Abstract
KcsA, the bacterial K+ channel from Streptomyces lividans, is the prototypical model system to study the functional and structural correlations of the pore domain of eukaryotic voltage-gated K+ channels (Kv channels). It contains all the molecular elements responsible for ion conduction, activation, deactivation and inactivation gating[1]. KcsA’s structural simplicity makes it highly amenable for structural studies. Therefore, it is methodological advantageous to produce large amount of functional and properly folded KcsA in a cost-effective manner. In the present study, we show an optimized protocol for the over-expression and purification of large amount of high-quality, fully functional and crystallizable KcsA using inexpensive detergents, which significantly lowered the cost of the purification process.
Keywords: ion channels, KcsA, biochemistry, over-expression, detergents, solubilization
1. Introduction
Potassium channels (K+ channels) are specialized membrane proteins that form an aqueous pore through which potassium ions cross the cell membrane at near diffusion-limited rates (~107 ions per second)[2], regulating the potassium homeostasis within the cells. K+ channels control the membrane potential of excitable cells such as neurons and muscle cells by opening and closing structural gates that regulate the flow of K+ across the cell membrane. The opening of a K+ channel gate (a process known as gating) is triggered by different type of stimuli (i.e., ligand binding, changes in the membrane voltage or mechanical deformation of the cell membrane). These multimeric membrane proteins have been classified by the number of transmembrane helices (TM) contained within their α-subunit (the pore forming subunit): among them the most common are the 2-transmembrane segments (2-TM) and the 6-TM subunits. Generally, four α-subunits assemble as a tetramer with four-fold symmetry around a central aqueous pore. Most of the cloned K+ channels contain a critical and highly-conserved amino-acid sequence known as the signature sequence, the TTVGYGD structural motif or selectivity filter[3]. The archetypal K+ channel from Streptomyces lividans, KcsA, belongs to the 2-TM group and displays high sequence identity with neuronal voltage-gated K+ channels. Despite its simplicity, KcsA is a pH-gated[4], highly selective potassium channel[5] that contains all the necessary elements to undergo activation, deactivation and inactivation gating[6–9]. The aforementioned features make KcsA the perfect functional and structural surrogate to study the pore domain of the more complex, poorly expressed and more difficult to crystallize eukaryotic voltage-gated K+ channels. To conduct structural-functional studies in any K+ channel, it is ideal to produce it in milligram quantities, since most of the mainstream techniques to determine protein structure require large amount of protein. In this study, we present an optimized protocol for the over-expression and purification of large amount of high-quality, fully functional and crystallizable KcsA. We have applied a systematic approach to optimize the production pipeline for KcsA by screening several E. coli strains for heterologous expression, growth media and inexpensive detergents for solubilization, which drastically lowered the production cost and yields around ~10 times more protein than any other published protocol.
2. Materials and methods
2.1 Materials
E. coli strains were purchased from Agilent Santa Clara, CA (XL-10 gold) and Lucigen Madison, WI (C41 and C43). Dodecyl β-D-maltoside (DDM), Triton X-100 and Anzergent 3–12, were obtained from Anatrace (Maumee, Ohio). Mouse anti-penta-Histidine antibody was from Qiagen (Chatsworth, CA). Alexa Fluor 680 conjugated goat anti-mouse secondary antibody, was obtained from Thermo Fisher Scientific (Valencia, CA). Pre-stained molecular weight markers were obtained from Bio-Rad (Richmond, CA). FAB antibody (for X-ray crystallography) was generously provided by Dr. Roderick Mackinnon. PD-10 desalting columns were acquired from GE Healthcare (Pittsburgh, PA). Soy Extract Polar lipids were purchased from Avanti Polar Lipids (Alabaster, AL). Spin-label (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl methanethiosulfonate) was ordered from Toronto Research Chemicals (Toronto, Canada). All other reagents were from Sigma, Difco or Fisher Scientific.
2.2 Expression of (His)6-KcsA in different E. coli strains
KcsA-pQE70 was transformed into different E. coli strains, freshly prepared competent cells of XL10-Gold (our control cells for protein expression), C41 and C43 (it has been documented these strains over-express membrane proteins), by the heat shock method and grown overnight at 37°C in the presence of 1% glucose and 0.4 mg/ml ampicillin. Next day, 1% of the overnight culture of each strain was used to start a 30 ml expression test culture using Luria-Bertani (LB) broth medium supplemented with 0.5% glycerol (as a chemical chaperone)[10], 0.2 % glucose and 0.4 mg/ml ampicillin at 37°C or CRAP based medium[11] ((NH4)2SO4 3.57 g/l, Na Citrate-2H2O 0.71g/l, KCl 1.07 g/l, Yeast extract 5.36g/l, HyCase 5.36 gr/l pH 7.3 and autoclaved; next day, 100 mM MOPS, 10.5 mM MgSO4 was added) supplemented with 0.5% glycerol (we have found that the addition of glycerol improves the expression and maximizes the final cell culture biomass) and 0.4 mg/ml ampicillin at 37°C. Once cells reached an optical density at 600 nm (O.D) of 0.6 they were cooled down to 29°C for 1 hour. Protein expression was started by the addition of 0.1 mM isopropyl thiogalactoside (IPTG), 10 mM BaCl2 (a well-known K+ channel blocker) and 0.4 mg/ml ampicillin. Cells were incubated overnight at constant agitation (250 rpm) at 29°C and harvested next day (since we have previously established that KcsA expression levels reach a plateau around ~ 18 hours) to quantify KcsA expression levels.
