Abstract
Previous studies based on single mitochondrial markers have shown that the common raven (Corvus corax) consists of two highly diverged lineages that are hypothesised to have undergone speciation reversal upon secondary contact. Furthermore, common ravens are paraphyletic with respect to the Chihuahuan raven (C. cryptoleucus) based on mitochondrial DNA (mtDNA). Here we explore the causes of mtDNA paraphyly by sequencing whole mitochondrial genomes of 12 common ravens from across the Northern Hemisphere, in addition to three Chihuahuan ravens and one closely related brown-necked raven (C. ruficollis) using a long-range PCR protocol. Our raven mitogenomes ranged between 16925–16928 bp in length. GC content varied from 43.3% to 43.8% and the 13 protein coding genes, two rRNAs and 22 tRNAs followed a standard avian mitochondrial arrangement. The overall divergence between the two common raven clades was 3% (range 0.3–5.8% in 16 regions including the protein coding genes, rRNAs and the control region). Phylogenies constructed from whole mitogenomes recovered the previously found mitochondrial sister relationship between the common raven California clade and the Chihuahuan raven (overall divergence 1.1%), which strengthens the hypothesis that mtDNA paraphyly in the common raven results from speciation reversal of previously distinct Holarctic and California lineages.
Introduction
In the era of genomics and high throughput sequencing, complete mitochondrial genomes (mitogenomes) are rapidly becoming available for many organisms [1]. In addition to increasing our knowledge of functional aspects of mitochondrial genes and molecular evolution [2, 3], complete mitogenomes offer increased resolution in phylogenetic and phylogeographic analyses [4–9]. Furthermore, obtaining complete mitogenomes reduces the likelihood of sequencing nuclear pseudogenes instead of mitochondrial DNA [10].
The number of published avian mitogenomes is increasing, and to date such genomes are available for more than 1530 specimens from more than 640 species in GenBank. Within the genus Corvus, there are complete mitogenomes available for seven species, but none from the clade of large-bodied raven species (clade V in [11]) distributed in the Afro-Holarctic region (Africa: 6 species; south-west North America: 1 species; Holarctic: 1 species). While the monophyly of the Afro-Holarctic raven clade is well established, relationships within the clade remain uncertain—especially those within and between the Corvus ruficollis species group (a complex of four morphologically variable crow-sized species in Africa: C. ruficollis, C. edithae, C. albus, C. rhipidurus), and the common raven species group (a complex of two larger-bodied species distributed across the Northern Hemisphere: the common raven C. corax, and Chihuahuan raven C. cryptoleucus) [11–14]. Here we focus on the common raven species group.
The common raven (Corvus corax L.) is one of the world’s largest passerines, with a wide breeding distribution in the Northern Hemisphere. Previous studies based on several mitochondrial markers (control region, cytochrome b, cytochrome c oxidase subunit 1 (COI)) show that this species harbours two highly diverged mtDNA lineages [15–17]. Structuring between California and Holarctic lineages is also apparent in microsatellite data [17], but not in the single nuclear intron studied to date (beta-fibrinogen intron,[12]). The Holarctic lineage is found across Eurasia and North America, while the California lineage is restricted to western North America where it co-occurs with the Holarctic lineage [15, 17]. Despite this deep divergence, there is no evidence that the two lineages are reproductively isolated as field studies suggest that they interbreed to a large extent and their offspring are viable [18]. Common ravens are therefore hypothesised to be an example of speciation reversal of divergent lineages upon secondary contact [18]. A further complication to the apparently reticulate history of the common raven is the fact that a second sympatric species, the Chihuahuan raven (Corvus cryptoleucus) restricted to south-western North America, is nested within the common raven based on mitochondrial DNA, having a closer relationship to the Californian clade, than either of them have to the Holarctic clade [12, 17]. However, these two species are phenotypically distinct, and there is no evidence of hybridization from either field or mtDNA studies [12, 17–19]. Overall, there appears to be good support for reproductive isolation between common and Chihuahuan ravens despite mtDNA paraphyly.
In this study, we present the first mitogenomes for Afro-Holarctic ravens. We sequence three raven species—the common raven, the Chihuahuan raven and the brown-necked raven (Corvus ruficollis)—and test whether mtDNA paraphyly results from reticulations in the speciation history of ravens. If complete mitogenomes and all constituent gene regions recover the deeply divergent California and Holarctic mtDNA clades and support paraphyly of common ravens with respect to Chihuahuan ravens, this offers evidence that paraphyly originates from a reticulate speciation history—most likely one where the mtDNA tree reflects an earlier divergence between the Holarctic lineage and the ancestor of California and Chihuahuan lineages followed by a more recent divergence of California and Chihuahua lineages. Alternatively, if complete mtDNA genomes reconstruct a different topology to that recovered in previous phylogenetic analyses of single mtDNA loci [15–17], this could suggest that apparent mtDNA paraphyly of common ravens resulting from a close sister relationship of California clade and Chihuahuan ravens could be caused by previous studies inadvertently sequencing nuclear pseudogenes.
Materials and methods
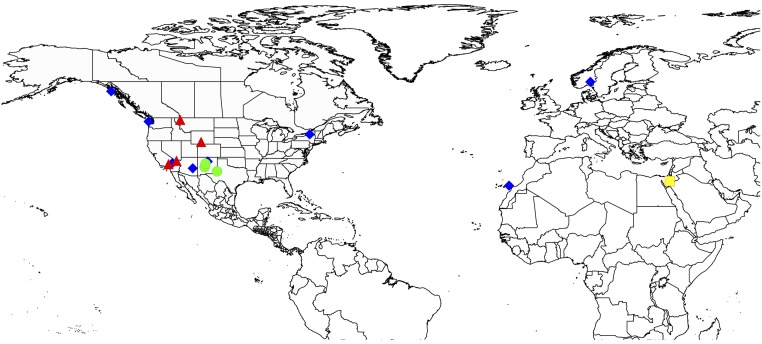
Blood samples were obtained from live birds (n = 8) and tissue samples from frozen tissue collections at official US natural history museums (n = 8). Seven of eight live-sampled birds were adults caught in mist nets/rocket nets, while the last one was a chick sampled in the nest. All birds were released in good condition immediately after blood sampling. All samples had been collected with the appropriate banding and collecting permits from relevant national authorities. Table 1 summarises specimen details and Fig 1 shows the geographic position of sampling locations. We analysed 12 samples of Corvus corax (10 from North America, 1 from Norway and 1 from the distinct Canary islands lineage that is nested within the Holarctic lineage [20]), three Corvus cryptoleucus (from New Mexico in south-west USA) and one Corvus ruficollis (from Israel). Of the 10 Corvus corax from North America, two were from northern and eastern regions with pure Holarctic mtDNA (Alaska, New York), two were from the state of California where the majority of ravens have California mtDNA, and six were from within the region of overlap of Holarctic and California mtDNA lineages in western North America (Arizona, New Mexico, Nevada, Washington, Wyoming, Montana) [18]. See Table 1 for mtDNA clade assignments of each sample based on single mtDNA loci from previous studies ([17, 18]; authors' unpubl. data).
