Abstract
The pharmacological effectiveness of oligonucleotides has been hampered by their tendency to remain entrapped in endosomes thus limiting their access to cytosolic or nuclear targets. We have previously reported a group of small molecules that enhance the effects of oligonucleotides by causing their release from endosomes. Here we describe a second novel family of oligonucleotide enhancing compounds (OECs) that is chemically distinct from the compounds reported previously. We demonstrate that these molecules substantially augment the actions of splice switching oligonucleotides (SSOs) and antisense oligonucleotides (ASOs) in cell culture. We also find enhancement of SSO effects in a murine model. These new compounds act by increasing endosome permeability and causing partial release of entrapped oligonucleotides. While they also affect the permeability of lysosomes, they are clearly different from typical lysosomotropic agents. Current members of this compound family display a relatively narrow window between effective dose and toxic dose. Thus further improvements are necessary before these agents can become suitable for therapeutic use.
Graphical abstract
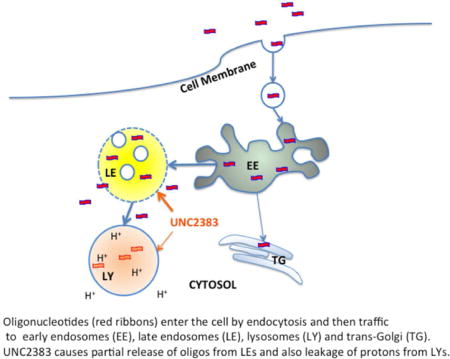
INTRODUCTION
Investigators have sought to utilize the precise effects of siRNA, antisense oligonucleotides (ASOs), and splice switching oligonucleotides (SSOs) for the therapy of cancer and other diseases(1, 2). However, even with massive investments in the medicinal chemistry and formulation of these molecules(3, 4), and despite initial advances in the clinic(5, 6) therapeutic use of oligonucleotides has thus far succeeded to only a limited degree. An important constraining factor is the ineffective delivery of oligonucleotides to their intracellular sites of action in the cytosol or nucleus, due in large part to trapping in endosomal compartments(7–9).
Oligonucleotides enter cells by various endocytotic pathways(10, 11). Initial uptake is followed by trafficking into multiple endomembrane compartments including early/sorting endosomes, late endosomes/multi-vesicular bodies, lysosomes and the Golgi complex(12, 13). During trafficking events, discontinuities in the lipid bilayer can occur thus potentially allowing for partial escape of vesicle contents(14, 15). Patterns of intracellular trafficking are regulated by a plethora of proteins that modulate the formation, movement and coalescence of membrane bound vesicles(16, 17). Thus it seems likely that small molecules could potentially regulate intracellular trafficking as well as the permeability properties of endomembrane vesicles. However, few such compounds have been described(18, 19).
Recently we conducted a high-throughput screen of multiple, diverse chemical libraries to identify small molecules that could enhance the pharmacological activities of oligonucleotides. We have previously reported a description of the screen as well as initial characterization of one group of oligonucleotide enhancing compounds (OECs) that act by partially releasing oligonucleotides from entrapment within endosomes(20). Here we describe a second family of compounds derived from a hit that emerged from that screen. As with the previously reported molecules, these compounds substantially enhance the effectiveness of oligonucleotides in the absence of any conventional transfection agents. The current compounds also act by increasing oligonucleotide release from endomembrane compartments. However, there is a relatively narrow gap between effective and toxic concentrations.
RESULTS & DISCUSSION
The original high throughput screen involved a HeLa cell line stably transfected with a luciferase reporter construct that is responsive to SSOs. Approximately 150,000 compounds were tested for their ability to enhance the luciferase induction effect of a SSO but not a mismatched oligonucleotide. The prototypical compound of the first family of small molecules discovered in the screen was termed UNC10217938 and has been described in detail elsewhere(20).
UNC2383, which emerged as another preliminary hit from our high throughput screen, is chemically distinct from previously published oligonucleotide enhancing compounds (Figure 1a). We tested UNC2383 for its ability to increase the effectiveness of a SSO using an assay that was similar to the initial high throughput screen. The cells were first incubated with the SSO followed by brief treatment with UNC2383; thereafter luciferase activity and cell protein were measured. As seen in Figure 1b exposure to increasing concentrations of UNC2383 resulted in progressive increases in luciferase induction. This result was highly specific since incubation with a mismatched oligonucleotide followed by UNC2383 had no effect. The onset of action of UNC2383 was quite rapid with effects first observed within 30 min. and reaching a plateau by 120 min. (Figure 1c). The cytotoxicity of 2383 in two cell lines is depicted in Figure 1d. As seen, the compound showed little toxicity at concentrations of 10uM or less, but toxicity was evident at higher concentrations.
Figure 1. Effects with SSOs.
