Abstract
MicroRNAs (miRNA) are 21-24 nucleotide RNAs present in many eukaryotes that regulate gene expression as part of the RNA-induced silencing complex. The sequence identity of the miRNA provides the specificity to guide the silencing effector Argonaute (AGO) protein to target mRNAs via a base pairing process1. The AGO complex either promotes translation repression and/or accelerated decay of this target mRNA2. There is overwhelming evidence both in vivo and in vitro that translation repression plays a major role3–7. However, there has been controversy about which of these three mechanisms is more significant in vivo, especially when effects of miRNA on endogenous genes cannot be faithfully represented by reporter systems in which, at least in metazoans, the observed repression vastly exceeds that typically observed for endogenous mRNAs8,9. Here, we provide a comprehensive global analysis of the evolutionarily distant unicellular green alga Chlamydomonas reinhardtii to quantify the effects of miRNA on protein synthesis and RNA abundance. We show that, similar to metazoan steady-state systems, endogenous miRNAs in Chlamydomonas can regulate gene-expression both by destabilization of the mRNA and by translational repression. However, unlike metazoan miRNA where target site utilization localizes mainly to 3'UTRs, in Chlamydomonas utilized target sites lie predominantly within coding regions. These results demonstrate the evolutionarily conserved mode of action for miRNAs, but details of the mechanism diverge between plant and metazoan kingdoms.
Keywords: miRNA, ribosome profiling, translation, Chlamydomonas, mRNA turnover
Recent in vivo studies in mammalian cells provide support for endogenous mRNA destabilization over translation repression as the dominant effect of miRNA under steady-state conditions9. However an inducible zebrafish embryo system in which miR430 is only expressed two hours post fertilization, reveals that translation repression occurs prior to accelerated mRNA decay10. This conclusion was further supported by findings in mouse liver, primary macrophages, primary B, T cells 8 and through reporter system in human HeLa cell-lines11 as well as Drosophila S2 cells12.
In contrast to the metazoan systems, there is a lack of comprehensive studies on the endogenous effects of miRNAs in plants and the question remains as to whether miRNA modulates by translation repression and/or promoting mRNA turnover. In plants miRNA-mediated gene regulation does occur13–15 but, unlike metazoan systems, the targets can be in the coding sequence as well as 3’UTR and the mechanism may involve endonucleolytic cleavage rather than accelerated decay or translation inhibition16,17. Most plant studies, however, are based on individual miRNAs or reporter assays that may not be informative about endogenous mRNA systems9,18,19. We therefore utilized the unicellular green alga Chlamydomonas reinhardtii, for which we have previously discovered and characterized its miRNAs20 and generated DCL3 mutants21.
Chlamydomonas is a particularly amenable experimental system because its unicellularity reduces complications with tissue-specific effects. Similar to higher plants, the machinery for miRNA-mediated translation regulation is also functional in Chlamydomonas, where the seed-region rule utilised by the metazoan system is adequate for translation repression, at least within reporter systems22. In this present study, we utilized two silencing mutants raised from our previous forward genetic screen at dcl321 and ago3 (Chung et. al. 2017 in preparation). The dcl3-1 mutant results in almost complete loss of miRNA as well as 21-nt small interfering (si)RNAs whereas ago3-25 is defective in AGO3 that binds to mRNA and is required for translation repression in the reporter system23. Neither mutant had obvious growth differences or morphological abnormalities under normal conditions21. Any effect seen in both dcl3-1 and ago3-25 on gene expression is likely, therefore, to be direct rather than an indirect secondary consequence of metabolic changes due to loss of miRNA-mediated regulation.
Here, through a combination of ribosome profiling, parallel RNA-Seq, sRNA-Seq and quantitative proteomics at mid-log phase of the dcl3-1 mutant and its corresponding complemented strain we have demonstrated that, in contrast to the metazoan system, the primary effect of miRNA in Chlamydomonas is through interaction with CDS regions instead of 3’ UTRs. However, similar to the metazoan system, miRNA in Chlamydomonas reinhardtii can also modulate gene expression via means of translational repression and mRNA turnover. Finally, and perhaps the most striking observation is that the translation apparatus itself is differentially regulated at the level of translation efficiency but not RNA abundance in the presence of the miRNA machinery.
Loss of DCL3 function does not affect the genome-wide RNA or translation profile
To explore the possibility that DCL3-dependent miRNA or siRNA regulates gene expression by either promoting mRNA turnover or through interfering with translation, we applied ribosome profiling, parallel RNA-Seq and quantitative N15 proteomics to biological triplicates of the vegetative mid-log phase dcl3-1 mutant and its corresponding complemented derivative (abbreviated as Cdcl3) carrying a wild type DCL3 allele introduced into the mutant strain. The experimental protocol is summarized in supplementary Figure 1 and supplementary Figure 2 illustrates the high degree of reproducibility between biological repeats in these data.