Table 1. Sample information.
Clade assignment of common ravens from North America to either California or Holarctic lineages are given for mitogenomes presented in this study and either mtDNA control region or COI sequenced in previous studies ([17, 18]; authors’ unpubl. data).
| Species | Accession number | Sampling locality | Coordinates (decimal degrees) | Collection date | Clade assignment | |
|---|---|---|---|---|---|---|
| Mito-genome | Single mtDNA locus | |||||
| C. corax | 2407–51899 | San Bernardino, CA, USA | 35.26N, 116.68W | 22.05.2001 | HOL | HOL |
| C. corax | USFWS 2327–69957 | Gila, AZ, USA | 33.64N, 110.52W | 25.04.2005 | HOL | HOL |
| C. corax | UAM30328 | Sitka, AK, USA | 57.05N, 135.33W | 2012 | HOL | HOL |
| C. corax | NYSM 11227 | Hamilton, NY, USA | 43.95N, 74.94W | 13.08.2011 | HOL | HOL |
| C. corax | MSB21677 | Santa Fe, NM, USA | 35.69N, 105.94W | 2009 | HOL | HOL |
| C. corax | 2387–36563 | Jefferson, WA, USA | 47.86N, 123.94W | 17.05.1997 | HOL | HOL |
| C. corax | NHMO-BI-23199 | Maridalen, Oslo, Norway | 60.00N, 10.79E | 04.05.2010 | HOL | HOL |
| C. corax | NHMO-BI-35585 | Fuerteventura, Spain | 28.35N, 14.03W | 2013 | HOL | HOL |
| C. corax | 1547–43719 | Wamsutter, WY, USA | 41.67N, 107.98W | 2013 | CAL | CAL |
| C. corax | UCSB 90–175 | Kern, CA, USA | 34.92N, 117.89W | 07.12.1994 | CAL | CAL |
| C. corax | 1807–88239 | Flathead, MT, USA | 48.39N, 114.33W | 23.04.2014 | CAL | CAL |
| C. corax | MBM 9200 | Clark, NV, USA | 35.93N, 115.47W | 18.05.2005 | CAL | CAL |
| C. cryptoleucus | MSB25417 | Socorro, NM, USA | 33.99N, 106.89W | 2005 | na | na |
| C. cryptoleucus | MSB40523 | Bernalillo, NM, USA | 35.02N, 106.63W | 2013 | na | na |
| C. cryptoleucus | MSB22405 | Lea, NM, USA | 32.70N, 103.14W | 1999 | na | na |
| C. ruficollis | NHMO-BI-18431 | Eilat, Israel | 29.55N, 34.93E | 21.03.2009 | na | na |
Fig 1. Map showing sampling locations.
Blue diamond = C. corax, Holarctic lineage; red triangle = C. corax, California lineage; green circle = C. cryptoleucus; yellow square = C. ruficollis.
Molecular analyses
DNA was extracted with EZNA blood/tissue kits (Omega Inc), following the protocol of the manufacturer. For sequencing of the complete mitogenomes, we followed the protocol recently described by Lifjeld et al. [21]. Briefly, mitochondrial DNA was amplified from high molecular weight genomic DNA using two primer pairs: mtCorvus531F (GGATTAGATACCCCACTATGC) & mtCorvus9431R (GTCTACRAAGTGTCAGTATCA) and mtCorvus8031F (CCTGAWCCTGACCATGAACCTA) & mtCorvus926R (GAGGGTGACGGGCGGTATGTA). These two primer pairs yielded respective amplicons with ~8,900 bp and ~9,800 bp. The primers were designed to anneal in conserved regions of the mitogenome, based on an alignment with genetic information from the following species: Corvus frugilegus, Corvus splendens, Corvus corax, Taeniopygia guttata, Molohthrus aeneus, Ficedula albicollis, Melamprosops phaeosoma and Gallus gallus. Annealing sites and overlapping regions are illustrated in Lifjeld et al. [21]. The following PCR conditions were utilized for amplification: 1X reaction buffer, 200 μM of each dNTP, 0.5 μM of each primer, ~20 ng template DNA, 0.02 U/μl Q5 High-Fidelity DNA polymerase (New England Biolabs) and dH2O to a final volume of 25 μl. The following thermal profiles were employed: Amplicon 1 –Initial denaturation 98°C in 30 seconds, 35 cycles with denaturation 98°C for 10 seconds, annealing 59°C for 20 seconds and elongation 72°C for 7.5 minutes, and a final elongation step for 2 minutes. Amplicon 2 –Initial denaturation 98°C in 30 seconds, 5 cycles with denaturation 98°C for 10 seconds following a touch down profile starting at 72°C with 1°C/cycle reduction, 30 cycles with denaturation 98°C for 10 seconds, annealing 67°C for 20 seconds and elongation 72°C for 7.5 minutes, and a final elongation step for 2 minutes.