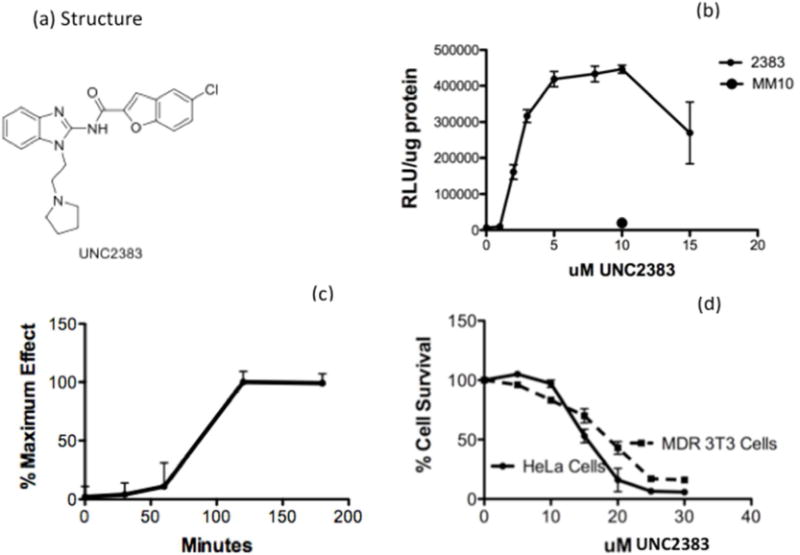
(a) Structure of UNC2383. (b) UNC2383 enhances SSO effects. HeLa Luc 705 cells were incubated in 24-well plates with 100 nM SSO623 or its mismatched control (MM) for 16 h in DMEM +10% FBS, rinsed and then treated with various concentrations of UNC2383 for 2 h. The cells were then rinsed and incubation continued for an additional 4 h in DMEM +10% FBS. Cells were rinsed twice in PBS and luciferase activity (RLU) and cell protein determined. The larger round symbol indicates the MM oligonucleotide. Means +/− SE. N = 3. Cell protein data for this assay is in Supporting Table 2. (c) Kinetics of UNC2383 action. Cells were preloaded with 100 nM SSO623 and then exposed to UNC2383 for various periods after which the compound was removed. The cells were further incubated and then luciferase and protein determined. The total time of incubation in each case was 6 h. (d) Cytotoxicity of UNC2383. HeLa Luc705 or NIH-3T3-MDR cells were exposed to UNC2383 as in (b) then incubated for 24 h in DMEM plus 10% FBS and tested using the Alamar Blue cytotoxicity assay. Means +/− SE. N = 3.
In an effort to improve the efficacy and reduce toxicity of UNC2383 we prepared several analogs. While some were inactive (namely those resulting from modification to the ethylpyrrolidine sidechain), several closely related analogs, which generally contain modifications to the substituents on the benzofuran and benzimidazole ring systems (see Supporting Information Figure 1), displayed activity in enhancing SSO effects. Typical dose-response curves for luciferase induction and cytotoxicity are shown in Supporting Figure 2. A summary of EC50 versus TC50 data for the analogs is given in Table 1. Interestingly, UNC4267 had somewhat reduced potency relative to UNC2383 but was also substantially less toxic. These results suggest that considerable alterations in effectiveness and toxicity may be possible through further modification. Chemical information regarding the synthesis of the various analogs is provided in the Supporting Information section.
Table 1.
TC50/EC50 Ratios for 2383 Analogs
| COMPOUND | EC50 (uM) | TC50 (uM) | TC50/EC50 |
|---|---|---|---|
| UNC2383 | 2 | 15 | 7.5 |
| UNC4425 | 6 | 12.5 | 2.1 |
| UNC4426 | 3.5 | 13 | 3.7 |
| UNC4428 | 10 | 50 | 5 |
| UNC4251 | 2 | 7 | 3.5 |
| UNC4253 | 11 | 48 | 4.4 |
| UNC4258 | 13 | 65 | 5 |
| UNC4267 | 8 | 68 | 8.5 |
These values were determined in experiments similar to Figure 1b,d.
We also tested UNC2383 for its ability to affect ASOs rather than SSOs. The ASO was designed to inhibit expression of the MDR1 gene and its product the P-glycoprotein multidrug transporter(21). Multi-drug resistant NIH-3T3-MDR cells were incubated with ASOs with or without further treatment with UNC2383 (Figure 2). One set of cells was treated with the ASO complexed with Lipofectamine 2000 as a positive control. Cell surface expression of P-glycoprotein was monitored using an Alexa 488 labeled anti-Pgp monoclonal antibody and flow cytometry. Treatment of the NIH-3T3-MDR cells with ASO alone had virtually no effect on Pgp levels, while treatment with ASO complexed with Lipofectamine 2000 caused a left shift of the flow cytometry profile indicating reduced Pgp expression. Similarly, incubation with ASO followed by treatment with UNC2383 resulted in a substantial reduction in Pgp expression thus indicating that UNC2383 can strongly enhance effects of ASOs as well as SSOs.
Figure 2. Effects with ASOs.
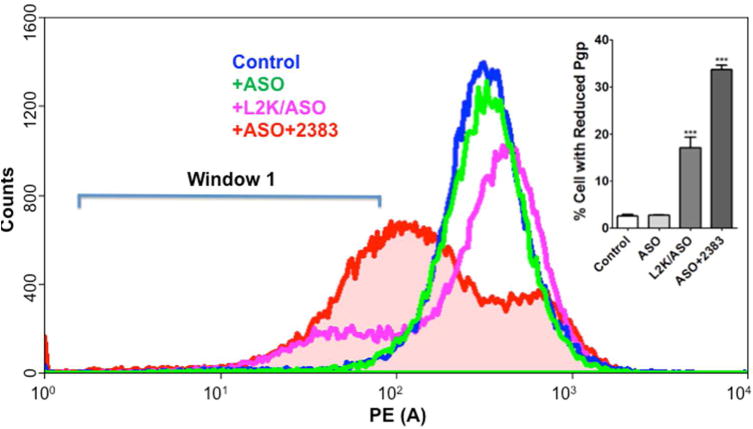
Reduction of Pgp expression. NIH 3T3-MDR cells were incubated with 100 nM anti-MDR1 ASO or a mismatched (MM) control for 16 h in DMEM +1% FBS. Cells were rinsed and then treated with 10 uM UNC2383 for 2 h. The compound was removed and the cells further incubated for 48 h. Expression of Pgp on the cell surface was determined by treating cells with Alexa 488 labeled anti-Pgp monoclonal antibody and binding quantitated by flow cytometry (PE(A)=units of fluorescence intensity). Treatment with ASO complexed with Lipofectamine 2000 (L2K) was a positive control. Y-axis, cell counts; X-axis, Alexa 488 fluorescence. In the inset the ordinate is the percentage of cells in Window 1 (reduced Pgp expression). Means +/− SE. N = 3.