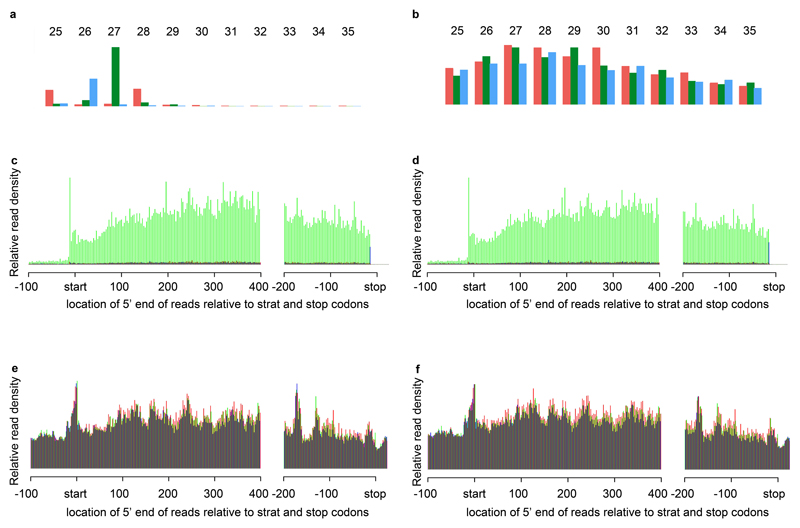
The slightly smaller footprint size of plant/algae ribosomes leads to differences in the phasing patterns compared to mammalian ribosome profiling studies24. In both the complemented strain Cdcl3 and the dcl3 mutant, the 5’ end of the 27-nt ribosome protected fragments (RPFs), mapped predominantly to the second codon position; in contrast and, as expected, RNA-Seq reads were uniformly distributed at all three codon positions (Figures 1A and B). The RPF 5’ end position distributions at start and stop codons were also similar in the dcl3-1 and Cdcl3 strains (Figures 1C and D respectively) in that there was a sharp 27-nt peak on the start codon (reflecting the rate-limiting initiation step of translation) and a sharp 28-nt peak on the stop codon (reflecting the conformation change from an elongating ribosome to a terminating ribosome, Supplementary figure 3B)24. In contrast, the RNA-seq reads are not limited to coding regions (Figures 1E, F and Supplementary Figures 3B).
Figure 1. Ribosome profiling data.
(A, B) Mapping the 5’ ends of ribosome protected fragments (RPFs) and corresponding RNA-Seq respectively, as a function of read size class (nt), within nucleus-encoded coding ORFs. Red, green and blue bars indicate the proportion of reads that map to codon positions 0, 1 and 2 (respectively).
(C, D) 5’ end positions of 27-nt RPFs relative to start and stop codons (nt). Reads were derived from strain Cdcl3 and dcl3-1 (respectively) and summed over all transcripts. Phasing is indicated using the same colors as in panels A and B.
(E, F) 5’ end positions of all RNA-seq reads relative to start and stop codons (nt). Reads were derived from strain Cdcl3 and dcl3-1 (respectively) and summed over all transcripts. Phasing is indicated using the same colors as in panels A and B.
The validity of these data was further confirmed with the analysis of DCL3. There were multiple DCL3 mRNA reads from three replicate samples of the Cdcl3 strain that were restricted to the open reading frame in the RPF datasets. In dcl3-1 the reads were from the region on the 5’ side of the mutagenic DCL3 insertion (Supplementary Figures 3C). Finally, Ribosome protected fragments (RPF), RNA abundance (RA), and translational efficiencies (TE) for expressed genes are well correlated between dcl3-1 and Cdcl3 (R2 = 0.95, 0.97 and 0.98 for TE, RPF and RNA, respectively, Supplementary Figure 3E). From these data, we conclude that any global effect of DCL3 on the translatome is minor but we could not rule out quantitative effects on a subset of RNAs.
To explore this possibility, we refined our analysis by dividing the mRNA profiles into those with or without predicted targets of the DCL3-dependent miRNAs. The first stage in this analysis was to re-evaluate the miRNA precursors in C. reinhardtii that we had previously identified as being both coding and non-coding RNAs. Now, however, with the use of the RPF data to identify translated open reading frames, we find that all miRNAs in this alga derive from introns or the exons (3’UTR or coding) of mRNAs. Supplementary Figure 4 and Table 2 is an updated summary of the 42 miRNA precursors in C. reinhardtii described in Valli et. al. 201621.
Our subsequent analysis differentiated mRNAs with miRNA targets in the 5’ UTR, CDS and 3’ UTR from those without targets. The CDS regions were defined by the R software Bioconductor package – riboSeqR - that utilizes the triplet periodicity of ribosome profiling for the de novo inference of AUG-initiated coding sequences that are supported by RPFs24 and we used the seed-sequence rule to identify miRNA target motifs25,26. This rule requires base-pairing of the first 8 nucleotides of miRNA and it is supported by direct assay of miRNA targeting and structural studies of human AGO227 and by experimental tests in higher plants28 and C. reinhardtii22.
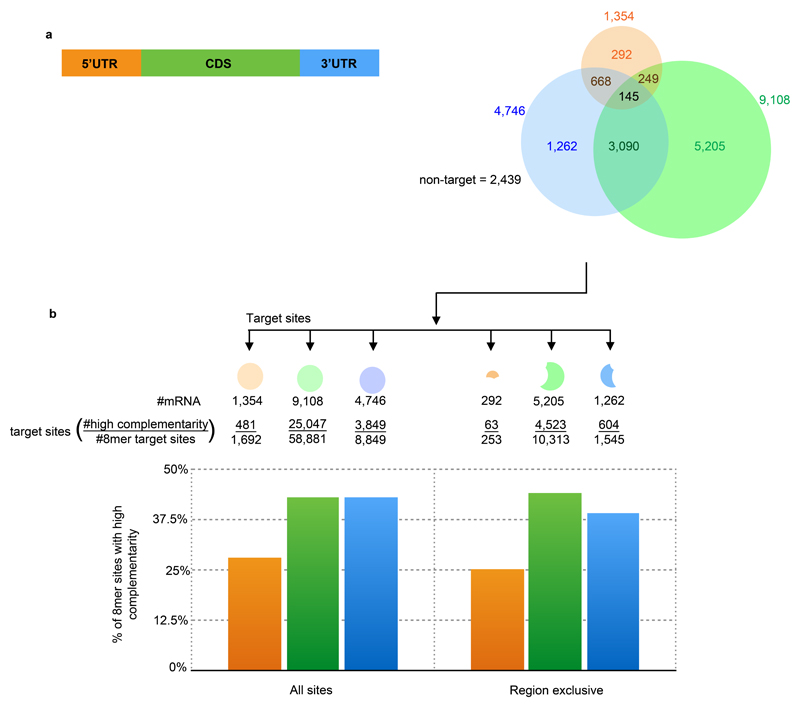
To identify the miRNA-target mRNAs we first filtered for the 19 most-abundant DCL3-dependent miRNAs in our sRNA-Seq data (Supplementary Figure 5; see also Materials and Methods). We then applied the TargetScan prediction algorithm25,26 to the mRNAs with RPF-validated ORFs. This criterion meant that the TargetScan algorithm was applied to 13,073 expressed transcripts (out of 17,741 annotated transcripts) of which 2,439 do not contain any predicted 8mer miRNA target sites. Of all the predicted target sites, a larger proportion (70%) are located in the CDS (Figure 2A) compared to UTRs (10% for 5'UTR and 36% for 3'UTR). This distribution is likely, at least in part, a reflection of greater length of the CDS compared to UTR regions. Using a more stringent miRNA targeting rule did not have a large change on these numbers: about half of the mRNA seed sequence targets also have >50% sequence complementarity to the relevant miRNA in the sequences upstream of the 3’ eight nucleotides (Figure 2B).
