Abstract
Extracellular matrix (ECM) is a system used to model the design of biomaterial matrices for tissue regeneration. Various biomaterial systems have been developed to mimic the composition or microstructure of the ECM. However, emulating multiple facets of the ECM in these systems remains a challenge. Here, a new strategy is reported which addresses this need by using silk fibroin and chitosan (CS) nanocomposite materials. Silk fibroin was first assembled into ECM-mimetic nanofibers in water and then blended with CS to introduce the nanostructural cues. Then the ratios of silk fibroin and CS were optimized to imitate the protein and glycosaminoglycan compositions. These biomaterial scaffolds had suitable compositions, hierarchical nano-to-micro structures, and appropriate mechanical properties to promote cell proliferation in vitro, and vascularization and tissue regeneration in vivo. Compared to previous silk-based scaffolds, these scaffolds achieved improvements in biocompatibility, suggesting promising applications in the future in tissue regeneration.
Graphical abstract
Introduction
Silk fibroin (SF) is a natural biomaterial that has been widely used as a matrix for tissue regeneration of bone, skin, blood vessels, nerves and other tissues.1–4 SF has a complex hierarchical microstructure and different compositions which depend on the source and processing. These features mean that it is feasible for silk-based materials to form various material types and shapes with tunable performance, (e.g., mechanical properties, degradation lifetime in vivo), thus satisfying the prerequisites for the regeneration of different tissues.5,6 While substantial developments have been achieved to date to optimize the biocompatibility of silk materials including the control of nanostructures and mechanical properties, new strategies are still required to further improve the bioactivity of silk-based materials for better tissue regeneration and to enrich silk-based biomaterial systems for various biomedical applications.
The extracellular matrix (ECM) is well-known for maintaining a preferred microenvironment for tissue regeneration and has been used as a guide for the design of artificial biomaterial matrices.7–9 ECM-like matrix systems could provide an appropriate niche and biological cues for accelerating the regeneration of tissues and organs.10–12 Matrices having a similar composition to ECM were first explored to give better tissue repair. Because the ECM contains proteins, glycoproteins, and glycosaminoglycans, different strategies have been developed to mimic the ECM compositions by blending natural proteins and polysaccharides.7,13,14 Although these ECM-biomimetic materials could provide suitable cues to induce tissue regeneration, their application is seriously limited because of insufficient mechanical properties, possible pathogen transfer risk and uncontrollable degradation behaviour.15–18 Because of its superior and tunable mechanical properties, controllable degradation and relatively low inflammatory response, SF has been used to replace collagen, and blended with chitosan (CS), a biocompatible natural biomaterial with some similarities with glycosaminoglycan, to mimic the ECM composition.4 Although many previous studies suggested better biocompatibility with silk rather than CS materials,19,20 better biocompatibility and cell growth in vitro and in vivo than pure silk and CS were achieved using SF-CS composite scaffolds, by tuning the ratio of SF and CS, suggesting that they have promising applications as matrices. 21–23
Besides the composition, the nanofibrous structure of the ECM was considered a critical factor in affecting cellular morphology and various cellular behaviours including cell proliferation, attachment and differentiation.24–26 Different nanofibrous biomaterials were fabricated using electrospinning and phase separation, and significant improvements in cytocompatibility, when compared with the controls without nanofibrous structures, were achieved.27,28 Recently, different self-assembly mechanisms were identified which indicated that the nanostructures of SF could be tuned in aqueous solutions by adjusting the molecular mobility, charge distribution and hydrophilic interactions.29–31 As well as the work of Jin and Kaplan,32 several feasible processes have been developed to form silk nanofibers of various diameters and secondary structures, and with enriched motifs, and these have been used to design silk-based materials with hierarchical microstructures.33,34 Although SF cannot resemble the hierarchical structure of natural collagen perfectly (diameter, 50–100 nm),35 the significantly improved cytocompatibility of these materials implied that the fabrication of nanofibrous structures was feasible for silk-based materials to achieve improved niches for cells and tissues.
Recently, nanofibrous SF-CS scaffolds were fabricated using dielectrophoresis, and these showed superior cytocompatibility to the SF-CS scaffolds that lacked nanofibrous structures.4 Therefore, it is reasonable to assume that simultaneously modifying the composition and microstructure in the SF-CS system, ECM-like niches will be generated. Although several studies have confirmed the feasibility,7, 36–40 the challenge remains to prepare SF-CS scaffolds with multiple ECM-mimetic cues, in part because of the difficulties in forming three-dimensional nanofibrous structures with SF and CS. Considering the finding, by Bai et al, of SF nanofibers in aqueous solutions,34 the goal of the present work was to introduce the SF nanofibers into CS for further development of novel SF-CS scaffolds with better performances. CS was utilized because of its similarity with the glycosaminoglycans of the ECM and its successful applications of closely mimicking natural ECM compositions by blending it with collagen.41,42 SF nanofibers in aqueous solution were blended with CS to form ECM-like scaffolds. The scaffolds were optimized by tuning the ratio of SF and CS. Both in vitro and in vivo studies confirmed compatibility and good performance of the scaffold materials when compared with the structures generated from pure SF nanofibers.