Based on the above results, wherein UNC2383 enhanced the effects of both SSOs and ASOs, molecules which have completely distinct mechanisms of action, it seemed likely that UNC2383 affects the trafficking and delivery of the oligonucleotides rather than their molecular mechanisms. Accordingly we tested UNC2383 for its ability to release material from endomembrane entrapment. Thus cells were incubated with a highly fluorescent dextran that is taken up via pinocytosis, then rinsed and treated with UNC2383, followed by observation using confocal fluorescence microscopy. As seen in Figure 3a,b control cells displayed an abundance of distinct, highly fluorescent vesicles containing entrapped dextran. However, there was no visible fluorescence in the cytosol itself. In contrast, in cells exposed to UNC2383 (Figure 3c,d) the fluorescence intensity of vesicles was reduced and there was clear evidence of a diffuse cytosolic fluorescence. This indicates that UNC2383 caused partial release of the dextran from vesicular entrapment. We also examined effects on the distribution of a SSO labeled with the fluorophore TAMRA. As seen in Figure 3e,f, treatment with UNC2383 caused a partial but substantial re-localization of the labeled oligonucleotide to the nucleus, which was delineated using a Hoechst dye. Quantitation of the increase in TAMRA fluorescence in the nucleus due to UNC2383 is shown in Figure 3g. Thus UNC2383 seems to act in a manner similar to previously described oligonucleotide enhancing compounds(18, 20) in that it increases the permeability of endomembranes which allows oligonucleotides to have greater access to the cytosol and nucleus. Encouragingly, similar results on oligonucleotide redistribution to the nucleus were seen with other UNC2383 analogs (Supporting Figure 3).
Figure 3. Effect on Endomembrane Permeability.
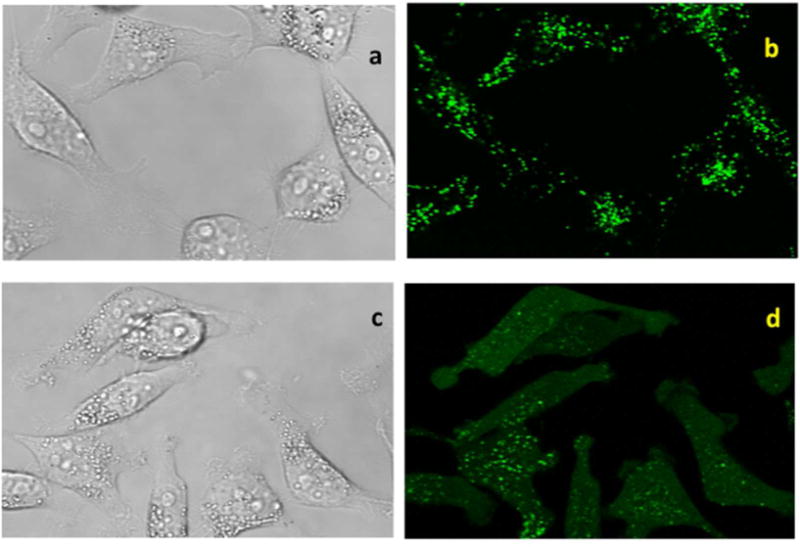
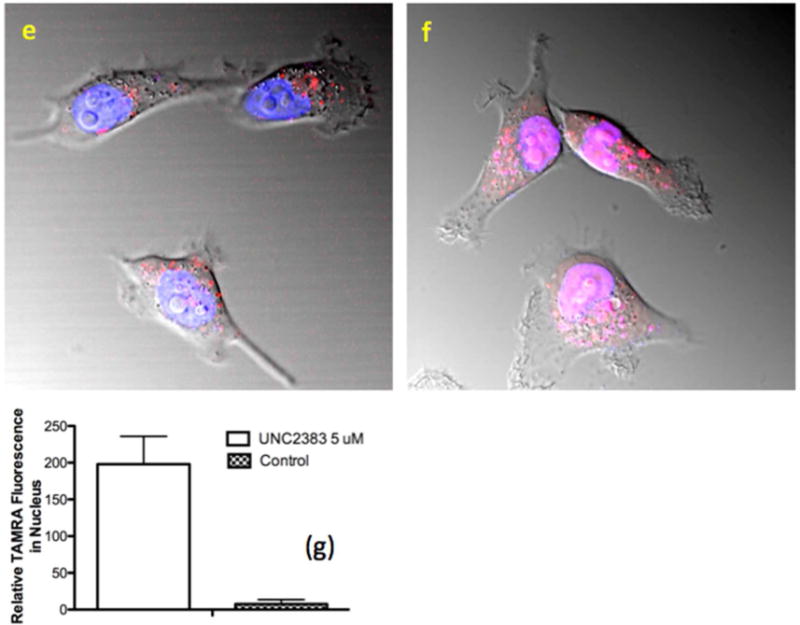
HeLa Luc 705 cells (50,000) were seeded into glass-bottom dishes and incubated at 37°C for attachment. Alexa 488-Dextran 10K (200μg/ml) or TAMRA SSO 623 (2.5 uM) were added into medium and the cells incubated for 24 hours. Cells were washed with PBS and then placed back in medium and treated with UNC2383 for 2 h or maintained as controls. Cells were rinsed after drug treatment. Live cells were imaged with an Olympus FV1200 or a Zeiss LSM710 confocal microscope. For the studies with Alexa 488 Dextran, the controls are shown in images (a,b) while cells treated with 10 uM UNC2383 are in images (c,d). For studies with TAMRA SSO 623 the control is image (e) while image (f) shows cells treated with 5uM UNC2383. Images (e,f) are composites of TAMRA (red), Hoechst (blue) and DIC images and the pink coloration in (f) indicates overlap of the TAMRA and Hoechst fluorescence. Images are typical of three independent assays. Intensity settings were identical for fluorescence images with or without UNC2383. Panel (g) quantitates the TAMRA fluorescence in the nucleus for controls versus cells treated with 5 uM UNC2383, as measured using Fiji software, with Hoechst stain used to delineate the nucleus (N=12).
