Abstract
Mitotic clonal expansion has been suggested as a prerequisite for adipogenesis in murine preadipocytes, but the precise role of cell proliferation during human adipogenesis is unclear. Using adipose tissue-derived human mesenchymal stem cells as an in vitro cell model for adipogenic study, a group of cell cycle regulators, including Cdk1 and CCND1, were found to be downregulated as early as 24 h after adipogenic initiation and consistently, cell proliferation activity was restricted to the first 48 h of adipogenic induction. Cell proliferation was either further inhibited using siRNAs targeting cell cycle genes or enhanced by supplementing exogenous growth factor, basic fibroblast growth factor (bFGF), at specific time intervals during adipogenesis. Expression knockdown of Cdk1 at the initiation of adipogenic induction resulted in significantly increased adipocytes, even though total number of cells was significantly reduced compared to siControl-treated cells. bFGF stimulated proliferation throughout adipogenic differentiation, but exerted differential effect on adipogenic outcome at different phases, promoting adipogenesis during mitotic phase (first 48 h), but significantly inhibiting adipogenesis during adipogenic commitment phase (days 3–6). Our results demonstrate that cellular proliferation is counteractive to adipogenic commitment in human adipogenesis. However, cellular proliferation stimulation can be beneficial for adipogenesis during the mitotic phase by increasing the population of cells capable of committing to adipocytes before adipogenic commitment.
Keywords: : human mesenchymal stem cells (hMSCs), adipogenesis, cell proliferation, cell cycle regulator, basic fibroblast growth factor (bFGF)
Introduction
Adipose tissue is a central component of whole-body energy homeostasis regulation. Obesity is characterized by excess body fat accumulation, as a result of increased number of adipocytes (fat cells) through adipogenesis and/or enlarged adipocytes due to increased lipid storage through lipogenesis [1]. Advancement in understanding adipose tissue biology may provide new strategies for the intervention of obesity and obesity-related diseases. Adipogenesis is the process in which uncommitted stem cells differentiate into mature adipocytes. Much of our current understanding of adipogenesis is based on in vitro studies using mouse preadipocyte cell line 3T3-L1 cells [2] and on a more limited scale, mesenchymal stem cells [3].
Human mesenchymal stem cells (hMSCs) are a type of adult stem cell that exists in multiple tissues in the body, including adipose tissue, bone marrow, and peripheral blood, and play important roles in maintaining normal tissue homeostasis. They can be isolated, expanded, and differentiated in vitro into a number of specialized cell types, including adipocytes, which make them an excellent in vitro cell model for studying human adipogenesis [4].
Using a simple cocktail of adipogenic inducing media (AIM) containing dexamethasone (DEX), 3-isobutyl-1-methylxanthine (IBMX), and insulin, hMSCs can be induced to differentiate into mature adipocytes [5]. Due to their ability to differentiate into a variety of mature cell types, low allogeneic immune response, and low tumorigenicity in graft recipients, hMSCs have been of great interests to researchers exploring cell-based therapies as well, and is the most prevalent cell type used in ongoing stem cell-based clinical trials [6]. In addition to advancing our basic understanding of adipose tissue biology, the potential therapeutic application of hMSCs in adipose tissue engineering makes it even more relevant to use these cells for studying human adipogenesis [7].
Over the past two decades or so, many individual adipogenic regulators have been independently uncovered, including specific signaling pathways, growth factors or cytokines, transcription factors, GTPase protein and its regulator, epigenetic regulators, microRNAs, and so on. [3,8,9]. Most significantly, C/EBPα (CCAAT/enhancer binding protein alpha) and PPARγ (peroxisome proliferator-activated receptor gamma) were identified as two key transcriptional factors, which when overexpressed could dictate adipogenic cell fate in both 3T3-L1 and hMSCs [10–14]. Despite the discovery of new players involved in adipogenesis, there is a lack of systematic understanding of the sequence of events underlying human adipogenesis on both the molecular and subcellular levels.
One of the questions that remain to be addressed is as follows: Do cells stop proliferating or do they undergo a period of mitotic clonal expansion before undergoing adipogenic commitment and/or terminal differentiation? It has been suggested that mitotic clonal expansion is a prerequisite for adipogenic differentiation in 3T3-L1 cells [15], although in a separate study, the opposite conclusion was drawn [16]. In both studies, cell proliferation activities were observed during the first 60 h of adipogenic induction, resulting in a threefold to fourfold increase of total cell numbers.
To our knowledge, no study so far has thoroughly addressed this question in human adipogenesis. One previous study briefly examined total cell counts before and after adipogenic induction in hMSCs and concluded that hMSCs derived from bone marrow did not undergo mitotic clonal expansion during adipogenic differentiation [11]. Another study examined the expression of Cyclin D1, which oscillated with interval insulin treatments following periodic AIM treatments [17]. In addition, it demonstrated that araC, a replication blocker, did not affect adipogenic differentiation at 1–3 pg/cell, although it was not clear whether araC had any effect on cell proliferation at those concentrations [17].
In this study, using adipose tissue-derived hMSCs as an in vitro model for adipogenic differentiation, we examined the temporal expression patterns of a group of cell cycle regulators identified through microarray analysis, as well as cellular proliferation activities of hMSCs undergoing continuous AIM treatment for 12 days. In addition, the role of cell proliferation during adipogenic differentiation was assessed by inhibiting or promoting cellular proliferation at different time intervals of AIM treatment.
Our results demonstrated that hMSCs do undergo a mitotic clonal expansion phase during the first 48 h of AIM induction. Inhibiting proliferation by knocking down the expression of cell cycle master regulator, Cdk1, resulted in an increased number of adipocytes, likely by enhancing cells' ability to commit to adipogenic lineage, even though total cell number was reduced. Supplementing basic fibroblast growth factor (bFGF), a fibroblast growth factor (FGF), during days 3–6 after the initiation of adipogenic induction that corresponds to adipogenic commitment phase, significantly inhibited adipogenesis. The above evidences suggest that cellular proliferation antagonizes adipogenic commitment of hMSCs. On the other hand, stimulating cellular proliferation by bFGF can be beneficial to adipogenic differentiation, but only if it is applied before adipogenic commitment, by increasing the number of cells capable of committing to adipogenesis.
Materials and Methods
hMSC culture and differentiation
Adipose tissue-derived hMSCs were purchased from Fisher Scientific (SV3010201) and cultured in Hyclone Advance STEM expansion media (SH30875KT; Fisher Scientific). Passage 4 hMSCs were used in all experiments described. For osteogenic differentiation induction, cells were exposed to osteogenic inducing media (OIM) composed of 0.05 mM ascorbic acid 2-phosphate (NC9445523; Fisher Scientific), 10 mM β-glycophosphate (NC9960188), and 0.2 or 1 μM DEX (NC9756434; Fisher Scientific) in Hyclone Advance STEM expansion media. For adipogenic differentiation induction, cells were either exposed to AIM cocktail composed of 0.45 mM IBMX (NC9875083; Fisher Scientific), 10 μg/mL insulin (I9278-5ML; Sigma), and 0.2 or 1 μM DEX, or IBMX+DEX only.
All adipogenic differentiation media were made in control growth media (CM) composed of high-glucose Dulbecco's modified Eagle's medium (DMEM), 10% heat-inactivated fetal bovine serum, 0.1 μM β-mercaptoethanol (Cat. No. ES-007-E; Millipore), and 1% MEM non-essential amino acids (Cat. No. 11140050; Fisher Scientific). Cells were grown in either Napco 8000wj CO2 incubator thermal conductivity CO2 sensor/relative humidity sensor or Heracell CO2 incubator with infrared CO2 sensor/relative humidity sensor and handled in Labconco biosafety cabinet.
Cell death assay
Cell death assay was carried out by following instructions in Annexin-V-FLUOS staining kit from Roche (Cat. No. 11 858 777 001). Cells stained positive for propidium iodide (PI; red) from each sample well were imaged and counted using an Olympus IX50 microscope. Cells were then fixed and stained with nuclear dye DAPI (Cat. No. P36931; Invitrogen), followed by nuclei counting using Cell Profiler software.
Resazurin assay
Bioreduction of the dye by viable cells reduces the amount of its oxidized form (blue) and concomitantly increases the amount of its fluorescent intermediate (red). Reduction in blue can be measured spectrophotometrically by monitoring decrease in absorbance at 600 nm (see instructions in Sigma TOX8-1KT). Briefly, 10% resazurin dye solution (diluted with cell culture media) was added to cells and incubated for 2 h in CO2 incubator. The solution was then transferred to a new empty plate for optical density (OD) reading at both 600 and 690 nm (background) using a Biotek ELx800 plate reader and Gen5 ELISA software.
Cell proliferation assay
The cell proliferation rate was determined by following the instructions of the Click-iT EdU Alexa Fluor 488 imaging kit (Cat. No. C10337; Fisher Scientific). Cells were incubated with thymidine analogue, 5-Ethynyl-2′-deoxyuridine (Edu), for 8 h before fixation and subsequent staining. Images were taken using Olympus IX50 fluorescence microscope and both total nuclei and EdU-labeled nuclei were counted using Cell Profiler software.
siRNA reverse transfection
The received 1 nmol lyophilized siRNA was first constituted in 100 μL of RNA-free water to obtain a stock concentration of 10 μM, which was further diluted to 2 μM working stock (1 μL of 2 μM siRNA working stock equals to about 28 ng of siRNA). To prepare transfection complex for each well of 96-well plate, dilute 0.28 μL of XtremeGENE siRNA transfection reagent (Cat. No. 04476093001; Roche) in 28 μL DMEM basal media in a tube, and then add 1 μL of 2 μM siRNA (28 ng) within 5 min. Incubate at room temperature for 25–30 min before transferring to the well, during which cells were trypsinized and counted. A total of 8,000 cells in 80 μL of growth media were then added to each well containing the roughly 28 μL of Xtreme/siRNA mix. Cells and transfection reagents used were increased proportionally when transfection was carried out in larger wells based on the relative growth areas. After 20 h, cells were changed into AIM.
SiControl (SiCon): AllStars Negative siRNA (Cat. No. 1027284; Qiagen); siCdk1, functionally validated siRNA directed against human Cdk1 (Cat. No. SI00299712; Qiagen); siCdk2, functionally validated siRNA directed against human Cdk2 (Cat. No. SI00299775; Qiagen); siCdk4, functionally validated siRNA directed against human Cdk4 (Cat. No. SI00299789; Qiagen); siCCND1, functionally validated siRNA directed against human CCND1 (Cat. No. SI02654540; Qiagen); and siC/EBPα, functionally validated siRNA directed against human C/EBPα (Cat. No. GS1050; Qiagen).
OilRedO and DAPI staining
Oil droplets in differentiated cells were stained by OilRedO solution (Cat. No. NC9773107; Fisher Scientific). Briefly, cells were fixed in 10% formalin for 20 min, rinsed with distilled water thrice, washed in 100% isopropylene glycol for 5 min, incubated in OilRedO solution for 30 min, washed with 85% isopropylene glycol for 5 min, and rinsed with distilled water thrice. Cells were then counterstained with 1 μg/mL DAPI solution in phosphate-buffered saline for 5 min before additional rinsing with water, followed by imaging using Olympus IX50 microscope (OilRedO: green light–red fluorescence; DAPI: ultraviolet light–blue fluorescence). Whole well images were taken with Leica EZD40 Stereoscope after staining.
