Abstract
We present RobotReviewer, an open-source web-based system that uses machine learning and NLP to semi-automate biomedical evidence synthesis, to aid the practice of Evidence-Based Medicine. RobotReviewer processes full-text journal articles (PDFs) describing randomized controlled trials (RCTs). It appraises the reliability of RCTs and extracts text describing key trial characteristics (e.g., descriptions of the population) using novel NLP methods. RobotReviewer then automatically generates a report synthesising this information. Our goal is for RobotReviewer to automatically extract and synthesise the full-range of structured data needed to inform evidence-based practice.
1 Introduction and Motivation
Decisions regarding patient healthcare should be informed by all available evidence; this is the philosophy underpinning Evidence-based Medicine (EBM) (Sackett, 1997). But realizing this aim is difficult, in part because clinical trial results are primarily disseminated as free-text journal articles. Moreover, the biomedical literature base is growing exponentially (Bastian et al., 2010). It is now impossible for a practicing clinician to keep up to date by reading primary research articles, even in a narrow specialty (Moss and Marcus, 2017). Thus healthcare decisions today are often made without full consideration of the existing evidence.
Systematic reviews (SRs) are an important tool for enabling the practice of EBM despite this data deluge. SRs are reports that exhaustively identify and synthesise all published evidence pertinent to a specific clinical question. SRs include an assessment of research biases, and often a statistical meta-analysis of trial results. SRs inform all levels of healthcare, from national policies and guidelines to bedside decisions. But the expanding primary research base has made producing and maintaining SRs increasingly onerous (Bastian et al., 2010; Wallace et al., 2013). Identifying, extracting, and combining evidence from free-text articles describing RCTs is difficult, time-consuming, and laborious. One estimate suggests that a single SR requires thousands of person hours (Allen and Olkin, 1999); and a recent analysis suggests it takes an average of nearly 70 weeks to publish a review (Borah et al., 2017). This incurs huge financial cost, particularly because reviews are performed by highly-trained persons.
To keep SRs current with the literature then we must develop new methods to expedite evidence synthesis. Specifically, we need tools that can help identify, extract, assess and summarize evidence relevant to specific clinical questions from free-text articles describing RCTs. Toward this end, this paper describes RobotReviewer (RR; Figure 1), an open-source system that automates aspects of the data-extraction and synthesis steps of a systematic review using novel NLP models.1
Figure 1.
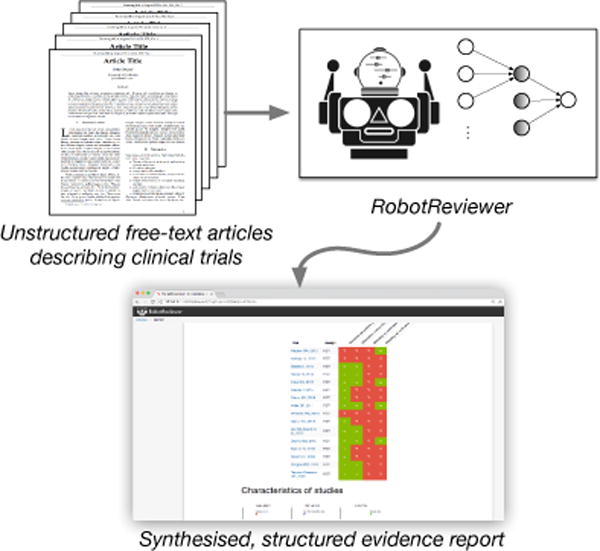
RobotReviewer is an open-source NLP system that extracts and synthesises evidence from unstructured articles describing clinical trials.
2 Overview of RobotReviewer (RR)
RR is a web-based tool which processes journal article PDFs (uploaded by end-users) describing the conduct and results of related RCTs to be synthesised. Using several machine learning (ML) data-extraction models, RR generates a report summarizing key information from the RCTs, including, e.g., details concerning trial participants, interventions, and reliability. Our ultimate goal is to automate the extraction of the full range of variables necessary to perform evidence synthesis. We list the current functionality of RR and future extraction targets in Table 1.
Table 1.
