Abstract
Living cells and tissues experience physical forces and chemical stimuli in a human body. The process of converting mechanical forces into biochemical activities and gene expression is mechanochemical transduction or mechanotransduction. Significant advances have been made in understanding mechanotransduction at cellular and molecular levels over the last two decades. However, major challenges remain in elucidating how a living cell integrates signals from mechanotransduction with chemical signals to regulate gene expression and to generate coherent biological responses in living tissues in physiological conditions and diseases.
For many years, researchers focus their studies on the influence and mechanisms of individual soluble molecules such as growth factors, cytokines, and chemotactic molecules on biological functions of living cells and tissues. However, during the last two decades, increasing evidence demonstrates that local microenvironment (e.g. matrix stiffness) and physical forces of cells and tissues play critical roles in controlling and regulating responses and behaviors in embryonic development, adult physiology, and various diseases like cancer, cardiovascular diseases, and pulmonary diseases. In order to elicit biological responses, cells must convert these physical signals into chemical processes and/or changes in gene expression. The conversion of mechanical signals into chemical signals or gene expression is called mechanochemical transduction, or mechanotransduction [1]. Despite significant progress over the last decades, the underlying molecular mechanisms of cellular mechanotransduction are not well understood and therefore more efforts are needed to address many open fundamental questions in this area.
Force-dependent integrin adhesion at cell-matrix interface
While some body cells are often in suspension (e.g., circulating cells in blood), most body cells need to attach to a polymer-like structure called the extracellular matrix (ECM) to function properly in a tissue. In 1984, Erkki Ruoslahti et al. discovered that the arginylglycylaspartic acid (Arg-Gly-Asp, or RGD) tripeptide was part of fibronectin [2], a matrix protein polymer, which interacts with a cell surface receptor, which was cloned by Richard Hines lab in 1986 and named as integrin [3], the primary transmembrane molecule to mediate cell-matrix adhesion. Thirty years later, 24 integrin subtypes have been discovered that are made of different combinations of α and β integrin heterodimers (e.g., α5β1 for binding to matrix protein fibronectin) [4]. Integrin subtypes αMβ2 (Mac-1) or αLβ2 (LFA-1), known to mediate adhesion between an immune cell and a target cell, have been cloned a year after cloning of β1 integrin [5]. When an integrin is in its inactive form, its ectodomains are in the bend configuration and its hybrid domain is in the closed configuration. ECM protein fibronectin or cytoplasmic protein talin can induce integrin activation. When the integrin is activated, its ectodomains become extended and upright [6] and its hybrid domain swings open away from the α-subunit [7], although the details of these processes are still under debate [8]. However, integrin activation alone is necessary but not sufficient for many vital cellular functions such as cell spreading, cell growth, and proliferation. For example, an early paper in late 1990’s on the role of integrin and cell function finds that it is the degree of cell spreading and not the total number of clustered integrins that dictate whether a normal endothelial cell enters the cycle of DNA synthesis [9]. During the early days of the integrin study, researchers focused their attention on chemical signaling of integrins (e.g., control of pH and Na/H antiporter) after integrins clustered and immobilized onto ECM [10, 11]. While chemical processes are certainly at play, it turns out that mechanical forces play a critical role in integrin-mediated adhesion and cellular responses. The first experimental evidence that integrins and focal adhesions (FAs) (a protein complex consists of clustered integrins and other cytoplasmic molecules) mediate mechanical force transmission to the cytoskeleton was shown in 1993 [12]. Several years later 3 groups independently show that the mechanical force signaling via FAs are bi-directional: Outside-In [13] and Inside-Out signaling [14, 15]. The details of the molecular mechanisms of activation are still being actively studied, however. For example, a molecular dynamics model has been used to explore how integrin αIIbβ3 is activated [16]. In addition to change the conformation of integrins, matrix forces can unfold fibronectin [17] and tune the interactions of fibronectin and collagen-1 in the ECM of fibroblasts [18]. It is now well accepted in the field that the primary function of the integrins is to mediate mechanical signaling, to recruit cytoplasmic proteins to the clustered integrins and FAs, and to propagate mechanical forces to alter activities of other cytoplasmic proteins and cellular responses, since integrins themselves do not have enzymatic activities. Molecular mechanisms of force-induced integrin activation and cell-ECM adhesion are discussed in a couple of recent reviews [19, 20]. To learn about the evidence for a force-reinforced bond (a catch bond) in α5β1 integrins [21], one can read a recent review on the topic [22]. In contrast to integrins, only relative recently cell-cell adhesion molecule E-cadherin, critical for formation of an intact epithelial monolayer, is shown to be mechanosensors [23]. In response to fluid shear stress, a complex consisting of PECAM-1 (platelet-endothelial cell adhesion molecule 1), VE-cadherin (vascular endothelial cadherin) and VEGF (vascular endothelial growth factor) receptor 2 (VEGFR2) is identified as a mechanosensor in endothelial cells [24] and G protein-coupled receptors is discovered as a mechanosensor in neutrophils [25]. Activation of IkappaB kinase or Flk-1 by fluid shear stress is mediated by integrins [26, 27]. In this review, we focus on integrin-mediated ECM force transmission and force transduction, i.e., mechanochemical transduction. Fort those who are interested in cellular mechanoelectrical transduction, they are suggested to a review on mechanosensitive ion channels on the plasma membrane [28].