We also examined the effects of UNC2383 on the co-localization of the SSO with markers for specific endomembrane compartments by using baculovirus vectors that express GFP chimeras of marker proteins (Figure 4). In control cells there was substantial co-localization of the TAMRA-SSO with GFP-LAMP1, a lysosome marker (4a), and with GFP-Rab7, a late endosome marker (4c). Treatment with UNC2383 caused a partial re-localization of the TAMRA-SSO to the nucleus and affected its co-localization with marker proteins (4b,d). Additional examples of these effects are shown in Supporting Figure 4. The extent of co-localization of oligonucleotide with the endomembrane marker proteins was quantitated using the Manders’ Correlation Coefficient(22) and is shown in (4e). Treatment with UNC2383 substantially reduced the degree of co-localization of the TAMRA-SSO with both late endosomes and lysosomes, as would be expected given the re-localization of oligonucleotide to the nucleus using this compound.
Figure 4. Effects on Subcellular Localization of Oligonucleotide.
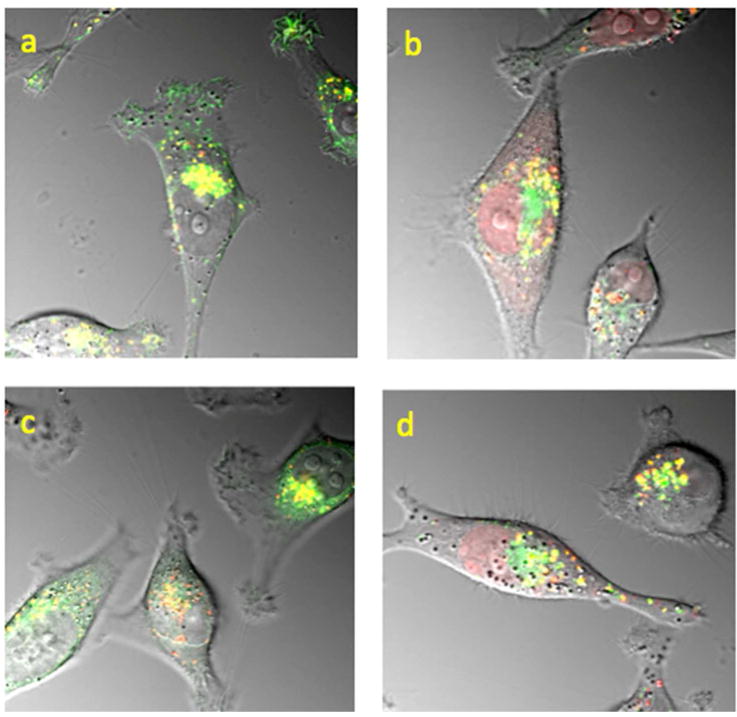
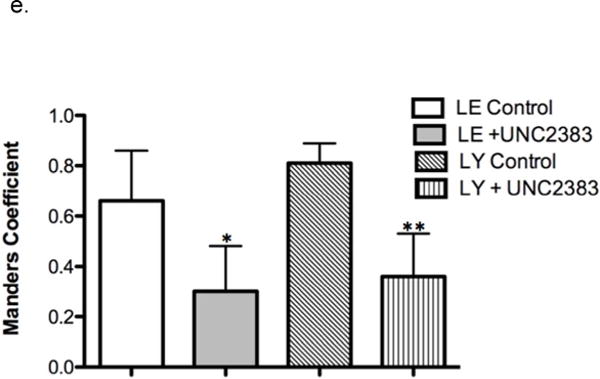
HeLa Luc 705 cells were dually labeled with TAMRA SSO and, by using baculovirus vectors, with GFP-Rab7 or GFP-LAMP1 as markers for late endosomes (LE) or lysosomes (LY) respectively. Subsequently cells were treated for 2h with 5 uM UNC2383 or maintained as controls. Composite images showing TAMRA fluorescence (red), GFP (green) and DIC are provided. Overlap is indicated in yellow/orange. (a) GFP-LAMP1 plus TAMRA, control; (b) GFP-LAMP1 plus TAMRA with 5 uM UNC2383; (c) GFP-Rab7 plus TAMRA, control; (d) GFP-Rab7 plus TAMRA with 5 uM UNC2383. Panel (e) shows a plot of the Manders’ Correlation Coefficients for TAMRA versus GFP in control cells and cells treated with 5 uM UNC2383 (N=12). The differences in Manders’ Coefficient in control versus treated cells were significant at the 5% level for LE and at the 1% level for LY, both using the paired t-Test.
We also investigated the effects of our compounds on lysosomes since such interactions may have toxic consequences. The effects of our compounds on the integrity of lysosomes were evaluated using the fluorescent probe Lysotracker Red ® which accumulates in low pH compartments, particularly lysosomes. Thus perturbation of the integrity of lysosome membrane would lead to leakage of protons, increased intra-lysosomal pH, and reduced lysotracker accumulation. As seen in Figure 5a, low concentrations of UNC2383 or its analogs that were effective in luciferase induction, but non-toxic in the Alamar Blue cytotoxicity assay, had limited effects on lysotracker accumulation, while higher concentrations of the compounds strongly affected this parameter. For example, 2uM UNC2383 or 10 uM of UNC4267, UNC4258 or UNC4428, concentrations that quite strongly enhanced SSO actions (see Figure 1b and Supporting Figure 2), had little effect on lysotracker accumulation. This suggests that, at low compound concentrations, some oligonucleotide is released from higher pH endomembrane compartments that do not strongly accumulate lysotracker dye, while at high compound concentrations low pH compartments such as lysosomes are also affected.
Figure 5. Effects on Lysosomes.
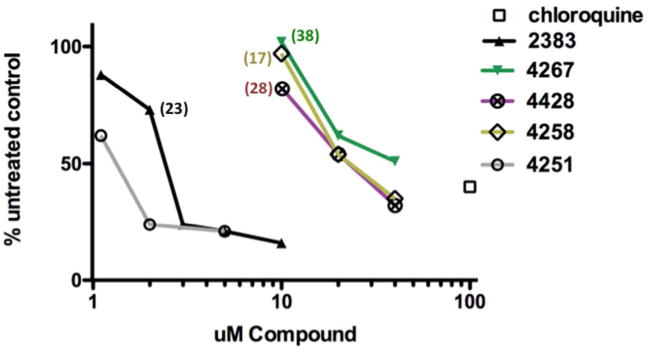
(a) Lysotracker Uptake. Accumulation of Lysotracker Red in HeLa Luc 705 cells was measured after a 2h exposure of cells to the indicated concentrations of various analogs. The lysosomotropic compound chloroquine was included as a positive control. Means of triplicate determinations are shown. The color-coded numbers refer to the fold increase in luciferase induction attained by the concentration of analog associated with the adjacent symbol on the graph. The numbers are taken from the data of Fig 1b for UNC2383 and Supporting Fig 2 for UNC4267, UNC4258 and UNC4428.