2.3 Assessing the expression of (His)6-KcsA in the presence of known K+ channel blockers
KcsA-pQE70 was transformed into the C41 E. coli strain (the best E. coli strain for KcsA overexpression identified in this work) by the heat shock method and grown overnight at 37°C in the presence of 1% glucose and 0.4 mg/ml ampicillin. Next day, 1% of the overnight culture was used to start a 30-ml culture in LB medium supplemented with 0.2 % glucose, 0.5 % glycerol and 0.4 mg/ml ampicillin, at 37°C. Once the cells reached an O.D of 0.6, the culture was cooled down to 29°C for 1 hour. KcsA expression was started by the addition of 0.1 mM IPTG, 10 mM of a subset of known potassium channel blockers (BaCl2, CsCl2, Tetraethyl ammonium, or Tetrabutyl ammonium Cl) and 0.4 mg/ml ampicillin. Then each culture was incubated overnight at constant agitation (250 rpm) at 29 °C. Next day, the cells were harvested to quantify KcsA expression levels.
2.4 Detergent-solubilization tests of (His)6-KcsA
Small scale KcsA solubilization tests were performed using different types of detergents[12]. A KcsA-containing E. coli membrane preparation was extracted with one of several detergents of interest, at the specified detergent concentration in mM or times the CMC (critical micellar concentration): 30 mM (~17 xCMC) Decyl Maltoside (DM), 20 mM (~118 xCMC) Dodecyl Maltoside (DDM), 1.5% (~250 xCMC) Thesit, 1.5% (~16 xCMC) Anzergent 3–12, 1.5% (~4 xCMC) Sodium Dodecanoyl Sarcosine or 1.5% (~93 xCMC) Triton X-100) in Buffer-A (Buffer-A: 50 mM Tris-HCl pH 8.0, 150 mM KCl, and protease inhibitors) for 1 h at room temperature. The insoluble material was spun down at 100,000 g and the supernatant was electrophoresed on a 15% acrylamide protein gel and electro-transferred to a polyvinylidene difluoride (PVDF) membrane according to the manufacturer’s instructions. Membranes were blocked for 1 h at room temperature in 5% non-fat milk in PBS, pH 7.4, and then incubated overnight in a mouse anti-pentahistidine antibody (1:3000 dilution) at 4 °C. After three washes with phosphate buffered saline solution (PBS) containing 0.05% Tween-20 (TPBS), the membranes were incubated with Alexa fluor-conjugated goat anti-mouse antibody (1:5000 dilution) for 1 h and after extensive washes with TPBS KcsA bands were visualized in a Licor Odyssey CLx.
2.5 Salt concentration effect on the detergent mediated solubilization of (His)6-KcsA
Small scale solubilization tests were performed to assess the effect of the salt concentration on KcsA solubilization. The inexpensive Triton X-100 was used to extract KcsA by titrating KCl or NaCl concentration. The solubilization was performed in Buffer A with increasing salt concentrations (150, 300, 600, 1200 mM KCl or NaCl) for 1 h at room temperature. The insoluble material was spun down at 100,000 g and the supernatant was electrophoresed and immunoblotted as described above.
2.6 (His)6-KcsA expression in optimized conditions and membrane preparation
KcsA-pQE70 was transformed in C41 E. coli strain by the heat shock method and grown overnight at 37°C. Next day, 1% of the overnight culture was used to inoculate 1 liter of CRAP medium (phosphate limiting media) containing 0.2 % glucose, 0.5 % glycerol and 0.4 mg/ml ampicillin at 37°C. Once the cells reached an O.D. of 0.6, they were cooled down to 29°C for 1 hour. Afterward 10 mM BaCl2, 0.4 mg/ml ampicillin and 0.1 mM IPTG were added to induce the overnight expression of KcsA. Next day, cells were harvested and resuspended in the following buffer: 50 mM Tris-HCl, 150 mM KCl, 170 ug/ml phenylmethylsulfonyl fluoride and 1 mg/mL of egg lysozyme and incubated at room temperature for 1 h under constant rotation. The mixture was homogenized on an Emulsiflex C3 and spun down at 100,000g for 1 h and the supernatant was discarded. The membrane pellet was resuspended with Buffer A containing protease inhibitors and aliquots were stored at −80 °C.
2.7 Large scale purification of (His)6-KcsA in new and optimized conditions
A KcsA containing E. coli membrane preparation was extracted with the detergent of interest (20 mM DDM, 1.5% Anzergent 3–12 or 1.5 % Triton X-100) in Buffer-A supplemented with 1M KCl and protease inhibitors for 1h at room temperature. The insoluble material was spun down at 100,000 g and the supernatant was loaded into a cobalt resin column. Once the KcsA channel was bound to the cobalt resin, the column was washed with 10 column volumes (CV) of Buffer-A, 10 mM imidazole and 1 mM DDM (this step cleans the sample form impurity and exchange the detergent of the bound-KcsA to 1 mM DDM, irrespective of the detergent used in the solubilization step). Finally, the protein was eluted in Buffer-A, 1 mM DDM and 400 mM imidazole. The analysis of properly-folded channels, i.e., oligomeric state, was determined by size-exclusion chromatography on an ENrich SEC 650 10 × 300 column (Bio-Rad) equilibrated with Buffer-A, 1mM DDM.