It is increasingly evident that almost all living cells generate endogenous forces, although the magnitudes of the forces vary among different cell types. Various types of cells appear to exhibit different sensitivities to changes in force magnitudes, frequencies, and durations, but the underlying mechanisms are not well understood. Inside the cytoplasm, different types of molecular motors generate forces and a single molecular motor can generate a force of a few picoNewtons (~pNs), but nonmuscle myosin II are particularly interesting because many of these molecules can align along actin microfilaments to significantly increase the total magnitude of forces in actin bundles to the nanoNewton (~nN) range at a single FA [15]. Forces exerted at single integrins are estimated from a few pNs to several dozens of pNs [29–31]. Recent measurements with tension gauge tethers reveal that a single integrin molecule mediates ~40 pN force to initiate cell spreading before FA formation and is independent of actomyosin [32, 33] and that a >54 pN molecular tension is transmitted by clustered integrins in motile actomyosin-dependent FAs [34]. Since estimates for the single integrin force are still considered by some to be a point of debate, future studies are needed to reconcile these estimates obtained in various conditions. More importantly, a FA has hundreds of clustered integrins and other cytoplasmic proteins to connect with hundreds of filamentous actins (F-actins) and myosin IIs such that the force at a single FA can be tens of nNs and can propagate along the cytoskeleton to distances ranging from several micrometers (μms) to tens of μms across the whole cell length.
Mechanotransduction in the cytoplasm
In the early 80’s, Don Ingber [35] and Mina Bissell [36] independently proposed that solid-state ECM could impact on cell/tissue organization/function and gene expression, but at the time the experimental evidence is scarce and the mechanism is not clear. A fundamental question in biology is how a living cell integrates forces sensed, say, at different FAs, and responds in a coherent manner. Ingber first proposed the model of cellular tensegrity (tensional integrity) in early 1980’s to explain the tension-integrated cytoskeletal structure and cellular responses to mechanical forces [35, 37]. While this model emphasizes the importance of cytoskeletal tension in integrating cytoskeletal structures and cellular responses, it should be viewed in the context of the dynamic processes of the cytoskeleton (e.g., assembly and disassembly of cytoskeletal filaments) and local biochemical processes in the cell. Early experimental results in living cells are consistent with the cellular tensegrity model [12, 38], but rigorous experimental tests and support for the tensegrity model with microtubules as the compression-bearing element in the cell [39, 40] and the prestress model (a model where the prestress is balanced from outside of the model) came several years later [41]. Here inherent cytoskeletal tension and pre-tensile stress (prestress) are equivalent and are used interchangeably.
Many labs have searched for intracellular mechanosensors downstream of integrins over the last decade. Molecular dynamics analyses that show the potential role of FA protein talin [42] and of FA protein vinculin [43] in mechanosensing are consistent with the experimental evidence for recombinant talin rod molecule [44] and for vinculin in living cells [45] in mechanosensing. Recent work reveals that differential ECM stiffness (i.e. ECM force) sensing by talin requires its actin-binding site 2 (ABS2) but not vinculin or actin-binding site 3 (ABS3) [46]. The force transmission capability of talin and integrins is downregulated by another focal adhesion protein Kank that binds close to the ABS2 site of talin [47]. These studies reveal the mechanosensing capacity of some FA proteins and are consistent with the notion that any molecules that are in the pathway of the force propagation should be in principle candidates for mechanosensors. However, for many years a prevailing view in the field of mechanotransduction is that force transmission in living cells are “short-ranged” and thus a local force can only exert its significant effects at the periphery of the cell such as a FA. This prevailing view dominates the field of mechanotransduction as studies have focused on the activation of proteins at or near FAs by force. For example, pulling on integrins activates Src protein near the applied force and Src activation slowly propagates along the plasma membrane via cytoskeletal dependent mechanisms [48], mechanical stretch results in phosphorylation of p130Cas by Src kinases [49], and elevation of substrate rigidity activates focal adhesion kinase (FAK) on fibronectin-coated substrates [50]; all demonstrate a local impact by a local force. From a material’s point of view, this “short-ranged” force prediction would be reasonable if the material was isotropic and homogeneous and therefore a local stress would decay as ~1/R2 where R is the distance between the site of local force application and a point in the material. But the cytoplasm of a living spread cell is neither isotropic nor homogeneous.
Since early 2000’s, evidence of long-distance force propagation (~tens of μms) in living cells has been demonstrated [51, 52]. The long-range force propagation becomes short-ranged (a few μms) when either the prestress in the actin bundles (i.e., stress fibers) are inhibited or the actin bundles are disrupted [51, 52], suggesting the critical roles by the prestressed actin bundles in long-distance force propagation. Importantly, it has been revealed that intracellular mechanotransduction is long-distant because Src or Rac1 can be directly activated within 300 ms of force application at a distance that is more than 30–60 μm away from the site of the local force application [53, 54] (Fig. 1). Mechanotransduction in the cytoplasm is also much faster (>40-fold) than a soluble growth factor induced signaling [53]. These findings of the anisotropic cytoskeleton mediating rapid long-distant force transmission have been predicted by a theoretical prestressed actin bundle model [55] and are consistent with the cellular tensegrity model [37]. The reason for the long-distant force propagation of the stress fibers is because they are much stiffer than the surrounding cytoplasm and thus can concentrate the stresses and propagate them to long distances throughout the whole cell length. Anisotropic cytoskeleton mediating long-distance force transmission in the cytoplasm has been demonstrated in several other cell types [56, 57] and tension-driven anisotropic alignment of ECM fibers is responsible for long-distance force transmission in the ECM [58]. Recently it is shown that collective cell durotaxis toward stiffer matrices during cell migration is a result of long-range intercellular force transmission [59], which would not be possible without the long-distance force transmission first within an individual cell, since cellular forces are first generated locally at cell-ECM interface in an integrin-dependent manner.