Experimental
Preparation of aqueous SF solutions
Bombyx mori SF solutions were prepared according to previously published procedures.43–45 Silk was boiled for 20 min in an aqueous solution of 0.02 M sodium carbonate, and then rinsed thoroughly with distilled water to extract the sericin proteins. The extracted silk was dissolved in 9.3 M lithium bromide solution at 60 °C for 4 h, yielding a 20% (w/v) solution. This solution was dialyzed against distilled water, using dialysis tubes (MWCO 3,500) for 72 h to remove the salt. The solution was optically clear after dialysis and was centrifuged at 9,000 rpm for 20 min at 4 °C to remove the silk aggregates formed during the process. The final concentration of aqueous SF solution was about 6 wt%, which was determined by weighing the remaining solid after drying.
Fabrication of SF nanofibers
The SF nanofibers were assembled as reported in a recent study.46,47 Fresh SF solution was treated by a concentration-dilution process. The solution (6 wt%) was slowly concentrated to about 20 wt% over 24 h at 60 °C to form metastable nanoparticles, and then diluted to 0.5 wt% with distilled water. The diluted SF solution was incubated for over 24 h at 60 °C to induce the nanofiber formation.
Fabrication of SF-CS scaffolds
SF scaffolds were prepared via a modified lyophilization method.44,48,49 The fresh SF solution was blended with SF nanofibers at dry weight ratios (SF:SF nanofibers) of 15:1, whereas CS (carboxymethyl CS, MW 100,000–300,000, Yuanye Biological Company, Shanghai, China) was dissolved directly in pure water to achieve a 2% CS solution. Then the SF and CS solutions were mixed at different ratios (2:1, 4:1, 6:1 and 8:1), following dilution to 2% with pure water. The uniform solution was poured into cylindrically shaped containers and frozen at −20 °C for 24 h, and then lyophilized for about 48 h. After lyophilization, the dried scaffolds were immersed in crosslinking agent (25 mM 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) and 10 mM N-hydroxysuccinimide in 80% methanol) for about 5 h. The crosslinked porous scaffolds were immersed in pure water to remove unreacted crosslinker. Then, the scaffolds were freeze dried again before characterization. According to the ratios of SF and CS, the scaffolds were termed as S2-C, S4-C, S6-C and S8-C. As controls, scaffolds derived from the same SF-CS blended solution were also prepared using the same lyophilization process but without the crosslinking process, or with the methanol treatment but without the crosslinking agent.
Characterization of SF-CS scaffolds
The morphology of the scaffolds was observed using scanning electron microscopy (SEM) (S-4800, Hitachi, Tokyo, Japan) at 3 kV. Samples were mounted on a copper plate and sputter coated with gold for 2 min to achieve a layer with a thickness of about 5 nm for better imaging.48 Fourier transform infrared spectroscopy (FTIR) analysis of the SF scaffolds was performed with an ATR-FTIR spectrometer (Nicolet FTIR 5700, Thermo Scientific, FL, USA), equipped with a MIRacle attenuated total reflection (ATR) accessory (germanium crystal).48 For each measurement, 64 scans were co-added with a resolution of 4 cm−1 with a wave number ranging from 800 to 1800 cm−1. The crystalline structure of the scaffolds obtained was measured using X-ray diffraction (XRD) (X’Pert Pro MPD, PANalytical BV, Almelo, The Netherlands) using monochromated Cu Kα radiation (30 mA, 40 kV) with a scanning speed of 6° min−1.48 The thermal properties of the scaffolds were measured using differential scanning calorimetry (DSC; Q2000, TA Instruments, New Castle, DE, USA) under a dry nitrogen flow of 50 mL min−1. Temperature modulation differential scanning calorimetry (TMDSC) measurements were performed on a instrument equipped with a refrigerated cooling system (Q2000, TA, Instruments). After baseline correction, 5 mg of sample was sealed inside the aluminum pans for further measurement. The samples were heated at 2 °C min−1 with a modulation period of 60 s and a temperature amplitude of 0.318.50 All these measurements were repeated three times.
Because soluble SF could be released and achieve a state of equilibrium within 0.5 h,51 samples were soaked in pure water and kept at 37 °C in a shaking water bath for 3 h to determine the water stability of the scaffolds. At each time point, five samples for each group were dried and weighed. The residual mass (%) was obtained by dividing the residual weight by the initial weight. To measure the compressive properties of the scaffolds (11 mm in diameter and 22 mm in height) in hydrated conditions, the scaffolds were first hydrated in water for 4 h and then measured with a cross head speed of 2 mm min−1 at 25 °C using a universal testing frame (3366, Instron, Norwood, MA, USA) with a 10 N loading cell.49,52 The load was applied until the cylinder was compressed by more than 30% of its original length.53 The compressive modulus was calculated as the slope of the linear elastic region of the stress-strain curve. Five samples were tested for each group.27 The porosity of the scaffolds was determined using a liquid displacement method with 100% ethanol, as previously reported.54 The lyophilized scaffolds were immersed in a graduated cylinder of known volume of ethanol (V1). The scaffold was immersed in ethanol for 5 min until it became saturated (V2). Then the ethanol soaked scaffold was removed from the graduated cylinder, and the remaining volume was recorded as (V3). The porosity of the scaffolds was expressed as:
In vitro cytocompatibility of scaffolds
Bone marrow mesenchymal stem cells (BMSCs) derived from Sprague Dawley (SD) rats were cultured in vitro to assess the biocompatibility of the crosslinked SF-CS scaffolds. Use of SD rats was granted by the Animal Ethics Committee of Soochow University. Briefly, BMSCs were cultured on the sterile scaffolds (diameter of 8 mm and height of 2 mm) in Dulbecco’s modified Eagle’s medium (low glucose) supplemented with 10% fetal bovine serum, and 1% IU mL−1 of streptomycin-penicillin (Invitrogen, Carlsbad, CA, USA) at 37 °C.