2.8 Functional evaluation of KcsA purified with optimized conditions
KcsA containing liposomes were used for electrophysiological studies according to the method of Delcour et al.[13] and Cortes et al[14], with some modifications. In brief, KcsA was reconstituted at a protein-to-lipid weight/weight ratio of 1:100 and incubated overnight with bio-beads. Next day, the proteoliposome suspension was centrifuged at 100,000 g for 1 h, and the pellet was resuspended in 60 ul of rehydration buffer (5 mM MOPS, 200 mM KCl, pH 7.0). Usually, three drops of the proteoliposome suspension were dried overnight in a desiccation chamber under vacuum for around 24 h. Next day, 20 μl of rehydration buffer were applied to each dried drop. The samples were rehydrated for 5 to 24 hours, producing giant liposomes suitable for patch clamp. All patch-clamp measurements were done in symmetrical conditions: 200 mM KCl and 5 mM MOPS-buffer at pH 4.0, at room temperature. Single-channel currents were recorded with a patch-clamp amplifier, and currents were sampled at 40 kHz with analogue filter set to 10 kHz. Pipette resistances were 2.0 MΩ after fire polishing (filled with 200 mM KCl and 5 mM MOPS-buffer at pH 4.0).
2.9 KcsA activation gating assessment by CW-EPRs
To test whether extraction with Anzergent 3–12 or Triton affected the gating properties of KcsA, we tracked the structural changes of the KcsA activation gate by using Continuous Wave Electron Paramagnetic Resonance Spectroscopy (CW-EPRs)[9]. KcsA G116C mutant (an excellent reporter for KcsA’s activation gating) was purified in the aforementioned detergents, concentrated to 10 mg/ml and reduced by adding 5 mM tris(2-carboxyethyl) phosphine (TCEP). A PD-10 desalting column, pre-equilibrated with a degassed buffer (50 mM Tris-HCl, 150 mM KCl at pH 7) + 1 mM DDM, was used to eliminate the excess of the reducing agent. Reduced KcsA-G116C mutant was incubated with a 10-fold molar excess of the thiol-specific spin-label (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl methanethiosulfonate) overnight at 4°C. The unreacted spin labels were removed using a desalting PD-10 column and labeled channels were reconstituted into preformed Asolectin liposomes at a 1:1600 (KcsA tetramer:lipid) molar ratio. Finally, liposomes were harvested by centrifugation at 100,000 g for CW-EPRs analysis.
2.10 X-ray Crystallography of Wild Type KcsA solubilized with Triton
We crystallized the KcsA channel extracted with Triton X-100 in the presence of antibody Fab fragment by the sitting-drop method as described previously[15]. After mixing C-terminal truncated KcsA with an antibody fragment (FAB) in a protein-to-Fab weight/weight ratio of 1:3, the KcsA-Fab complex was further purified by size exclusion chromatography in the crystallization buffer (150 mM KCl, 50 mM Tris-HCl and 5 mM decyl maltoside). Crystals of the KcsA–Fab complex were obtained in the following experimental conditions: 20–25% PEG400 (v/v), 50 mM magnesium acetate, 50 mM sodium acetate (pH 4.8–5.4) at 19 °C. Next, the PEG concentration in the reservoir was gradually augmented until it reached 40%, which produced better diffracting crystals. Crystals diffracted X-rays to 2.3 Å Bragg spacing at the synchrotron and the structure of the channel was solved by molecular replacement. Data were collected at the Stanford Synchrotron Radiation Laboratory Beamline (SSRL) 14-1 and processed with the HKL2000 package.
3. Results
3.1 Strategies to enhance KcsA protein expression levels
Our strategy to maximize the production of recombinant KcsA by E. coli consisted in improving the following critical steps: 1) to screen KcsA expression levels in different E. coli strains, 2) to screen different expresion media and additives to maximize protein expression, 3) to identify K+ channels blockers that could mitigate any cytotoxic effect caused by KcsA overexpression levels in preparations with the growth media and E. coli strain identified in steps 1 and 2 and 4) to search for affordable alternative detergents that could extract larger amounts of pure, fully functional and properly folded KcsA in a cost-effective manner.
Toward this end, we tested: a) the effect on KcsA expression level of 3 different E. coli strains (XL10-Gold, C41 and C43 cells), b) the presence of a chemical chaperone (glycerol) and c) the use of 2 different growth media (LB and CRAP media) (Fig. 1). 1 out of the 9 tested conditions seemed to significantly increase KcsA expression levels. The combination of C41, CRAP media and 0.5 % glycerol produced the largest amount of KcsA per cell (Fig. 1) in addition to yielding the largest biomass, which is highly convenient when very large amounts of recombinant protein are needed. Next, we decided to further optimize KcsA expression levels using the experimental conditions identified earlier (i.e., C41 cells, CRAP media and 0.5 % glycerol). Hence, we evaluated the effect of different known K+ channel blockers (i.e., BaCl2, CsCl2, TEA and TBA) on KcsA expression. Our experimental results showed that irrespective of the K+ channel blocker tested, KcsA’s expression levels were larger in the presence of a blocker (data not shown), which led us to use for all our following experiments the conventional and inexpensive blocker Barium Chloride.
Figure 1. Western blot analysis of KcsA expression levels.