Figure 1. Long range mechanotransduction in the cytoplasm and the nucleus.
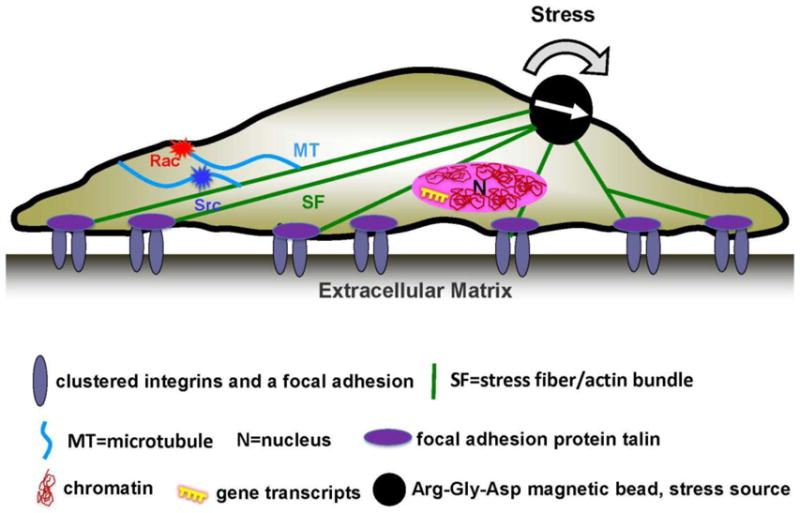
A local shear stress of physiologic magnitudes and frequency is applied via integrins with a Arg-Gly-Asp tripeptide coated magnetic bead. The applied force concentrates at the stress fibers and propagates to long distances in the cytoplasm to directly activate enzyme Src [53] and Rac1 [54] and into the nucleus to stretch chromatin to upregulate transcription [73].
Viscoelastic mechanics of the cell and mechanotransduction
The stress fibers are viscoelastic, just like the rest of the cytoplasm. The viscoelastic nature of the cell has been investigated in detail over the last decade. It is discovered that as the frequency of force application increases, the change of cell stiffness with the force frequency follows a weak power law, as if the cell behaves like a soft glass material [60]. Although the exact molecular mechanism underlying this unique behavior of the cell remains unclear up to this day, rupture of protein-protein noncovalent bonds in living cells has been proposed as a possible source of the weak power law [61]. Recently, it is shown that the plastic deformation in a living cell after removal of mechanical load is a constant fraction of the total deformation, originates from bond ruptures within the cytoskeleton, and also follows a weak power law with loading frequency [62]. Whether the weak power law behaviors during both loading and after loading conditions emerge from the same noncovalent protein-protein bond rupture mechanism needs to be examined in the future.
At the present time, it is not clear how much cell mechanics, which focuses on the viscoelastic mechanical properties of the living cells, relates to mechanotransduction. While some of the viscoelastic behaviors of living cells originates solely from the material properties of the cells, it is conceivable that some of the temporal changes of the viscoelastic properties is a result of cellular remodeling, which would depend, at least in part, on the processes of mechanotransduction. A recent report shows that during differentiation, nuclei in bovine and human mesenchymal cells stiffen and the stiffening response sensitizes the cells to calcium signaling [63]. However, more research is needed to understand how mechanotransduction processes in the cytoplasm (and the nucleus) vary with changes in viscoelastic properties of the cell and vice versa.