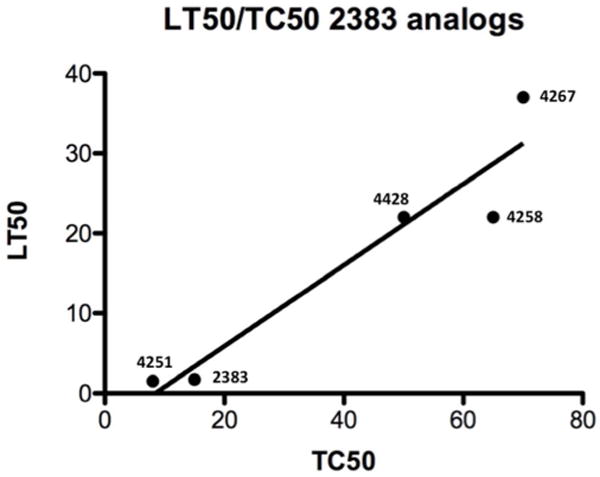
(b) LT50 vs TC50. The plot shows the ratio of the concentration of analog required for 50% inhibition of Lysotracker Red uptake (LT50) to the concentration required for 50% cell killing (TC50) under the same conditions of exposure. Nonlinear regression was calculated using Prism® software. The R2 value for the plot is 0.91.
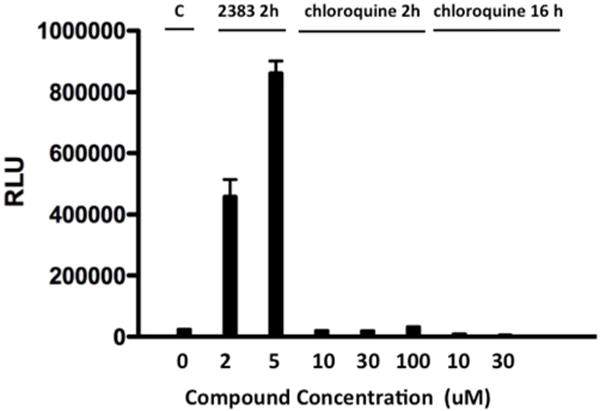
(c) 2383 vs chloroquine; luciferase induction. HeLa Luc705 cells were exposed to SSO623 and then treated with various concentrations of UNC2383 or chloroquine for 2h and then tested for luciferase induction following the methods described in Figure 1. In a sub-set of the experiment cells were exposed to chloroquine for 16 h rather than 2h. Means +/− SE. N=3.
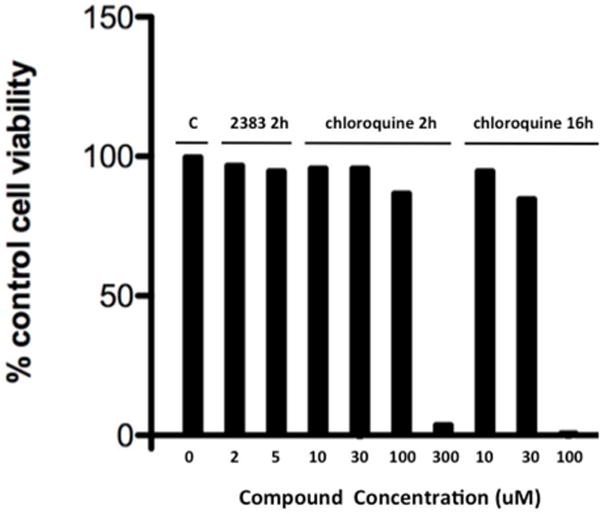
(d) UNC2383 vs chloroquine; cytotoxicity. After treatment as in 5c the cells were tested for viability using the Alamar Blue assay. Means, N=3.
A plot of the concentration for 50% toxicity versus the concentration for 50% inhibition of lysotracker accumulation for 5 analogs showed an approximately linear relationship (Figure 5b). Thus part of the toxicity of these compounds may be ascribed to permeabilization of lysosomes, although other effects cannot be ruled out. There is an enormous literature on the benzimidazole pharmacophore with various compounds described as having multiple therapeutic or toxic effects(23); thus it is difficult to specify additional toxicities. Interestingly, substantial reductions in lysotracker accumulation were observed at compound concentrations that did not affect cell viability, indicating that some degree of permeabilization of lysosomes can be tolerated. This is also suggested by the slope of Figure 5b that is less than 1 (~0.5). Importantly, although UNC2383 and its analogs affect lysosomes, they are not typical lysosomotropic compounds. This is clearly shown in Figure 5c & d where we compare the effects of UNC2383 to those of chloroquine, a classic lysosomotrope. Chloroquine had little effect on SSO-mediated luciferase induction even at very high concentrations or long duration of exposure, whereas UNC2383 was very active at non-toxic concentrations.
We have also examined the effects of UNC2383 in vivo. We used a transgenic mouse model termed EGFP654 that incorporates an EGFP reporter whose coding sequence is interrupted by an intron that is aberrantly spliced, resulting in failure to produce mature EGFP mRNA and protein(24). However, successful delivery of an appropriate SSO will correct splicing leading to restoration of message and protein expression in tissues(20). In these studies we pretreated mice with the SSO and subsequently administered UNC2383. As seen in Figure 6 (a–e), treatment with the SSO plus the small molecule resulted in an increase in correctly spliced EGFP message above that provided by the SSO alone. Results are shown for liver, kidney, lung and intestine, but splice correction was observed in other tissues as well. The magnitudes of the enhancing effects observed in vivo were much less than those observed in cell culture. However, this is not uncommon and at this point nothing is known about the pharmacokinetics or biodistribution of these compounds; thus the route and schedule of administration may have been suboptimal.