Typical variables required for an evidence synthesis (Centre for Reviews and Dissemination, 2009), and current RR functionality. Text: extracted text snippets describing the variable (e.g. ‘The randomization schedule was produced using a statistical computer package’). Structured: translation to e.g., standard bias scores or medical ontology concepts.
| Extraction type | Text | Structured | Extraction type | Text | Structured |
|---|---|---|---|---|---|
| General | Intervention and setting | ||||
| Record number | ✓ | ✓ | Setting | ✓ | |
| Author | ✓ | ✓ | Interventions and controls | ✓ | |
| Article title | ✓ | ✓ | co-interventions | ✓ | |
| Citation | ✓ | ✓ | Outcome data/results | ||
| Type of Publication | ✓ | Unit of analysis | |||
| Country of origin | Statistical techniques | ||||
| Source of funding | Outcomes reported? | ✓ | |||
| Study characteristics | Outcome definitions | ✓ | |||
| Aims/objectives | Measures used | ✓ | |||
| Study design | ✓ | ✓ | Length of follow up | ||
| Inclusion criteria | ✓ | N participants enrolled | ✓ | ||
| Randomization/blinding | ✓ | ✓ | N participants analyzed | ✓ | |
| Unit of allocation | Withdrawals/exclusions | ✓ | |||
| Participants | Summary outcome data | ||||
| Age | ✓ | Adverse events | |||
| Gender | ✓ | ||||
| Ethniticy | ✓ | ||||
| Socio-economic status | ✓ | ||||
| Disease characteristics | ✓ | ✓ | |||
| Co-morbidities | ✓ | ✓ |
RR comprises several novel ML/NLP components that target different sub-tasks in the evidence synthesis process, which we describe briefly in the following section. RR provides access to these models both via a web-based prototype graphical interface and a REST API service. The latter provides a mechanism for integrating our models with existing software platforms that process biomedical texts generally and that facilitate reviews specifically (e.g., Covidence2). We provide a schematic of the system architecture in Figure 2. We have released the entire system as open source via the GPL v 3.0 license. A live demonstration version with examples, a video, and the source code is available at our project website.3
Figure 2.
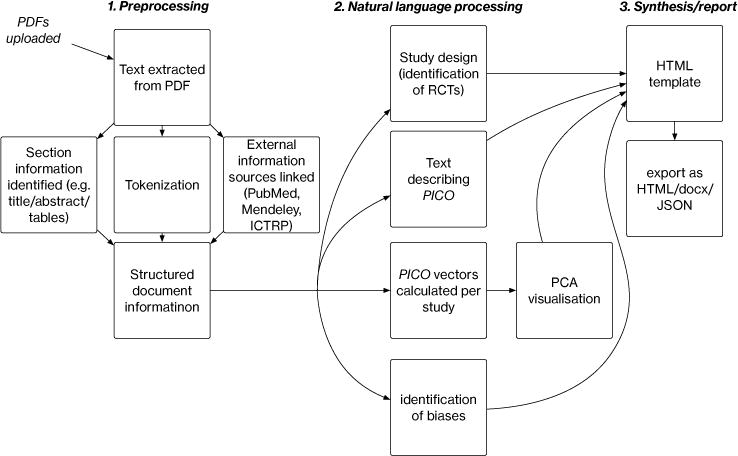
Schematic of RR document processing. A set of PDFs are uploaded, processed and run through models; the output from these are used to construct a summary report.
3 Tasks and Models
We now briefly describe the tasks RR currently automates and the ML/NLP models that we have developed and integrated into RR to achieve this.
3.1 Risks of Bias (RoB)
Critically appraising the conduct of RCTs (from the text of their reports) is a key step in evidence synthesis. If a trial does not rigorously adhere to a well-designed protocol, there is a risk that the results exhibit bias. Appraising such risks has been formalized into the Cochrane4 Risk of Bias (RoB) tool (Higgins et al., 2011). This defines several ‘domains’ with respect to which the risk of bias is to be assessed, e.g., whether trial participants were adequately blinded.