Figure 2. Distribution of 8mer target sites.
(A) Venn diagram showing number of transcripts predicted to be targeted with the 8mer rule.
(B) Proportion of 8mer target sites that also have at least 50% complementarity from nucleotides 11-21 of the miRNA
Next, we excluded the RNAs with predicted target sites in more than one region (5'UTR/CDS/3'UTR) because for these it would have not been possible to differentiate the effects of miRNA acting in the different regions. In addition, we also excluded mRNAs with miRNA precursors because they are unstable in the presence of DCL3 as a consequence of miRNA processing (see supplementary Figure 4 and21). Following application of these filters our further analysis was based on 292 mRNAs with 5’ UTR targets, 5,205 with CDS targets, 1,262 with targets in the 3’ UTR and the 2,439 without predicted targets.
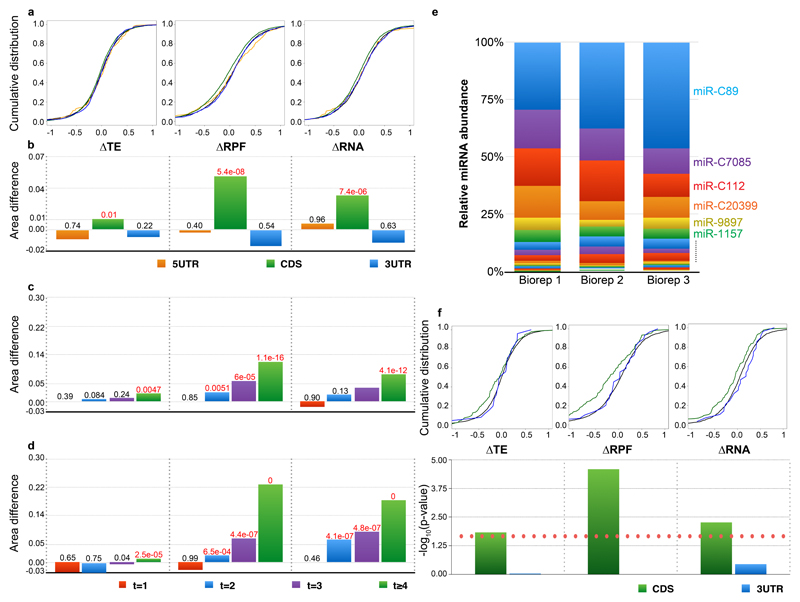
Similar to studies by the Bartel and Giraldez groups10,8,9 we plotted cumulative distributions of differential translation efficiency, total RPF and RA for target and non-target mRNAs in the dcl3-1 mutant and Cdcl3 to assess the miRNA-mediated effects of DCL3 (Figure 3A and B). Differential TE is computed as (RPFC/RNAC)/(RPFdcl3/RNAdcl3). The analysis revealed that, similar to the analysis of zebrafish10, the major effects of Dicer loss of function (dcl3-1 vs Cdcl3) were on mRNAs containing target sites within the CDS and the effect is more significant in the RPF than the RNA data, contributing to its significant but small effect in TE. The effects were evident as a shift to increased RNA abundance for mRNAs with target sites in dcl3-1 and they are consistent with the canonical role of miRNAs as negative regulators.
Figure 3. miRNA downregulates gene expression primarily through mRNA destabilization by CDS targeting.
(A) Cumulative distributions of ΔTE (left), ΔRPF (middle) and ΔRA (right) log2 fold changes in dcl3-1 relative to Cdcl3. Colors correspond to genes containing predicted 8mer miRNA target sites exclusively in the 5’UTR (orange), CDS (green), 3’UTR (blue), or no targets (black).
(B) Bar graph of differences between area under cumulative distribution of mRNA containing target sites and non-target containing mRNAs (5'UTR, CDS and 3'UTR in orange, green and blue, respectively). Significance (K.S. test) of the differences are indicated above each bar; p-values less than or equal to 0.01 are highlighted in red.
(C-D) Bar graph of differences between area under cumulative distribution of mRNA containing 1 (red), 2 (blue), 3 (purple) or 4 or more (green) CDS-exclusive target sites and non-target containing mRNAs. Significance (K.S. test) of the differences are indicated above each bar; p-values less than or equal to 0.01 are highlighted in red.
(E) Normalised miRNA abundances of Cdcl3 (in three biological replicates).
(F) Cumulative distributions (top) and significance (bottom; the red dotted line indicates p-value of 0.01) of ΔTE (left), ΔRPF (middle) and ΔRA (right) log2 fold changes for mRNAs containing miR-C89 target sites exclusively within the CDS (green) or 3’UTR (blue) (sample sizes 141 and 25, respectively). 5’UTR-exclusive targets were omitted due to low sample size.