For consistency across all treatment groups within each experimental set, the same exposure time was used for taking OilRedO images or DAPI images at high magnification (200 × ). Imaging was carried out from the top of the well to the bottom of the well, with the top edge of next imaging field juxtaposed with the bottom edge of the previous image field, resulting in 14 images/well for 24-well plates. For quantification, cells were air-dried overnight in fume hood, extracted with pure isopropyl alcohol (Cat. No. A426P; Fisher Scientific; 100 μL/well for 24-well plate), transferred to 96-well plate, and OD was measured at 510 and 690 nm using Biotek Elx800 plate reader.
bFGF treatment
Frozen bFGF stock solution (Cat. No. 100-18B; PeproTech) at 2 μg/mL was freshly diluted into 2 ng/mL working solution in adipogenic differentiation media upon each use. After desired duration of treatment, cells were rinsed twice with basal media before changing into adipogenic differentiation media. To avoid well positional effect, 3 different bFGF treatment groups plus a control treatment group are tested in each 24-well plate, with 6 wells/treatment group and all groups symmetrically positioned in the plate. It took three plates to complete the whole set of bFGF treatments at nine different time intervals, D0–1, D1–2, D0–2, D2–3, D3–4, D2–4, D4–6, D6–8, and D8–10.
bFGF was applied at the beginning of indicated treatment window and removed at the end of the treatment window by washing it off twice with basal media before switching to non-bFGF containing adipogenic differentiation media. For example, for D0–2 treatment, bFGF was added along with adipogenic differentiation media on the first day of differentiation (D0) and 48 h later (D2), media were removed and treatment wells were rinsed with basal media twice before regular differentiation media were added for remaining days of culture.
Gene expression by reverse transcription–polymerase chain reaction analysis
Total RNA was isolated from cells with the RNeasy kit (74104; Qiagen). SUPERSCRIPT II reverse transcriptase (Cat. No. 11752050; Fisher Scientific) was used for reverse transcription (RT; the same amount of RNA was used in each RT reaction). Polymerase chain reaction (PCR) was carried out using the HotStarTaq plus master mix kit (Cat. No. 203645; Qiagen) or the Fast cycling PCR kit (Cat. No. 203741; Qiagen).
Primer sequences used are as follows: CDK1-forward: GGATCTACCATACCCATTGACT, CDK1-reverse: CCATGTACTGACCAGGAGG G; CDC6-forward: GTTTGCTGCTGCCGCTGTGC, CDC6-reverse: GCCCAGACGTTTCCTGGGGC; CDC25B-forward: ATCGCGCCCTGTAGCCTGGA, CDC25B-reverse: GCTCTCCCCCAGCCTCAGCT; CCNA2-forward: GACGAGACGGGTTGCACCCC, CCNA2-reverse: GGCCAGCTTTGTCCCGTGACT; CCNE1-forward: GGAGCGGGATGCGAAGGAGC, CCNE1-reverse: TACAGGCAGCGGGGAGCCTC; CCND1-forward: AGCCTCCAGAGGGCTGTCGG, CCND1-reverse: CTTCTCGGCCGTCAGGGGGA; CDK2-forward: TGACTCGCCGGGCCCTATTCC, CDK2reverse: CCCAAGGCCAAGCCTGGTCA; PPARγ-forward: AAG CCC TTC ACT ACT GTT GA, PPARγ-reverse: ACC TGA TGG CAT TAT GAG AC; C/EBPα-forward: CCT AAG GTT GTT CCC CTA GT, C/EBPα-reverse: GAG AGT CTC ATT TTG GCA AG; PLK2-forward: ACTCGGGGCCGGAGATCTCG, PLK2-reverse: TGCTTGGCAGGAGCCAGCAA; and FBXO5-forward: AGGCAGACTCCACGTCGGCT, FBXO5-reverse: AGGGCTCACAATCGGTGACCCAA.
RNA quantification was carried out using a Nanodrop spectrophotometer, PCR was conducted using MJ mini 48w thermo cycler and Invitrogen StepOne real time PCR machine, and gel images were obtained using Bio Rad Gel Doc imager and Image Lab software 5.2.1.
Image area measurement using Image Pro software
Area measurement of OilRedO stain was carried out by analyzing images using Image Pro Premier v9.2 Software (Media Cybernetics, Rockville, MD).
Statistical analysis
Unpaired Student's t-test was used to evaluate the statistical differences between control and treatment groups.
Results
Temporal expression patterns of cell cycle genes during adipogenic differentiation of hMSCs
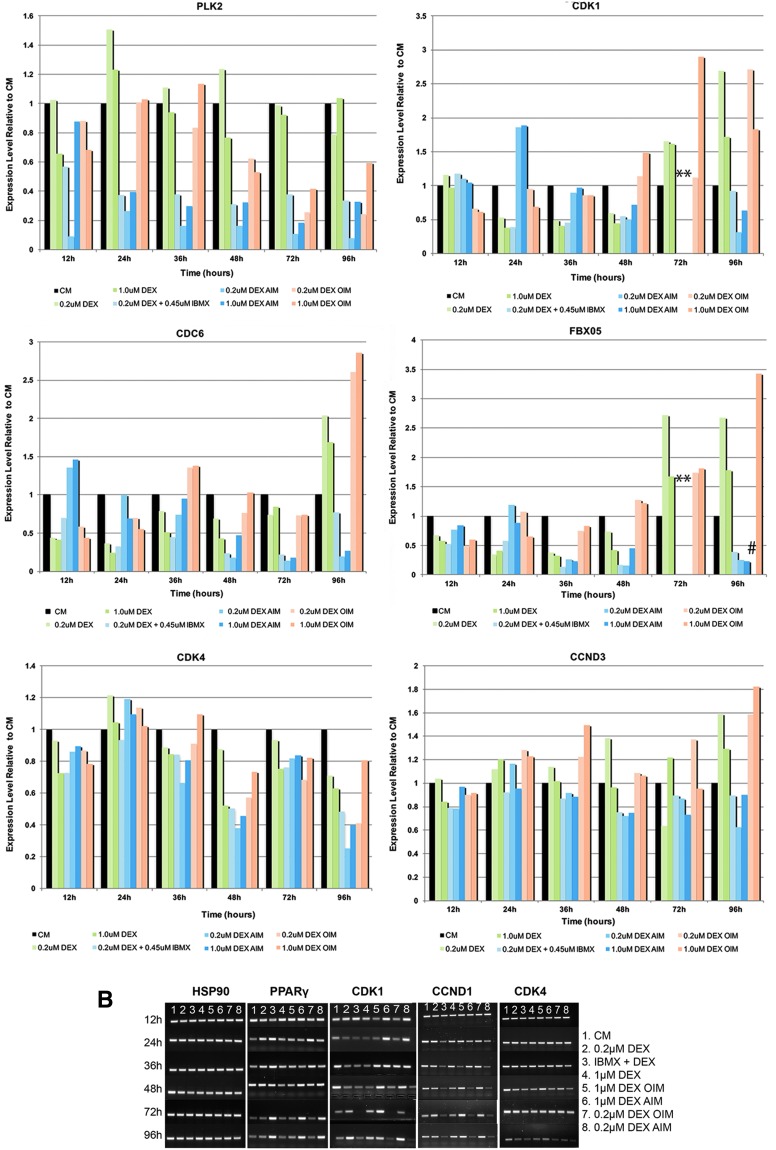
Through a microarray study comparing gene expression profiles between hMSCs treated with AIM induction cocktail in growth media and hMSC control cells treated with CM only at 36 and 72 h postinduction, a number of cell cycle regulators, including CDC6, CCND1, Cdk1, Cdk4, CCNA2, CCND3, PLK2, and FBX05, were found to be significantly downregulated in AIM-treated cells at both time points. To further examine the expression patterns of identified cell cycle genes and two additional selected cell cycle genes, Cdk1 and CDK4, RT-PCR was carried out at 12, 24, 36, 48, 72, and 96 h after initial treatment in eight different treatment conditions, CM, DEX alone at 0.2 or 1.0 μM, IBMX (0.45 mM)+DEX (0.2 μM), AIM with 0.2 μM DEX or with 1.0 μM DEX, and OIM with 0.2 or 1.0 μM DEX, with media change at every 48-h intervals. As control, expression of PPARγ and C/EBPα was also analyzed.
It has been suggested that human adipogenesis and osteogenesis are two inverse processes, with one process inhibiting the other [18–20]. Our previous study also identified a host of osteogenic suppressor genes that play opposite roles during these two processes [21]. To see whether such opposing expression pattern exists in the identified cell cycle genes, gene expression was also examined in cells treated with OIM composed of 0.05 μM ascorbic acid 2-phosphate, 10 mM beta-glycerophosphate, and 1.0 μM DEX. Since DEX is often used at a lower concentration in AIM (0.2 μM) and higher in OIM (1.0 μM) in the literature, both concentrations were examined to dissect its potential effect on the expression of interested genes.
Last, IBMX+DEX could induce mature adipocyte formation similar to AIM in murine 3T3-L1 cells [16] and hMSCs ([21] and unpublished data), and was therefore included in this study to eliminate the effect of insulin on cell proliferation during adipogenic differentiation.
As expected, PPARγ was upregulated by DEX, DEX+IBM and AIM starting at 24 h after initial media treatment, followed by a greater induction in IBMX+DEX or AIM only at 72 h after treatment initiation (Fig. 1). The expression of C/EBPα followed a similar pattern in DEX, IBMX+DEX, and AIM (Fig. 1).
FIG. 1.
Gene expression patterns of adipogenic markers and cell cycle regulators during adipogenic differentiation. (A) Expression of PPARγ, CDC6, CCND1, Cdk1, Cdk4, CCNA2, CCND3, PLK2, and FBX05 were examined at 12, 24, 36, 48, 72, and 96 h after initial treatment in eight different treatment conditions, CM, DEX alone at 0.2 or 1.0 μM, IBMX (0.45 mM)+DEX (0.2 μM), AIM with 0.2 μM DEX or with 1.0 μM DEX, and OIM with 0.2 or 1.0 μM DEX. Expression of C/EBPα was examined in CM, DEX, IBMX+DEX, and AIM at 12, 24, 36, and 72 h after treatment initiation, as well as at 7 and 18 days postinitiation. PPARγ and C/EBPα were both upregulated by DEX, DEX+IBMX, and AIM starting at 24 h after initial media treatment, followed by a greater induction in IBMX+DEX or AIM at 72 h after treatment initiation. Expression of all cell cycle regulators was downregulated in IBMX+DEX and AIM relative to its expression in CM at 48, 72, and 96 h after treatment initiation. Expression of cell cycle genes was normalized to those of HSP90, a housekeeping gene used as internal loading control, and set relative to its level in CM (100%) at each time point. *Expression level was deemed undetectable in IBMX+DEX and AIM samples; #sample was missing. (B) Agrose gel images of RT-PCR products for HSP90, PPARγ, CCND1, Cdk1, and Cdk4, whose expression was examined at the selected time points after treatment initiation in eight treatment media conditions as described in (A) (n = 2). AIM, adipogenic inducing media; C/EBPα, CCAAT/enhancer binding protein alpha; CM, control growth media; DEX, dexamethasone; IBMX, 3-Isobutyl-1-methylxanthine; PPARγ, peroxisome proliferator-activated receptor gamma; RT-PCR, reverse transcription–polymerase chain reaction. Color images available online at www.liebertpub.com/scd
On the other hand, expression of all examined cell cycle regulators was downregulated in IBMX+DEX- and AIM-treated cells relative to its expression in CM-treated cells at 48 h and later time points after treatment initiation, with some detected at as early as 24 h (PLK2, CCND1, and CCNA2) (Fig. 1). Expression of Cdk1, CDC6, and FBX05 was downregulated by DEX or DEX+IBMX starting at 24 h, but not by AIM until at 36 h (FBX05) or 48 h (Cdk1 and CDC6) time points (Fig. 1). In addition, in many cases such as with CCND1, CCNA2, Cdk1, CDC6, and FBX05, their expression was inversed in response to IBMX+DEX or AIM (downregulated) versus OIM (upregulated) treatment starting at the 48-h time point (Fig. 1). Last, neither the two different concentrations of DEX used nor the absence of insulin seemed to have any determinant effect on the expression of examined genes.