EBM aims to make evidence synthesis transparent. Therefore, it is imperative to provide support for one’s otherwise somewhat subjective appraisals of risks of bias. In practice, this entails extracting quotes from articles supporting judgements, i.e. rationales (Zaidan et al., 2007). An automated system needs to do the same. We have therefore developed models that jointly (1) categorize articles as describing RCTs at ‘low’ or ‘high/unknown’ risk of bias across domains, and, (2) extract rationales supporting these categorizations (Marshall et al., 2014; Marshall et al., 2016; Zhang et al., 2016).
We have developed two model variants for automatic RoB assessment. The first is a multitask (across domains) linear model (Marshall et al., 2014). The model induces sentence rankings (w.r.t. to how likely they are to support assessment for a given domain) which directly inform the overall RoB prediction through ‘interaction’ features (interaction of n-gram features with whether identified as rationale [yes/no]).
To assess the quality of extracted sentences, we conducted a blinded evaluation by expert systematic reviewers, in which they assessed the quality of manually and automatically extracted sentences. Sentences extracted using our model were scored comparably to those extracted by human reviewers (Marshall et al., 2016). However, the accuracy of the overall classification of articles as describing high/unclear or low risk RCTs achieved by our model remained 5–10 points lower than that achieved in published (human authored) SRs (estimated using articles that had been independently assessed in multiple SRs).
We have recently improved overall document classification performance using a novel variant of Convolutional Neural Networks (CNNs) adapted for text classification (Kim, 2014; Zhang and Wallace, 2015). Our model, the ‘rationale-augmented CNN’ (RA-CNN), explicitly identifies and up-weights sentences likely to be rationales. RA-CNN induces a document vector by taking a weighted sum over sentence vectors (output from a sentence-level CNN), where weights are set to reflect the predicted probability of sentences being rationales. The composite document vector is fed through a softmax layer for overall article classification. This model achieved gains of 1–2% absolute accuracy across domains (Zhang et al., 2016).
RR incorporates these linear and neural strategies using a simple ensembling strategy. For bias classification, we average the predicted probabilities of RCTs being at low risk of bias from the linear and neural models. To extract corresponding rationales, we induce rankings over all sentences in a given document using both models, and then aggregate these via Borda count (de Borda, 1784).
3.2 PICO
The Population, Interventions/Comparators and Outcomes (PICO) together define the clinical question addressed by a trial. Characterising and representing these is therefore an important aim for automating evidence synthesis.
3.2.1 Extracting PICO sentences
Past work has investigated identifying PICO elements in biomedical texts (Demner-Fushman and Lin, 2007; Boudin et al., 2010). But these efforts have largely considered only article abstracts, limiting their utility: not all clinically salient data is always available in abstracts. One exception to this is a system called ExaCT (Kiritchenko et al., 2010), which does operate on full-texts, although assumes HTML/XML inputs, rather than PDFs. ExaCT was hindered by the modest amount of available training data (∼160 annotated articles).
Scarcity of training data is an important problem in this domain. We have thus taken a distant supervision (DS) approach to train PICO sentence extraction models, deriving a corpus of tens of thousands of ‘pseudo-annotated’ full-text PDFs. DS is a training regime in which noisy labels are induced from existing structured resources via rules (Mintz et al., 2009). Here, we exploited a training corpus derived from an existing database of SRs using a novel training paradigm: supervised distant supervision (Wallace et al., 2016).
Briefly, the idea is to replace the heuristics usually used in DS to derive labels from the available structured resource with a function that maps from instances and DS derived labels to higher precision labels . Crucially, the representations include features derived from the available DS; such features will thus not be available for test instances. Parameters are to be estimated using a small amount of direct supervision. Once a higher precision label set is induced via , we can train a model as usual, training the final classifier fθ using . Further, we can incorporate the predicted probability distribution over true labels estimated by directly in the loss function used to train fθ. This approach results in improved model performance, at least for our case of PICO sentence extraction from full-text articles (Wallace et al., 2016).
Text describing PICO elements is identified in RR using this strategy; the results are displayed both as tables and as annotations on individual articles (see Figures 3 and 4, respectively).
Figure 3.