The cell morphology on the scaffolds was evaluated using confocal microscopy. After culturing for 1, 6, and 12 d, the cell seeded scaffolds were washed three times with phosphate buffered saline (PBS) and fixed in 4% paraformaldehyde for 30 min, followed by further washing. The cells were permeabilized with 0.1% Triton X-100 for 30 min and incubated with fluorescein isothiocyanate (FITC)-phalloidin (Sigma-Aldrich, St. Louis, MO, USA) for 20 min at room temperature, followed by washing with PBS and, finally, staining with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO, USA) for 10 min. The images of the stained samples were obtained using a confocal microscope (FV10 inverted microscope, Olympus, Nagano, Japan).
The proliferation behavior of the BMSCs was determined using the deoxyribonucleic acid (DNA) content on days 1 to 12. Before measurement, the samples were treated with proteinase K overnight at 56 °C to digest the remaining scaffolds.28 The DNA content was measured using the PicoGreen DNA assay according to the protocol of the manufacturer (Invitrogen, Carlsbad, CA, USA). A standard curve over a range of concentrations was prepared using λ-phage DNA in 10 × 10−3 M Tris-hydrochloride (pH 7.4), 5 × 10−3 M sodium chloride, and 0.1 × 10−3 M ethylenediaminetetraacetic acid (EDTA). Then the DNA spectra were collected at an excitation wavelength of 480 nm and an emission wavelength of 530 nm using a spectrofluorometer (BioTek Synergy 4 BioTek, Winooski, VT, Canada) and the DNA concentration was calculated based on the standard curves.55
In vivo study
The use of SD rats (Nine 8-week-old male rats, from the Animal Resource Center, Soochow University) was approved by the Animal Ethics Committee of Soochow University. Four subcutaneous pockets, approximately 2 cm apart, were created along the central line of the shaved dorsal area with. The scaffolds were implanted into the pockets. At weeks 1, 2, and 4 post implantation, the animals were euthanized and the implanted scaffolds together with the adjacent tissues were collected. The specimens were fixed in 4% paraformaldehyde in PBS at pH 7.4 for further histological and immunohistochemistry examination. For histological morphology, the samples were embedded in paraffin, sectioned into 4 µm slices, and stained with hematoxylin and eosin (H&E), and Masson’s trichrome stain to visualize the cell nuclei and cytoplasm. Sections were stained for the primary antibody against the endothelial cell marker (CD34) to visualize the formation of newly formed blood vessels.55
Statistical analysis
SPSS v.16.0 software was used for statistical analyses. Comparison of the mean values of the data sets was performed using a one-way analysis of variance (ANOVA). Measurements are presented as mean ± standard deviation, unless otherwise specified. P < 0.05 was considered to be significant.
Results and discussion
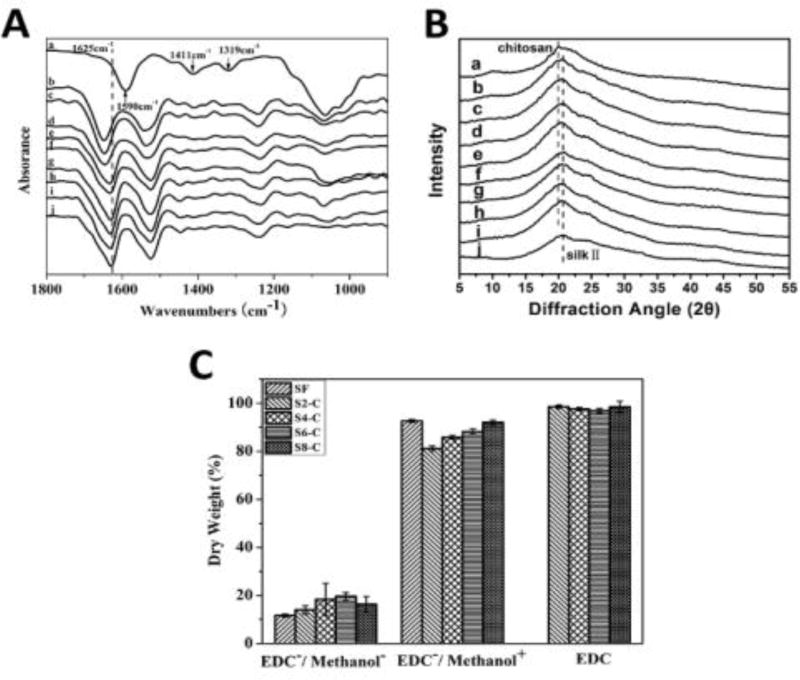
A modified nanofiber-assisted lyophilization process was used to form nanofiber-based microporous SF-CS scaffolds. SF nanofibers prepared using the prior self-assembly method were blended with fresh SF and CS solution sequentially to form a homogeneous aqueous solution [Figure S1; Electronic Supplementary Information (ESI)]. The porous scaffolds were then prepared after lyophilization and stabilized in methanol solution where EDC was used to crosslink SF and CS in order to restrain the release of CS. The FTIR spectra were used to assess the structural changes of the scaffolds. Before crosslinking treatment, the amide I peak (1600–1700 cm−1) of the scaffolds showed a red shift with the increase of CS,35 indicating that the interaction between CS and SF might be able to