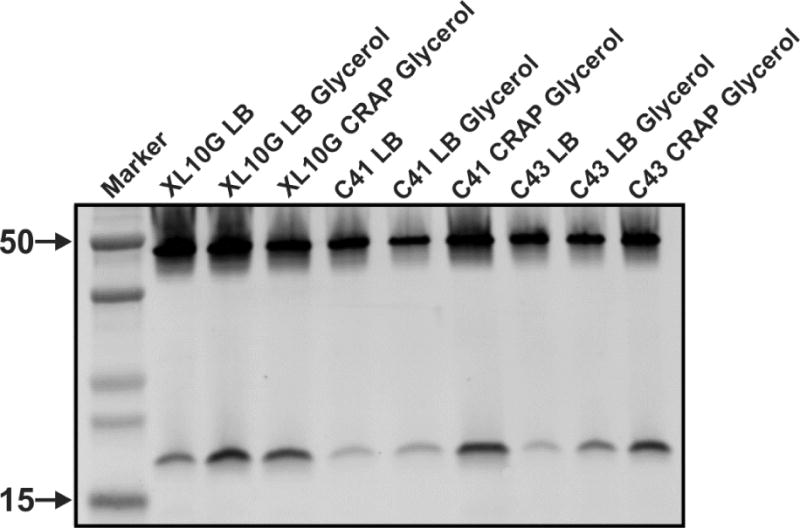
A Western blot (mouse anti-His5 and Alexa Fluor 680 conjugated goat anti-mouse) shows KcsA migrating as a 50 kDa oligomer (tetramer) and as a monomer at around 19 kDa (the predicted KcsA molecular weight is 19,725 Da). The different lanes in the Western blot show the normalized expression of KcsA in different E. Coli strains (XL10Gold, C41 or C43), in two types of media (LB or CRAP) with or without 0.5% glycerol.
3.2 Optimizing the detergent-mediated extraction of KcsA from E. coli membranes
To optimize the amount of KcsA extracted from E. coli membranes we reasoned that the following two aspects should be considered: 1) find a detergent and/or additives that can maximize the extraction of large amounts of properly folded and functional KcsA and 2) if possible, the detergent should be an inexpensive one, since most structural studies require large amounts of protein and that imposes a serious financial burden. Toward this end, we tested 5 different detergents to solubilize a KcsA containing membrane preparation (Fig 2A). To assess quantitatively the amount of KcsA extracted by each detergent type, the relative integrated intensities of the extracted KcsA bands were measured on a Western blot membrane by measuring the fluorescence signal with a Licor Odyssey CLx (Fig 2D).
Figure 2. Detergent screening for KcsA solubilization.
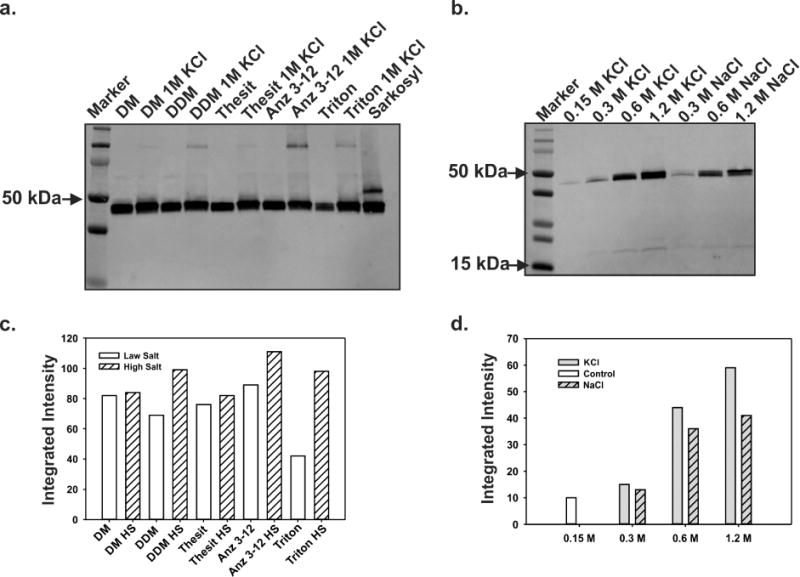
A Western blot analysis shows tetrameric KcsA running as a 50 kDa molecular weight complex. A. The different lanes are showing KcsA extracted from E. coli membranes using various detergents. B. Western blot showing the extraction of KcsA using Triton X-100 while increasing either NaCl or KCl concentration. C. Quantification by densitometric analysis of the effect of increasing salt concentration on KcsA solubilization per detergent type (Fig. 2A). D. Densitometric analysis to quantify the effect on KcsA extraction from E. coli membrane by Triton X-100 while increasing the NaCl or KCl concentration (Fig 2B).
An initial solubilization screen of KcsA containing membranes was performed with five detergents at physiological KCl concentration (Decyl Maltoside, Dodecyl Maltoside, Thesit, Anzergent 3–12, Triton X100 and Sarkosyl), at concentrations ranging from 10–30× the detergent critical micellar concentration (CMC). The amount of extracted KcsA in this experiment showed the following sequence: DDM>DM>Sarkosyl>Anzergent-3–12>Thesit>>>Triton-X100. (Fig 2A). Our experimental results showed that Anz 3–12, Sarkozyl and Thesit, were as effective extracting large amounts of KcsA in its native oligomeric state (a tetramer), as the more expensive detergents DM and DDM.