The complete PCR reactions were transferred to a 0.8% agarose gel and ran at 90V. When completely separated, the respective amplicons were cut from the gel and purified using the GenJet Gel Extraction Kit (ThermoFischer Scientific). Concentrations of the purified amplicons were measured on a Qubit instrument (ThermoFischer Scientific) and equimolar amounts of each amplicon were pooled. Approximately 20 ng of pooled amplicons from each individual where sheared using a Covaris M220 Focused-ultrasonicator (Covaris, Inc.), running the pre-programmed DNA shearing protocol for 800 bp twice. Size selection was performed using a BluePippin (Sage Science) instrument. We size selected DNA in the 350–450 bp range using a 2% agarose gel cassette (Sage Science). To generate barcoded libraries for sequencing, we employed the NEBNext library prep kit for Ion Torrent (New England Biolabs) on the sheared size selected amplicons using the IonXpress barcode adapter kit (ThermoFischer Scientific). Barcoded libraries were pooled and concentration of the final library was measured on a Fragment Analyzer (Advanced Analytical) using the DNF-474 High Sensitivity NGS Fragment Analysis kit. The size selected, barcoded, sheared amplicons were sequenced on a 314 chip using an IonPGM sequencing instrument (ThermoFischer Scientific).
Bioinformatics
Trimming, removal of low quality reads and demultiplexing were performed on the Torrent Suite™ software (ThermoFisher Scientific). The Corvus splendens mitogenome (GenBank acc. NC024607; [22]) was used as reference in the Torrent Suite ™ software for coverage estimates, using the plugin coverageAnalysis (v4.4.2.2). Additional trimming to improve quality of the data was performed using Trimmomatic v0.33 [23] with the following settings: Sliding window 4:20. Minimum read length:100. Mitogenomes were reconstructed by iterative mapping using MITObim v1.8 [24]. The complete mitogenome of Corvus splendens was used as reference in the mapping.
Duplications in the region between the cytB gene and the 12S rRNA gene have frequently been observed among bird taxa (e.g. [4, 25]) and such duplications may be indicated by increased coverage in this region [26]. We addressed this issue by inspecting coverage plots, emphasizing the area where duplications are likely to occur. This was performed in the software Tablet v1.14.10.20 [27]. We observed a drop in coverage in the start of the complement strand of the NAD6 gene. Hence, we employed a second iterative mapping using the assembled NAD6 gene as short sequence bait. Using this strategy, we avoided having a reference that covers the complete problematic region where duplications have been shown to occur. If the reference sequence covers this area, one may expect a bias in assembled reads based on similarity to the reference. In other words, if the true sequence includes a duplicated region and the reference sequence does not, the duplication may be masked since only the most similar reads will map to the reference. When using a short sequence bait, the assembly will be generated from the iterative mapping and accordingly independently from a large reference sequence. We found that the region where the coverage dropped had a local GC content of 72% and contained two stretches of C(n) homopolymers, which have been shown to introduce errors in sequence data generated by the Ion Torrent platform [28]. Such local high GC content may lead to systematic coverage drop in vertebrate mitochondria assemblies [29]. This may result from DNA polymerase introducing premature elongation stops in the GC rich region during the library amplification or from incomplete nucleotide incorporation in the sequencing reaction. We observed a major overrepresentation of reverse reads in the region with the coverage drop, indicating incomplete sequencing of forward reads through the respective region. We also observed a similar coverage drop in the same region in Illumina generated data [30]. Furthermore, our contig sequences were identical to the first iterative mapping when a short NAD6 sequence was used as bait. Hence, we conclude that the observed drop in coverage in our dataset is likely explained by the biochemical properties of the template DNA and/or the chemical reagents and not because of gene order alterations.
We were not able to cover the complete control region in all individuals in our assemblies, but for both of the common raven clades and the Chihuahuan raven we obtained the complete control region from at least one individual. For the single individual of brown-necked raven, we lack ~140 bp from the end of the control region, and the total mitogenome length for this individual was estimated based on the alignment of the other study specimens.
Mitochondrial genes were first automatically annotated using MITOs [31], and thereafter manually inspected.
Phylogenetic analyses
Genetic distances for nucleotides were measured as the number of base substitutions divided by the respective sequence length, averaging over all sequence pairs between groups (uncorrected p-distance). Standard error estimates were obtained by a bootstrap procedure (100 replicates). Genetic distance analyses were conducted using the maximum composite likelihood model [32]. The coding data was translated assuming a vertebrate mitochondrial genetic code. Genetic distance estimates were calculated in MEGA7 [33].
Given that accumulation of substitutions may bias phylogenetic interpretations, we performed substitution saturation tests on five different data sets: (i) all codon positions in protein coding genes (11,381 bp), (ii) first and second codon position in all protein coding genes (7,586 bp), (iii) first codon position (3,793 bp), (iv) second codon position and (v) third codon position. We followed Xia & Lemey [34] to estimate the index of substitution saturation (ISS). The ISS statistics were performed on the gap-free sites in the specified data sets using the software DAMBE 6.3.17 [34, 35]. The ISS values were significantly lower (p <0.0001) than the critical ISS values for all the data sets (S1 Table), indicating no saturation of substitutions. Accordingly we included all codon positions and non-coding nucleotide position in our phylogenetic analyses.
ClustalW was used to align our raven mitogenome dataset with previously published mitogenomes available for seven other species of Corvus–C. brachyrhynchos, C. cornix, C. frugilegus, C. hawaiiensis, C. macrorhynchos, C. moriorum, and C. splendens–and Pica pica, which were obtained from GenBank (accession numbers appear in the figures). The alignment was manually inspected in MEGA7. Maximum likelihood (ML) trees were constructed in MEGA7 using Pica pica as an outgroup. ML trees used the best fit substitution model according to Bayesian information criterion and Akaike information criterion (general time reversible + G + I) as estimated by MEGA7. Initial tree for the heuristic search was obtained by applying the maximum parsimony method [36]. The tree with the highest log likelihood (-52,715) was chosen for visualization. Test of node support in the phylogenetic tree was performed using 1,000 bootstrap replicates [37]. All positions with less than 95% coverage in the alignment were eliminated. ML trees were also constructed from nucleotide subsets containing the minimum (16S rRNA) and the maximum (ATP6) genetic distance between the clades (S1 and S2 Figs), and from subsets containing only protein coding genes (S3 Fig) and the control region (S4 Fig), which has been the region used most extensively to study the geographic distribution of Holarctic and California clades in western North America [18, 38].