Nuclear mechanotransduction
In contrast to the understanding of mechanics of the cytoplasm, we know very little about nuclear mechanics and mechanotransduction. A mammalian cell nucleus is a highly organized structure where gene expression is thought to be regulated temporally and spatially [64], but the role of force and mechanics in gene regulation is far from clear. The nuclear envelope is physically tethered to the actin cytoskeleton via the LINC (Linker of Neucleoskeleton and Cytoskeleton) complex that consists of KASH-domain proteins and SUN-domain proteins [65]. The evidence of force-carrying connections that reach from the plasma membrane to the genome was first shown late 1990’s [66]. However, precise control and application of physiologically relevant magnitudes of force and simultaneous quantitation of synchronous intranuclear deformation were only achieved later [67]. In addition, it is shown that intranuclear protein-protein complexes (coilin-SMN (survival of motor neuron) in Cajal bodies) can be directly dissociated by force at the cell surface and this force propagation into the nucleus depends on the tensed cytoskeleton as well as presence of Lamin A/C [68]. The nuclear lamina lie underneath the nuclear envelope. Lamins, a major protein network in the nuclear lamina, are known to function as a mechanosensor for differentiation and regulation of transcription factors [69–71]. Nevertheless, the fundamental question in the field of mechanotransduction that whether a physiologically-relevant force or deformation can directly deform a chromatin structure in a living cell to regulate specific gene expression has not been answered during the last 2 decades. It has been known for a long time that chromatin decondensation is associated with transcription, but it is not clear whether it is the chromatin decondensation that drives the gene expression or vice versa. A method has been developed using bacterial artificial chromosome (BAC) to insert multiple green fluorescent proteins (GFPs) and dihydrofolate reductase (DHFR) gene in the same chromatin domain [72]. Direct evidence has been demonstrated that externally applied forces via integrins alone can directly unfold and stretch a chromatin and elevate DHFR transcription within the same chromatin domain [73] (Fig. 1). Inhibiting cytoskeletal tension or disrupting the force transmission pathways from the cell surface to the nuclear structural proteins that connect to the chromatin inhibited or downregulated the DHFR expression. DHFR transcription processes can be quantified rapidly (within 15 sec, after RNA polymerase II binding) and are sensitive to the angle and direction of loading relative to the actin bundles: the higher the stress angle, the bigger the chromatin stretching, the greater the transcription. The finding of matrix force induced direct gene expression in the nucleus is consistent with the prestressed actin bundle model and the cellular tensegrity model in long-distance force propagation. It would be difficult to interpret this force-induced rapid transcription of DHFR by models of diffusion or translocation of FA proteins or other cytoplasmic molecules such as Yes-associated Protein (YAP)/transcriptional coactivator with PDZ-binding motif (TAZ) [74] or TWIST1 [75] into the nucleus, which can certainly regulate matrix rigidity/force dependent gene expression at longer time scales (~tens of minutes). Matrix force dependent cytoplasmic YAP translocation into the nucleus is indeed necessary for gene upregulation of nuclear protein 1 (Nupr1), whose role as a tumor suppressor is just recently discovered [76]. Histone modifications in the nucleus are one of the major means of epigenetic alterations to regulate gene expression. Stiff ECM or applied forces via integrins have been shown to increase methylation of histone 3 at the site of lysine 9 (H3K9) [77], but it is not clear at this time whether H3K9 methylation is a direct result of force propagation and impact in the nucleus or via indirect pathways of cytoplasmic biochemical activities.
The nuclear envelope is punctuated by many nuclear pore complexes (NPCs) that varies among cell types. The NPCs control the traffic of molecules into (e.g., YAP/TAZ or TWIST1) and out of the nucleus (e.g., nuclear actin [78]) and hence regulate gene expressions to influence mechanotransduction. However, how the NPCs are related to the LINC complexes is still elusive [79] and should be investigated in the future.
Mechanobiology in mechanobiomedicine
Owing largely to advances in mechanotransduction, mechanobiology, a study of biology using mechanical and engineering approaches, is becoming increasingly relevant to stem cell biology and cancer in the last decade or so. For many years, researchers have cultured cells on top of rigid plastic or glass. However, it is well known that various types of living cells in soft tissues attach to matrices of stiffness varying from 0.1 kPa to tens of kPa [80]. Tuning the substrate stiffness in a controlled manner, it is demonstrated that FAs and migration of living cells are dramatically different on substrates of various rigidity [14]. Later it is reported that mesenchymal stem cell differentiation can be directed by ECM stiffness [81], which is driven by myosin II dependent processes. A few years later, it is demonstrated that an applied local force can differentiate a single embryonic stem cell [82]. The impact of mechanical microenvironment on living cells can be long-lasting; when stem cells [83] or cancer cells [77] are plated on substrates of different stiffness, they maintain their behaviors on the previous substrate for several days to exhibit features of memory. It turns out that mechanics and forces are not only important in stem cell differentiation but also critical in normal patterning and correct organization of germ layers during early mammalian embryonic development [84].
Mechanical forces are even making an impact in cancer biology. Despite decades of efforts and much progress in research, many aspects of cancer remain poorly understood. For example, it is not clear why only a few cancer cells, out of thousands of cancer cells, are able to form metastatic colonization, an irreversible and critical step in malignant tumor progression to clinically detectable macroscopic metastases [85]. The concept of cancer stem cells (CSCs) has been proposed in 1990’s to explain the propensity of a few primitive cells of leukemia to proliferate [86], but others fail to find an association between tumorigenicity and cell surface stem cell markers in solid tumor cells [87, 88]. In primary tumors, high mechanical tension and matrix stiffening are shown to be important in cancer progression [89] and high fluid/solid pressure in the primary tumor often accompanies tumor growth [90], but secondary metastatic sites of tumors appear to be softer (suggesting lower forces) than the surrounding normal tissues [91], suggesting that low forces and low matrix stiffness might play be important in the tumorigenicity and metastasis of CSC-like tumor cells. Creating a 3D soft matrix made of fibrin gels, a small population of tumor cells that are highly tumorigenic and malignant can be selected and grown from the general cancer cell population in several murine or human cancer cell lines [92]. Since these cells are not selected by cell surface stem cell markers and appear to be different from the conventional CSCs or tumor-initiating cells (TICs) that exhibit three distinct subtypes, they are called tumor-repopulating cells (TRCs). Interestingly, those melanoma TRCs in 3D soft matrices are less differentiated than the melanoma cells cultured in 3D stiff matrices or on rigid plastic [77], suggesting that it is the low forces and low matrix stiffness in the 3D microenvironment that drive undifferentiated, tumorigenic TRC growth. These soft melanoma TRCs also extravasate (move out of the blood vessels) to secondary sites more efficiently (Fig. 2) than those stiff differentiated melanoma cells [93]. These findings suggest a common thread in metastatic colonization of malignant tumors: a few tumorigenic TRCs are able to metastasize and grow at the secondary sites of soft matrices because these cells exert low forces, are soft and undifferentiated.