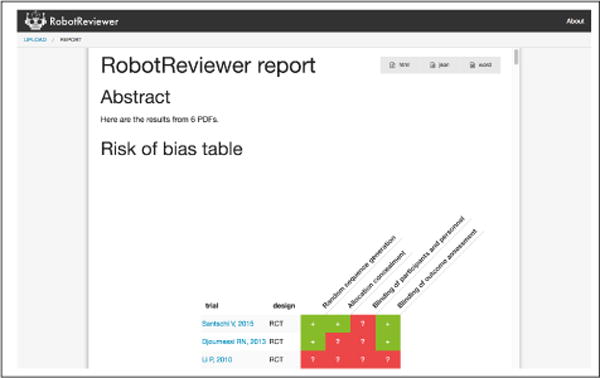
Report view. Here one can see the automatically generated risk of bias matrix; scrolling down reveals PICO and RoB textual tables
Figure 4.
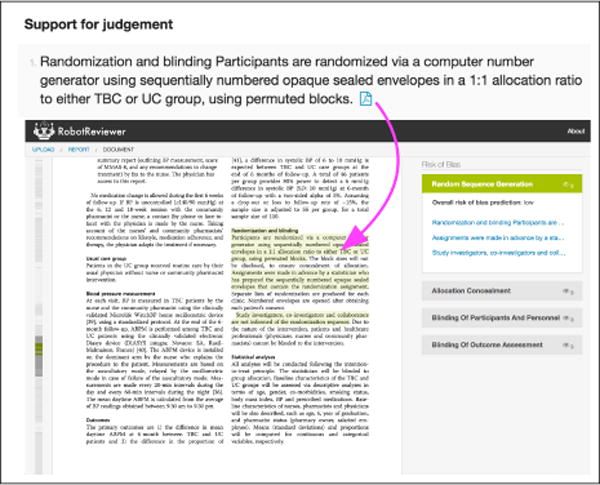
Links are maintained to the source document. We show predicted annotations for the risk of bias w.r.t. random sequence generation. Clicking on the PDF icon in the report view (top) brings the user to the annotation in-place in the source document (bottom).
3.2.2 PICO embeddings
We have begun to explore learning dense, low-dimensional embeddings of biomedical abstracts specific to each PICO dimension. In contrast to monolithic document embedding approaches, such as doc2vec (Le and Mikolov, 2014), PICO embeddings are an example of disentangled representations.
Briefly, we have developed a neural approach which assumes access to manually generated free-text aspect summaries (here, one per PICO element) with corresponding documents (abstracts). The objective is to induce vector representations (via an encoder model) of abstracts and aspect summaries that satisfy two desiderata. (1) The embedding for a given abstract/aspect should be close to its matched aspect summary; (2) but far from the embeddings of aspect summaries for other abstracts, specifically those which differ with respect to the aspect in question.
To train this model, we used data recorded for previously conducted SRs to train our embedding model. Specifically we collected 30,000+ abstract/aspect summary pairs stored in the Cochrane Database of Systematic Reviews (CDSR). We have demonstrated that the induced aspect representations improve performance an information retrieval task for EBM: ranking RCTs relevant to a given systematic review.5
For RR, we incorporate these models to induce abstract representations and then project these down to two dimensions using a PCA model pre-trained on the CDSR. We then present a visualisation of study positions in this reduced space, thus revealing relative similarities and allowing one, e.g., to spot apparently outlying RCTs. To facilitate interpretation, we display the uni and bigrams most activated for each study by filters in the learned encoder model on mouse-over. Figure 5 shows such an example. We are actively working to refine our approach to further improve the interpretability of these embeddings.
Figure 5.
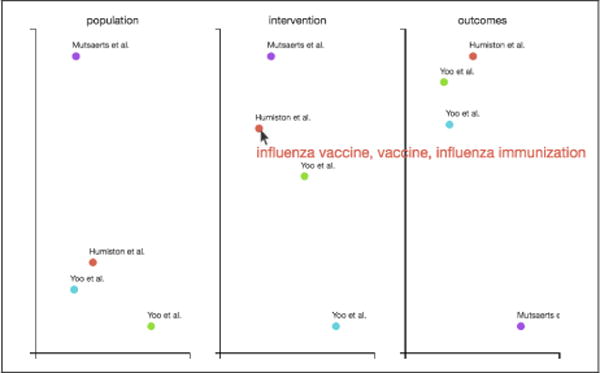
PICO embeddings. Here, a mouse-over event has occurred on the point corresponding to Humiston et al. in the intervention embedding space, triggering the display of the three uni-/bigrams that most excited the encoder model.