The difference in dcl3-1 versus Cdcl3 was greater in transcripts with CDS rather than UTR target sites and this effects appears to be dosage dependent, where mRNAs with four or more CDS targets were affected to a greater extent than those with fewer target sites (Figure 3C). However, this dosage-dependent effect was not observed for mRNAs containing target sites in the UTRs (Supplementary Figure 6A). Furthermore, these effects are also consistent at the protein level for mRNAs with supportive proteomics data (Supplementary Figure 6B).
As the key AGO in Chlamydomonas known to be associated with miRNA is AGO3 which mediates translational repression in a reporter system23, we also performed ribosome profiling as well as corresponding RNA-seq on an AGO3 mutant (ago3-25), raised from the same forward genetic screen as dcl3-121, as well as the corresponding parental strain and the wild type cc-1883 (Chung et. al. 2017, in preparation) in order to further validate whether these effects are truly due to the miRNA machinery. Supporting this, we also observed the dosage-dependent effect only for mRNAs containing target sites within the CDS in the ago3-25 mutant background (Figure 3D and Supplementary Figure 6A).
The global effect of mRNA repression is not likely due to target RNA cleavage as there are only 85 potential CDS target sites (83 mRNAs) complying with the plant targeting rule in Chlamydomonas20. Moreover, of these potential CDS cleavage site mRNAs, only 18/83 were expressed in our dataset, albeit at very low level (Supplementary Figure 6C). We also investigated potential targets for expressed miRNA where the base-pairing is between positions 2-15 (allowing one mismatch) and, similar to the plant-rule potential targets, there were very few candidates (47 in total), of which only 31 are expressed in our dataset and the expression level for all 31 mRNAs is low (Supplementary Figure 6C). Thus, well expressed genes are unlikely to be cleaved under steady-state conditions, consistent with the lack of phenotype for both dcl3-1 and ago3-25 mutants. A recent degradome study is also consistent with there being minimal miRNA target site cleavage in Chlamydomonas. The study involved miR-910, an miRNA also expressed in our sample, that cleaved only two mRNAs upon salt-stress29. The endogenous miRNA-mediated RNA down-regulation by CDS-targeted miRNA is not, therefore, likely to be mainly through target cleavage.
Finally, we tested the effect of miRNA abundance on TE, RPF and RA by focusing on the most abundant miRNA in our corresponding sRNA-Seq datasets: miR-C89 (Figure 3E, F and supplementary Figure 5; 5’UTR and protein data excluded due to small sample size). MiR-C89 correlated with a larger shift in TE and RA than other miRNAs consistent with magnitude of the effect being influenced by miRNA abundance.
From these findings we conclude that, similar to metazoan systems8,9, Chlamydomonas miRNA generally fine tunes gene expression through an effect on both RNA abundance and translation efficiency (Figure 3). The global effect on translation efficiency was significant although smaller than the effect on RNA abundance (Figures 3A and B), as in metazoans9. Unlike metazoans, however, the primary targets of miRNAs in Chlamydomonas are in the CDS instead of 3’UTRs (Figure 3). This difference may reflect differences between Chlamydomonas and metazoans in the ways in which miRNAs may influence elongating ribosomes.
Translation efficiency of 80S ribosomal proteins is higher in the DCL3 mutant
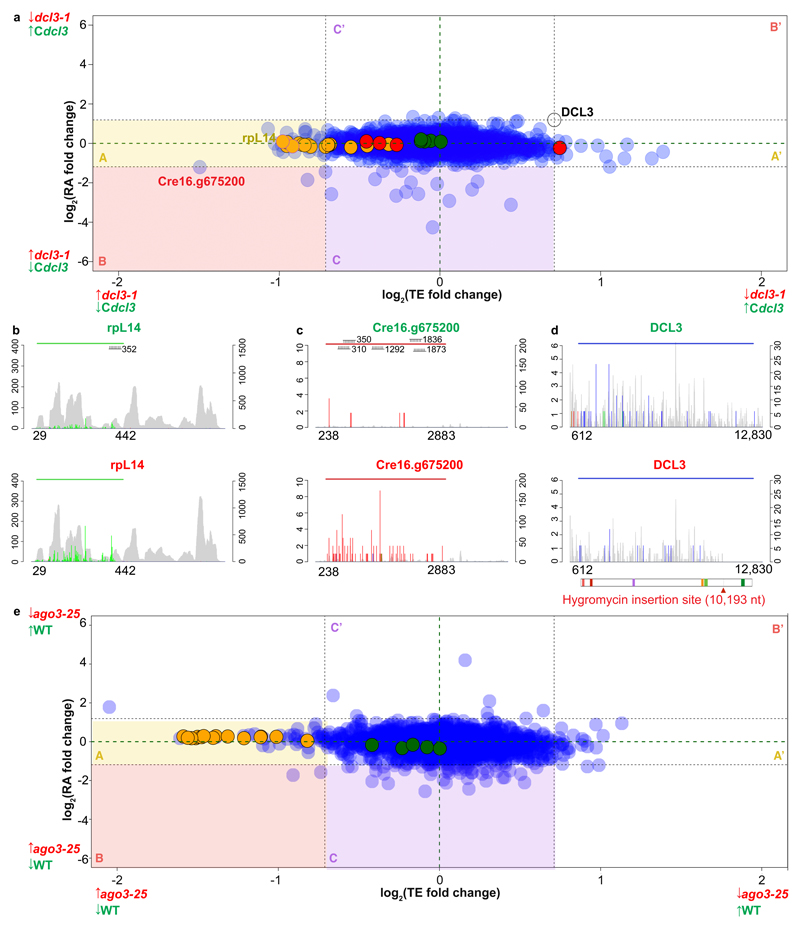
Our finding that miRNA targeting in Chlamydomonas is influenced by miRNA abundance and the number of target sites (Figure 3) implies that some mRNAs may be affected more than others. Therefore, to detect possible changes in individual mRNAs, we plotted the dcl3-1 versus Cdcl3 differences in TE and RA for all mRNAs with CDS-exclusive target sites (Figure 4). Using the dcl3-1 mutation as a benchmark (log2FC(TE) = 0.7 and log2FC(RNA) = 1.18), individual RNAs that are negatively regulated by miRNAs would distribute in field A of this figure if TE is affected (i.e. log2FC(TE) ≤ -0.7, yellow shaded area), field C if RA is affected but not TE (i.e. log2FC (RA) ≤ -1.18, -0.7 ≤ log2FC(TE) ≤ 0.7, purple shaded area) and in field B if there was a double effect on both TE and RA (log2FC(RA) ≤ -1.18, log2FC(TE) ≤ -0.7, red shaded area). Corresponding positive regulation would be indicated by distribution in fields A’, B’ and C’ respectively (Figure 4A).