Figure 2. Soft melanoma tumor-repopulating cells extravasate efficiently to secondary sites of metastasis.
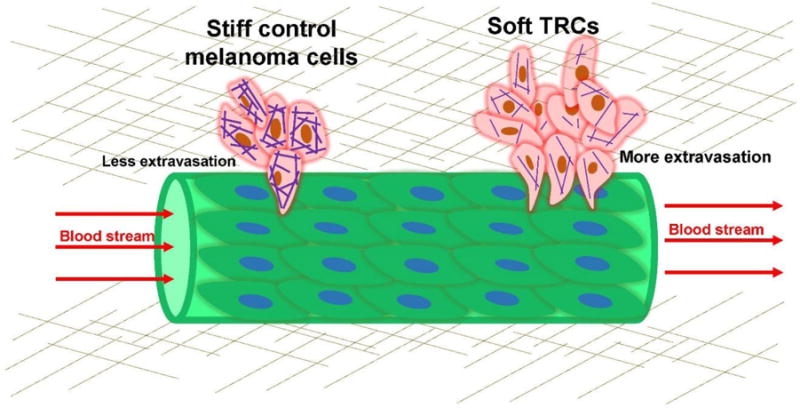
The melanoma tumor-repopulating cells (TRCs) are injected into the pericardial cavity (to the left of the image, not shown) of a zebrafish. The undifferentiated soft TRCs arrest at the tail and squeezed out of the small blood vessel (green color) more efficiently than the differentiated stiff control melanoma cells. The efficient extravasation of the soft TRCs and the ensuing micrometastasis formation and metastatic colonization is inhibited by differentiating the TRCs with retinoic acid, stiffening F-actin with a polymerizing drug, or promoting F-actin via overexpressing small GTPase Cdc42 (from [93]).
Mechanical forces and mechanotransduction are shown to be important not just in basic research in biology. Advances in life sciences and medicine in the next 20 years will likely depend much on the development of novel mechanical and engineering technologies. Human organs-on-a-chip for novel drug screening [94], shear force-activated cleaning of thrombosis [95], bioinspired surface coating to prevent thrombosis and biofouling [96], mechanically-tuned hydrogels for bone formation [97], and tumor cell membrane-derived therapeutic microparticles for reversing cancer drug resistance [98], are a few examples of applications of mechanobiology-based technologies in medicine. Mechanobiology-based medicine (Mechanobiomedicine or Mechanomedicine) is poised to emerge as an exciting branch of medicine that uses mechanics- and engineering-based technologies for better diagnostics and more effective therapeutics of diseases than currently commercially available tools can provide.
Outlook of mechanotransduction
Despite significant progress in understanding the cellular and molecular mechanisms of mechanotransduction (Table 1), the current research is limited by the scope and the tools of study. For example, we know very little why various cell types have vastly different sensitivities to ECM stiffness and forces. We also do not know the underlying mechanisms and the significance of why a particular cell type express different integrin types and why several cell types express similar integrin subtypes. While many research groups have focused on the molecular details of FA components and their force sensing dynamics, it is rather challenging to visualize and to quantify two or more molecules at the same time to understand the intricacies of physical interactions among these molecules at FAs and in the depth of the cytoplasm. Combining super resolution fluorescence microscopy such as STED (stimulated emission depletion) and PALM (photo-activated localization microscopy)/STORM (stochastic optical reconstruction microscopy) [99] or the ultra-super resolution fluorescence nanoscopy (called MINFLUX, a method that combines STED with PALM/STORM to resolve 1-nm precision and spatial resolution of 6 nm) [100] with biophysical probes [101, 102] may help advance the understanding in molecular mechanisms of biophysical interactions among molecules in a living cell. We need to know better how transcription or gene expression in the nucleus is controlled and regulated epigenetically by mechanical forces and by biochemical signals. More importantly, we need to understand how a living cell or a groups of interacting cells, whether normal or abnormal (e.g., a malignant tumor cell), integrate all the stimuli (physical, chemical, and electrical) to respond biologically in a coherent manner in living tissues.
Table 1.