A known strategy that maximizes the solubilization and stabilization of membrane proteins is the addition of large amount of salts in the detergent-mediated solubilization step[16]. To further increase the amount of KcsA extracted from E. coli membranes, we decided to evaluate the effect of increasing salt concentration during the solubilization step. With this end in view, 5 of the detergents previously tested, DM, DDM, Anzergent 3–12, Triton X100 and Thesit were reevaluated in their KcsA extracting capabilities in the presence of 1M KCl. Since KcsA is a K+ channel the excess of K+ could potentially stabilize and preserve the channel native structure as well as its biological function (Sarkozyl was not tested with high salt concentration since it is an ionic detergent).
Interestingly, the amount of KcsA extracted by each detergent is significantly larger when compared to the control at physiological salt concentration (Fig 2B and 2D). While the expensive DDM and Anzergent 3–12 showed a clear increase of the amount of KcsA extracted in the presence of 1M KCl, Triton X100 displayed the largest increase in the amount of extracted KcsA (~ 300% increment). Since Triton X100 is a very affordable detergent, we decided to perform a titration of the salt concentration effect on the solubilization of KcsA (KCl or NaCl) to further optimize the solubilization strategy (Figure 2B). The relative integrated intensities of the extracted KcsA bands, on the Western blot shown in Fig. 2C, displayed a remarkably increase in the intensity concomitant to the increment in salt concentration, irrespective whether KCl or NaCl was used. However, the effect of high salt concentration on KcsA Triton-X100 mediated-solubilization was larger when using KCl (Fig. 2D). It is also important to indicate that in the presence of 1M KCl, Anzergent-312, although significantly more expensive than Triton X100, extracted the largest amount of KcsA in our solubilization tests (Fig. 2D).
3.3 A large scale preparation of KcsA using optimized conditions for expression and purification
The best conditions for KcsA expression (C41 + 0.5% glycerol + CRAP media) were used to made a large-scale expression and a membrane preparation according to the procedure described on the Material and Methods section. Based on our solubilization test results, we proceeded to perform a large scale purification by solubilizing a KcsA containing membrane preparation with DDM (control), Triton X-100 and Anzergent 3–12 in the presence 1M KCl. KcsA was purified by cobalt affinity chromatography, and its hydrodynamic properties were evaluated by size-exclusion chromatography (Figure 3A). All the detergent tested in this work preserved KcsA’s hydrodynamic behavior based on its elution volume from a size exclusion chromatography column. Remarkably, Sarcosyl and Anzergent 3–12, which are very harsh detergents, and the mild and inexpensive Triton X100 preserved the tetrameric conformation of the channel as well as the most commonly used but more expensive detergents DDM and DM did (Fig 3A and 3B).
Figure 3. KcsA purification using a new and improved protocol. A. Size-exclusion chromatogram on an ENrich SEC 650 10×300 column of KcsA purified by metal-chelate chromatography.
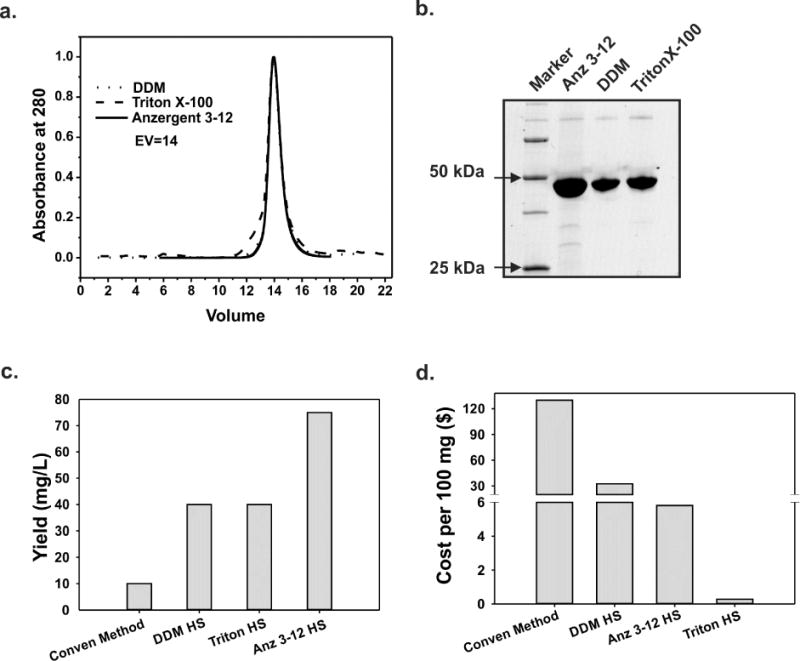
KcsA extracted by DDM (control, dotted line), Triton X-100 (dashed line) or Anzergent 3–12 (solid line) eluted as monodispersed peaks with elution volumes = 14 ml. B. SDS-PAGE analysis of the eluted fractions containing KcsA’s monodispersed peaks in A. C. KcsA yield in milligrams per liter of culture when extracted with different detergents. D. Cost of solubilizing 100 mg of KcsA per detergent type tested in this study.
When KcsA was solubilized with DDM or Triton X100, the final yield of pure, tetrameric and monodispersed channel was about 40mg/L, which is about 4 times larger than the yield obtained by using typical previous published methods for non-isotope enriched preparations[12, 17] (we attribute the yield increment to the combined effects of improved protein expression and the use of high salt concentration during solubilization). However, the solubilization of KcsA using Anzergent 3–12 resulted in the outstanding final yield of 75 mg/L of pure, functional and properly folded channel (Fig 3C). The use of Anzergent 3–12 doubled the yield of purified KcsA (i.e., mg/L of culture), compared to DDM or Triton X100 (Fig 3C). Nonetheless, the use of Triton X100 to purify KcsA represents the best compromise between yield and cost when compared to DDM and Anzergent 3–12. The cost of producing 100 mg of high quality KcsA using Triton X100, Anzergent 3–12 or DDM was $0.28, $5 and $30, respectively.