Figure 4. Effects of miRNAs on TE and RA.
(A) Correspondence between TE and RA fold-changes between dcl3-1 and Cdcl3 for nuclear-encoded genes containing miRNA target sites exclusively within the CDS (except DCL3, which was included as a marker). 80S, chloroplast and mitochondria ribosomal proteins are in orange, green and red, respectively.
(B-C) Histograms of 5' end positions of normalized RPF (colored, left-axis) and RNA-Seq (grey, right-axis) 27-nt reads mapped to genes with high differential TE: ribosomal proteins rpL14 and Cre16.g675200. The top (green title) and bottom (red title) graphs are derived from either Cdcl3 or dcl3-1, respectively. The colored horizontal line indicates the riboSeqR de novo-defined ORF; positions of potential miRNA target sites are annotated.
(D) Histogram of 5’ end positions of normalized RPF (colored, left-axis) and RNA-Seq (grey, right-axis) 27-nt reads mapped to DCL3 transcripts. The blue horizontal line indicates the CDS (612-12,830 nt). The schematic below the plot shows the domain organization of DCL3 which contains two DEAD/DEAH box helicase domains (light and dark red boxes), a helicase domain (purple box), a proline-rich domain (orange box) and two ribonuclease III domains a and b (light and dark green boxes, respectively). The thick grey line and the corresponding red arrow below indicate the hygromycin insertion site (nt 10,193).
(E) Correspondence between TE and RA fold-changes between ago3-25 and wild type CC-1883 for nuclear-encoded genes containing miRNA target sites exclusively within the CDS. Nuclear-encoded 80S and chloroplast ribosomal proteins are in orange and green, respectively. Mitochondrial ribosomal proteins are not shown due to low level of detection in the dataset.
The distribution of mRNA in this plot is consistent with a higher degree of negative rather than positive regulation on a few mRNAs: there were 32 and 16 targets in A and A’ respectively, 3 and 0 in B and B’, and 15 and 3 in C and C’. From this analysis we conclude that there may be up to 32 mRNAs that are subject to strong translational regulation by miRNAs (from the A and B fields), 15 subject to strong regulation of RNA abundance (from the B and Cdcl3 fields) and 3 subject to strong regulation at both levels. The RNA-Seq and RPF data for DCL3 mRNA and selected miRNA targets including rpL14 and Cre16.g67520 from field A are presented in Figure 4 C-E.
To assess whether the mRNAs in field C could either be miRNA targets or they could have DCL3 cleavage sites we monitored their level in ago3-25 and the wild-type (Supplementary Figure 6D). Repression of RNAs that are targeted by DCL3 would be relieved in dcl3-1 but not ago3-25 whereas those that are targeted by miRNAs would be depressed in both mutants.
The data are consistent with miRNA targeting for most of the field C RNAs of Figure 4 because their repression was relieved in both mutants although Cre15.g643503.t1.1 was an exception (perhaps related to it having an unusually long CDS - 7884 nt, cf. average CDS length for expressed genes = 2429 nt; Supplementary Figure 7D). We therefore conclude that the RA effect we observe is genuinely directed by the miRNA-AGO complex. Further, in order to distinguish whether reduced expression in Cdcl3 relative to dcl3-1 was a global effect or merely due to a small number of strongly repressed genes (i.e. fields A, B, C, A’, B’ and C’ of Figure 4A), we repeated the analysis with the strongly repressed candidates excluded and found a similar pattern of global mRNA repression as with all mRNAs (Supplementary Figure 8A). Similarly, with the targets of miR-C89 the repression of TE or RA primarily results from small changes in the expression of many genes (Supplementary Figure 8A and B).
It is striking that mRNAs subject to either strong translational or RNA stability regulation (i.e. field A and C) are enriched with those encoding RNA-interacting proteins (e.g. translation, transcription and rRNA processing) (Supplementary Table 3). Of the mRNAs subject to translational regulation a gene ontology analysis revealed the enriched pathway of “translation and ribosome” with the mRNAs for 80S ribosomal proteins being particularly prominent (Figure 4A and Supplementary Table 3). These candidates also contribute to the outlier group for TE and RPF but not RA in the cumulative distributions for transcripts with supporting proteomic data (Supplementary Figure 6B). Furthermore, the same enrichment is also observed in the ago3-25 mutant (Figure 4E and Supplementary Figure 7C). However, we do not observe enrichment for this pathway in previously published mammalian datasets9 of miR-233 knockout cultured neutrophils compared with wild-type culture neutrophils, and HeLa cells after transfection with miR-1 or miR-155 (Supplementary Figure 8).
The enrichment of “translation and ribosome” function in fields A and C of Figure 4A and E is specific for 80S ribosomal proteins; the nucleus-encoded 70S ribosomal proteins for both chloroplasts and mitochondria were an internal control and cluster around the 0-fold change axis for both TE and RNA (Figure 4A and E). It is likely therefore that the specific effect for the 80S factors reflects the targeting specificity of miRNAs in Chlamydomonas or that it is a compensatory mechanism for the loss of a layer of regulation in the dcl3-1 and ago3-25 mutants.