Force-induced changes of molecules or protein aggregates
| Molecule | Probe | Detection | Cell type | Time | Distance | Reference |
|---|---|---|---|---|---|---|
| RGD-Integrin | MTC | CSK disrupt stiffening | endothelial cells | ~1 min | <1 μm | 12 |
| FN7-10-Integrin | OT | stiffening | fibroblasts | 10–20 sec | <2 μm | 13 |
| Col-1-Integrin | Flexible PA gel | FA dynamics | fibroblasts | hrs | ~ μm | 14 |
| Integrin/FA | Flexible PDMS | PDMS strain | fibroblasts | hrs | <1 μm | 15 |
| FN-integrin | Strain device | FRET | fibroblasts | hrs | <1 μm | 17 |
| Src | OT | FRET | HUVECs | 1 min | <1 μm | 48 |
| p130Cas | Biaxial strain | IP | 293 cells | 1 min | ~1–5 μm | 49 |
| Src | MTC | FRET | HASM cells | 100–300 ms | >30 μm | 53 |
| Rac1 | MTC | FRET | HASM cells | 300 ms | >30 μm | 54 |
| Talin | MT/AFM | TIRF | Recomb. talin | a few sec | <1 μm | 44 |
| Vinculin | Tension probe | FRET | BAECs | ~100 sec | <1 μm | 45 |
| Coilin-SMN | MTC | FRET | Hela cells | 300 ms | >20 μm | 68 |
| Chromatin | 3DMTC | GFP spots strain | CHO cells | <100 ms | <5 μm | 73 |
| DHFR transcripts | 3DMTC | FISH | CHO cells | <15 sec | <5 μm | 73 |
Abbreviations. Distance, the distance between the force (strain) probe and the molecule(s) of interest; Time, the time between measurements and initiation of force application; MTC, magnetic twisting cytometry; RGD, Arg-Gly-Asp containing peptides; CSK disrupt., disruption of cytoskeleton with specific drugs; FN, fibronectin; FN7-10, a fragment of fibronectin type III, domain 7–10 that contains RGD peptides; OT, optical tweezer; Col-1, type 1 collagen; PA, polyacrylamide; FA, focal adhesion; PDMS, polydimethylsiloxane; FRET, fluorescence resonance energy transfer; HUVECs, human umbilical vein cells; IP, immunoprecipitation; 293 cells, human embryonic 293 cells; HASM cells, human airway smooth muscle cells; MT, magnetic tweezer; AFM, atomic force microscopy; TIRF, total internal reflection fluorescent microscopy; Recomb. talin, recombinant talin rod; BAECs, bovine arterial endothelial cells; SMN, survival of motor neuron protein; Hela cells, human cervical cancer cells; 3DMTC, three-dimensional MTC; CHO cells, Chinese hamster ovary cells; GFP spots, inserted multiple green fluorescent protein spots on the same chromatin; DHFR, dihydrofolate reductase; FISH, fluorescence in situ hybridization.
Acknowledgments
The author thanks collaborators, former and current lab members for help and discussion. This work is supported by NIH grant GM 072744 and Ministry of Science and Technology of China grant 2016YFA0101100.
References
- 1.Mofrad MR, Kamm R. Cellular Mechanotransduction: Diverse Perspectives From Molecules to Tissues. Cambridge University Press; 2014. [Google Scholar]
- 2.Li F, Redick SD, Erickson HP, Moy VT. Biophys J. 2003;84:1252–1562. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamkun JW, DeSimone DW, Fonda D, Patel RS, Buck C, Horwitz AF, Hynes RO. Cell. 1986;46:271–82. doi: 10.1016/0092-8674(86)90744-0. [DOI] [PubMed] [Google Scholar]
- 4.Humphries JD, Byron A, Humphries MJ. J Cell Sci. 2006;119:3901–3. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kishimoto TK, O’Connor K, Lee A, Roberts TM, Springer TM. Cell. 1987;48:681–90. doi: 10.1016/0092-8674(87)90246-7. [DOI] [PubMed] [Google Scholar]
- 6.Takagi J, Petre BM, Walz T, Springer TA. Cell. 2002;110:599–11. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 7.Campbell ID, Humphries MJ. Cold Spring Harb Perspect Biol. 2011;3(3) doi: 10.1101/cshperspect.a004994. pii: a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong JP, Stehle T, Goodman SL, Arnaout MA. Blood. 2003;102:1155–9. doi: 10.1182/blood-2003-01-0334. [DOI] [PubMed] [Google Scholar]
- 9.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 10.Ingber DE, Prusty D, Frangioni JV, Cragoe EJ, Jr, Lechene C, Schwartz MA. J Cell Biol. 1990;110:1803–11. doi: 10.1083/jcb.110.5.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz MA, Lechene C, Ingber DE. Proc Natl Acad Sci U S A. 1991;88:7849–53. doi: 10.1073/pnas.88.17.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang N, Butler JP, Ingber DE. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 13.Choquet D, Felsenfeld DP, Sheetz MP. Cell. 1997;88:39–48. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- 14.Pelham RJ, Wang YL. Proc Natl Acad Sci U S A. 1997;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balaban NQ, et al. Nat Cell Biol. 2001;3:466–72. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 16.Mehrbod M, Trisno S, Mofrad MR. Biophys J. 2013;105:1304–15. doi: 10.1016/j.bpj.2013.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubow KE, Vukmirovic R, Zhe L, Klotzsch E, Smith ML, Gourdon D, Luna S, Vogel V. Nat Commun. 2015;6:8026. doi: 10.1038/ncomms9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphrey JD, Dufresne ER, Schwartz MA. Nat Rev Mol Cell Biol. 2014;15:802–12. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Z, Guo SS, Fässler R. J Cell Biol. 2016;215:445–456. doi: 10.1083/jcb.201609037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong F, García AJ, Mould AP, Humphries MJ, Zhu C. J Cell Biol. 2009;185:1275–84. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Lee H, Zhu C. Exp Cell Res. 2016;349:85–94. doi: 10.1016/j.yexcr.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.le Duc Q, et al. J Cell Biol. 2010;189:1107–15. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, Engelhardt B, Cao G, DeLisser H, Schwartz MA. Nature. 2005;437:426–31. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 25.Makino A, Prossnitz ER, Bünemann M, Wang JM, Yao W, Schmid-Schönbein GW. Am J Physiol Cell Physiol. 2006;290:C1633–9. doi: 10.1152/ajpcell.00576.2005. [DOI] [PubMed] [Google Scholar]