3.4 Functional evaluation of KcsA solubilized by Triton X100
An essential aspect when using any detergent to extract and purify a membrane protein is the preservation of its biological function. Since we found that for KcsA purification, Triton X-100 is an alternative cost-effective detergent to the conventional and very expensive DM and DDM, we carried out a functional evaluation of purified KcsA by patch clamp methodology, after extraction with Triton X-100 and reconstitution in giant liposomes. In Figure 4a we show representative KcsA single-channel recordings measured under symmetric K+ concentrations (200 mM, pH 4.0). The conduction properties of KcsA channels extracted with the conventional DDM or Triton X100 were undistinguishable, displaying chord conductances at 150 mV of 125 pS and 131 pS, respectively.
Figure 4. Functional analysis of KcsA extracted with Triton X-100 and 1M KCl.
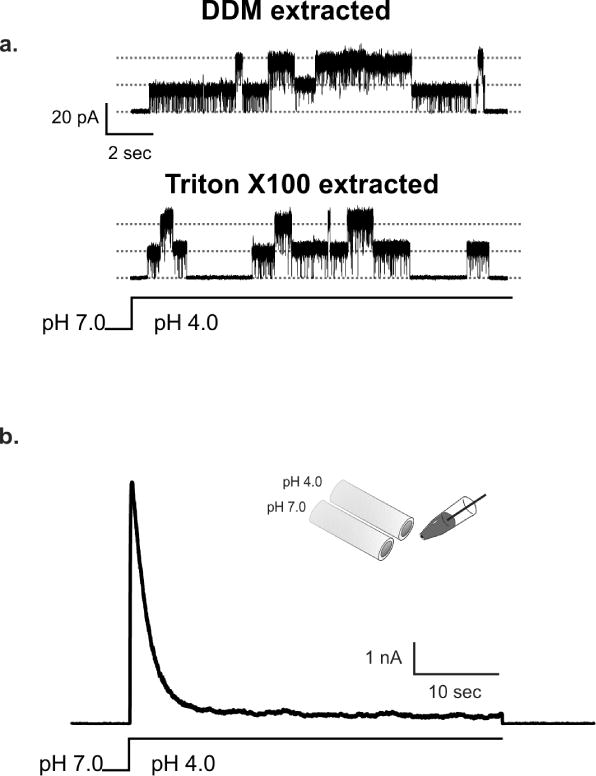
A. Top panel shows representative single-channel recordings of KcsA extracted by DDM (upper trace) or by Triton X-100 (lower trace). KcsA single-channel activity was recorded in symmetrical solutions containing 200 mM KCl, pH 4.0 and at 100 mV. B. Macroscopic current of KcsA extracted by Triton X-100 displayed intact pH-dependent activation and C-type inactivation gating, which are indistinguishable from those observed with channels purified by conventional methods. The inactivation rate constant for KcsA extracted Triton X-100 was 2.5±0.1 sec, which is very similar to the one reported for KcsA-extracted by conventional methods.
We also studied KcsA’s activation and C-type inactivation properties by measuring the kinetic behavior of macroscopic currents elicited by a rapid change in solution pH from 7.0 to 3.0. Satisfactorily, the activation and inactivation properties of KcsA extracted with Triton X-100 were fully preserved with our new purification scheme, as seen in Figure 4b. The channel at 100 mV and after a swift solution exchange to pH 3.0, reached rapidly full activation (T0.5 ~20 ms) and inactivated slowly (Tinact=~2.5 sec), which is the typical behavior of KcsA extracted and purified using the standard DDM [18, 19].
3.5 KcsA activation gating assessed by CW-EPRs
An essential behavior of a properly folded and functional KcsA biochemical preparation is the pH-dependent conformational change reported by a nitroxide group attached at position G116C[9]. KcsA-G116C mutant extracted with Triton X-100 was spin labeled and reconstituted in Asolectin liposomes. In Figure 5a we show an ensemble of CW-EPR spectra displaying the typical change in dipolar coupling displayed by the nitroxide group attached at position G116C. The KcsA activation gate is closed at pH 7.0 and spin labels attached at position G116C display a large dipolar coupling due to their close proximity in space. As the channel undergoes pH-dependent activation, those spin labels move away from each other and the G116C-SL dipolar coupling decreases as a function of the opening of KcsA’s activation gate, reaching maximum opening at pH 3.0. The KcsA-Triton activation pKa measured by this approach is 4.3 ± 0.1, which is the same measured in our control experiments using DDM.
Figure 5. Spectroscopic assessment of gating and structural evaluation by X-ray Crystallography of KcsA extracted by Triton X-100.