Figure 6.

In Vivo Effects-RNA. EGFP654 mice were treated with SSO623 or a mismatched control (MM) and then received UNC2383 or diluent (N=3). RT=PCR with gel analysis was used to detect correctly or incorrectly spliced EGFP mRNA. The gel images were quantitated using Fiji software. Gel image for liver (6a). The lower band (87bp) is correctly spliced EGFP mRNA while the upper band (160bp) is uncorrected. (b–e) Quantitation of splice correction in liver (b), kidney (c), intestine (d) and lung (e). The bars indicate the ratio of correctly spliced to incorrectly spliced mRNA ×100. The differences between the SSO623 only samples and the SSO623 plus UNC2383 samples are statistically significant.
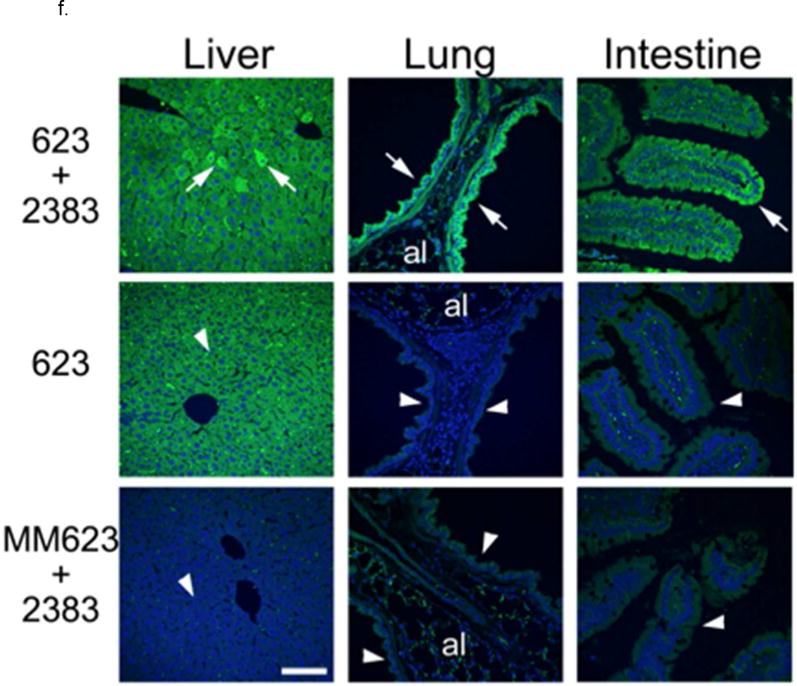
f. In Vivo Effects-Protein. EGFP immunostaining was performed in liver, lung and intestine tissue and analyzed by confocal microscopy. Increased EGFP signal (green) was seen in groups of liver cells, epithelial cells lining the bronchi, and epithelial cells in the colonic crypts in mice treated with both SSO623 and UNC2383 (arrows) compared to matching tissues from control mice (arrowheads in control mouse tissues point to equivalent structures indicated by arrows in tissues from the SSO623 plus UNC2383 treated mice). Some non-specific fluorescence signal was observed in the alveoli (al) in the lung tissue sections of all mouse groups. Nuclei were counter stained with DAPI (blue). Bar =70 um.
We also examined correction at the protein level by using an anti-EGFP antibody. Figure 6f shows that the epithelial cells of bronchi and intestinal crypts expressed EGFP in mice that received both SSO623 and UNC2383, as compared to control mice that received mismatched oligo plus UNC 2383, or SSO623 in the absence of the small molecule. In intestine the most proximal region of the crypt exhibited the highest expression probably due to the effect of epithelial regeneration in the time frame of treatment. As expected, liver exhibited degrees of expression with all SSO treatments but EGFP immunostaining was highest in mice treated both with the SSO and the small molecule.
Airway and intestine are difficult therapeutic targets for oligonucleotides in genetic disorders such as cystic fibrosis(25, 26); thus it is interesting that effects were observed in these tissues. In these experiments toxicity was monitored by obtaining blood samples and analyzing parameters that reflect renal, hepatic and hematotoxicity (Supporting Table 1). The only indication of toxicity was a moderately elevated level of the liver enzyme ALT, although this was not significant at the 5% level.
The experiments described above indicate that UNC2383 can substantially enhance the pharmacological effects of oligonucleotides in cell culture and provide in vivo enhancement as well, accompanied by limited toxicity. Our studies suggest that UNC2383 acts similarly to previously described OECs in that it increases the permeability of endomembrane compartments(18, 20). This allows partial release of oligonucleotides from non-productive endomembrane entrapment and provides access to targets in the cytosol or nucleus. While it is clear that UNC2383 and its closely related analogs act on endomembranes, we have not yet pursued the precise molecular target since the affinity of these compounds is likely too low to permit identification by proteomic or lipidomic techniques. Although higher concentrations of UNC2383 and various analogs can affect lysosomal pH, data presented here clearly shows that their action is quite distinct from typical lysosomotropic compounds such as chloroquine. Thus simple pH-driven drug accumulation and subsequent osmotic swelling of endomembranes cannot account for the oligonucleotide enhancing effects observed here. The observations of Figures 4 and 5 suggest that low concentrations of UNC2383 and its analogs primarily affect endosomes while higher concentrations can affect both endosomes and lysosomes. Although we have examined only SSOs and ASOs in this report, we anticipate that UNC2383 and related OECs will also affect the actions of siRNA, as well as various types of oligonucleotide conjugates, since these molecules are also restricted by endomembrane trapping(27).