- 26.Bhullar IS, Li YS, Miao H, Zandi E, Kim M, Shyy JY, Chien S. J Biol Chem. 1998;273:30544–9. doi: 10.1074/jbc.273.46.30544. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Miao H, Li S, Chen KD, Li YS, Yuan S, Shyy JY, Chien S. Am J Physiol Cell Physiol. 2002;283:C1540–7. doi: 10.1152/ajpcell.00222.2002. [DOI] [PubMed] [Google Scholar]
- 28.Arnadóttir J, Chalfie M. Annu Rev Biophys. 2010;39:111–37. doi: 10.1146/annurev.biophys.37.032807.125836. [DOI] [PubMed] [Google Scholar]
- 29.Thoumine O, Kocian P, Kottelat A, Meister JJ. Eur Biophys J. 2000;29:398–408. doi: 10.1007/s002490000087. [DOI] [PubMed] [Google Scholar]
- 30.Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Nature. 2003;424:334–337. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 31.Li F, Redick SD, Erickson HP, Moy VT. Biophys J. 2003;84:1252–1562. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Ha T. Science. 2013;340:991–4. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chowdhury F, et al. Integr Biol (Camb) 2015;7:1265–71. doi: 10.1039/c5ib00080g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Sun J, Xu Q, Chowdhury F, Roein-Peikar M, Wang Y, Ha T. Biophys J. 2015;109:2259–67. doi: 10.1016/j.bpj.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingber DE, Madri JA, Jamieson JD. Proc Natl Acad Sci U S A. 1981;78:3901–5. doi: 10.1073/pnas.78.6.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bissell MJ, Hall HG, Parry G. J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 37.Ingber DE, Wang N, Stamenovic D. Rep Prog Phys. 2014;77:046603. doi: 10.1088/0034-4885/77/4/046603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pourati J, Maniotis A, Spiegel D, Schaffer JL, Butler JP, Fredberg JJ, Ingber DE, Stamenovic D, Wang N. Am J Physiol Cell Physiol. 1998;274:C1283–9. doi: 10.1152/ajpcell.1998.274.5.C1283. [DOI] [PubMed] [Google Scholar]
- 39.Wang N, Naruse K, Stamenović D, Fredberg JJ, Mijailovich SM, Tolić-Nørrelykke IM, Polte T, Mannix R, Ingber DE. Proc Natl Acad Sci U S A. 2001;98:7765–70. doi: 10.1073/pnas.141199598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamenović D, Mijailovich SM, Tolić-Nørrelykke IM, Chen J, Wang D. Am J Physiol Cell Physiol. 2002;282:C617–24. doi: 10.1152/ajpcell.00271.2001. [DOI] [PubMed] [Google Scholar]
- 41.Wang N, Tolić-Nørrelykke IM, Chen J, Mijailovich SM, Butler JP, Fredberg JJ, Stamenović D. Am J Physiol Cell Physiol. 2002;282:C606–16. doi: 10.1152/ajpcell.00269.2001. [DOI] [PubMed] [Google Scholar]
- 42.Lee SE, Kamm RD, Mofrad MR. J Biomech. 2007;4:2096–106. doi: 10.1016/j.jbiomech.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Golji J, Mofrad MR. Biophys J. 2010;99:1073–81. doi: 10.1016/j.bpj.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Science. 2009;323:638–41. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grashoff C, et al. Nature. 2010;466:263–6. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar A, Ouyang M, Van den Dries K, McGhee EJ, Tanaka K, Anderson MD, Groisman A, Goult BT, Anderson KI, Schwartz MA. J Cell Biol. 2016;213:371–83. doi: 10.1083/jcb.201510012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Z, et al. Nat Cell Biol. 2016;18:941–53. doi: 10.1038/ncb3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Nature. 2005;434:1040–5. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 49.Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Cell. 2006;127:1015–26. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seong J, et al. Proc Natl Acad Sci U S A. 2013;110:19372–7. doi: 10.1073/pnas.1307405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu S, Chen J, Fabry B, Numaguchi Y, Gouldstone A, Ingber DE, Fredberg JJ, Butler JP, Wang N. Am J Physiol Cell Physiol. 2003;285:C1082–C1090. doi: 10.1152/ajpcell.00159.2003. [DOI] [PubMed] [Google Scholar]
- 52.Hu S, Eberhard L, Chen J, Love JC, Butler JP, Fredberg JJ, Whitesides GM, Wang N. Am J Physiol Cell Physiol. 2004;287:C1184–C1191. doi: 10.1152/ajpcell.00224.2004. [DOI] [PubMed] [Google Scholar]
- 53.Na S, Collin O, Chowdhury F, Tay B, Ouyang M, Wang Y, Wang N. Proc Natl Acad Sci USA. 2008;105:6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poh YC, Na S, Chowdhury F, Ouyang M, Wang Y, Wang N. PLoS One. 2009;4:e7886. doi: 10.1371/journal.pone.0007886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang N, Suo Z. Biochem Biophys Res Commun. 2005;328:1133–1138. doi: 10.1016/j.bbrc.2005.01.070. [DOI] [PubMed] [Google Scholar]