The results above indicate that examined cell cycle genes were specifically downregulated during adipogenic differentiation, but not during osteogenic differentiation, and such downregulation was induced by IBMX+DEX, with insulin as a dispensable factor.
Cell proliferation is restricted to the first 48 h of adipogenic induction
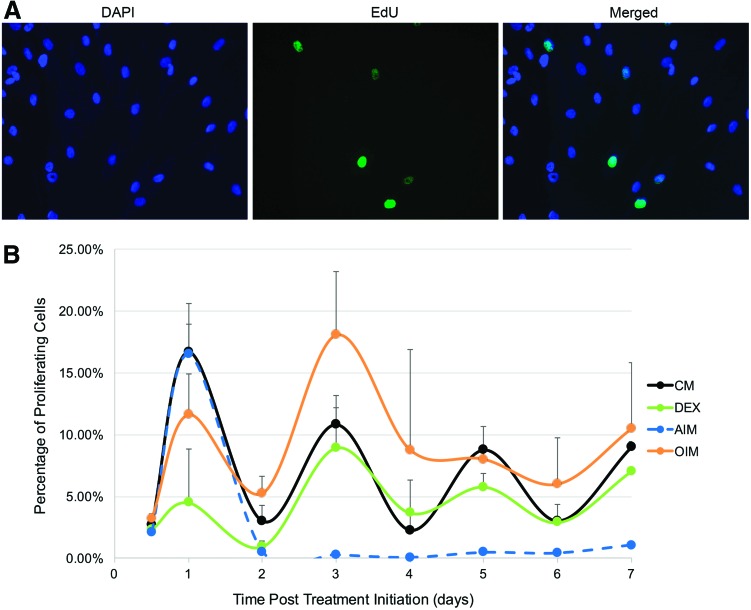
The overall downregulated expression of examined cell cycle genes during adipogenic treatment suggests that the cell proliferation activity may be inhibited. To examine cell proliferation activity, hMSCs were plated at equal density (50,000 cells/well in 24-well plates, around 70%–75% confluent) and treated the next day (day 0) with CM, DEX, AIM, or OIM, followed by media change at precisely 48-h intervals. The percentage of proliferating cells was determined at 0.5, 1, 2, 3, 4, 5, 6, and 7 days after initial exposure by adding EdU, a nucleoside analog of thymidine that is incorporated into DNA during active DNA synthesis, at 8 h before examined time points, followed by fixation, detecting EdU-labeled cells (green), and Hoechst stain that labels all cells (blue) (Fig. 2A). Cells in each treatment group (two wells) were imaged and the number of EdU-labeled green nuclei and total nuclei in each well were quantified, with the percentage of green over blue cells calculated.
FIG. 2.
Cell proliferation activity was limited to the first 48 h of adipogenic induction. (A) Cells were plated at equal density across different treatment groups (triplicate wells per treatment group) before media treatment. Proliferating cells were labeled by EdU, a thymidine analog, which was added 8 h before the designated fixation hours, and detected by Alexa Fluor 488 tagged Azide-containing detection reagent (green). All cells were stained with DAPI (blue). (B) Percentage of proliferating cells was determined at 12, 24, 48, 72, 96, 120, 144, and 168 h after treatment initiation in CM, DEX, AIM, or OIM (n = 3). EdU, 5-ethynyl-2′-deoxyuridine; OIM, osteogenic inducing media. Color images available online at www.liebertpub.com/scd
Interestingly, while the number of dividing cells oscillated with each media change at 48-h intervals in CM, DEX, and OIM treatments, it was only induced during the first 48 h of AIM treatment and subsequently diminished despite additional AIM changes (Fig. 2B). The results indicate that cell proliferation is restricted to the first 48 h of adipogenic differentiation.
Total cell numbers are reduced in response to adipogenic induction
Changes in cell proliferation rate would predict changes in total cell numbers. To examine total cell numbers, a resazurin-based cell number and viability assay were used to measure the relative changes in total live cells in DEX-, DEX+IBMX-, AIM-, or OIM-treated cells compared to control, CM-treated cells, at 24 h, 72 h, 7 days, and 10 days after treatment initiation (media change at 48-h interval) [22].
hMSCs were plated at equal density in 24-well plates followed by treatment with different media for 10 days, with media change at 48-h intervals. At designated time points, resazurin OD readings at 600 nm in each treatment group were determined and then subtracted from the average of control resazurin OD readings (resazurin solution incubated with no cells). The calculated change in absorbance readings positively correlates with total live cell numbers in its corresponding sample. Its values in DEX, DEX+IBMX, AIM, or OIM were subsequently normalized against those in CM to compare their relative cell numbers and graphed.
At 24 h after treatment initiation, no significant differences were observed across all treatment groups (Fig. 3). At 72 h and later time points, however, adipogenic induction by DEX+IBMX or AIM resulted in significantly decreased total live cells compared to CM, whereas OIM and DEX treatments resulted in significantly increased total live cells compared to CM starting at 7 and 10 days, respectively (Fig. 3).
FIG. 3.
Total cell numbers were significantly reduced in adipogenic induction medium-treated wells compared to CM-, DEX-, or OIM-treated wells. Cells were plated at equal density across different treatment groups (triplicate wells per treatment group) before media treatment and incubated with resazurin solution for 2 h at designated time point before OD readings of incubated resazurin solution at 600 nm were obtained. The graphed percentage value for each treatment group was calculated by dividing the difference between its OD reading and control resazurin solution (incubated with no cells) against the difference obtained for the CM group. P value was calculated based on comparing raw OD readings between control CM and other treatment groups. *P < 0.01; **P < 0.05 (n = 3). OD, optical density. Color images available online at www.liebertpub.com/scd
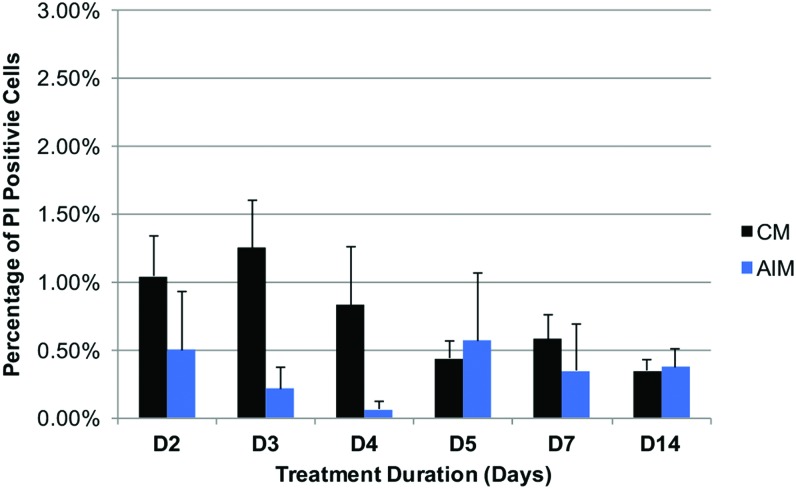
To find out whether cell death might be a contributing factor to the reduced total cell numbers in adipogenic medium-treated groups, cell death rate was compared between CM and AIM treatments. hMSCs were plated at equal density and continuously cultured for 14 days with media change at 48-h intervals. At 2, 3, 4, 5, 7, and 14 days after initial CM or AIM treatment, live cells were stained with PI, followed by fixation and staining with DAPI solution. PI (red) labels both apoptotic and necrotic cells, whereas DAPI labels all cells. The percentage of dead cells was calculated after quantifying the number of red and blue cells in each group. Cell death appeared reduced in AIM-treated group compared to CM on D3 and D4, but overall, no significant differences were observed between the two groups (Fig. 4).
FIG. 4.
Cell death rate between CM- and AIM-treated cells was overall similar. Cells were plated at equal density across different treatment groups (triplicate wells per treatment group) before media treatment initiation. Cell death rate was determined by counting PI-labeled cells and total cells for each treatment group at designated time points after initial treatment (n = 3). PI, propidium iodide. Color images available online at www.liebertpub.com/scd
The results above indicate that, consistent with decreased cellular proliferation activities, adipogenic differentiation results in reduced total cell numbers compared to CM, DEX, or OIM treatment after 48 h of treatment, and such reduction is not linked to cell death.
Expression knockdown of cell cycle genes by siRNA promotes adipogenic differentiation
To examine how further knockdown of identified cell cycle regulators would affect adipogenic differentiation, siRNAs that have been commercially validated to specifically target Cdk1 or Cdk2 were transfected into hMSCs. It has been shown that Cdk1 was the only essential cell cycle Cdk that was sufficient to drive the mammalian cell cycle, whereas all other Cdks were dispensable [23]. Cdk2 was chosen due to its structural and functional similarity to Cdk1 as a cell cycle regulator, even though it was not identified in our microarray study. siCon made of scrambled siRNAs was used as the negative control.
A previous protocol developed in the laboratory allowed over 90% transfection efficiency with bone marrow-derived hMSCs based on live cell counts [21]. A similar protocol was tested in adipose-derived hMSCs using both cell death inducing siDeath and nonsilencing siRNAs with Alex Fluor 488 modification that labels transfected cells, and the results indicated around 85%–90% transfection efficiency (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd).
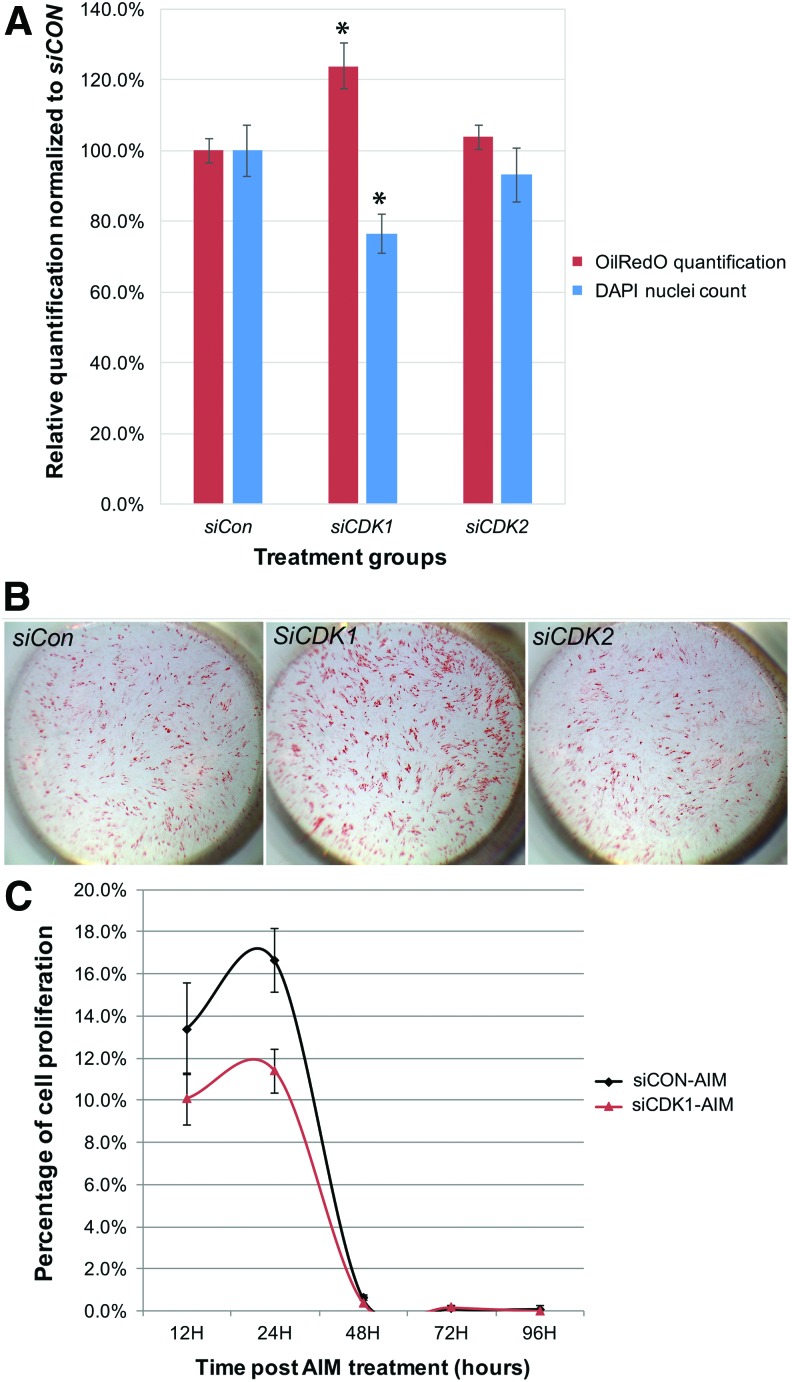
Cells were reverse transfected with siCon, siCdk1, and siCdk2 in hMSC growth media at 20 nM, with equal cell density plating across all treatment wells, followed by AIM treatment at 20 h posttransfection. After 12 days of AIM treatment with media change at 48-h intervals, cells were fixed, stained with OilRedO (stains fat droplets) and DAPI solution, and subsequently imaged before OilRedO stain was extracted for OD reading.