It is possible that the distribution of ribosomes on the mRNA would be affected by absence of miRNAs (see Figures 4B and C for example rpL14 and Cre16.g675200). However, we did not observe any significant correlation between the position of the miRNA target sites and the distribution of RPF or RNA reads for the mRNAs of fields A and C of Figure 4A either individually or through a global analysis of multiple RNAs. In contrast, in the mRNA for DCL3 there was an effect: the RPFs in the Cdcl3 sample extended to the stop codon and the RNA-Seq reads covered the full length mRNA whereas, in dcl3-1, the RPF and RNA-Seq data were more sparse than in Cdcl3 and they stopped at the site of the mutagenic hyg insert (Figure 4D and Supplementary 3C). Clearly, from this DCL3 analysis, the RPF and RNA-Seq data can reflect both the quantitative and qualitative aspects of ribosome distribution and RNA accumulation.
We hypothesized that CDS-targeting of the miRNA-AGO complex should result in road-blocking of elongating ribosomes, resulting in ribosome pile-up and/or drop-off 5’ and 3’ end of miRNA target sites respectively. However we did not observe any significant changes in RPF density around miRNA target sites, indicating that RISC does not induce ribosome pileup within CDS regions. Presumably the efficient RNA helicase activity of the ribosomes is able to overcome the steric hindrance by the RISC in Chlamydomona 30,31. There may, however, be a transient effect on ribosome translocation. Having now identified these RNAs with the greatest effect on TE and RNA we will be able to explore the factors affecting the two modes of RNA regulation and the conditions under which miRNAs have the greatest effect on their mRNA targets.
Materials and Methods
Culturing and harvesting Chlamydomonas
Three independent fresh single colonies of Chlamydomonas reinhardtii cells were sub-cultured as biological triplicates. Cells where grown in 50 ml Tris-acetate-phosphate (TAP) medium at 23 ºC in baffled flasks on a rotatory shaker (140 rpm) under constant illumination with white light (70 µE m2 sec-1) to mid-log phase (OD750 ~ 0.6), followed by inoculation into 750 ml TAP in 2 L baffled flasks at OD750 = 0.2. These were cultured in the same conditions until mid-log phase prior to harvesting by filtering off the media, after which the cell paste was immediately flash frozen and pulverized in liquid nitrogen with 5 mL of pre-frozen buffer (20 mM Tris-Cl pH 7.5, 140 mM KCl, 5 mM MgCl2, 10 µg/ml cycloheximide, 100 µg/mL chloramphenicol, 0.05 mM DTT, 0.5% NP40, 1% Triton X-100 and 5% sucrose). The frozen powder was gradually thawed on ice and clarified by centrifugation for 30 min at 4700 rpm at 4 ºC followed by adjustment of A254 = 10 before further treatment, or snap frozen in liquid nitrogen and stored at −80 ºC. The extraction efficiency was monitored by polysome profiling (Supplementary Figure 3F). The flash freezing method was preferred as methods involving pretreatment with translational inhibitors such a cycloheximide or chloramphenicol can introduce various biases, in particular in artificially enhancing the initiation peak of the profile32, which we also observed in Chlamydomonas reinhardtii when we compared flash-freezing with cycloheximide pretreatment (Supplementary Figure 3G).
Metabolic labelling and LC-MS/MS
For metabolic labelling, ammonia chloride (14N) was replaced with ammonia chloride-15N (Cambridge Isotope Laboratories Inc) in the TAP media used to maintain dcl3-1. There were no obvious differences in growth rates between algae maintained in N14 and N15. dcl3-1-N15 and Complement-N14 were mixed equally prior to protein extraction via TCA-acetone precipitation followed by resuspension in resuspension buffer (8 M urea, 500 mM NaCl, 10 mM Tris-Cl pH 8, 5 mM DTT) and resolved in 1.5 mm 10% bis-tris Novex Gel (Thermo Fisher Scientific Inc, Waltham, MA, USA). The experiment was performed in biological triplicate.
1D gel bands (12 per lane) were transferred into a 96-well PCR plate. The bands were cut into 1 mm2 pieces, de-stained, reduced (DTT), alkylated (iodoacetamide) and subjected to enzymatic digestion with trypsin overnight at 37 °C. After digestion, the supernatant was pipetted into a sample vial and loaded onto an autosampler for automated LC-MS/MS analysis.
All LC-MS/MS experiments were performed using a Dionex Ultimate 3000 RSLC nanoUPLC (Thermo Fisher Scientific Inc, Waltham, MA, USA) system and a QExactive Orbitrap mass spectrometer (Thermo Fisher Scientific Inc, Waltham, MA, USA). Separation of peptides was performed by reverse-phase chromatography at a flow rate of 300 nL/min and a Thermo Scientific reverse-phase nano Easy-spray column (Thermo Scientific PepMap C18, 2 µm particle size, 100 Å pore size, 75 µm i.d. x 50 cm length). Peptides were loaded onto a pre-column (Thermo Scientific PepMap 100 C18, 5 µm particle size, 100 Å pore size, 300 µm i.d. x 5 mm length) from the Ultimate 3000 autosampler with 0.1% formic acid for 3 min at a flow rate of 10 µL/min. After this period, the column valve was switched to allow elution of peptides from the pre-column onto the analytical column. Solvent A was water + 0.1% formic acid and solvent B was 80% acetonitrile, 20% water + 0.1% formic acid. The linear gradient employed was 2-40% B in 30 min (total run time including a high organic wash step and requilibration was 60 min).