- 56.del Alamo JC, Norwich GN, Li YS, Lasheras JC, Chien S. Proc Natl Acad Sci U S A. 2008;105:15411–6. doi: 10.1073/pnas.0804573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abhilash AS, Baker BM, Trappmann B, Chen CS, Shenoy VB. Biophys J. 2014;107:1829–40. doi: 10.1016/j.bpj.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, Abhilash AS, Chen CS, Wells RG, Shenoy VB. Biophys J. 2014;107:2592–603. doi: 10.1016/j.bpj.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sunyer R, et al. Science. 2016;353:1157–61. doi: 10.1126/science.aaf7119. [DOI] [PubMed] [Google Scholar]
- 60.Fabry B, Maksym GN, Butler JP, Glogauer M, Navajas D, Fredberg JJ. Phys Rev Lett. 2001;87:148102. doi: 10.1103/PhysRevLett.87.148102. [DOI] [PubMed] [Google Scholar]
- 61.Chowdhury F, Na S, Collin O, Tay B, Li F, Tanaka T, Leckband DE, Wang N. Biophys J. 2008;95:5719–27. doi: 10.1529/biophysj.108.139832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonakdar N, Gerum R, Kuhn M, Spörrer M, Lippert A, Schneider W, Aifantis KE, Fabry B. Nat Mater. 2016;15:1090–4. doi: 10.1038/nmat4689. [DOI] [PubMed] [Google Scholar]
- 63.Heo SJ, Driscoll TP, Thorpe SD, Nerurkar L, Baker BM, Yang MT, Chen CS, Lee DA, Mauck RL. Elife. 2016;5 doi: 10.7554/eLife.18207. pii: e18207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dekker J, Misteli T. Cold Spring Harb Perspect Biol. 2015;7:a019356. doi: 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang N, Tytell JD, Ingber DE. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 66.Maniotis AJ, Chen CS, Ingber DE. Proc Natl Acad Sci USA. 1997;94:849–54. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu S, Chen J, Butler JP, Wang N. Biochem Biophys Res Commun. 2005;329:423–428. doi: 10.1016/j.bbrc.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 68.Poh YC, Shevtsov SP, Chowdhury F, Wu DC, Na S, Dundr M, Wang N. Nat Commun. 2012;3:866. doi: 10.1038/ncomms1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Proc Natl Acad Sci U S A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swift J, Harada T, Buxboim A, Shin JW, Tang HY, Speicher DW, Discher DE. Science. 2013;341:1240104. [Google Scholar]
- 71.Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Nature. 2013;497:507–11. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS. J Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N. Nat Mater. 2016;15:1287–1296. doi: 10.1038/nmat4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dupont S, et al. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 75.Wei SC, et al. Nat Cell Biol. 2015;17:678–88. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jia Q, et al. Oncogenesis. 2016;5:e220. doi: 10.1038/oncsis.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan Y, et al. Nat Commun. 2014;5:4619. doi: 10.1038/ncomms5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.HQ Le, et al. Nat Cell Biol. 2016;18:864–75. doi: 10.1038/ncb3387. [DOI] [PubMed] [Google Scholar]
- 79.Jahed Z, Soheilypour M, Peyro M, Mofrad MR. J Cell Sci. 2016;129:3219–29. doi: 10.1242/jcs.184184. [DOI] [PubMed] [Google Scholar]
- 80.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1673–7. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 82.Chowdhury F, Na S, Li D, Poh YC, Tanaka TS, Wang F, Wang N. Nat Mater. 2010;9:82–8. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang C, Tibbitt MW, Basta L, Anseth KS. Nat Mater. 2014;13:645–52. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poh YC, et al. Nat Commun. 2014;5:4000. doi: 10.1038/ncomms5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valastyan S, Weinberg RA. Cell. 2011;147:275–92. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 87.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Nature. 2008;456:593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quintana E, Shackleton M, Foster HR, Fullen DR, Sabel MS, Johnson TM, Morrison SJ. Cancer Cell. 2010;18:510–23. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paszek MJ, et al. Cancer Cell. 2005;8:241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 90.Jain RK. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 91.Plodinec M, et al. Nat Nanotechnol. 2012;7:757–65. doi: 10.1038/nnano.2012.167. [DOI] [PubMed] [Google Scholar]
- 92.Liu J, Tan Y, Zhang H, Zhang Y, Xu P, Chen J, Poh YC, Tang K, Wang N, Huang B. Nat Mater. 2012;11:734–41. doi: 10.1038/nmat3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen J, et al. Sci Rep. 2016;6:19304. doi: 10.1038/srep19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science. 2010;328:1662–8. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Korin N, et al. Science. 2012;337:738–42. doi: 10.1126/science.1217815. [DOI] [PubMed] [Google Scholar]
- 96.Leslie DC, et al. Nat Biotechnol. 2014;32:1134–40. doi: 10.1038/nbt.3020. [DOI] [PubMed] [Google Scholar]
- 97.Huebsch N, et al. Nat Mater. 2015;14:1269–77. doi: 10.1038/nmat4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma J, et al. Cell Res. 2016;26:713–27. doi: 10.1038/cr.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hell SW, et al. J Phys D: Appl Phys. 2015;48:44301. [Google Scholar]
- 100.Balzarotti F, Eilers Y, Gwosch KC, Gynnå AH, Westphal V, Stefani FD, Elf J, Hell SW. Science. 2017;355:606–612. doi: 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]
- 101.Heller I, et al. Nat Methods. 2013;10:910–916. doi: 10.1038/nmeth.2599. [DOI] [PubMed] [Google Scholar]
- 102.Zhang Y, et al. Nat Proc 2017 [Google Scholar]