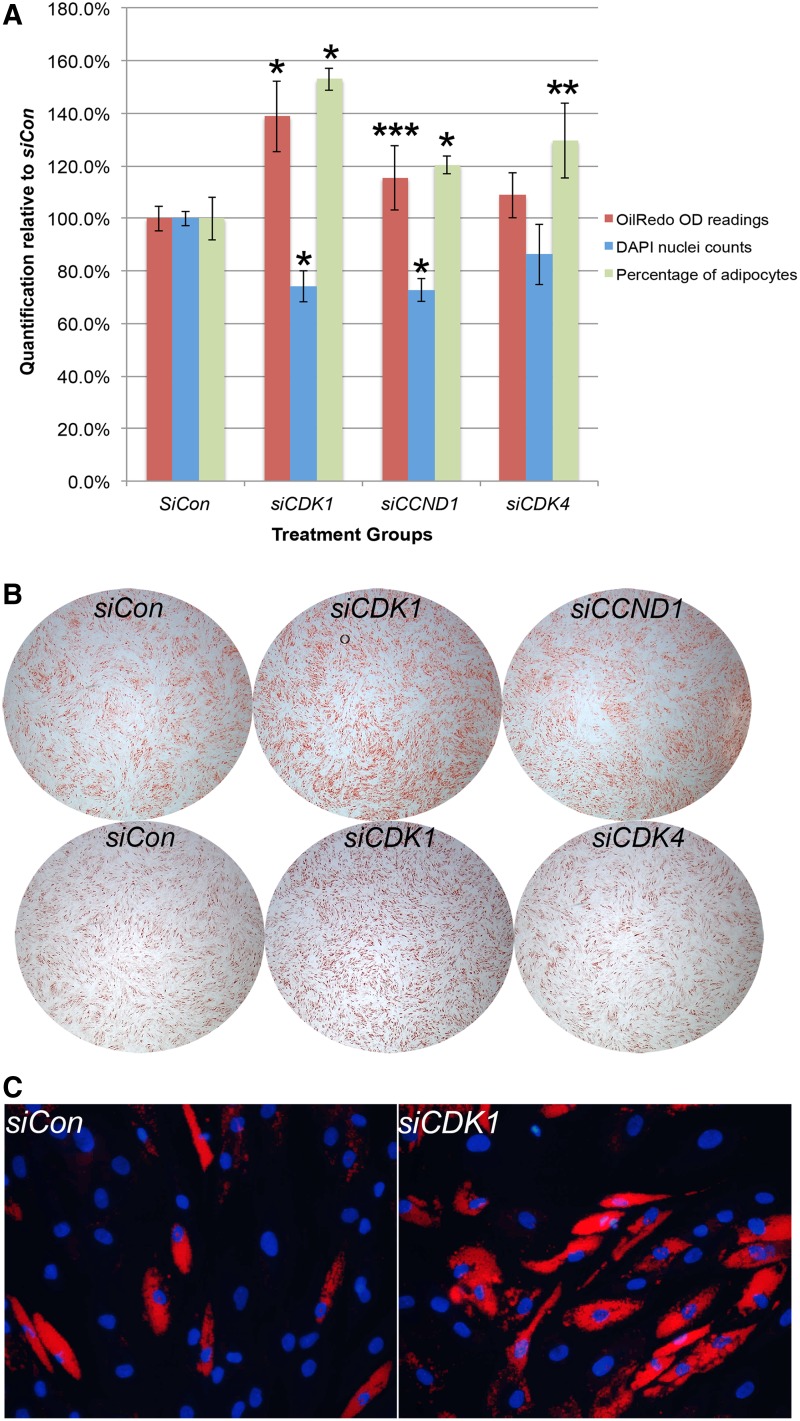
Consistently, siCdk1-transfected wells resulted in stronger OilRedO staining intensity and significantly greater OilRedO OD readings, but significantly reduced total cell counts compared to the siCon group (Fig. 5A, B). The overall effect of siCdk2, however, is insignificant, even though its nuclei count always trended lower than siCon (Fig. 5A, B).
FIG. 5.
SiCdk1 significantly enhanced adipogenic differentiation of hMSCs, while inhibiting its proliferation. (A) Equal number of cells was transfected with siCon, siCDK1, or siCDK2 at 20 h before AIM treatment initiation, followed by AIM change at 48-h intervals. OilRedO quantification and DAPI cell counts were determined after 12 days of AIM treatment and values for each treatment group were graphed after they were normalized against those of the siCon group. (B) After 12 days of AIM treatment, whole well images of siCon, siCdk1, or siCdk2 stained with OilRedO solution were taken. (C) Cell proliferation rate in siCon and siCdk1 transfected cells was determined by EdU labeling at the designated time points after AIM treatment initiation. *P < 0.01 (n = 3). hMSCs, human mesenchymal stem cells; siCon, SiControl. Color images available online at www.liebertpub.com/scd
To confirm that total cell count reduction in siCdk1 cells was a result of decreased cellular proliferation, EdU-based cell proliferation assay was performed at 32, 44, 68, 92, and 116 h post-siRNA transfection (equivalent of 12, 24, 48, 72, and 96 h after AIM treatment initiation, with media change at 48-h intervals). At 24 h AIM post treatment initiation, the proliferation rates of siCdk1 transfected cells were considerably lower than those of siCon-transfected cells (P = 0.056) (Fig. 5C). To also confirm that the enhanced adipogenic differentiation by siCdk1 was not a random event, cells were transfected by two different siRNAs against C/EBPα or siCon, followed by AIM treatment at 24, 48, or 72 h posttransfection for 12 days. As anticipated, siC/EBPα resulted in a significantly decreased differentiation in comparison to siCon, in contrary to siCdk1 treatment (Supplementary Fig. S2).
To see whether a similar effect would be observed for siRNAs targeting the other identified cell cycle genes, the effect of siCCND1 and siCdk4 was also examined, along with siCdk1 and siCon as controls. CCND1 and Cdk4 were chosen due to their well-established role in regulating G1/S phase transition [24].
SiCCND1 treatment resulted in significantly reduced total cell numbers than those of siCon, with OilRedO OD readings trending higher as well (P = 0.1), whereas the effect of siCdk4 appeared insignificant, with total nuclei count trending lower than siCon in general (Fig. 6A, B). To determine adipogenic differentiation efficiency, 14 sequential images of OilRedO stain and 14 overlaying images of DAPI stain were taken from each of triplicate wells of individual treatment group. Adipocytes were manually counted in merged OilRedO and DAPI images and percentage of adipocytes over total cell counts in each well was determined (Fig. 6C). The percentages of adipocytes were significantly higher in both siCdk1- and siCCND1-treated wells than in siCon wells, but only trended higher in siCdk4-treated wells (Fig. 6A).
FIG. 6.
The effect of siCCND1 and siCdk4 on adipogenic differentiation of hMSCs was weaker than those of siCdk1. (A) Equal number of cells was transfected with siCon, siCdk, or siCCND1 separately, or siCon, siCdk1, or siCdk4 separately, at 20 h before AIM treatment initiation, followed by AIM change at 48-h intervals. OilRedO quantification, adipocyte counts, and DAPI cell counts were determined after 12 days of AIM treatment, and values for each treatment group were graphed after they were normalized against those of the siCon group. (B) Representative whole well images of different treatment groups stained with OilRedO solution were taken after 12 days of AIM treatment. (C) Images merging both OilRedO and DAPI stain allowed better visual distinction of individual adipocytes. Images were taken from the same positions within siCon- and siCdk-transfected wells, depicting a significantly greater percentage of adipocytes forming in siCdk wells (24/40) compared to those in siCon wells (14/49). The average differentiation efficiency of the former is about 1.5-fold over the latter. *P < 0.01; **P ≤ 0.05; ***P ≤ 0.1 (n = 3). Color images available online at www.liebertpub.com/scd
The results above demonstrated that both siCdk1 and siCCND1 significantly promoted adipogenic differentiation efficiency in hMSCs, while inhibiting its proliferation, with siCdk1 conferring a greater effect.
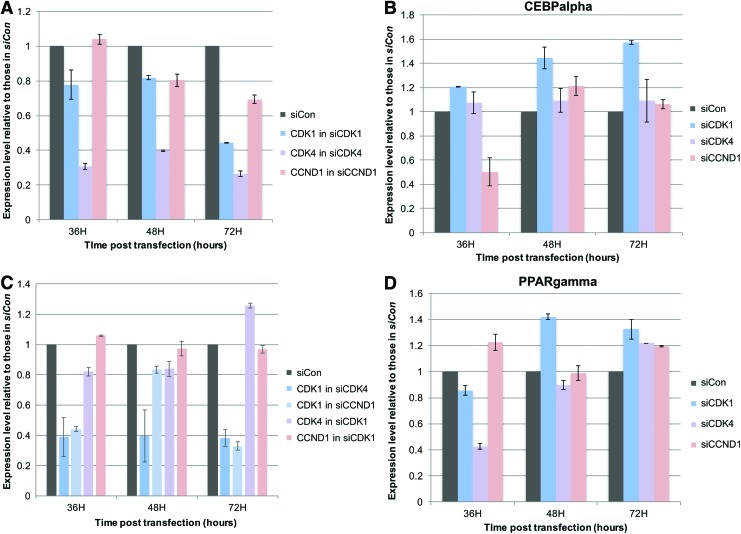
Gene expression knockdown by siRNAs was confirmed by semiquantitative RT-PCR in RNA samples isolated at 36, 48, and 72 h posttransfection. In siCdk1, siCdk4, and siCCND1 cells, the expression of Cdk1, Cdk4, and CCND1 were downregulated to about 40%, 30%, and 70% of their control levels in siCon cells, respectively (Fig. 7A and Supplementary Fig. S3). Interestingly, expression of Cdk1 and Cdk4 appeared to be reciprocally regulated, with Cdk1 downregulated to about 40% of its control level in siCdk4 cells at all three time points examined and Cdk4 downregulated to about 80% of its control level in siCdk1 cells at 36 and 48 h posttransfection (Fig. 7B). In addition, expression of Cdk1 was also downregulated by siCCND1, although CCND1 expression appeared unchanged in siCdk1 cells (Fig. 7B).