Results
The Ion PGM run yielded 1,022,295 reads. In total, 45.2% was filtered as polyclonals and 9.3% as low quality reads in the Torrent Suite software. Sample specific information regarding total number of reads, trimmed reads, mapped reads, coverage and GenBank accessions numbers are provided in S2 Table.
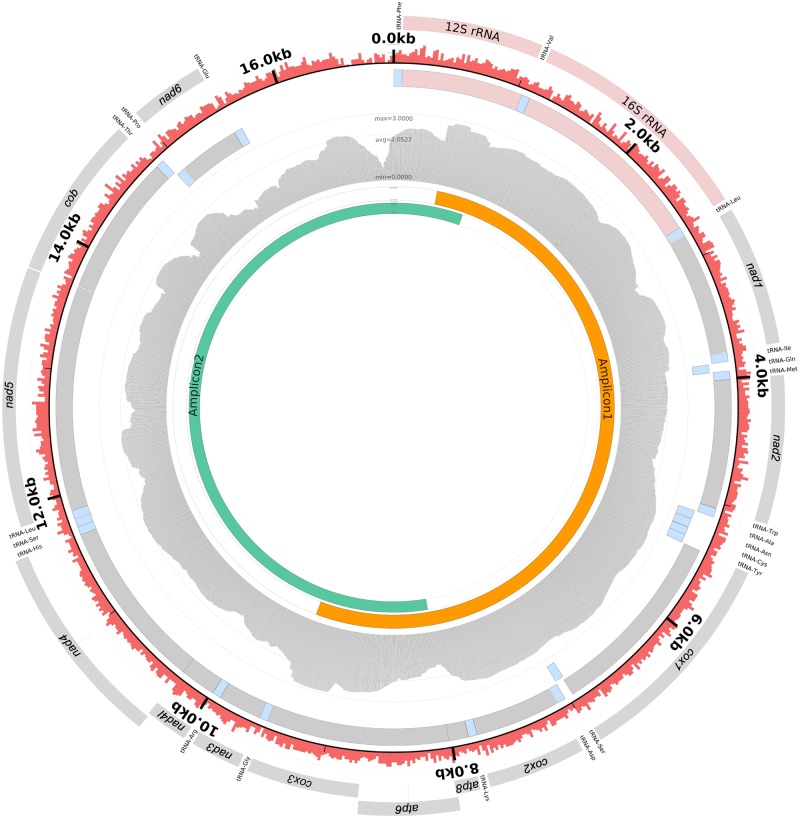
The total length of the raven mitogenomes ranged between 16,925 bp (Californian clade) and 16,928 bp (Holarctic clade), while that of the Chihuahuan raven was 16,928 bp and the brown-necked raven 16,927 bp. Gene annotation analyses revealed 13 protein coding genes, 2 rRNAs and 22 tRNAs (Fig 2). The gene arrangement followed a standard avian mitogenome model [39], similar to published mitogenomes from other Corvus species [22, 40, 41]. Analyses of base composition revealed a GC content of 43.8% (Holarctic clade), 43.6% (Californian clade), 43.7% (Chihuahuan raven) and 43.3% (brown-necked raven).
Fig 2. Graphical overview of the Corvus corax mitogenome.
The figure contains the following information, from the outermost to the innermost layer: (1) Gene products for annotated genes. (2) Genome position, minor ticks for every kb. Red bars illustrate GC-content in 20 bp windows. (3) Forward and (4) reverse genes. Blue colours show tRNAs, red colours show rRNAs and grey colours illustrate protein coding genes. (5) Coverage plot with log transformed coverage. (6) Position for amplicon 1 and (7) position for amplicon 2. The figure was created from GenBank accession KX245135 in the software Circleator [42].
There was full agreement between the clade assignment and phylogenetic relationships based on previous single-marker mtDNA studies ([17, 18]; authors' unpubl. data) and that based on the complete mitogenome (Fig 3; Table 1). The overall sequence divergence across the alignment between birds belonging to the Holarctic and the Californian clade of common ravens was 3%. However, there was a large degree of variation across the 13 protein coding genes, the two rRNA genes and the control region, ranging from 0.3% in 16S rRNA to 5.8% in ATP6 (Table 2). The divergence between the Holarctic clade and the Chihuahuan raven was very similar to that between the Holarctic and the Californian Clade (overall: 3.1%; range across regions: 0.3% - 5.5%; Table 2), while the Californian clade and the Chihuahuan raven was on average around two percentage points less diverged (overall: 1.1%; range across regions: 0.2% - 2.8%; Table 2).
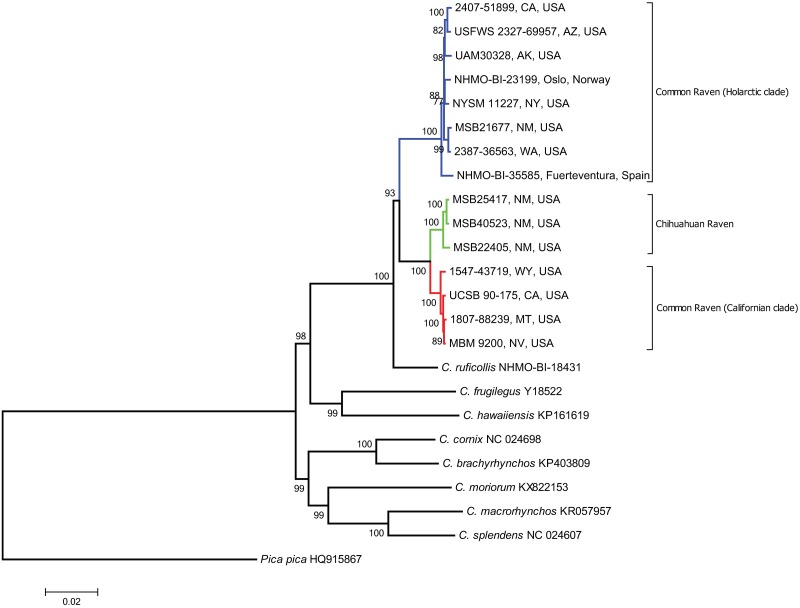
Fig 3. Maximum likelihood phylogeny inferred from whole mitogenomes of three species of ravens (common raven C. corax, Chihuahuan raven C. cryptoleucus, brown-necked raven C. ruficollis), and six other Corvus species with published mitogenomes (labeled with GenBank acc. no.).