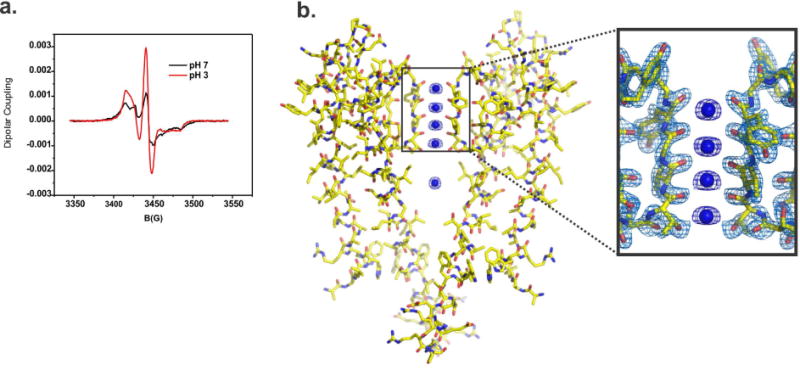
A. pH dependent structural changes at the activation gate monitored by CW-EPR spectroscopy of KcsA extracted by Triton X-100. Black spectrum shows typical dipolar coupling of spin labels at position 116 when the channel is closed at pH 7 (black line) or open at pH 4. B. On the left, X-ray crystal structure of KcsA extracted by Triton X-100 @ 2.3 Å resolution, the high-quality of the electron density map allowed us to build a complete KcsA model. On the right, a view of the structural model of KcsA’s selectivity filter, the 2Fo-Fc electron density map (contoured @ 2σ for the protein and 3σ for the K+) is surrounding amino acids that form KcsA’s selectivity filter (only two subunits are depicted).
3.6 Crystal structure of KcsA extracted with Triton X-100
To understand the function of an ion channel at atomic resolution very often we need to use large amounts of protein to conduct significantly different but complementary biophysical studies. They all require that during the purification process the atomic structure of the protein studied is preserved. To prove that the structure of KcsA remains unaltered after being solubilized with Triton X-100, we crystallized it in the presence of antibody fragment (Fab) by the sitting-drop method, as previously described Fab-Kcsa complex crystallized in the I4 space group and diffracted X-rays to 2.3 Å resolution. The phases of the structure were solved by molecular replacement methods using as a search model the antibody fragment molecule. The resulting structural model of the Fab-KcsA complex, after several refinement cycles, showed excellent refinement statistics (Table 1). Under the present conditions the structure of the selectivity filter of KcsA-extracted with Triton X-100 is indistinguishable from that of KcsA purified and crystallized in DM (1K4C) [15] (Figure 5)
Table 1.
| Statistic | KcsA-Triton |
|---|---|
| Space Group | I4 |
| Cell Dimension | |
| a=b, c (Å) | 156.53, 74.61 |
| α=β= ϒ (°) | 90 |
| Resolution (Å) | 26.09-2.3 (2.1-2.0) |
| Rmerge (Å) (%) | 35 |
| I/σI | 3.7 |
| Completeness (%) | 98.80 (93.70) |
| Redundancy | 4.9 |
| Refinement | |
| No. reflections | 39844 |
| Rwork/Rfree | 0.2106/0.2382 |
| No. atoms | 4089 |
| Protein | 3964 |
| Ligand/ion | 7 |
| Waters | 77 |
| Other ligands | 41 |
| Protein residues | 531 |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.933 |
| Wilson B-factor | 49.18 |
| Ramachandran Favored (%) | 97.7% |
| Ramachandran outliers (%) | 0.6% |
Highest resolution shell is show in parenthesis.
Data sets were collected from a single crystal.
4. Discussion
The low protein expression levels and challenging purification processes represent the major obstacles to overcome, when pursuing structural-functional studies of membrane proteins. Most of the biophysical and structural techniques (i.e., isothermal titration calorimetry, X-ray crystallography, solid-state and solution nuclear magnetic resonance and CW-EPR spectroscopies) used nowadays to determine the structures and dynamics of membrane proteins require the continuous supply of large amounts of pure, properly folded and fully functional test protein. Producing large amounts of any membrane protein represents currently a huge financial burden for the mainstream structural-biology laboratory, and in many cases drastically limits the number and/or diversity of studies that can be performed on a test protein.
Aiming to overcome these limitations we have successfully applied a systematic approach to optimize the expression and purification of a prototypical ion channel. This approach can be applied to other membrane proteins. In this study we have significantly improved the expression and purification of the archetypal K+ channel, KcsA, and made it highly cost effective. KcsA is a well-studied K+ channel[12, 20–22], which has become the archetypal pore domain for the superfamily of voltage-gated K+ channels. The previously reported yield for pure and properly folded KcsA is less than 7 mg/L and the vast majority of these studies used very expensive detergents, like DM[15, 17] and DDM[12, 21].
In this study we simultaneously increased the KcsA yield per liter of culture while reducing the production cost. To achieve these improvements, the following strategies were systematically applied to the production pipeline: 1) testing different E. coli growth media to maximize protein expression in the presence of a well-known chemical chaperone, glycerol, which seems to help in the proper folding of membrane proteins[10], 2) testing inexpensive detergents for membrane proteins solubilization and 3) maximizing the extraction capabilities of a given detergent by increasing the ionic strength of the buffer.
The use of a modified version of CRAP. media, the C41 strain and 0.5% Glycerol represented the best conditions for KcsA overexpression. Additionally, we established that Triton X-100 supplemented with 1 M KCl was a highly cost-effective alternative for KcsA solubilization, compared to the most expensive detergents DDM and DM. The cost of extracting 100 mg of KcsA with Triton X-100 was 28 cents. On the other hand, the cost of extracting the same amount of KcsA with DDM or DM was $33, which represents a 100-fold reduction in the solubilization cost. KcsA extracted by Triton X-100 was fully functional and amenable to crystallographic studies yielding high-quality crystals.