restrain beta sheet formation in various transformational processes. After crosslinking in methanol, the scaffolds showed a typical beta sheet structure (main peak at 1625 cm−1) because methanol induced beta sheet formation in the SF during the crosslinking process.56,57 However, other significant structural changes after the treatment were not shown in the FTIR spectra partly because the new peaks overlapped with those from SF. Although the main peaks of CS and SF were overlapped in the XRD curves, the XRD results revealed beta-sheet formation after methanol treatment, and this was in good agreement with the FTIR results (Figure 1B). The water stability of the scaffolds under different conditions was studied to confirm the crosslinking reaction between SF and CS.58–60 The untreated scaffolds easily dissolved in water at 37 °C, because CS and SF with a low beta sheet content are water soluble (Figure 1C). When the scaffolds were immersed in methanol without EDC for 3 h, pure CS scaffolds remained soluble in water, while the weight loss was 19%, 15%, 12%, 8% and 4% for S2-C, S4-C, S6-C, S8-C, and pure SF, respectively, because the CS was soluble after methanol treatment. The weight loss decreased to below 5% for all the crosslinked scaffolds, which was a similar results to that obtained for the methanol treated pure SF scaffolds. Considering that SF became mostly insoluble after methanol treatment but showed a slight weight loss in water, the results indicated that CS without EDC crosslinking cannot be stabilized in water. Therefore, although there was no direct evidence, the improved water insolubility of the scaffolds after the crosslinking treatment suggested that CS has been stabilized in the scaffolds. Other crosslinking agents such as genipin might achieve a better crosslinking effect, and this will be studied in the future.
Figure 1.
Structures of SF-CS scaffolds: (A) FTIR spectra and (B) XRD spectra. The samples were as follows: (a) pure CS, (b–e) SF-CS scaffolds without crosslinking treatment with SF-CS ratios of 2:1, 4:1, 6:1, and 8:1, respectively, (f–i) crosslinked SF-CS scaffolds with SF-CS ratios of 2:1, 4:1, 6:1, and 8:1, respectively, and (j) methanol treated SF scaffolds. (C) Water stability of various scaffolds. The samples of SF, S2-C, S4-C, S6-C and S8-C indicate that the ratio of SF and CS is 1:0, 2:1, 4:1, 6:1 and 8:1, respectively, while EDC−/methanol− indicated scaffolds without crosslinking, EDC−/methanol+ indicates methanol-treated scaffolds without crosslinking treatment, and EDC shows the crosslinked scaffolds in methanol.
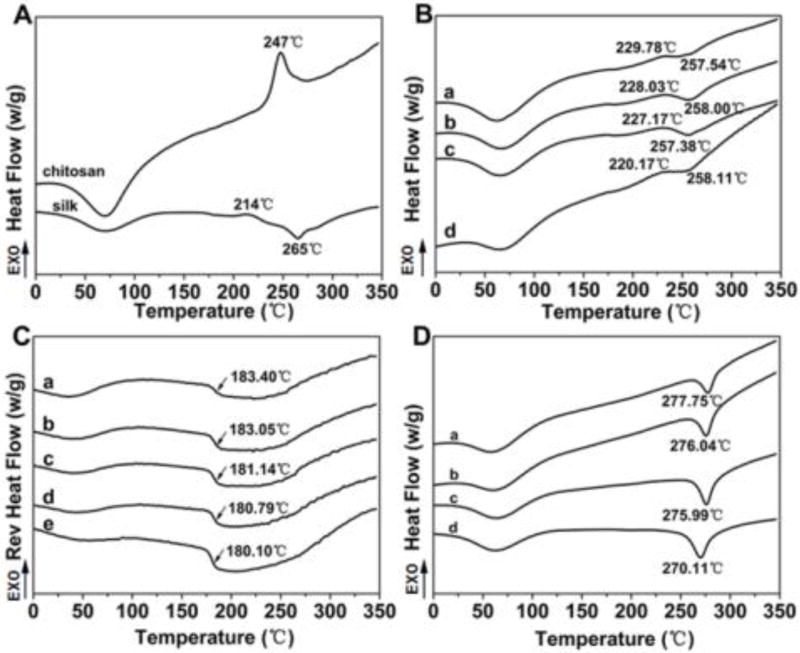
The structural changes as well as the interactions of SF and CS were further investigated using DSC. The DSC peaks of SF strongly depend on the measurement conditions such as the heating rate.61 Based on previous studies under similar measurement conditions,55,62 the degradation peak at 250–260 °C belongs to a random/silk I structure whereas the peaks at 260–280 °C are the beta-sheet formation.63 The increase in temperature of the degradation peaks suggests a higher beta-sheet content.62,63 Figure 2A illustrates the standard DSC curves for pure CS and SF scaffolds. Besides the water evaporation peaks at 50–100 °C, the CS scaffolds showed a significant nonisothermal crystallization peak at 247 °C whereas the SF scaffolds showed a nonisothermal crystallization peak at 214 °C and a degradation peak at 265 °C. Similar to previous studies,62,63 the crystallization peak at 214 °C suggested beta-sheet formation that then resulted in a degradation peak at 265 °C, which was higher than that of the random/silk I structure (250–260 °C). In comparison, only one nonisothermal