3.3 Study design
RCTs are regarded as the gold standard for providing evidence on of the effectiveness of health interventions (Chalmers et al., 1993) Yet these articles form a small minority of the available medical literature. We employ an ensemble classifier, combining multiple CNN models, Support Vector Machines (SVMs), and which takes account of meta-data obtained from PubMed. Our evaluation on an independent dataset has found this approach achieves very high accuracy (area under the Recevier Operating Characteristics curve = 0.987), outperforming previous ML approaches and manually created boolean filters.6
4 Discussion
We have presented RobotReviewer, an open-source tool that uses state-of-the-art ML and NLP to semi-automate biomedical evidence synthesis. RR incorporates the underlying trained models with a prototype web-based user interface, and a REST API that may be used to access the models. We aim to continue adding functionality to RR, automating the extraction and synthesis of additional fields: particularly structured PICO data, outcome statistics, and trial participant flow. These additional data points would (if extracted with sufficient accuracy) provide the information required for statistical synthesis.
For example, for assessing bias, RR is competitive with, but modestly inferior to the accuracy of a conventional manually produced systematic review (Marshall et al., 2016) We therefore recommended that RR be used as a time-saving tool for manual data extraction, or that one of two humans in the conventional data-extraction process be replaced by the automated process.
However, there is an increasing need for methods that trade a small amount of accuracy for increased speed (Tricco et al., 2015). The opportunity cost of maintaining current rigor in SRs is vast: reviews do not exist for most clinical questions (Smith, 2013), and most reviews are out of date soon after publication (Shojania et al., 2007).
RR used in a fully automatic workflow (without manual checks) might improve upon relying on the source articles alone, particularly given those in clinical practice are unlikely to have time to read the full texts. To explore how automation should be used in practice, we plan to experimentally evaluate RR in real-world use: in terms of time saved, user experience, and the resultant review quality.
Acknowledgments
RobotReviewer is supported by the National Library of Medicine (NLM) of the National Institutes of Health (NIH) under award R01LM012086. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. IJM is supported of the Medical Research Council (UK), through its Skills Development Fellowship program (MR/N015185/1).
Footnotes
We described an early version of what would become RR in (Kuiper et al., 2014); we have made substantial progress since then, however.
Cochrane is a non-profit organization dedicated to conducting SRs of clinical research: http://www.cochrane.org/.
Under review: preprint available at http://www.byronwallace.com/static/articles/PICO-vectors-preprint.pdf.
Under review; pre-print available at https://kclpure.kcl.ac.uk/portal/iain.marshall.html
References
- Allen IE, Olkin I. Estimating time to conduct a meta-analysis from number of citations retrieved. The Journal of the American Medical Association (JAMA) 1999;282(7):634–635. doi: 10.1001/jama.282.7.634. [DOI] [PubMed] [Google Scholar]
- Bastian H, Glasziou P, Chalmers I. Seventy-five trials and eleven systematic reviews a day: how will we ever keep up? PLoS medicine. 2010;7(9) doi: 10.1371/journal.pmed.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah R, Brown AW, Capers PL, Kaiser Kathryn A. Analysis of the time and workers needed to conduct systematic reviews of medical interventions using data from the prospero registry. BMJ open. 2017;7(2):e012545. doi: 10.1136/bmjopen-2016-012545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudin F, Nie J-Y, Dawes M. Positional language models for clinical information retrieval; EMNLP; 2010. pp. 108–115. [Google Scholar]
- Centre for Reviews and Dissemination. Systematic reviews: CRD’s guidance for undertaking reviews in health care. University of York; York: 2009. [Google Scholar]