The final dataset used in the phylogenetic analysis contained 15,392 bp after indels and positions in the alignment with less than 95% coverage were eliminated. The tree was rooted with Pica pica. USA = United States of America, CA = California, AZ = Arizona, AK = Alaska, NY = New York, NM = New Mexico, WA = Washington, WY = Wyoming, MT = Montana, NV = Nevada.
Table 2. Genetic distances (uncorrected p-distances) among Holarctic (HOL) and Californian (CAL) lineages of the common raven and the Chihuahuan (CHI) raven, for different partitions, the whole alignment and protein coding genes concatenated.
| Gene/Region | Comparison | Distance | SE | Length (bp)a |
|---|---|---|---|---|
| 12S rRNA | HOL-CAL | 0.0046 | 0.0022 | 980 |
| 12S rRNA | HOL-CHI | 0.0045 | 0.0021 | 980 |
| 12S rRNA | CAL-CHI | 0.0020 | 0.0014 | 980 |
| 16S rRNA | HOL-CAL | 0.0027 | 0.0013 | 1,601 |
| 16S rRNA | HOL-CHI | 0.0023 | 0.0013 | 1,601 |
| 16S rRNA | CAL-CHI | 0.0025 | 0.0013 | 1,601 |
| NAD1 | HOL-CAL | 0.026 | 0.008 | 975 |
| NAD1 | HOL-CHI | 0.026 | 0.008 | 975 |
| NAD1 | CAL-CHI | 0.007 | 0.003 | 975 |
| NAD2 | HOL-CAL | 0.031 | 0.005 | 1,041 |
| NAD2 | HOL-CHI | 0.032 | 0.006 | 1,041 |
| NAD2 | CAL-CHI | 0.021 | 0.004 | 1,041 |
| COXI | HOL-CAL | 0.026 | 0.004 | 1,551 |
| COXI | HOL-CHI | 0.026 | 0.004 | 1,551 |
| COXI | CAL-CHI | 0.004 | 0.002 | 1,551 |
| COXII | HOL-CAL | 0.019 | 0.005 | 681 |
| COXII | HOL-CHI | 0.020 | 0.005 | 681 |
| COXII | CAL-CHI | 0.011 | 0.003 | 681 |
| ATP8 | HOL-CAL | 0.033 | 0.013 | 153 |
| ATP8 | HOL-CHI | 0.023 | 0.012 | 153 |
| ATP8 | CAL-CHI | 0.010 | 0.004 | 153 |
| ATP6 | HOL-CAL | 0.058 | 0.009 | 684 |
| ATP6 | HOL-CHI | 0.055 | 0.009 | 684 |
| ATP6 | CAL-CHI | 0.028 | 0.007 | 684 |
| COXIII | HOL-CAL | 0.023 | 0.005 | 738 |
| COXIII | HOL-CHI | 0.025 | 0.005 | 738 |
| COXIII | CAL-CHI | 0.008 | 0.003 | 738 |
| NAD3 | HOL-CAL | 0.036 | 0.010 | 348 |
| NAD3 | HOL-CHI | 0.038 | 0.010 | 348 |
| NAD3 | CAL-CHI | 0.009 | 0.005 | 348 |
| NAD4l | HOL-CAL | 0.048 | 0.012 | 290 |
| NAD4l | HOL-CHI | 0.046 | 0.012 | 290 |
| NAD4l | CAL-CHI | 0.012 | 0.005 | 290 |
| NAD4 | HOL-CAL | 0.034 | 0.008 | 1,377 |
| NAD4 | HOL-CHI | 0.031 | 0.007 | 1,377 |
| NAD4 | CAL-CHI | 0.011 | 0.003 | 1,377 |
| NAD5 | HOL-CAL | 0.046 | 0.006 | 1,815 |
| NAD5 | HOL-CHI | 0.048 | 0.006 | 1,815 |
| NAD5 | CAL-CHI | 0.013 | 0.002 | 1,815 |
| COB | HOL-CAL | 0.039 | 0.007 | 1,093 |
| COB | HOL-CHI | 0.045 | 0.007 | 1,093 |
| COB | CAL-CHI | 0.017 | 0.004 | 1,093 |
| NAD6 | HOL-CAL | 0.043 | 0.010 | 518 |
| NAD6 | HOL-CHI | 0.042 | 0.010 | 518 |
| NAD6 | CAL-CHI | 0.013 | 0.005 | 518 |
| CR | HOL-CAL | 0.039 | 0.005 | 1,198 |
| CR | HOL-CHI | 0.043 | 0.005 | 1,198 |
| CR | CAL-CHI | 0.022 | 0.004 | 1,198 |
| Whole mt-genome | HOL-CAL | 0.029 | 0.001 | 16,553 |
| Whole mt-genome | HOL-CHI | 0.030 | 0.001 | 16,553 |
| Whole mt-genome | CAL-CHI | 0.011 | 0.001 | 16,553 |
| Protein coding genes | HOL-CAL | 0.037 | 0.002 | 11,275 |
| Protein coding genes | HOL-CHI | 0.038 | 0.002 | 11,275 |
| Protein coding genes | CAL-CHI | 0.013 | 0.001 | 11,275 |
a All alignment positions with missing data (gaps or ambiguous bases) were eliminated.