crystallization peak appeared between 220 °C and 230 °C for all the SF-CS scaffolds, indicating that there was a strong interaction between SF and CS (Figure 2B).63,64–66 The crystallization peak moved to a lower temperature for the samples with a higher SF content, confirming that there was a strong SF-CS interaction. A significantly lower peak intensity than pure SF and CS indicated that both the crystallization transformations of SF and CS were restrained by the strong interaction. The change of degradation peaks further confirmed the existence of a strong SF-CS interaction. Compared to pure SF scaffolds, the degradation peaks at the lower temperature (below 260 °C) appeared in the curves of all the SF-CS scaffolds, suggesting that SF was in a more amorphous state.63 A higher glass transition temperature (Tg) also appeared in the TMDSC curves for the SF-CS scaffolds (Figure 2C). Unlike the Tg at lower temperatures (10–100 °C) that was related to SF-water interaction,62,63 the Tg temperature was influenced by the SF-CS interaction and further increased following the increase of CS content, confirming the interaction between SF and CS. After the crosslinking process in methanol solution, all the SF-CS scaffolds had a higher degradation temperature (above 270 °C) than the pure SF scaffolds (Figure 2D), because of beta sheet formation and also because of the crosslinking between the SF and CS. In addition, both the crystallization peaks of SF and CS disappeared from the curves of the crosslinked scaffolds, because SF was transformed into beta sheet structures whereas CS was immobilized with SF by the crosslinking process.
Figure 2.
The DSC curves of SF-CS scaffolds: (A) the standard DSC curves of pure CS and SF scaffolds without methanol treatment; (B) the standard DSC curves of SF-CS scaffolds without crosslinking, (a–d) indicates S2-C, S4-C, S6-C and S8-C, respectively; (C) the TMDSC curves of SF-CS scaffolds without crosslinking, (a–d) indicates S2-C, S4-C, S6-C and S8-C, respectively, whereas (e) was pure silk scaffolds without crosslinking; (D) the standard DSC curves of crosslinked SF-CS scaffolds, (a–d) indicate S2-C, S4-C, S6-C and S8-C, respectively.
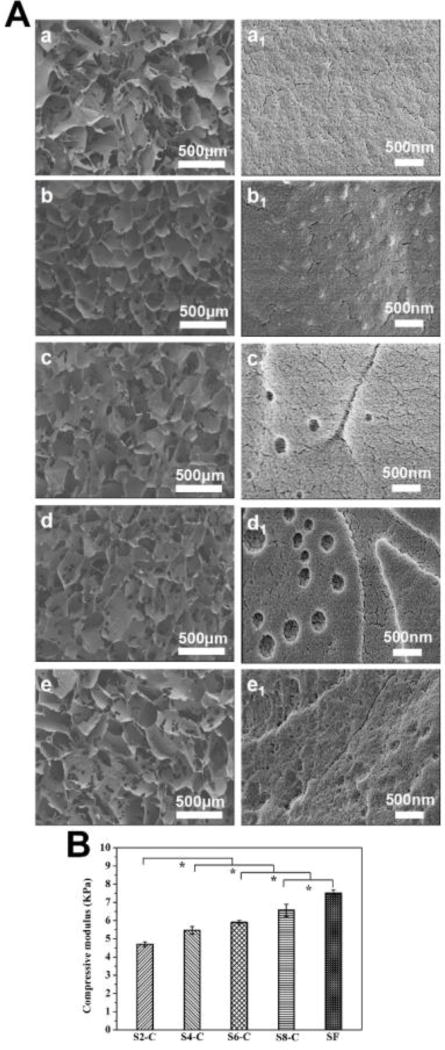
The strong interaction between SF and CS facilitated the formation of better porous structures in the lyophilization process. The hierarchical microstructure of the scaffolds was investigated using SEM. All the SF-CS scaffolds showed better interconnected porous morphologies, than those composed of pure SF, with a pore size of above 100 µm and they also had a high porosity of above 85% (Figure S2; ESI), which is suitable for cell and tissue ingrowth. Various phase separated structures appeared on the porous walls partly because of the different hydrophilic-hydrophobic properties of SF and CS.28 Higher magnification images revealed nanofibrous structures for all the SF-CS scaffolds, similar to the SF scaffolds prepared using similar lyophilization processes.30 The results revealed better micro-nanofibrous hierarchical morphologies for the SF-CS scaffolds, suggesting the formation of ECM-mimetic structures that were considered to be favorable for cell adhesion and growth. As expected, the SF-CS scaffolds also showed tunable mechanical properties which were obtained by varying the ratios of SF and CS in the wet state. A previous study revealed that biomaterials with an elastic modulus of 1–7 kPa could induce the differentiation of stem cells into endothelial cells.55 The compressive modulus changed from 6.5 kPa to 4.7 kPa for these scaffolds, suggesting the potential for a stem cell support for differentiation into endothelial cells and for vascularization.