FIG. 7.
Gene expression of Cdk1, Cdk4, CCND1, PPARγ, and C/EBPα in siRNA-transfected cells. RNA was isolated at 36, 48, or 72 h post-siRNA transfection. Expression level for each gene was normalized against those of HSP90. Percentage value graphed was derived by comparing the average value of triplicates for each gene in each treatment group to its value in siCon cells at the same time point. (A) Expression of Cdk1, Cdk4, and CCND1 was downregulated in siCdk1-, siCdk4-, siCCND1-transfected cells, respectively. (B) Expression of Cdk1 was downregulated in both siCdk4- and siCCND1-transfected cells. Expression of Cdk4 was transiently downregulated in siCdk1 cells before it was upregulated, while expression of CCND1 was unchanged in siCdk1 cells. (C) Expression of C/EBPα was upregulated in siCdk1 cells, but transiently downregulated in siCCND1-transfected cells. (D) Expression of PPARγ was upregulated in siCdk1 cells, but transiently downregulated in siCdk4-transfected cells (n = 3). Color images available online at www.liebertpub.com/scd
To determine how expression knockdown of Cdk1, Cdk4, and CCND1 could affect the expression of early adipogenic markers, RT-PCR was carried out for PPARγ and C/EBPα at 36, 48, and 72 h posttransfection. SiCdk1 slightly, but significantly enhanced the expression of both C/EBPα and PPARγ at 48- and 72-h time points. On the contrary, siCdk4 did not have a significant effect on C/EBPα at any time point, but significantly downregulated the expression of PPARγ at 36 h, which was then upregulated at 72 h, whereas siCCND1 slightly upregulated the expression of PPARγ, but significantly downregulated the expression of C/EBPα at 36 h, which was then elevated to control or higher levels at 48 and 72 h (Fig. 7C, D).
The transient downregulation of PPARγ and C/EBPα by siCdk4 and siCCND1, respectively, may partially account for the overall less prominent enhancement effect by siCdk4 and siCCND1 on adipogenic differentiation compared to siCdk1, as shown in Fig. 6 (see Discussion section).
Exogenous bFGF inhibits adipogenic differentiation after mitotic phase
Since inhibiting cell proliferation during the first 48 h of adipogenic induction enhanced adipogenic differentiation, we examined how promoting cellular proliferation would affect adipogenic differentiation by applying exogenous bFGF at 2 ng/mL during different time intervals that span across the 12-day adipogenic induction, 0–1D, 1–2D, 0–2D, 2–3D, 3–4D, 2–4D, 4–6D, 6–8D, and 8–10D.
For each treatment group, except for during the specified time when bFGF was applied along with adipogenic induction media (AIM or IBMX+DEX), cells were treated with adipogenic induction media alone on the remaining days. At the end of treatment, cells were double stained with OilRedO and DAPI solution and imaged, followed by OilRedO extraction for quantification. Total cell numbers and adipocytes were counted based on images taken from 6 wells of each treatment group, with 14 overlapping images of DAPI and OilRedO stain taken from each well. Results from using AIM (two independent repeats) or IBMX+DEX (three independent repeats) as adipogenic induction media were identical and results are therefore presented as the mean value of both.
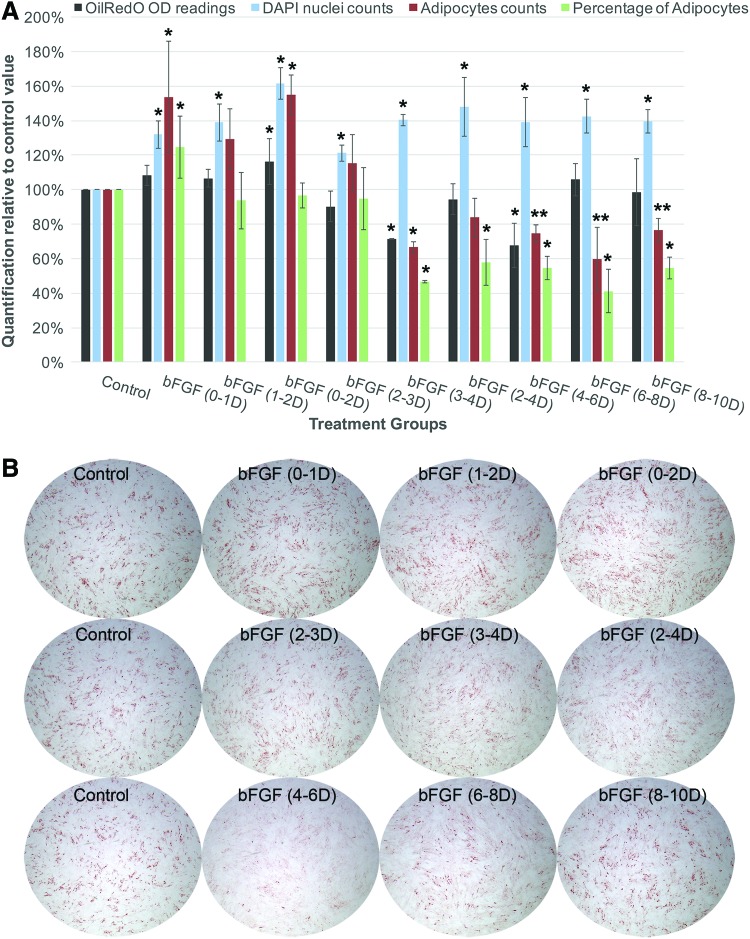
Regardless of when bFGF was applied, it significantly enhanced total cell numbers compared to control treated with adipogenic media alone (Fig. 8, blue columns). However, bFGF only increased total number of adipocytes when it was applied during the first 48 h of adipogenic induction and significantly reduced it when applied after day 3 (Fig. 8, red columns). Adipogenic differentiation efficiency based on the calculation of percentage of adipocytes was only significantly enhanced when bFGF was applied during the first 24 h of adipogenic induction (0–1D), and remained unchanged or significantly reduced when bFGF was applied during day 1–3 or after day 3, respectively (Fig. 8, green columns).
FIG. 8.
Exogenous bFGF exerted differential effect on the adipogenic differentiation of hMSCs depending on the timing of its application. Cells were plated at equal density across all treatment groups (six replicate wells per treatment group) as well as before treatment initiation in each experimental set. For each treatment group, bFGF was applied in AIM at the indicated time window (rinsed off at the end of treatment), and cells were in AIM alone on the remaining days. Control group was treated with AIM only. OilRedO quantification, adipocyte counts, and DAPI nuclei counts were determined at the end of the 12-day AIM treatment. (A) bFGF enhanced cellular proliferation across all stages of adipogenic differentiation of hMSCs, but only promoted adipogenesis during days 0–2 by increasing the total number of cells committing to adipocytes. Between days 3 and 6, bFGF inhibited adipogenesis by reducing the total number of adipocytes, while after day 6, bFGF had insignificant effect in terms of total accumulation of fat oil droplets based on OilRedO quantification, but the number of recognizable adipocytes was significantly reduced. Data were derived from the average of four independent replicates of the whole experimental set, with 6 wells per group and a total of 256 OilRedO and 256 DAPI images processed for each treatment group. (B) Representative whole well images of each treatment group taken at the end of 12-day adipogenic induction. *P < 0.01, **P < 0.05 (n = 4). bFGF, basic fibroblast growth factor. Color images available online at www.liebertpub.com/scd
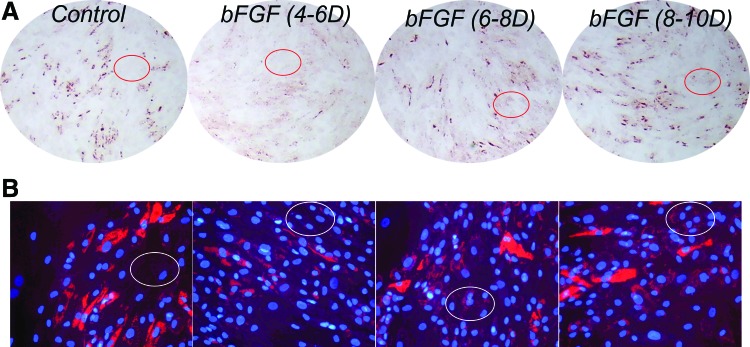
Quantification of OilRedO stain by isopropanol extraction followed by OD reading at 515 nm was overall highly consistent with the manual counting of adipocytes in all treatment groups, except for groups bFGF (6–8D) and bFGF (8–10D), which, despite having significantly lower counts of adipocytes compared to the control group, had similar level of OilRedO quantification (Fig. 8, black columns). One likely explanation was that in bFGF (6–8D) and bFGF (8–10D) treatment wells, there was noticeably stronger “background” stain compared to the control or bFGF (4–6D) wells (Fig. 9A, circled areas), which could be cells that contain small and faint oil droplets, but were not recognized as well-defined adipocytes during manual counting (Fig. 9B).
FIG. 9.
Application of bFGF after day 6 of adipogenic initiation resulted in greater number of cells containing small and faint oil droplets. (A) Images demonstrating the different background intensity (representative area is circled in each image) across different treatment groups at 8× magnification. Images were taken under the same exposure time and adjusted with identical parameters in Adobe Photoshop. (B) Images demonstrating differences in the abundance of cells containing small and faint oil droplets that cannot be easily recognized as individual adipocytes (representative area is circled in each image) across different treatment groups at 200× magnification. Images were taken under the same exposure time and adjusted with identical parameters in Adobe Photoshop (n = 4). Color images available online at www.liebertpub.com/scd
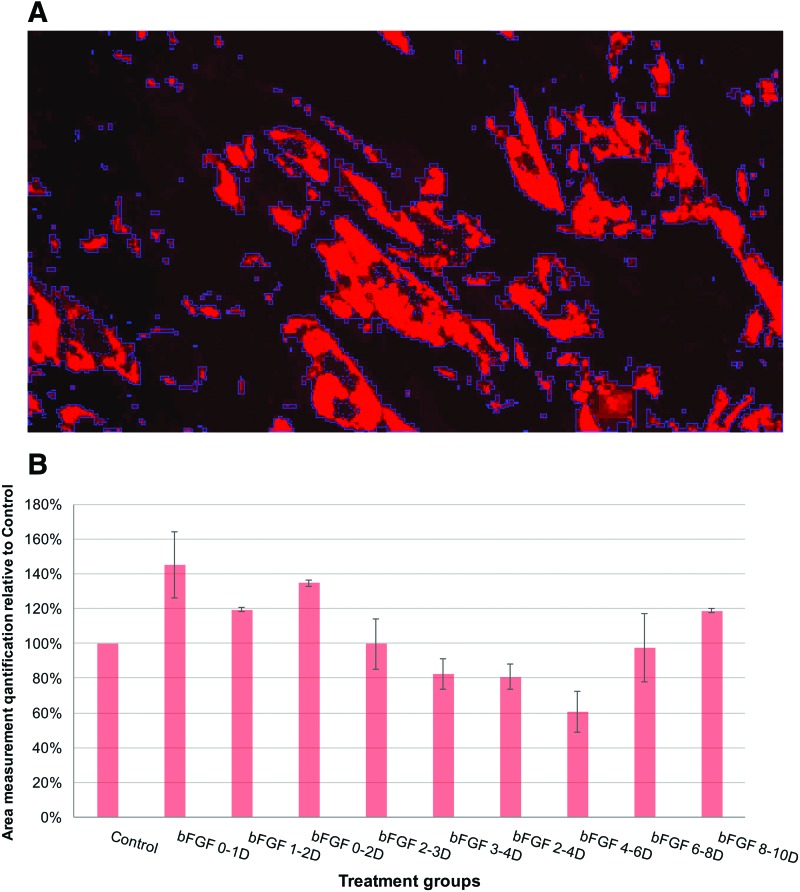
To confirm that, ImagePro, an image analysis software, was used to analyze the same sets of images used in manual counting by measuring the area of all fluorescent stain in each image (Fig. 10A). The total area measurement for each treatment group was derived from the average of three independent sets of experiments and graphed based on its relative quantification against the control (Fig. 10B). The distribution pattern of the area measurements across different treatment groups was similar to the pattern of the OilRedO OD readings in Fig. 8, indicating that the speckled faint oil droplets observed in groups bFGF (6–8D) and bFGF (8–10D) were indeed the underlying cause for the discrepancy between the individual adipocyte count quantification and OilRedO quantification observed in these two treatment groups.