The LC eluant was sprayed into the mass spectrometer by means of an Easy-Spray source (Thermo Fisher Scientific Inc.). All m/z values of eluting ions were measured in an Orbitrap mass analyzer, set at a resolution of 70000 and was scanned between m/z 380-1500. Data dependent scans (Top 20) were employed to automatically isolate and generate fragment ions by higher energy collisional dissociation (HCD, NCE:25%) in the HCD collision cell and measurement of the resulting fragment ions was performed in the Orbitrap analyser, set at a resolution of 17500. Singly charged ions and ions with unassigned charge states were excluded from being selected for MS/MS and a dynamic exclusion window of 20 s was employed.
Protein identification and relative quantitation
Data were recorded using Χcalibur™ software version 2.1 (Thermo Fisher Scientific, San Jose, CA). Files were converted from .raw to .mzXML using MSConvert and then .mzXML files to .mgf using the in-house software iSPY33,34. The .mgf files were submitted to the Mascot search algorithm. The following parameters were employed: carbamidomethyl as a fixed modification, and oxidation on methionine (M) residues and phosphorylation on serine (S), threonine (T), and tyrosine (Y) residues as variable modifications; 20 ppm for peptide tolerance, 0.1 Da of MS/MS tolerance; a maximum of two missed cleavages, a peptide charges of +2, +3, or +4; and selection of a decoy database. Mascot .dat output files were imported into iSPY for 14N/15N quantitation and analysed through Percolator for improved identification35. The 14N and 15N peptide isotopic peaks from the MS1 dataset were used to compare the theoretical mass difference between the heavy and light peptides, and the typical isotopic distribution patterns. Only unique peptides with a posterior error probability (PEP-value) of ≤ 0.05 were considered for further analysis. Spectra were merged into peptides and proteins based on their median intensity in MS1, meaning the more intense the signal of the spectrum, the more weight it added to quantitation. The statistical programming environment R was used to process iSPY output files to check for the 15N incorporation rate and to confirm that the data were normally distributed. After normalization, only peptides detected in at least two biological replicates, with a fold change ≥ 1.5 and a p-value ≤ 0.05 were considered for further analysis. Relative protein expression values were computed as (ProteinC/Proteindcl3) using the average of the triplicates for all follow-up analysis.
Nuclease footprinting
Lysates (200 µL) were slowly thawed on ice and treated with 6000 units RNase I (Thermo Fisher Scientific Inc.). in a thermo-mixer at 28 ºC, 400 rpm for 30 min. The reaction was stopped by mixing the digest reaction with 120 units of SUPERase-In RNase inhibitor (Thermo Fisher Scientific Inc.) followed by centrifugation for 2 min at 14000 rpm at 4 ºC to further clarify any remaining debris. The supernatant was layered onto a 1 M sucrose cushion prepared in Chlamydomonas polysome buffer, and RNA were purified as described in Ingolia et. al.36. Polysome integrity for the lysate and digestion conditions were assessed via polysome profiling (Supplementary Figure 3F).
Ribosome profiling and RNA-Seq
The methodologies were largely based on the protocols of Ingolia et. al. and Guo et. al.9,36 with modifications (i) mRNA for corresponding RNA-Seq was enriched by removal of rRNA using the ribo-zero kit (plant seed and root kit), (ii) RNA-Seq size selection was in parallel with ribosome profiling (i.e. between 26 and 34 nt), and (iii) for ribosome profiling, ribosomal RNA contamination was removed by two rounds of treatment with duplex specific nuclease (DSN) for 30 min as described in (Chung et. al. 2015).
Preparation for sRNA libraries
Small RNA from total RNA samples used for RNA-Seq were size excluded in 15% TBU gel for miRNA enrichment (Thermos Scientific). The sRNA were further prepared according to the NEXTflex small RNA-Seq kit v2 (Bio Scientific), followed by sequencing on the NextSeq500 platform.
Computational analysis of ribosome profiling and RNA-Seq data
After removal of adaptor sequences, Illumina sequencing reads were mapped to the reference transcriptome (Phytozome 281) or miRNA precursor sequences described in Valli et. al. 201621 using bowtie-1 and processed as described in Chung et. al. 201524. Only mRNAs with more than 50 RPF reads of size 27 or 28 nt uniquely mapped to more than 10 positions were considered. Corresponding RNA-Seq reads within coding regions de novo defined by ribosome profiling were extracted for differential RA as well as TE analysis using riboSeqR as described in Chung et. al. 201524. Further filtering was applied for fold change analyses where mRNAs were only considered if they had (i) at least 10 normalised RPF and 10 normalised RNA counts, and (ii) the sum of all RPF or RNA counts over the three biological replicates for both dcl3-1 and complement combined is at least 200. Normalisation was based on BaysSeq output 37. Cumulative distributions for TE, RPF and RA fold changes were calculated based on the average of all three replicates. Differential analyses for the mouse data in Guo et. al. 20109 were obtained from the Gene expression Omnibus in NCBI (accession:GSE220001 and GSE21992).
Target prediction
Target prediction was done using TargetScan26 using the same transcriptome input as for the ribosome profiling analysis. As there are no conserved sites available due to lack of miRNA data from the green algae phylum, we could not calculate context and scores; thus we only utilized the part of the software to detect all possible miRNA target sites. Further, as the efficacy between 8mer-A1 and 8mer-m8 sites are similar, we combined both types of target sites in the 8mer prediction, similar to Guo et. al. 2010 and Agarwal et. al. 20159,26. Target prediction based on the plant rule was performed via TAPIR38.