The cytocompatibility of the scaffolds was evaluated using BMSC attachment and proliferation. Because CS scaffolds were soluble, only nanofibrous SF scaffolds were used as controls. The cells showed different proliferation behaviours on the scaffolds. Cell numbers increased without reaching a plateau in the 12 d of the culture period, suggesting good cytocompatibility of all scaffolds. Compared to the SF scaffolds, cell proliferation was lower on the S2-C scaffolds. Several studies have suggested better cytocompatibility on SF than CS.67,68 However, significantly higher cell numbers were achieved on the other scaffolds with low CS contents and optimized on the S8-C scaffolds. Previous studies revealed that tuning SF and CS content to mimic ECM composition resulted in improved cell compatibility.69,70 The present DNA results confirmed that optimizing the ratios of SF and CS provided a preferable microenvironment for cell proliferation. A recent study by Han et al also showed that the nanofibrous SF scaffolds had better cytocompatibility than that those without nanofibrous structures.55 Cell compatibility was further improved by optimizing the ratio of SF and CS, which implied that multiple ECM simulation was feasible for designing biomaterial systems with improved cytocompatibility. The confocal microscopy results further supported the different cell behaviours on the SF-CS scaffolds. From day 1 to day 12, fewer cell aggregates formed on the S2-C scaffolds than on the silk scaffolds, while cell proliferation was significantly enhanced on the S4-C and S6-C scaffolds and the most cell aggregates were found in the S8-C scaffolds.
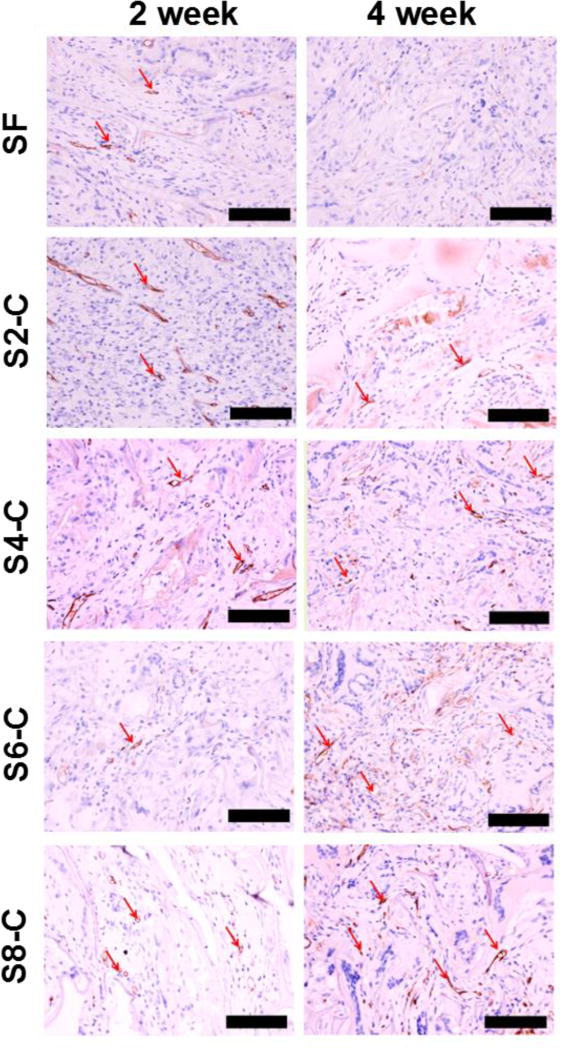
To further evaluate the biocompatibility, the different SF-CS scaffolds were implanted subcutaneously in rats. The nanofibrous SF scaffolds with vascularization capacity were used as a control.37 The histology of the tissue sections from the implanted scaffolds is shown in Figure 5. Extensive vasculature was observed visually in all the scaffolds after implantation, because the scaffolds had a suitable stiffness range to induce vascularization. Significantly better vascularization appeared in the S4-C, S6-C and S8-C scaffolds, implying more rapid neovascularization because of the better ECM mimetic microenvironment.71–73
Figure 5.
In vivo vascularization of all the scaffolds: CD34 immunohistochemical staining of sections of the scaffolds implanted subcutaneously in rats for two and four weeks. The red arrows indicate the blood vessels inside the scaffolds, the scale bar is 100 µm. *Statistical significance P < 0.05.
Immunohistochemistry analysis was used to quantify the number of vessels inside the different scaffolds (Figure 6). After 28 days of implantation, the number of newly formed vessels per mm2 increased by 8% in the pure silk scaffolds, and by 28%, 29% and 42% for the S4-C, S6-C and S8-C scaffolds, respectively. Previous work demonstrated the higher vascularization capacity of the nanofibrous scaffolds,53 thus the present results implied further improvement by using the ECM simulation strategy.
Figure 6.