FIG. 10.
Area measurements of stained oil droplets across different bFGF treatment groups demonstrated similar pattern as OilRedO quantification. (A) An exemplary image demonstrating how the ImagePro software identifies and outlines stained oil droplets and outlined areas was measured quantitatively. (B) The average value of area measurements from three independent experimental sets was calculated for each treatment group and graphed by comparing to the measurements of control group (n = 3). Color images available online at www.liebertpub.com/scd
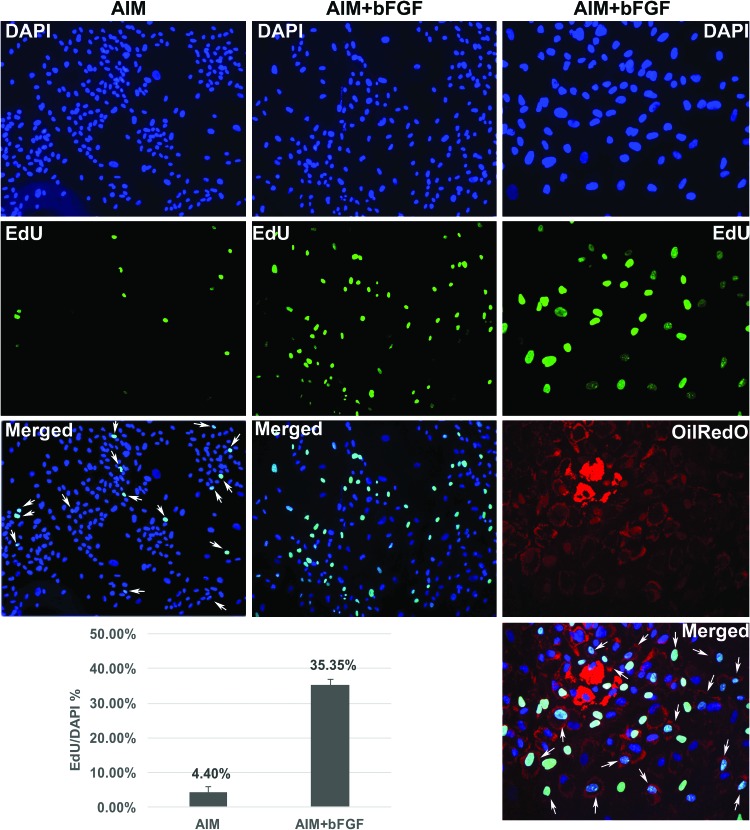
One potential explanation for the abundant speckled faint oil droplets in groups bFGF (6–8D) and bFGF (8–10D) was that premature adipocytes might have been dividing in response to bFGF stimulation. To test that, cells treated with AIM for 6 or 8 days were changed to fresh AIM with or without bFGF and EdU was added 24 h later (at days 7 and 9, respectively) for 12 h before cells were fixed and cell proliferation was analyzed. Compared to cells treated with AIM alone, cells treated with AIM plus bFGF had s significantly greater cell proliferation rate (35.4% vs. 4.4%) (Fig. 11, panels in left hand and middle columns). Interestingly, many cells committed to adipogenic lineage, as indicated by OilRedO-stained oil droplets, were EdU positive, indicating that they had reentered cell cycle and would likely split accumulated oil droplets into daughter cells (Fig. 11, panels in right hand column).
FIG. 11.
Premature adipocytes reenter cell cycle in response to bFGF stimulation. After 6 or 8 days of AIM treatment, cells were treated with AIM or AIM+bFGF for 24 h before EdU labeling for 12 h, followed by DAPI (blue), EdU (green), and OilRedO (red) costaining. The percentage of EdU-positive cells over total cells was graphed. Arrows in bottom right “Merged” panel point to premature adipocytes that were also stained positive for EdU labeling (n = 3). Color images available online at www.liebertpub.com/scd
In summary, the results above demonstrated that bFGF promoted cellular proliferation at all stages of adipogenic differentiation of hMSCs, which enhanced total adipogenesis during the first 48 h of adipogenic induction, but exerted strong inhibitory effect during days 3–6 after adipogenic initiation. After day 6, however, the effect of bFGF appeared to be “diffusive,” with similar amount of fat oil droplets dispersed into greater number of cells, resulting in fewer easily recognizable adipocytes with prominent oil droplets.
Summary
In summary, we present evidence that cellular proliferation antagonizes adipogenic differentiation of hMSCs. However, cellular proliferation stimulation can be beneficial for adipogenesis during the mitotic phase by increasing the population of cells capable of committing to adipogenic cell fate before the onset of adipogenic commitment.
We demonstrated that (1) cell cycle regulators, including Cdk1, CCND1, CCNA2, CDC6, FBX05, PLK2, and Cdk4, were downregulated during adipogenic differentiation, with some starting as early as 24 h after initiation of induction; (2) during the process of 12-day continuous adipogenic induction by AIM, the mitotic activity is limited to the first 48 h; (3) expression knockdown by siRNAs against Cdk1 and CCND1 inhibited cellular proliferation, but promoted adipogenic differentiation, with siCdk1 conferring a greater effect than siCCND1; and last, (4) application of exogenous growth factor bFGF promoted cellular proliferation across all stages of adipogenic differentiation, which enhanced total adipogenesis during the first 48 h of adipogenic induction, a phase that coincides with the mitotic activity, but exerted strong inhibitory effect during days 3–6 after adipogenic initiation, a phase that coincides with adipogenic commitment marked by the significant upregulation of C/EBPα and PPARγ.
Discussion
Adipogenesis has long been studied in vitro using murine 3T3-L1 cells subcloned initially from mouse embryonic fibroblast cell line 3T3-M [2]. A fraction of total cell population would automatically undergo adipogenesis even in growth media alone supplemented with 10%–30% of calf serum when saturation density was reached [2]. When stimulated with AIM consisting of DEX, insulin, and IBMX, these cells would undergo about two rounds of cell division during the first 2–3 days of adipogenic induction (a period referred to as mitotic clonal expansion) before growth arrest and terminal differentiation [16].
The question of whether mitotic clonal expansion is a prerequisite for adipogenic differentiation, however, has been debatable, with evidence supporting both sides of the arguments presented in the literature.
Evidence supporting this theory has largely been derived from studies that demonstrated dual negative effect of molecules on both cell proliferation and differentiation in 3T3-L1 cells [16,25–30]. Ectopic expression of Mad1 [30], an antagonizer of C-Myc, overexpression of Nur77 [27], a member of the nuclear receptor 4A subgroup, or ectopic expression of A-C/EBP [26], a C/EBP-specific dominant negative protein that could form stable inactive heterodimer with C/EBPβ, inhibited adipogenesis as well as proliferation. Similarly, applying DNA polymerase alpha inhibitor aphidicolin [28], rapamycin [29], an immunosuppressant with an antiproliferative activity, U0126 [15], an inhibitor of mitogen-activated protein kinase (MEK), roscovitine [15], a Cdk inhibitor, or N-acetyl-Leu-Leu-norleucinal [25], an inhibitor of calpain, which is a calcium-activated protease that was shown to degrade p27 (Cdk inhibitor), also demonstrated dual suppressing effect on both adipogenesis and proliferation.
However, to conclude that proliferation is a prerequisite for adipogenesis based on such dual effect, one would need to assume that these agents affected adipogenic differentiation indirectly by inhibiting cell proliferation first. Evidence of such causal effects, however, has been lacking and there remains the possibility that many of the agents may play parallel roles in both proliferation and differentiation, especially for the compound inhibitors, which often have pleiotropic effects other than what they are intended for when used at high concentrations.
For example, we have tested roscovitine on adipogenic differentiation of hMSCs and found that at 1 or 2 μM, it did not have any significant effect on either proliferation or adipogenic differentiation (unpublished data). At 5 μM, it significantly inhibited adipogenesis without affecting cellular proliferation, regardless if it was applied during the first 4 days of adipogenic induction or throughout the 12-day course (Supplementary Fig. S4). In this case, roscovitine appeared to have a direct effect on adipogenesis without affecting proliferation. When it was used at 20 μM or higher as shown in a previous study [15], it is likely that it could affect both adipogenesis and proliferation simultaneously.
Similar arguments could be made for A-C/EBP. A-C/EBP could partner with C/EBP members to form stable inactive heterodimers [31]. C/EBPβ is one of the earlier genes being induced by AIM that subsequently induces the transcription of PPARγ [32]. In addition, it has been shown to play an important role in cell cycle reentry and mitotic clonal expansion [33]. Given the dual roles of C/EBPβ, it is not surprising that in the A-C/EBP study, A-C/EBP could inhibit both adipogenesis and proliferation by “sequestering” C/EBPβ [26], and it does not necessarily establish proliferation inhibition as an underlying cause for adipogenic inhibition. A-C/EBP could also potentially partner with other C/EBP members, including C/EBPα, to inhibit adipogenesis. Similar caveats might have confounded the interpretation of the relationship between mitotic clonal expansion and adipogenesis in other previous studies.
Furthermore, there is evidence published in the literature that argues against the notion that mitotic clonal expansion is a prerequisite for adipogenesis [2,16,34–37]. In contrast to MEK1 inhibitor U0126, PD98059, another MEK1 inhibitor, was shown to exert enhancement effect on adipocyte differentiation in 3T3-L1 cells, even though it inhibited cellular proliferation [16,38,39]. We have tested both compounds in the past on bone-marrow derived hMSCs and observed that at 10 μM, PD98059 promoted adipogenesis, while U0126 inhibited it (unpublished), suggesting that these two compounds may differ in their mode of actions.
It has been argued that the differential outcome of these two compounds are due to the fact that U0126 was a more specific and potent inhibitor of MEK1 that could completely prevent mitotic clonal expansion, unlike PD98059, which only partially inhibited mitotic clonal expansion [15]. However, in a separate study, it was shown that troglitazone, which activated PPARγ as its ligand, could rescue the U0126-associated block in adipogenesis without reactivating clonal expansion [34], indicating that the effect of U0126 on adipogenesis and proliferation was independent of each other.
Additional arguments were provided by overexpression of C-Myc, which is an oncogene that precludes cells from exiting cell cycle, inhibited adipogenesis [36]. Similarly, treatment with tumor growth factor beta (TGFβ), which promotes proliferation and activates MEK1, inhibited adipogenesis in 3T3-L1 cells [37]. In addition, cells grown in suspension in media containing methyl cellulose were growth arrested, but were still able to differentiate into adipocytes [2], and finally, single hMSCs plated on surfaces that promoted rounded morphology could differentiate into adipocytes without undergoing cell division [35]. These data suggest that mitotic clonal expansion and adipogenic differentiation can be uncoupled. Last, even if we assume that the two are tightly coupled events, little is known about why or how cell cycle reentry leads to adipogenesis.