In comparing to previously described oligonucleotide enhancing compounds including Retro-1(18) and UNC10217938(20), it seems that compound UNC2383 is far more effective than Retro-1 and approximately equivalent to UNC10217938 in terms of actions in cells. However, the window between effective and toxic concentrations is narrower for UNC2383 than for UNC10217938. We hypothesize that UNC10217938 may be more selective for endosomes rather than lysosomes as compared to UNC2383, but this remains to be determined. The OECs described here are all closely related to the initial hit from a high throughput screen and are thus still far removed from being mature drug candidates. However, further analysis of structure activity relationships may lead to the development of new compounds with greater efficacy and reduced toxicity. It is interesting to note that other groups have begun to describe oligonucleotide enhancing small molecules(28, 29). The structures of those molecules are very different from ones described here or previously by us(20), suggesting that there may be a variety of mechanisms by which small molecules can augment effects of oligonucleotides.
METHODS
Cells and Culture Methods
HeLaLuc705 cells are stably transfected with a firefly luciferase reporter whose coding sequence is interrupted by an abnormal intron. Effective delivery of an appropriate SSO, such as the 2′-O Me phosphorothioate SSO623 (5′-GTTATTCTTTAGAATGGTGC-3′), to the nucleus of these cells will correct splicing and allow luciferase expression. NIH-3T3-MDR cells are stably transfected with a human MDR1 cDNA coding for the P-glycoprotein (Pgp). We have described maintenance and use of these cell lines elsewhere(18, 20).
Functional Assays in Cell Models
SSO-mediated luciferase induction assays were conducted as previously described(18). In short, HeLaLuc705 cells in 24 well culture plates were pre-incubated with SSO623 followed by brief treatment with the small molecule; at intervals thereafter luciferase activity and protein content were determined. ASO mediated inhibition of MDR1 gene expression utilized an Alexa 488 labeled anti-P-glycoprotein antibody and flow cytometry to measure Pgp expression as described(20). The anti-MDR1 sequence is 5′-CCATCccgacctcgcGCTCC-3′ (all phosphorothiopate with 2-O-Me residues in capitals). Cytotoxicity of 2383 was monitored using the Alamar Blue assay(30).
Confocal Microscopy and Immunostaning
Effects of UNC2383 on endosome stability were monitored in cell culture models as previously described(20). Briefly, HeLaLuc705 cells were preincubated overnight with Alexa 488 labeled dextran or with TAMRA-labeled SSO 623. Live cells were imaged by fluorescence confocal microscopy with or without treatment with UNC2383 or its analogs. In some cases the location of the nucleus was delineated by treating the cells with Hoechst 33342 (ThermoFisher Scientific) at 5 ug/ml in PBS after exposure to analogs. In some cases cells were transfected with baculovirus expression vectors (Cell Lights, Life Technologies) for GFP-Rab7a (late endosome marker) or GFP-LAMP-1 (lysosome marker). Live cell imaging utilized a Zeiss LSM710 confocal microscope with environmental stage. Co-localization of TAMRA labeled SSO 623 with marker proteins was quantitated using Fiji (Image J) software and expressed as a Manders’ Correlation Coefficient(22). For the in vivo effects of UNC2383, EGFP protein expression was examined in formalin-fixed, paraffin embedded tissues harvested from treated or control mice. EGFP protein expression was monitored by immunostaining with an anti-EGFP antibody (Abcam) using techniques previously described(31). EGFP immunostaining was analyzed using a Leica SP2 confocal microscope with acquisition parameters constant throughout the study: images were processed with Adobe Photoshop software.
Lysotracker Assays
Integrity of lysosomes was evaluated using the fluorescent probe Lysotracker Red ® (Thermo Fisher). Cells in 24 well culture plates were incubated with various concentrations of UNC2383 or its analogs for 60 min. At this point 200 nM Lysotracker was added and the incubation continued for an additional 15 min. Cells were thoroughly rinsed in PBS, lysed in 0.2% TX 100, centrifuged briefly at 4000 rpm, and 100 ul of the supernate was distributed to black 96 well plates for analysis of fluorescence using a FLUOstar Omega 96 well microplate reader.
In Vivo Studies
All animal procedures were conducted in compliance with guidelines of the UNC Laboratory Animal Medicine Department and with federal guidelines. The EGFP654 transgenic mouse contains a reporter gene comprised of the EGFP coding sequence interrupted by an aberrantly spliced intron(24). Correct splicing and EGFP production can be restored by delivery of an appropriate SSO to the nucleus of tissue cells(20, 32). EGFP654 mice were administered 35 mg/kg SSO623 or mis-matched oligonucleotide in buffer by intra-peritoneal injection on two consecutive days. One day later mice received either 5 mg/kg of UNC2383 in a 1/1 DMSO/PEG400 solution, or only the diluent, via intra-peritoneal injection. After 5h one cohort of mice was euthanized and tissue samples collected for RNA analysis. These were quick frozen on dry ice and RT-PCR performed as we have previously described(18, 20) (additional details in the Supporting Information). After 24 h the second cohort was euthanized and tissue and cardiac blood samples were obtained. Tissues were fixed in 10% formalin and processed for EGFP antibody staining as described(31). Blood samples were analyzed by the UNC Animal Clinical Chemistry Core facility.
Additional Methods
A summary of additional methods and use of statistics is provided in the Supporting Information.
Supplementary Material
Acknowledgments
The authors thank P. Ariel of the UNC Microscopy Services Laboratory, C. Santos and her staff of the UNC Animal Studies Core, and the UNC Animal Clinical Chemistry Core for their outstanding assistance, F. Chung and K. Burns for their assistance with mouse tissue processing, and E. Hull-Ryde for chemical sample management. They also thank S. Frye for his helpful guidance.
Funding. This work was supported by NIH grants R01CA151964 and 1 R41 TR 001330 to RLJ and by NIH grant R01CA194064 to XM and by Cystic Fibrosis Foundation grant RDP15RO and NIH Grant P01 HL 110873 (R Boucher, PI) to SMK.