The analysis of in vivo vascularization of all the scaffolds: Vessel density within the scaffolds after subcutaneous implantation for four weeks on the dorsal region. *Statistical significance P < 0.05.
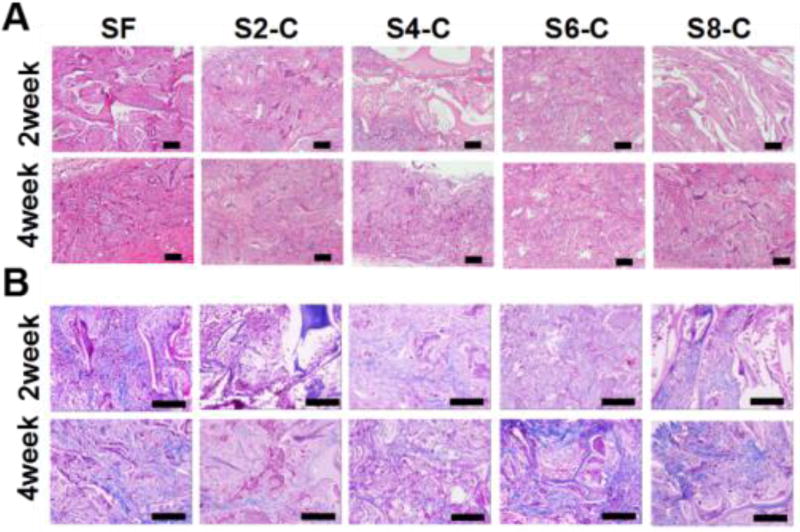
Better granulation tissue regeneration was also found inside the SF-CS scaffolds (Figure 7). At 28 days after implantation, only about 60% of the nanofibrous scaffolds were occupied by tissue ingrowth, but more than 80% of the SF4-C, SF6-C and SF8-C scaffolds were filled with new tissue. Therefore, compared to previously reported SF-CS scaffolds,69,70,74 the present findings suggest that biocompatibility could be improved using ECM simulation, including composition and nanostructure. The new strategy offers improved biomaterial systems because of the control of the biomaterial microenvironment.
Figure 7.
In vivo compatibility of all the scaffolds: (A) H&E staining of sections of the scaffolds implanted subcutaneously in rats for two and four weeks; (B) Masson’s trichrome staining of sections of the scaffolds implanted subcutaneously in rats for two and four weeks, the scale bar is 100 µm.
Conclusions
The present study demonstrates a multiple ECM simulation strategy for constructing natural scaffolds with improved biocompatibility. The nanostructures were introduced to simulate ECM microstructures and the content of SF and CS was optimized to achieve simulation of ECM compositions and morphology. The improved cell proliferation in vitro, and better neovascularization and tissue regeneration in vivo suggest promising applications for these scaffolds in tissue regeneration.
Supplementary Material
Figure 3.
Morphology of SF-CS scaffolds: (A) SEM images of scaffolds: (a–e) indicate the porous structures of SF, S2-C, S4-C, S6-C and S8-C samples, while a1-e1 show the nanotopography of the porous walls; (B) compressive modulus of the scaffolds in wet conditions. *Statistically significant P < 0.05.
Figure 4.
Cytocompatibility of the scaffolds in vitro: (A) confocal microscopy images of BMSCs cultivated on day 1, day 6 and day 12 on different SF-CS scaffolds. (B) BMSC proliferation behavior on different SF-CS scaffolds on day 1, 3, 6, 9, and 12. The blue color (DAPI) represents the cell nucleus and SF whereas the green color (FITC labeled phalloidin) indicates the cytoskeleton of cells. The scale bar is 200 µm. *Statistical significance P < 0.05.
Acknowledgments
The authors wish to thank National Basic Research Program of China (973 Program, 2013CB934400), the National Natural Science Foundation of China (NSFC) (81272106, 81671028) and the US National Institute of Health (NIH) (R01 DE017207, P41 EB002520). We also thank the Natural Science Foundation of Jiangsu Province (Grants No BK20140397, BK20140401) and the China National Textile and Apparel Council (J201405) for support of this work.