On the contrary, evidences have pointed to molecular coupling between growth arrest and differentiation. Both PPARγ and C/EBPα have been shown to promote cell cycle arrest by upregulating Cdk inhibitors p18 and p21 and repressing E2F-dependent transcription, respectively [40,41]. TGFβ treatment was shown to prevent the upregulation of p18, which could have led to the failure of the cells exiting cell cycle, resulting in inhibited adipogenesis [42]. It has also been demonstrated that before the expression of a differentiated phenotype, preadipocytes must arrest their growth at a distinct state in the G1 phase of the cell cycle [43].
Limited studies have been done so far to address the relationship between cellular proliferation and differentiation in hMSCs [10,17]. In this study, we demonstrated that similar to 3T3-L1 cells, hMSCs induced by AIM also undergo active proliferation for about 48 h before entering growth arrest. This process is insulin independent, as cells treated with IBMX and DEX only also behaved identical to those treated with AIM. Interestingly, a host of cell cycle regulators, including Cdk1 and CCND1, were downregulated starting from as early as 24 h after initiation of induction, which appears to coincide with the upregulation of C/EBPα. Furthermore, downregulation of Cdk1 or CCND1 by gene-specific siRNA resulted in greater adipogenesis, even though total cell numbers were reduced. This indicates that decreased proliferative activity is more permissive for adipogenic commitment.
This is consistent with previous studies demonstrating that CCND1 inhibits PPARγ-dependent activity, and CCND1−/− murine embryonic fibroblasts have increased PPARγ activity and increased propensity to undergo differentiation into adipocytes [44]. In addition, knockdown of REX1/ZFP42, a zinc finger protein, was shown to decrease cell proliferation and increase adipogenic potential in hMSCs, accompanied with suppressed expression of Cdk2 and CCND1 [45]. Interestingly, of all three siRNAs tested, there appeared to be a correlation between the degree of proliferation inhibition and the degree of adipogenic enhancement: siCdk4 conferred insignificant inhibitory effect on both proliferation and adipogenesis, while siCdk1 consistently had the strongest inhibitory effect on proliferation and the most significant effect on adipogenic enhancement among the three. The effect of siCCND1 lied in between those of siCdk4 and siCdk1.
It is not surprising to see the strongest inhibitory effect on proliferation by siCdk1, as Cdk1 has been shown to be the only Cdk protein that was sufficient to drive mammalian cell cycle, while all the other Cdks, including Cdk2, Cdk3, Cdk4, and Cdk6, were dispensable [23]. Furthermore, the changing levels of Cdk1 activity throughout cell cycle progression was shown to coordinate cell cycle transitions and govern periodic expression of critical genes to bring about proper cell cycle progression [46]. Perturbation of its expression levels, even at 40%–80% of its normal expression level as demonstrated in this study, could therefore significantly impact cell cycle progression and overall cellular proliferation.
It is also interesting to note that expression of Cdk1 is downregulated the most at 72 h after adipogenic initiation during the normal course of adipogenesis induced by AIM or IBMX+DEX, which correlates to a postmitotic stage when expression of both C/EBPα and PPARγ was drastically upregulated in cells treated with AIM or IBMX+DEX compared to their expression in cells treated with CM or DEX alone. On the other hand, the expression of Cdk4 was the least affected during the normal course of adipogenesis compared to Cdk1 or CCND1.
One intriguing observation was that the expression of Cdk1 was also downregulated in siCdk4- or siCCND1-treated cells, yet their effect on cellular proliferation and adipogenesis was not nearly as strong as that of siCdk1. The fact that expression change in Cdk4 or CCND1 could affect the expression of Cdk1 may not be surprising as Cdk2 and Cdk4 were shown to regulate Cdk1 expression [47]. However, downregulation of Cdk4 or CCND1 might also involve other molecular changes that somehow counteracted the effect of Cdk1 downregulation in siCdk4- or siCCND1-treated cells. Indeed, we found that siCdk4 and siCCND1 also transiently downregulated the expression of PPARγ and C/EBPα, respectively, while siCdk1 enhanced the expression of both.
A previous study established that Cdk4 positively regulated the expression and activation of PPARγ in mouse embryonic fibroblasts [48]. It is plausible that downregulation of Cdk4 by siCdk4 could affect adipogenic differentiation of hMSCs through its impact on cellular proliferation, growth arrest, and activation of PPARγ. Therefore, there are likely underlying complex molecular interplays that rendered the differential outcomes by the three different siRNAs. Nevertheless, unlike previous studies based on compound inhibitors or overexpression of genes that interfere with cell cycle regulation in 3T3-L1 cells, we believe that direct inhibition of cell proliferation by transiently downregulating players of the core cell cycle control machinery, the Cdk/Cyclin complexes, offers a more straightforward interpretation of the relationship between cell proliferation and adipogenesis.
FGF2 (bFGF) belongs to a large family of FGFs and has long been used in human pluripotent stem cell growth media to maintain cell self-renewal and inhibit differentiation (reviewed in the introduction of Quang et al. [49]). Bone marrow-derived MSCs isolated from bFGF−/− knockout mice demonstrated increased potency in adipogenic differentiation and reduced potency in osteogenic differentiation compared to wild-type MSCs, indicating that bFGF inhibits adipogenesis [50]. Our bFGF study provided additional confirmation and insights into the role of bFGF during the different stages of adipogenesis.
When applied during the first 48 h of adipogenic induction, which correlates to the mitotic phase, bFGF promoted adipogenesis. This was most likely by increasing the number of cells capable of committing to adipocytes later on. Indeed, at this stage, we demonstrated that bFGF significantly increased the total number of adipocytes, especially when it was applied during the first 24 h of adipogenic induction. It is plausible that cells responding to bFGF treatment did not undergo their usual transition to growth arrest until after bFGF withdrawal and additional exposure to AIM alone for another 2 days. In other words, these cells might have delayed adipogenic commitment, which would require additional future studies.
When applied between day 2 and 3 of induction, bFGF had minimum effect on adipogenesis, even though the total number of cells was still significantly increased. This stage appeared to correspond to a cell growth arrest stage when proliferation has stopped, but adipogenic markers, including C/EBPα and PPARγ, are yet to be substantially induced. At this stage, it appears that cells capable of committing to adipocytes were responding to the proliferative stimulus of bFGF on a minimum level (the total adipocytes always trended higher, although insignificantly), and their cell fate was not deterred by bFGF either. On the other hand, cells not capable of committing were still dividing in response to bFGF, contributing to the increase in the total cell number.
When applied between day 3 and 6 of induction, bFGF had a strong inhibitory effect on adipogenic differentiation, while also significantly increasing total cell numbers. This stage correlates to the adipogenic commitment stage when expression of C/EBPα and PPARγ were both substantially upregulated in AIM- or IBMX+DEX-treated cells compared to cells treated with DEX alone (Fig. 1A). It is worth noting that even though both genes were upregulated in response to AIM or IBMX+DEX starting as early as 24 h after adipogenic initiation, such induction was solely due to the effect of DEX, as both were induced at a similar level in cells treated with DEX alone or cells treated with OIM, indicating that at this lower induction level, cells were still very fluid in their cell fate commitment, as they were still capable of committing to osteogenic lineage in the presence of ascorbic acid-2-phosphate and beta-glycerophosphate (Fig. 1A).
It is therefore highly likely that the soaring expression of C/EBPα and PPARγ starting at 72 h after adipogenic initiation was the pivotal event that tipped the cells' commitment toward adipogenic lineage. At this stage, the total number of adipocytes was significantly reduced, while total cell numbers were significantly increased in the presence of bFGF, indicating that adipogenic commitment was hindered or reversed by bFGF.
An alternative explanation for the observed inhibition on adipogenesis imposed by bFGF at this stage is that cells might have merely been “arrested” in differentiation, instead of active suppression on the molecular machinery driving the differentiation. If cells were simply transiently arrested in differentiation in the presence of bFGF, one may expect a mere delay in the progression of differentiation. In another word, cells exposed to bFGF during D3−4 of adipogenic differentiation should have similar level of differentiation on D12 as control-treated cells on D11, more or less. This, however, did not appear to be the case. The absolute number of total adipocytes in bFGF D3−4 treatment group was only about 65% of those in the control group by D12 of differentiation, whereas in the control treatment group, its total adipocytes on D11 are about the same as on D12.
Therefore, during the commitment phase, bFGF did not merely arrest cells in differentiation, rather it actively inhibited differentiation. It is plausible that by forcing cells to reenter cell cycle, it somehow deterred/derailed the progression of differentiation.
When applied after day 6 of adipogenic induction, bFGF appeared to have a very different effect from the other stages: it did not affect the total fat accumulation in the whole population, but reduced fat accumulation in some individual adipocytes, making their oil droplets less prominent, while increasing a significant number of cells carrying small faint oil droplets that were not easily recognizable as adipocytes.
There are two possible explanations for this observation: (1) bFGF stimulated existing adipocytes to divide, splitting the oil droplets into two different cells, with one inheriting fewer than the other and (2) bFGF inhibited lipogenesis in existing adipocytes, making their oil droplets less prominent as they would otherwise have been in the absence of bFGF. In addition, bFGF stimulated additional cells that previously did not commit to adipogenic cell fate to undergo early stage of adipogenesis somehow and accumulated small speckled oil droplets. Considering the strong inhibitory effect of bFGF on adipogenic commitment during day 3–6, scenario 1 is more plausible.
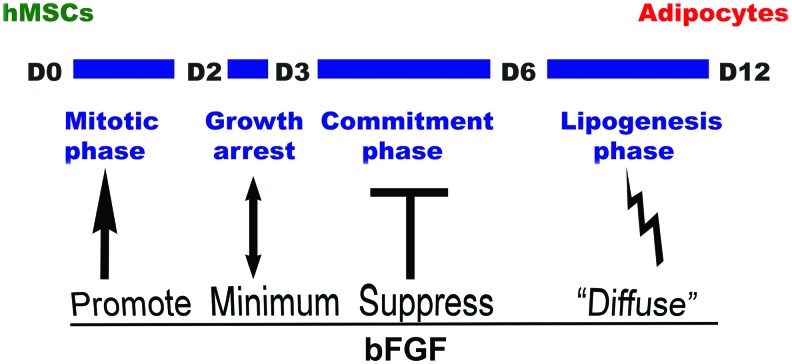
Together, based on our above findings, we propose a model of adipogenic differentiation of hMSCs that consists of four different stages: mitotic phase, growth arrest phase, commitment phase, and lipogenesis phase, with each phase demonstrating different response to bFGF signaling (Fig. 12).
FIG. 12.
A simple model illustrating the four different phases during adipogenic differentiation of hMSCs and how cells respond to bFGF differently in different phases, resulting in distinct outcomes in adipogenic differentiation. During mitotic phase, bFGF help expand cells capable of committing to adipocytes; during growth arrest, the effect of bFGF is minimum with overall effect trending slightly upward; during adipogenic commitment phase, bFGF strongly inhibits adipogenic differentiation; and during lipogenesis phase, bFGF appears to “diffuse” total oil droplets into greater number of cells. Color images available online at www.liebertpub.com/scd
Finally, our combined assays using OilRedO staining and quantification, individual adipocyte counting, as well as ImagePro software for measuring total areas of stained oil droplets in the bFGF experiments helped to elucidate the pros and cons of each assay tool.
The conventionally used approach to quantify adipogenesis is by OilRedO stain quantification, which is fast and convenient, but is relatively crude and has low sensitivity level due to the nature of the assay itself. In addition, it does not distinguish whether any level of OilRedO quantification change was due to changed total adipocytes (hyperplasia/hypoplasia) or change in the amount of fat accumulation in individual fat cells (hypertrophy/hypotrophy). To distinguish those, one would need to rely on the other two approaches.