- Chalmers I, Enkin M, Keirse MJNC. Preparing and updating systematic reviews of randomized controlled trials of health care. Milbank Q. 1993;71(3):411. [PubMed] [Google Scholar]
- de Borda J. A paper on elections by ballot. In: Sommerlad F, McLean I, editors. The political theory of Condorcet. 1784. pp. 122–129. (1989, eds) [Google Scholar]
- Demner-Fushman D, Lin J. Answering clinical questions with knowledge-based and statistical techniques. Computational Linguistics. 2007;33(1):63–103. [Google Scholar]
- Higgins JPT, Altman DG, Gøtzsche PC, Jüni P, Moher D, Oxman AD, Savović J, Schulz KF, Weeks L, Sterne JAC. The Cochrane Collaborations tool for assessing risk of bias in randomised trials. BMJ. 2011;343:d5928. doi: 10.1136/bmj.d5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. Convolutional neural networks for sentence classification. 2014 arXiv preprint arXiv:1408.5882. [Google Scholar]
- Kiritchenko S, de Bruijn B, Carini S, Martin J, Sim I. ExaCT: automatic extraction of clinical trial characteristics from journal publications. BMC medical informatics and decision making. 2010;10(1):56. doi: 10.1186/1472-6947-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper J, Marshall IJ, Wallace BC, Swertz MA. Spá: A web-based viewer for text mining in evidence based medicine; ECML-PKDD; Springer. 2014. pp. 452–455. [Google Scholar]
- Le QV, Mikolov T. Distributed representations of sentences and documents; ICML; 2014. pp. 1188–1196. [Google Scholar]
- Marshall IJ, Kuiper J, Wallace BC. Automating risk of bias assessment for clinical trials; ACM-BCB; 2014. pp. 88–95. [DOI] [PubMed] [Google Scholar]
- Marshall IJ, Kuiper J, Wallace BC. RobotReviewer: Evaluation of a System for Automatically Assessing Bias in Clinical Trials. Journal of the American Medical Informatics Association (JAMIA) 2016;23(1):193–201. doi: 10.1093/jamia/ocv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz M, Bills S, Snow R, Jurafsky D. Distant supervision for relation extraction without labeled data; IJCNLP; 2009. pp. 1003–1011. [Google Scholar]
- Moss AJ, Marcus FI. Changing times in cardiovascular publications: A commentary. Am J Med. 2017 Jan;130(1):11–13. doi: 10.1016/j.amjmed.2016.06.052. [DOI] [PubMed] [Google Scholar]
- Sackett DL. Evidence-based medicine: how to practice and teach EBM. WB Saunders Company; 1997. [Google Scholar]
- Shojania KG, Sampson M, Ansari MT, Ji J, Garritty C, Rader T, Moher D. Updating Systematic Reviews Technical Review No 16. Agency for Healthcare Research and Quality (US); 2007. Sep 1, [PubMed] [Google Scholar]
- Smith Richard. The Cochrane collaboration at 20. BMJ. 2013 Dec 18;347:f7383. doi: 10.1136/bmj.f7383. [DOI] [PubMed] [Google Scholar]
- Tricco Andrea C, Antony Jesmin, Zarin Wasifa, Strifler Lisa, Ghassemi Marco, Ivory John, Perrier Laure, Hutton Brian, Moher David, Straus Sharon E. A scoping review of rapid review methods. BMC Med. 2015 Sep 16;13224 doi: 10.1186/s12916-015-0465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BC, Dahabreh IJ, Schmid CH, Lau J, Trikalinos TA. Modernizing the systematic review process to inform comparative effectiveness: tools and methods. Journal of Comparative Effectiveness Research (JCER) 2013;2(3):273–282. doi: 10.2217/cer.13.17. [DOI] [PubMed] [Google Scholar]
- Wallace BC, Kuiper J, Sharma A, Zhu M, Marshall IJ. Extracting PICO Sentences from Clinical Trial Reports using Supervised Distant Supervision. Journal of Machine Learning Research. 2016;17(132):1–25. [PMC free article] [PubMed] [Google Scholar]
- Zaidan O, Eisner J, Piatko CD. Using “annotator rationales” to improve machine learning for text categorization; NAACL; 2007. pp. 260–267. [Google Scholar]
- Zhang Y, Wallace B. A sensitivity analysis of (and practitioners’ guide to) convolutional neural networks for sentence classification. 2015 arXiv preprint arXiv: 1510.03820. [Google Scholar]
- Zhang Y, Marshall IJ, Wallace BC. Rationale-Augmented Convolutional Neural Networks for Text Classification; EMNLP; 2016. pp. 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]