The monophyly of the Afro-Holarctic raven clade was also supported (bootstrap support = 100; Fig 3), as per previous studies based on single mtDNA loci or concatenation of nuclear introns and mtDNA [11–14]. The maximum likelihood tree estimated from whole mitogenomes strongly supported a sister relationship between the Californian clade of the common raven and the Chihuahuan raven (bootstrap support = 100; Fig 3). There was strong support for a sister relationship between the Holarctic clade and the clade containing both the Chihuahuan raven and the California clade (bootstrap support = 93; Fig 3). ML trees constructed from four different subsets of the mitogenome dataset (16S rRNA, ATP6, all protein coding genes, control region; S1–S4 Figs) recovered similar topologies as the whole mitogenome tree (Fig 3). As expected 16S rRNA showed the least resolution with generally low bootstrap values. For the other three subsets, we found high support for the monophyly of the Afro-Holarctic raven clade (98–100) and each of its three multi-individual terminal clades (Holarctic clade, California clade, Chihuahuan raven; 97–100), as well as for the sister relationship between the California clade and the Chihuahuan raven (94–100). The single brown-necked raven was placed outside these three clades in the ATP6 and all protein coding genes subsets, with moderate support (72–79), but came out together with the Holarctic clade in the control region tree, again with moderate support (75). Uncertainty with respect to the monophyly of the common raven species group in the different subsets could be explained by ambiguity stemming from variable levels of divergence with respect to the brown-necked raven.
Discussion
Whole mitogenome sequences confirmed previous findings based on single mitochondrial markers; a deep split between Holarctic and California clades within the common raven and a sister relationship between the California clade and the Chihuahuan raven (Fig 3) [12, 17]. The deep split was present in all parts of the mitogenome, although the different regions showed different levels of divergence (S1–S4 Figs), reflecting variable mutation rates among mitochondrial regions [43]. The sister relationship between the Californian clade and the Chihuahuan raven was also confirmed in all but the least diverged region (16S rRNA; S1 Fig). The apparent conflict between species boundaries supported by phenotypic and behavioural traits and the mtDNA tree can be explained by the speciation reversal hypothesis, wherein the mtDNA tree reflects the “original” divergence history of three previously distinct raven lineages (Holarctic, Californian and Chihuahuan) before secondary contact and speciation reversal of the Holarctic and California lineages into a single admixed species, the common raven [18]. An alternative hypothesis is that the mtDNA sister relationship of California and Chihuahuan ravens results from ancient introgressive hybridization between the two, which resulted in the capture and replacement of one species’ original mitochondrial lineage with the others’ (see [44] for a similar case in Emberiza buntings), with subsequent divergence. Differentiating between these three hypotheses—(1) mtDNA phylogeny reflects the original speciation history prior to speciation reversal of California and Holarctic lineages, (2) Chihuahuan ravens have mtDNA captured from ancient introgressive hybridization with California clade common ravens, (3) California clade common ravens have mtDNA captured from ancient introgressive hybridization with Chihuahuan ravens—would require the addition of nuclear genomic data and is beyond the scope of this study. However, the consistent signal of divergence and paraphyly combined with a lack of stop codons and frameshift mutations throughout the mitogenome, strongly supports a true mitochondrial origin of the highly diverged raven lineages.
Our study illustrates the advantages of using whole mitogenomes to resolve phylogenetic relationships. The high resolution of our whole mitogenome phylogeny suggests that uncertainty about relationships between the eight species of Afro-Holarctic ravens could be resolved with additional sequencing of mitogenomes from the species missing from our mitogenome dataset—i.e., C. albus, C. edithae, C. rhipidurus, C. albicollis, and C. crassirostris. Inclusion of the remaining members of the C. ruficollis species group is particularly critical given that some studies have supported these species as closer to the Holarctic common raven clade than the Holarctic clade is to either the California common raven clade and the Chihuahuan raven (i.e., paraphyly of the “common raven species group”; [12, 13]). This conflicts with our strong support for the monophyly of the “common raven species group” to the exclusion of C. ruficollis in the analysis based on whole mitogenomes (Fig 3). Note, however, that a sister relationship between C. ruficollis and the Holarctic clade was recovered in the control region (S4 Fig), albeit with rather weak support. Given the conflict between phylogenies based on mitogenomes (this study), single loci [13] and multilocus datasets [11, 12, 45], and considering reports that many of the species in the C. ruficollis species group hybridize with each other [19], we suggest that future attempts to produce a fully resolved phylogeny for the Afro-Holarctic ravens should include multiple individuals from the ranges of all species (including regions of allopatry and sympatry where possible), incorporate whole mitogenomes, and extensive sampling of the nuclear genome (e.g. sequence capture or whole genome re-sequencing approaches).
In conclusion, mitogenomes support previously found deep lineages and paraphyly in the common raven/Chihuahuan raven species complex. Our sequencing strategy provides 16 new mitogenomes with high coverage even from the smallest IonPGM chip, showing that our approach of long-range PCR amplification and IonPGM sequencing provides a cost efficient alternative to mitogenome assembly from whole genome sequencing data.
Supporting information
The tree was rooted with Pica pica. USA = United States of America, CA = California, AZ = Arizona, AK = Alaska, NY = New York, NM = New Mexico, WA = Washington, WY = Wyoming, MT = Montana, NV = Nevada.
(TIF)
The tree was rooted with Pica pica. USA = United States of America, CA = California, AZ = Arizona, AK = Alaska, NY = New York, NM = New Mexico, WA = Washington, WY = Wyoming, MT = Montana, NV = Nevada.
(TIF)
The tree was rooted with Pica pica. USA = United States of America, CA = California, AZ = Arizona, AK = Alaska, NY = New York, NM = New Mexico, WA = Washington, WY = Wyoming, MT = Montana, NV = Nevada.
(TIF)
C moriorum was excluded from the tree due to missing data. The tree was rooted with Pica pica. USA = United States of America, CA = California, AZ = Arizona, AK = Alaska, NY = New York, NM = New Mexico, WA = Washington, WY = Wyoming, MT = Montana, NV = Nevada.
(TIF)
The table provide index of substitution saturation (ISS) and critical values of ISS for symmetric and asymmetric tree topologies. The ISS values were significantly lower (p <0.0001) than the critical ISS in all data sets.