Footnotes
Supporting Information is available free of charge via the internet at at http://pubs.acs.org. This includes information on the synthesis of the compounds tested as well their structures. It also includes additional methods and statistical information. Additionally it includes the following data:
References
- 1.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annual Review of Pharmacology and Toxicology. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 2.Wu SY, Lopez-Berestein G, Calin GA, Sood AK. RNAi Therapies: Drugging the Undruggable. Sci Transl Med. 2014;6:240ps247. doi: 10.1126/scitranslmed.3008362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr Opin Chem Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Watts JK, Corey DR. Silencing disease genes in the laboratory and the clinic. J Pathol. 2011;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tse MT. Regulatory watch: Antisense approval provides boost to field. Nat Rev Drug Discovery. 2013;12:179. [Google Scholar]
- 6.Bennett CF, Baker BF, Pham N, Swayze E, Geary RS. Pharmacology of Antisense Drugs. Annual Review of Pharmacology and Toxicology. 2017;57:81–105. doi: 10.1146/annurev-pharmtox-010716-104846. [DOI] [PubMed] [Google Scholar]
- 7.Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Research. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juliano RL. The delivery of therapeutic oligonucleotides. Nucleic Acids Research. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Controlled Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Juliano RL, Ming X, Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjugate Chem. 2012;23:147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu S, Olenyuk BZ, Okamoto CT, Hamm-Alvarez SF. Targeting receptor-mediated endocytotic pathways with nanoparticles: rationale and advances. Adv Drug Delivery Rev. 2013;65:121–138. doi: 10.1016/j.addr.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 13.Mellman I, Emr SD. A Nobel Prize for membrane traffic: vesicles find their journey’s end. J Cell Biol. 2013;203:559–561. doi: 10.1083/jcb.201310134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kummel D, Ungermann C. Principles of membrane tethering and fusion in endosome and lysosome biogenesis. Curr Opin Cell Biol. 2014;29:61–66. doi: 10.1016/j.ceb.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Smith EA, Chapman ER, Weisshaar JC. Lipid mixing and content release in single-vesicle, SNARE-driven fusion assay with 1-5 ms resolution. Biophys J. 2009;96:4122–4131. doi: 10.1016/j.bpj.2009.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Pfeffer SR. Rab GTPase regulation of membrane identity. Curr Opin Cell Biol. 2013;25:414–419. doi: 10.1016/j.ceb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ming X, Carver K, Fisher M, Noel R, Cintrat JC, Gillet D, Barbier J, Cao C, Bauman J, Juliano RL. The small molecule Retro-1 enhances the pharmacological actions of antisense and splice switching oligonucleotides. Nucleic Acids Research. 2013;41:3673–3687. doi: 10.1093/nar/gkt066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Kleist L, Haucke V. At the crossroads of chemistry and cell biology: inhibiting membrane traffic by small molecules. Traffic. 2012;13:495–504. doi: 10.1111/j.1600-0854.2011.01292.x. [DOI] [PubMed] [Google Scholar]
- 20.Yang B, Ming X, Cao C, Laing B, Yuan A, Porter MA, Hull-Ryde EA, Maddry J, Suto M, Janzen WP, Juliano RL. High-throughput screening identifies small molecules that enhance the pharmacological effects of oligonucleotides. Nucleic Acids Research. 2015;43:1987–1996. doi: 10.1093/nar/gkv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang H, Fisher MH, Xu D, Miyamoto YJ, Marchand A, Van Aerschot A, Herdewijn P, Juliano RL. Inhibition of MDR1 gene expression by chimeric HNA antisense oligonucleotides. Nucleic Acids Research. 2004;32:4411–4419. doi: 10.1093/nar/gkh775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keri RS, Hiremathad A, Budagumpi S, Nagaraja BM. Comprehensive Review in Current Developments of Benzimidazole-Based Medicinal Chemistry. Chem Biol Drug Design. 2015;86:19–65. doi: 10.1111/cbdd.12462. [DOI] [PubMed] [Google Scholar]
- 24.Sazani P, Gemignani F, Kang SH, Maier MA, Manoharan M, Persmark M, Bortner D, Kole R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nature biotechnology. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 25.Kreda SM, Pickles RJ, Lazarowski ER, Boucher RC. G-protein-coupled receptors as targets for gene transfer vectors using natural small-molecule ligands. Nat Biotechnol. 2000;18:635–640. doi: 10.1038/76479. [DOI] [PubMed] [Google Scholar]
- 26.Quon BS, Rowe SM. New and emerging targeted therapies for cystic fibrosis. BMJ. 2016;352:i859. doi: 10.1136/bmj.i859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juliano RL, Ming X, Nakagawa O. The chemistry and biology of oligonucleotide conjugates. Acc Chem Res. 2012;45:1067–1076. doi: 10.1021/ar2002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilleron J, Paramasivam P, Zeigerer A, Querbes W, Marsico G, Andree C, Seifert S, Amaya P, Stoter M, Koteliansky V, Waldmann H, Fitzgerald K, Kalaidzidis Y, Akinc A, Maier MA, Manoharan M, Bickle M, Zerial M. Identification of siRNA delivery enhancers by a chemical library screen. Nucleic Acids Research. 2015;43:7984–8001. doi: 10.1093/nar/gkv762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osborn MF, Alterman JF, Nikan M, Cao H, Didiot MC, Hassler MR, Coles AH, Khvorova A. Guanabenz (Wytensin) selectively enhances uptake and efficacy of hydrophobically modified siRNAs. Nucleic Acids Research. 2015;43:8664–8672. doi: 10.1093/nar/gkv942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ming X, Ju W, Wu H, Tidwell RR, Hall JE, Thakker DR. Transport of dicationic drugs pentamidine and furamidine by human organic cation transporters. Drug Metab Dispos. 2009;37:424–430. doi: 10.1124/dmd.108.024083. [DOI] [PubMed] [Google Scholar]
- 31.Kreda SM, Gentzsch M. Imaging CFTR protein localization in cultured cells and tissues. Methods Mol Biol. 2011;742:15–33. doi: 10.1007/978-1-61779-120-8_2. [DOI] [PubMed] [Google Scholar]
- 32.Roberts J, Palma E, Sazani P, Orum H, Cho M, Kole R. Efficient and persistent splice switching by systemically delivered LNA oligonucleotides in mice. Mol Ther. 2006;14:471–475. doi: 10.1016/j.ymthe.2006.05.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