Another important detergent for KcsA solubilization identified by us in this study was Anzergent 3–12, which extracted the largest amount of properly folded KcsA. Anzergent 3–12 will be very useful for NMR studies, since typically this technique requires tenths of milligrams of isotopic labeled proteins, which is very expensive to produce, so it is highly desirable to extract as much protein as possible. Finally, we have shown that a straightforward systematic screening of the bacterial strain, growth media, chemical chaperones and detergent for membrane solubilization can dramatically increase the final yield of a pure, functional and properly folded membrane protein in a cost-effective fashion.
Highlights.
Improved protocol for high-expression levels of KcsA in E. coli.
Cost-effective method for the purification of large quantities of KcsA.
Cost-effective production of properly folded, fully functional and crystallizable KcsA.
Acknowledgments
L.G.C is grateful to L.G.G.F. and A.A.C. L.G.C and C.T. are grateful to L.R. for his continuous technical advice and proofreading the manuscript. Dr. Roderick Mackinnon provided the hybridoma cells for Fab production. We thank the member of the Cuello Laboratory for technical advice on this project. We thank Silvia Russi at the Stanford Synchrotron Radiation Laboratory Beamline (SSRL) 14-1. This work was supported in part by: CMPR-TTUHSC seeding grant, American Heart Association [11SDG5440003], National Institute of Health [1RO1GM097159-01A1] and Welch Foundation [BI-1757].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pan AC, Cuello LG, Perozo E, Roux B. Thermodynamic coupling between activation and inactivation gating in potassium channels revealed by free energy molecular dynamics simulations. The Journal of general physiology. 2011;138:571–580. doi: 10.1085/jgp.201110670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hille B. Ionic Channels of Excitable Membranes. Sinauer; Sunderland, MA: 1992. [Google Scholar]
- 3.Heginbotham L, Lu Z, Abramson T, MacKinnon R. Mutations in the K+ channel signature sequence. Biophysical journal. 1994;66:1061–1067. doi: 10.1016/S0006-3495(94)80887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuello LG, Romero JG, Cortes DM, Perozo E. pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry. 1998;37:3229–3236. doi: 10.1021/bi972997x. [DOI] [PubMed] [Google Scholar]
- 5.LeMasurier M, Heginbotham L, Miller C. KcsA: it’s a potassium channel. The Journal of general physiology. 2001;118:303–314. doi: 10.1085/jgp.118.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordero-Morales JF, Cuello LG, Zhao Y, Jogini V, Cortes DM, Roux B, Perozo E. Molecular determinants of gating at the potassium-channel selectivity filter. Nature structural & molecular biology. 2006;13:311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 7.Cuello LG, Jogini V, Cortes DM, Pan AC, Gagnon DG, Dalmas O, Cordero-Morales JF, Chakrapani S, Roux B, Perozo E. Structural basis for the coupling between activation and inactivation gates in K(+) channels. Nature. 2010;466:272–275. doi: 10.1038/nature09136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuello LG, Jogini V, Cortes DM, Perozo E. Structural mechanism of C-type inactivation in K(+) channels. Nature. 2010;466:203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science (New York, NY) 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 10.Figler RA, Omote H, Nakamoto RK, Al-Shawi MK. Use of chemical chaperones in the yeast Saccharomyces cerevisiae to enhance heterologous membrane protein expression: high-yield expression and purification of human P-glycoprotein. Archives of biochemistry and biophysics. 2000;376:34–46. doi: 10.1006/abbi.2000.1712. [DOI] [PubMed] [Google Scholar]
- 11.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortes DM, Perozo E. Structural dynamics of the Streptomyces lividans K+ channel (SKC1): oligomeric stoichiometry and stability. Biochemistry. 1997;36:10343–10352. doi: 10.1021/bi971018y. [DOI] [PubMed] [Google Scholar]
- 13.Delcour AH, Martinac B, Adler J, Kung C. Modified reconstitution method used in patch-clamp studies of Escherichia coli ion channels. Biophys J. 1989;56:631–636. doi: 10.1016/S0006-3495(89)82710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortes DM, Cuello LG, Perozo E. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. The Journal of general physiology. 2001;117:165–180. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution.[see comment] Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 16.Stauffer KA, Kumar NM, Gilula NB, Unwin N. Isolation and purification of gap junction channels. J Cell Biol. 1991;115:141–150. doi: 10.1083/jcb.115.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhate MP, Wylie BJ, Thompson A, Tian L, Nimigean C, McDermott AE. Preparation of uniformly isotope labeled KcsA for solid state NMR: Expression, purification, reconstitution into liposomes and functional assay. Protein expression and purification. 2013;91:119–124. doi: 10.1016/j.pep.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakrapani S, Cordero-Morales JF, Perozo E. A quantitative description of KcsA gating II: single-channel currents. The Journal of general physiology. 2007;130:479–496. doi: 10.1085/jgp.200709844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakrapani S, Cordero-Morales JF, Perozo E. A quantitative description of KcsA gating I: macroscopic currents. The Journal of general physiology. 2007;130:465–478. doi: 10.1085/jgp.200709843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schrempf H, Schmidt O, Kummerlen R, Hinnah S, Muller D, Betzler M, Steinkamp T, Wagner R. A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. Embo J. 1995;14:5170–5178. doi: 10.1002/j.1460-2075.1995.tb00201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heginbotham L, Kolmakova-Partensky L, Miller C. Functional reconstitution of a prokaryotic K+ channel. The Journal of general physiology. 1998;111:741–749. doi: 10.1085/jgp.111.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science (New York, NY) 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]