(DOCX)
(DOCX)
Acknowledgments
We thank the following persons and institutions for generously sharing samples: John Marzluff, Marco Restani, Bryan Bedrosian, William Boarman, Eduardo García-del-Rey, Reuven Yosef, Carsten Lome, Mark Holmgren at the University of California, Santa Barbara, Chris Witt at the University of New Mexico, Museum of Southwestern Biology, Division of Birds, Kevin Winker at the University of Alaska, Bird Collection, John Klicka at the Burke Museum of Natural History and Culture and Department of Biology, University of Washington and Jeremy Kirchman at the New York State Museum. Thanks to Silje Hogner for lab assistance.
Data Availability
Mitogenomes were deposited in GenBank (accession no. KX245133-KX245148). The sequence data are available in the NCBI Sequence Read Archive in the Bioproject no. PRJNA321255.
Funding Statement
This study was financed by a grant to AJ from the Norwegian Research Council, https://www.forskningsradet.no/en/Home_page/1177315753906 (grant no 213592). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Smith DR. The past, present and future of mitochondrial genomics: have we sequenced enough mtDNAs? Brief Funct Genomics. 2015; 15: 47–54. 10.1093/bfgp/elv027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci USA. 2015; 112: 10177–10184. 10.1073/pnas.1422049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moray C, Lanfear R, Bromham L. Domestication and the mitochondrial genome: comparing patterns and rates of molecular evolution in domesticated mammals and birds and their wild relatives. Genome Biol Evol. 2014; 6: 161–169. 10.1093/gbe/evu005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol Biol Evol. 2007; 24: 269–280. 10.1093/molbev/msl158 [DOI] [PubMed] [Google Scholar]
- 5.Sloan DB, Havird JC, Sharbrough J. The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol Ecol. 2017; 26: 2212–2236. 10.1111/mec.13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allcock AL, Cooke IR, Strugnell JM. What can the mitochondrial genome reveal about higher-level phylogeny of the molluscan class Cephalopoda? Zool J Linn Soc. 2011; 161: 573–586. 10.1111/j.1096-3642.2010.00656.x [DOI] [Google Scholar]
- 7.Miya M, Nishida M. The mitogenomic contributions to molecular phylogenetics and evolution of fishes: a 15-year retrospect. Ichthyological Research. 2015; 62: 29–71. 10.1007/s10228-014-0440-9 [DOI] [Google Scholar]
- 8.Haran J, Timmermans MJTN, Vogler AP. Mitogenome sequences stabilize the phylogenetics of weevils (Curculionoidea) and establish the monophyly of larval ectophagy. Mol Phyl Evol. 2013; 67: 156–166. 10.1016/j.ympev.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Talavera G, Vila R. What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evol Biol. 2011; 11: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song H, Buhay JE, Whiting MF, Crandall KA. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci USA. 2008; 105: 13486–13491. 10.1073/pnas.0803076105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jønsson K, Fabre P-H, Irestedt M. Brains, tools, innovation and biogeography in crows and ravens. BMC Evol Biol. 2012; 12: 72 10.1186/1471-2148-12-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman CR, Omland KE. Phylogenetics of the common raven complex (Corvus: Corvidae) and the utility of ND4, COI and intron 7 of the β-fibrinogen gene in avian molecular systematics. Zool scripta. 2005; 34: 145–156. [Google Scholar]
- 13.Haring E, Däubl B, Pinsker W, Kryukov A, Gamauf A. Genetic divergences and intraspecific variation in corvids of the genus Corvus (Aves: Passeriformes: Corvidae)–a first survey based on museum specimens. J Zool Syst Evol Res. 2012; 50: 230–246. 10.1111/j.1439-0469.2012.00664.x [DOI] [Google Scholar]
- 14.Rutz C, Klump BC, Komarczyk L, Leighton R, Kramer J, Wischnewski S, et al. Discovery of species-wide tool use in the Hawaiian crow. Nature. 2016; 537: 403–407. 10.1038/nature19103 [DOI] [PubMed] [Google Scholar]
- 15.Omland KE, Baker JM, Peters JL. Genetic signatures of intermediate divergence: population history of Old and New World Holarctic ravens (Corvus corax). Mol Ecol. 2006; 15: 795–808. 10.1111/j.1365-294X.2005.02827.x [DOI] [PubMed] [Google Scholar]
- 16.Johnsen A, Rindal E, Ericson P, Zuccon D, Kerr K, Stoeckle M, et al. DNA barcoding of Scandinavian birds reveals divergent lineages in trans-Atlantic species. J Ornithol. 2010; 151: 565–578. [Google Scholar]
- 17.Omland KE, Tarr CL, Boarman WI, Marzluff JM, Fleischer RC. Cryptic genetic variation and paraphyly in ravens. Proc R Soc B. 2000; 267: 2475–2482. 10.1098/rspb.2000.1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webb WC, Marzluff JM, Omland KE. Random interbreeding between cryptic lineages of the Common Raven: evidence for speciation in reverse. Mol Ecol. 2011; 20: 2390–2402. 10.1111/j.1365-294X.2011.05095.x [DOI] [PubMed] [Google Scholar]
- 19.del Hoyo J, Elliot A, Christie D, editors. Handbook of the Birds of the World—Volume 14 Bush-shrikes to Old World Sparrows. Barcelona: Lynx Edicions; 2009. [Google Scholar]
- 20.Baker JM, Omland KE. Canary Island Ravens Corvus corax tingitanus have distinct mtDNA. Ibis. 2006; 148: 174–178. 10.1111/j.1474-919X.2006.00493.x [DOI] [Google Scholar]
- 21.Lifjeld JT, Anmarkrud JA, Calabuig P, Cooper JEJ, Johannessen LE, Johnsen A, et al. Species-level divergences in multiple functional traits between the two endemic subspecies of Blue Chaffinches Fringilla teydea in Canary Islands. BMC Zoology. 2016; 1: 1–19. 10.1186/s40850-016-0008-4 [DOI] [Google Scholar]
- 22.Krzeminska U, Wilson R, Rahman S, Song BK, Gan HM, Tan MH, et al. The complete mitochondrial genome of the invasive house crow Corvus splendens (Passeriformes: Corvidae). Mitochondrial DNA A DNA Mapp Seq Anal. 2016; 27: 974–975. 10.3109/19401736.2014.926512 [DOI] [PubMed] [Google Scholar]
