. doi: 10.1371/journal.pgen.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Driscoll MD, Huang J, Bhagat S, Hilf R, Bambara RA, Muyan M. The effects of estrogen-responsive element- and ligand-induced structural changes on the recruitment of cofactors and transcriptional responses by ERalpha and ERbeta. Mol Endocrinol. 2002;16(4):674–693. doi: 10.1210/mend.16.4.0810. [DOI] [PubMed] [Google Scholar]
- Young AP, Bandarian V. Radical mediated ring formation in the biosynthesis of the hypermodified tRNA base wybutosine. Curr Opin Chem Biol. 2013;17(4):613–618. doi: 10.1016/j.cbpa.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalucki YM, Shafer WM, Jennings MP. Directed evolution of efficient secretion in the SRP-dependent export of TolB. Biochim Biophys Acta. 2011;1808(10):2544–2550. doi: 10.1016/j.bbamem.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16(3):274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- Zhang G, Ignatova Z. Generic algorithm to predict the speed of translational elongation: implications for protein biogenesis. PLoS One. 2009;4(4):e5036. doi: 10.1371/journal.pone.0005036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12(9):835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wang T, Fu J, Xiao G, Liu Y. Nonoptimal codon usage influences protein structure in intrinsically disordered regions. Mol Microbiol. 2015;97(5):974–987. doi: 10.1111/mmi.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]