The list of miRNA used was based on the 19 DCL3-dependent miRNAs expressed based on the sRNA data, where the average reads within the complement is greater than 400 and the average ratio of complement to dcl3-1 reads is greater than 150. The selected DCL3-dependent miRNA used are: chromosome_5_3227666_3227753_+ (miR-C89), chromosome_6_6776108_6776193_+ (miR-cluster20399), chromosome_13_2001067_2001197_- (miR-cluster 7085), chromosome_10_3399870_3399999_- (miR9897), chromosome_13_3152367_3152452_- (miR-C112), chromosome_6_3067368_3067456_+ (miR1162), chromosome_12_6402226_6402307_- (miR1157), chromosome_9_6365928_6366014_- (miR912), chromosome_7_4386252_4386309_- , chromosome_17_6144120_6144204_+ (miR-cluster12551), chromosome_1_7070552_7070605_-, chromosome_16_185088_185174_-(miR1169), chromosome_2_8349161_8349264_+ , chromosome_2_9129508_9129593_- miR-cluster14712), chromosome_7_5926395_5926482_+ (miR-C59), chromosome_14_3218783_3218866_- (miR910), chromosome_6_7063792_7063881_- (miR1152), chromosome_4_3100624_3100751_+ (miR1153) and chromosome_1_5106349_5106475_+ (miR-C82). The miRNA precursor sequence used for mapping was based on Valli et. al. (2016). Only 8mer sites were utilized, and 8mer complementarity was verified via extraction of target sites followed by miRNA complementarity assessment using the Vienna RNA package program RNAduplex. The level of 3’ complementarity was similarly investigated where nt 9 to 21 of the target site 3’ of the seed region was extracted and the level of complementarity assessed with RNAduplex.
Data and code availability
All raw sequencing data are deposited on Arrayexpress with accessions E-MTAB-3852 and E-MTAG-3851. All code used for statistical analysis is available on request.
Supplementary Material
Acknowledgements
We thank J. Barlow for technical assistance and media preparation; T. J. Hardcastle and B. Santos for technical bioinformatic support; A. Valli for providing the silencing mutants and, A. Molnar and A. E. Firth for discussions. This work was supported by a Balzan Prize award and the European Research Council Advanced Investigator Grant ERC-2013-AdG 340642 TRIBE. B.Y.W.C. was supported by an EMBO long-term postdoctoral fellowship and a Sir Henry Wellcome Fellowship [096082]. D.C.B. is the Royal Society Edward Penley Abraham Research Professor.
Footnotes
Author contributions
B.Y.W.C. and D.C.B. conceived and designed the research. B.Y.W.C performed the experiments and analysed the data. M.J.D., A.J.G. and J.H. performed all the LC-MS/MS sample processing and iSPY analysis. B.Y.W.C. and D.C.B. wrote the manuscript.
Conflict of interest
The authors declared that they have no conflict of interest.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–88. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 3.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Iwakawa H oki, Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Bazzini AA, Lee MT, Giraldez AJ. Supporting Online Material for Supporting Online Material for. 2012 doi: 10.1126/science.1215704. [DOI] [Google Scholar]
- 7.Izaurralde BE. Breakers and blockers— miRNAs at work. Science (80-.) 2015;349:380–382. doi: 10.1126/science.1260969. [DOI] [PubMed] [Google Scholar]
- 8.Eichhorn SW, et al. MRNA Destabilization Is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bazzini Aa, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–7. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Béthune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–23. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–40. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brodersen P, Voinnet O. Target Recognition and Mode of Action. Nat Rev Mol Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 14.Reis RS, Hart-smith G, Eamens AL, Wilkins MR, Waterhouse PM. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nat Plants. 2015;1:1–6. doi: 10.1038/nplants.2014.27. [DOI] [PubMed] [Google Scholar]
- 15.Li S, et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in arabidopsis. Cell. 2013;153:562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brodersen P, et al. Widespread translational inhibition by plant miRNAs and siRNAs. TL - 320. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. VN- [DOI] [PubMed] [Google Scholar]
- 17.Iwakawa H, Tomari Y. Molecular Insights into microRNA-Mediated Translational Repression in Plants. Mol Cell. 2013;52:591–601. doi: 10.1016/j.molcel.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 18.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:25–29. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molnar a, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 21.Valli AA, et al. Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs. Genome Res. 2016;26:519–529. doi: 10.1101/gr.199703.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamasaki T, et al. Complementarity to an miRNA seed region is sufficient to induce moderate repression of a target transcript in the unicellular green alga Chlamydomonas reinhardtii. Plant J. 2013;76:1045–1056. doi: 10.1111/tpj.12354. [DOI] [PubMed] [Google Scholar]
- 23.Yamasaki T, Kim E-J, Cerutti H, Ohama T. Argonaute3 is a key player in miRNA-mediated target cleavage and translational repression in Chlamydomonas. Plant J. 2016;85:258–68. doi: 10.1111/tpj.13107. [DOI] [PubMed] [Google Scholar]
- 24.Chung BY, et al. The use of duplex-specific nuclease in ribosome profiling and a user-friendly software package for Ribo-seq data analysis. Rna. 2015;21:1731–1745. doi: 10.1261/rna.052548.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis BP, Shih I, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science (80-.) 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallory AC, et al. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5’ region. EMBO J. 2004;23:3356–64. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao X, et al. MicroRNAs modulate adaption to multiple abiotic stresses in Chlamydomonas reinhardtii. Sci Rep. 2016;6:38228. doi: 10.1038/srep38228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 31.Qu X, et al. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutteridge A, et al. Nutrient control of eukaryote cell growth: a systems biology study in yeast. BMC Biol. 2010;8:68. doi: 10.1186/1741-7007-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marondedze C, et al. A Quantitative Phosphoproteome Analysis of cGMP-Dependent Cellular Responses in Arabidopsis thaliana. Mol Plant. 2016;9:621–623. doi: 10.1016/j.molp.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Brosch M, Yu L, Hubbard T, Choudhary J. Accurate and sensitive peptide identification with mascot percolator. J Proteome Res. 2009;8:3176–3181. doi: 10.1021/pr800982s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardcastle TJ, Kelly KA. baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonnet E, He Y, Billiau K, van de Peer Y. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. 2010;26:1566–1568. doi: 10.1093/bioinformatics/btq233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data are deposited on Arrayexpress with accessions E-MTAB-3852 and E-MTAG-3851. All code used for statistical analysis is available on request.