- 23.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 2013; 41: e129–e129. 10.1093/nar/gkt371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh TR, Shneor O, Huchon D. Bird Mitochondrial Gene Order: Insight from 3 Warbler Mitochondrial Genomes. Mol Biol Evol. 2008; 25: 475–477. 10.1093/molbev/msn003 [DOI] [PubMed] [Google Scholar]
- 26.Gibb GC, England R, Hartig G, McLenachan PA, Taylor Smith BL, McComish BJ, et al. New Zealand passerines help clarify the diversification of major songbird lineages during the oligocene. Genome Biol Evol. 2015. 10.1093/gbe/evv196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, et al. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013; 14: 193–202. 10.1093/bib/bbs012 [DOI] [PubMed] [Google Scholar]
- 28.Bragg LM, Stone G, Butler MK, Hugenholtz P, Tyson GW. Shining a Light on Dark Sequencing: Characterising Errors in Ion Torrent PGM Data. PLoS Comput Biol. 2013; 9: e1003031 10.1371/journal.pcbi.1003031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekblom R, Smeds L, Ellegren H. Patterns of sequencing coverage bias revealed by ultra-deep sequencing of vertebrate mitochondria. BMC Genomics. 2014; 15: 467 10.1186/1471-2164-15-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anmarkrud JA, Lifjeld JT. Complete mitochondrial genomes of eleven extinct or possibly extinct bird species. Mol Ecol Res. 2017; 17: 334–341. 10.1111/1755-0998.12600 [DOI] [PubMed] [Google Scholar]
- 31.Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, et al. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phyl Evol. 2013; 69: 313–319. 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004; 101: 11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016; 33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia X, Lemey P. Assessing substitution saturation with DAMBE In: Lemey P, Salemi M, Vandamme A-M, editors. The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing. 2 ed Cambridge: Cambridge University Press; 2009. [Google Scholar]
- 35.Xia X, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Mol Phyl Evol. 2003; 26: 1–7. 10.1016/S1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- 36.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- 37.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985; 39: 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- 38.Fleischer RC, Boarman WI, Gonzalez EG, Godinez A, Omland KE, Young S, et al. As the raven flies: using genetic data to infer the history of invasive common raven (Corvus corax) populations in the Mojave Desert. Mol Ecol. 2008; 17: 464–474. 10.1111/j.1365-294X.2007.03532.x [DOI] [PubMed] [Google Scholar]
- 39.Desjardins P, Morais R. Sequence and gene organization of the chicken mitochondrial genome. J Mol Biol. 1990; 212: 599–634. 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- 40.Krzeminska U, Wilson R, Rahman S, Song BK, Seneviratne S, Gan HM, et al. Mitochondrial genomes of the jungle crow Corvus macrorhynchos (Passeriformes: Corvidae) from shed feathers and a phylogenetic analysis of genus Corvus using mitochondrial protein-coding genes. Mitochondrial DNA A DNA Mapp Seq Anal. 2016; 27: 2668–2670. 10.3109/19401736.2015.1043540 [DOI] [PubMed] [Google Scholar]
- 41.Li X, Lu J, Lu J, Hu X, Huang Z. The complete mitochondrial genome of the American crow, Corvus brachyrhynchos (Passeriformes, Corvidae). Mitochondrial DNA A DNA Mapp Seq Anal. 2016; 27: 4213–4214. 10.3109/19401736.2015.1022745 [DOI] [PubMed] [Google Scholar]
- 42.Crabtree J, Agrawal S, Mahurkar A, Myers GS, Rasko DA, White O. Circleator: flexible circular visualization of genome-associated data with BioPerl and SVG. Bioinformatics. 2014; 30: 3125–3127. 10.1093/bioinformatics/btu505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galtier N, Enard D, Radondy Y, Bazin E, Belkhir K. Mutation hot spots in mammalian mitochondrial DNA. Genome Res. 2006; 16: 215–222. 10.1101/gr.4305906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Irwin DE, Rubtsov AS, Panov EN. Mitochondrial introgression and replacement between yellowhammers (Emberiza citrinella) and pine buntings (Emberiza leucocephalos) (Aves: Passeriformes). Biol J Linn Soc. 2009; 98: 422–438. 10.1111/j.1095-8312.2009.01282.x [DOI] [Google Scholar]
- 45.Scofield RP, Mitchell KJ, Wood JR, De Pietri VL, Jarvie S, Llamas B, et al. The origin and phylogenetic relationships of the New Zealand ravens. Mol Phyl Evol. 2017; 106: 136–143. 10.1016/j.ympev.2016.09.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The tree was rooted with Pica pica. USA = United States of America, CA = California, AZ = Arizona, AK = Alaska, NY = New York, NM = New Mexico, WA = Washington, WY = Wyoming, MT = Montana, NV = Nevada.
(TIF)
The tree was rooted with Pica pica. USA = United States of America, CA = California, AZ = Arizona, AK = Alaska, NY = New York, NM = New Mexico, WA = Washington, WY = Wyoming, MT = Montana, NV = Nevada.
(TIF)
The tree was rooted with Pica pica. USA = United States of America, CA = California, AZ = Arizona, AK = Alaska, NY = New York, NM = New Mexico, WA = Washington, WY = Wyoming, MT = Montana, NV = Nevada.
(TIF)
C moriorum was excluded from the tree due to missing data. The tree was rooted with Pica pica. USA = United States of America, CA = California, AZ = Arizona, AK = Alaska, NY = New York, NM = New Mexico, WA = Washington, WY = Wyoming, MT = Montana, NV = Nevada.
(TIF)
The table provide index of substitution saturation (ISS) and critical values of ISS for symmetric and asymmetric tree topologies. The ISS values were significantly lower (p <0.0001) than the critical ISS in all data sets.
(DOCX)
(DOCX)
Data Availability Statement
Mitogenomes were deposited in GenBank (accession no. KX245133-KX245148). The sequence data are available in the NCBI Sequence Read Archive in the Bioproject no. PRJNA321255.