Counting individual adipocytes requires highly laborious imaging and manual counting due to the irregularity of cell shapes that are difficult to be automated using imaging analysis software, but it provides valuable insights that OilRedO quantification could not provide. And finally, automated area measurement of stained oil droplets using software like ImagePro also requires laborious imaging process, but is much faster than individual adipocyte counting and provides significantly greater sensitivity than OilRedO quantification. Bear in mind, however, area measurement by ImagePro does not help distinguish hyperplasia/hypoplasia versus hypertrophy/hypotrophy, and therefore adipocyte counting is still valuable. Overall, the three approaches complement each other and should be used with discretion.
In summary, our study demonstrated that similar to 3T3-L1 cells, hMSCs also undergo a mitotic clonal expansion stage at the beginning of their adipogenic differentiation, followed by growth arrest and adipogenic commitment. Attenuating early cellular proliferation by downregulating cell cycle genes, such as Cdk1, significantly promoted adipogenesis, indicating that cell proliferation antagonizes adipogenic differentiation. In addition, growth factor bFGF promotes cellular proliferation throughout all phases of adipogenesis, but only enhanced adipogenesis when applied during the mitotic phase by increasing the total number of cells capable of committing to adipocytes. During the commitment phase, however, bFGF significantly inhibited adipogenesis, further demonstrating an antagonizing relationship between cellular proliferation and adipogenic differentiation.
Supplementary Material
Acknowledgments
This work was supported by NIH grants no. 1SC3GM094078 and 1SC3GM116720-01, both awarded to Y.Z. No additional external funding was received for this study.
Author Disclosure Statement
The authors declare that there are no conflicts of interest.
References
- 1.Arner P. and Spalding KL. (2010). Fat cell turnover in humans. Biochem Biophys Res Commun 396:101–104 [DOI] [PubMed] [Google Scholar]
- 2.Green H. and Meuth M. (1974). An established pre-adipose cell line and its differentiation in culture. Cell 3:127–133 [DOI] [PubMed] [Google Scholar]
- 3.Cristancho AG. and Lazar MA. (2011). Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 12:722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peltz L, Gomez J, Marquez M, Alencastro F, Atashpanjeh N, Quang T, et al. (2012). Resveratrol exerts dosage and duration dependent effect on human xmesenchymal stem cell development. PLoS One 7:e37162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott MA, Nguyen VT, Levi B. and James AW. (2011). Current methods of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev 20:1793–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong WK. and Sugii S. (2013). Adipose-derived stem cells: fatty potentials for therapy. Int J Biochem Cell Biol 45:1083–1086 [DOI] [PubMed] [Google Scholar]
- 7.Tran TT. and Kahn CR. (2010). Transplantation of adipose tissue and stem cells: role in metabolism and disease. Nat Rev Endocrinol 6:195–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe CE, O'Rahilly S. and Rochford JJ. (2011). Adipogenesis at a glance. J Cell Sci 124(Pt 16):2681–2686 [DOI] [PubMed] [Google Scholar]
- 9.Zhang R, Wang D, Xia Z, Chen C, Cheng P, Xie H, et al. (2013). The role of microRNAs in adipocyte differentiation. Front Med 7:223–230 [DOI] [PubMed] [Google Scholar]
- 10.Qian SW, Li X, Zhang YY, Huang HY, Liu Y, Sun X, et al. (2010). Characterization of adipocyte differentiation from human mesenchymal stem cells in bone marrow. BMC Dev Biol 10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, et al. (1999). Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 3:151–158 [DOI] [PubMed] [Google Scholar]
- 12.MacDougald OA. and Lane MD. (1995). Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem 64:345–373 [DOI] [PubMed] [Google Scholar]
- 13.Tontonoz P, Hu E. and Spiegelman BM. (1994). Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 79:1147–1156 [DOI] [PubMed] [Google Scholar]
- 14.Freytag SO, Paielli DL. and Gilbert JD. (1994). Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev 8:1654–1663 [DOI] [PubMed] [Google Scholar]
- 15.Tang QQ, Otto TC. and Lane MD. (2003). Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc Natl Acad Sci U S A 100:44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu Z, Wei Y, Chen N, Jiang M, Wu J. and Liao K. (2001). DNA synthesis and mitotic clonal expansion is not a required step for 3T3-L1 preadipocyte differentiation into adipocytes. J Biol Chem 276:11988–11995 [DOI] [PubMed] [Google Scholar]
- 17.Janderova L, McNeil M, Murrell AN, Mynatt RL. and Smith SR. (2003). Human mesenchymal stem cells as an in vitro model for human adipogenesis. Obes Res 11:65–74 [DOI] [PubMed] [Google Scholar]
- 18.Beresford JN, Bennett JH, Devlin C, Leboy PS. and Owen ME. (1992). Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. J Cell Sci 102(Pt 2):341–351 [DOI] [PubMed] [Google Scholar]
- 19.Meunier P, Aaron J, Edouard C. and Vignon G. (1971). Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. A quantitative study of 84 iliac bone biopsies. Clin Orthop Relat Res 80:147–154 [DOI] [PubMed] [Google Scholar]
- 20.Justesen J, Stenderup K, Ebbesen EN, Mosekilde L, Steiniche T. and Kassem M. (2001). Adipocyte tissue volume in bone marrow is increased with aging and in patients with osteoporosis. Biogerontology 2:165–171 [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y. and Ding S. (2007). A high-throughput siRNA library screen identifies osteogenic suppressors in human mesenchymal stem cells. Proc Natl Acad Sci U S A 104:9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dienstknecht T, Ehehalt K, Jenei-Lanzl Z, Zellner J, Muller M, Berner A, et al. (2010). Resazurin dye as a reliable tool for determination of cell number and viability in mesenchymal stem cell culture. Bull Exp Biol Med 150:157–159 [DOI] [PubMed] [Google Scholar]
- 23.Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. (2007). Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448:811–815 [DOI] [PubMed] [Google Scholar]
- 24.Orford KW. and Scadden DT. (2008). Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet 9:115–128 [DOI] [PubMed] [Google Scholar]
- 25.Patel YM. and Lane MD. (2000). Mitotic clonal expansion during preadipocyte differentiation: calpain-mediated turnover of p27. J Biol Chem 275:17653–17660 [DOI] [PubMed] [Google Scholar]
- 26.Zhang JW, Tang QQ, Vinson C. and Lane MD. (2004). Dominant-negative C/EBP disrupts mitotic clonal expansion and differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci U S A 101:43–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao LC, Bensinger SJ, Villanueva CJ, Wroblewski K. and Tontonoz P. (2008). Inhibition of adipocyte differentiation by Nur77, Nurr1, and Nor1. Mol Endocrinol 22:2596–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reichert M. and Eick D. (1999). Analysis of cell cycle arrest in adipocyte differentiation. Oncogene 18:459–466 [DOI] [PubMed] [Google Scholar]
- 29.Yeh WC, Bierer BE. and McKnight SL. (1995). Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc Natl Acad Sci U S A 92:11086–11090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pulverer B, Sommer A, McArthur GA, Eisenman RN. and Luscher B. (2000). Analysis of Myc/Max/Mad network members in adipogenesis: inhibition of the proliferative burst and differentiation by ectopically expressed Mad1. J Cell Physiol 183:399–410 [DOI] [PubMed] [Google Scholar]
- 31.Greenwel P, Tanaka S, Penkov D, Zhang W, Olive M, Moll J, et al. (2000). Tumor necrosis factor alpha inhibits type I collagen synthesis through repressive CCAAT/enhancer-binding proteins. Mol Cell Biol 20:912–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saladin R, Fajas L, Dana S, Halvorsen YD, Auwerx J. and Briggs M. (1999). Differential regulation of peroxisome proliferator activated receptor gamma1 (PPARgamma1) and PPARgamma2 messenger RNA expression in the early stages of adipogenesis. Cell Growth Differ 10:43–48 [PubMed] [Google Scholar]
- 33.Zhang YY, Li X, Qian SW, Guo L, Huang HY, He Q, et al. (2011). Transcriptional activation of histone H4 by C/EBPbeta during the mitotic clonal expansion of 3T3-L1 adipocyte differentiation. Mol Biol Cell 22:2165–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prusty D, Park BH, Davis KE. and Farmer SR. (2002). Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor gamma (PPARgamma) and C/EBPalpha gene expression during the differentiation of 3T3-L1 preadipocytes. J Biol Chem 277:46226–46232 [DOI] [PubMed] [Google Scholar]
- 35.McBeath R, Pirone DM, Nelson CM, Bhadriraju K. and Chen CS. (2004). Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell 6:483–495 [DOI] [PubMed] [Google Scholar]
- 36.Freytag SO. (1988). Enforced expression of the c-myc oncogene inhibits cell differentiation by precluding entry into a distinct predifferentiation state in G0/G1. Mol Cell Biol 8:1614–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choy L, Skillington J. and Derynck R. (2000). Roles of autocrine TGF-beta receptor and Smad signaling in adipocyte differentiation. J Cell Biol 149:667–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Font de Mora J, Porras A, Ahn N. and Santos E. (1997). Mitogen-activated protein kinase activation is not necessary for, but antagonizes, 3T3-L1 adipocytic differentiation. Mol Cell Biol 17:6068–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boney CM, Gruppuso PA, Faris RA. and Frackelton AR., Jr. (2000). The critical role of Shc in insulin-like growth factor-I-mediated mitogenesis and differentiation in 3T3-L1 preadipocytes. Mol Endocrinol 14:805–813 [DOI] [PubMed] [Google Scholar]
- 40.Morrison RF. and Farmer SR. (1999). Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J Biol Chem 274:17088–17097 [DOI] [PubMed] [Google Scholar]
- 41.Slomiany BA, D'Arigo KL, Kelly MM. and Kurtz DT. (2000). C/EBPalpha inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol Cell Biol 20:5986–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang JW, Choi Y, Park JH, Kim JS, Park KD, Baek DH, et al. (2008). The effects of cyclin-dependent kinase inhibitors on adipogenic differentiation of human mesenchymal stem cells. Biochem Biophys Res Commun 366:624–630 [DOI] [PubMed] [Google Scholar]
- 43.Scott RE, Hoerl BJ, Wille JJ, Jr, Florine DL, Krawisz BR. and Yun K. (1982). Coupling of proadipocyte growth arrest and differentiation. II. A cell cycle model for the physiological control of cell proliferation. J Cell Biol 94:400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu M, Rao M, Bouras T, Wang C, Wu K, Zhang X, et al. (2005). Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem 280:16934–16941 [DOI] [PubMed] [Google Scholar]
- 45.Bhandari DR, Seo KW, Roh KH, Jung JW, Kang SK. and Kang KS. (2010). REX-1 expression and p38 MAPK activation status can determine proliferation/differentiation fates in human mesenchymal stem cells. PLoS One 5:e10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banyai G, Baidi F, Coudreuse D. and Szilagyi Z. (2016). Cdk1 activity acts as a quantitative platform for coordinating cell cycle progression with periodic transcription. Nat Commun 7:11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berthet C. and Kaldis P. (2006). Cdk2 and Cdk4 cooperatively control the expression of Cdc2. Cell Div 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abella A, Dubus P, Malumbres M, Rane SG, Kiyokawa H, Sicard A, et al. (2005). Cdk4 promotes adipogenesis through PPARgamma activation. Cell Metab 2:239–249 [DOI] [PubMed] [Google Scholar]
- 49.Quang T, Marquez M, Blanco G. and Zhao Y. (2014). Dosage and cell line dependent inhibitory effect of bFGF supplement in human pluripotent stem cell culture on inactivated human mesenchymal stem cells. PLoS One 9:e86031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao L, Sobue T, Esliger A, Kronenberg MS, Coffin JD, Doetschman T, et al. (2010). Disruption of the Fgf2 gene activates the adipogenic and suppresses the osteogenic program in mesenchymal marrow stromal stem cells. Bone 47:360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.