Notes and references
- 1.Cestari M, Muller V, Rodrigues JH, Nakamura CV, Rubira AF, Muniz EC. Biomacromolecules. 2014;15:1762. doi: 10.1021/bm500132g. [DOI] [PubMed] [Google Scholar]
- 2.Karageorgiou V, Tomkins M, Fajardo R, Meinel L, Snyder B, Wade K, Chen G, Vunjak-Novakovic G, Kaplan DL. J.Biomed. Mater. Res. 2006;78A:324. doi: 10.1002/jbm.a.30728. [DOI] [PubMed] [Google Scholar]
- 3.Lovett M, Cannizzaro C, Daheron L, Messmer B, Vunjak-Novakovic G, Kaplan DL. Biomaterials. 2007;28:5271. doi: 10.1016/j.biomaterials.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunne LW, Iyyanki T, Hubenak J, Mathur AB. Acta Biomater. 2014;10:3630. doi: 10.1016/j.actbio.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rockwood DN, Preda RC, Yücel T, Wang X, Lovett ML, Kplan DL. Nature Protocols. 2011;6:1612. doi: 10.1038/nprot.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vepari CC, Kaplan DL. Prog Polym Sci. 2007;32:991. doi: 10.1016/j.progpolymsci.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu Y, Zhu JB, Xue CB, Li Z, Ding F, Yang YM, Gu XS. Biomaterials. 2014;35:2253. doi: 10.1016/j.biomaterials.2013.11.087. [DOI] [PubMed] [Google Scholar]
- 8.Langer R, Vacanti JP. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 9.Shalumon KT, Lai GJ, Chen CH, Chen JP. ACS Appl. Mater. Interfaces. 2015;7:21170. doi: 10.1021/acsami.5b04962. [DOI] [PubMed] [Google Scholar]
- 10.Calle EA, Hill RC, Leiby KL, Le AV, Gard AL, Madri JA, Hansen KC, Niklason LE. Acta Biomater. 2016;46:91. doi: 10.1016/j.actbio.2016.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atala A, Kasper FK, Mikos AG. Sci. Transl. Med. 2012;4:160. doi: 10.1126/scitranslmed.3004890. [DOI] [PubMed] [Google Scholar]
- 12.Chen G, Chen JL, Yao B, Li L, Luo XY, Zhang XX, Feng L, Jiang ZT, Yu M, Guo WH, Tian WD. Biomaterials. 2015;52:56. doi: 10.1016/j.biomaterials.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Badylak SF. Biomaterials. 2007;28:3587. doi: 10.1016/j.biomaterials.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 14.Lee JS, Shin J, Park HM, Kim YG, Kim BG, Oh JW, W S. Biomacromolecules. 2014;15:206. doi: 10.1021/bm4015039. [DOI] [PubMed] [Google Scholar]
- 15.Mao Z, Shi H, Guo R, Ma L, Gao C, Han C, Shen J. Acta Biomater. 2009;5:2983. doi: 10.1016/j.actbio.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Bell E, Ivarsson B, Merrill C. Proc Natl Acad Sci USA. 1979;76:1274. doi: 10.1073/pnas.76.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng KW, Khor HL, Hutmacher DW. Biomaterials. 2004;25:2807. doi: 10.1016/j.biomaterials.2003.09.058. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, You C, Hu X, Zheng Y, Q Li, Feng Z, Sun H, Gao C, Han C. Acta Biomater. 2013;9:7822. doi: 10.1016/j.actbio.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 19.Li DR, Jiao GL, Zhang W, Chen XF, Ning RJ, Du C. RSC Adv. 2016;6:19887. [Google Scholar]
- 20.Xu ZP, Shi LY, Yang MY, Zhang HP, Zhu LJ. J. Mater. Chem. B. 2015;3:3634. doi: 10.1039/c5tb00226e. [DOI] [PubMed] [Google Scholar]
- 21.Gobin AS, Froude VE, Mathur AB. J. Biomed. Mater. Res. 2005;74A:465. doi: 10.1002/jbm.a.30382. [DOI] [PubMed] [Google Scholar]
- 22.She ZD, Liu WQ, Feng QL. Front. Mater. Sci. Chin. 2009;3:241. [Google Scholar]
- 23.Silva SS, Motta A, Rodrigues MT, Pinheiro AFM, Gomes ME, Mano JF, Reis RL, Migliaresi C. Biomacromolecules. 2008;9:2764. doi: 10.1021/bm800874q. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, Gao HL, Shen LL, Pan Z, Mao LB, Wu T, He JC, Zhou DH, Zhang ZY, Yu SH. Nanoscale. 2016;8:309. doi: 10.1039/c5nr06876b. [DOI] [PubMed] [Google Scholar]
- 25.Li XT, Zhang Y, Chen GQ. Biomaterials. 2008;29:3720. doi: 10.1016/j.biomaterials.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Hashemi SM, Soudi S, Shabani I, Naderi M, Soleimani M. Biomaterials. 2011;32:7363. doi: 10.1016/j.biomaterials.2011.06.048. [DOI] [PubMed] [Google Scholar]
- 27.Armentano I, Dottori M, Fortunati E, Mattioli S, Kenny JM. Polym. Degrad. Stab. 2010;95:2126. [Google Scholar]
- 28.Nogueira GM, Swiston AJ, Beppu MM, Rubner MF. Langmuir. 2010;26:8953. doi: 10.1021/la904741h. [DOI] [PubMed] [Google Scholar]
- 29.Rabotyagova OS, Cebe P, Kaplan DL. Biomacromolecules. 2009;10:229. doi: 10.1021/bm800930x. [DOI] [PubMed] [Google Scholar]
- 30.Gomes S, Numata K, Leonor IB, Mano JF, Reis RL, Kaplan DL. Biomacromolecules. 2011;12:1675. doi: 10.1021/bm2000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong J, Ma M, Li W, Zhou J, Yan ZQ, He DN. Biopolymers. 2014;101:1181. doi: 10.1002/bip.22532. [DOI] [PubMed] [Google Scholar]
- 32.Jin HJ, Kaplan DL. Nature. 2003;424:1057. doi: 10.1038/nature01809. [DOI] [PubMed] [Google Scholar]
- 33.Lu Q, Zhu HS, Zhang CC, Zhang F, Zhang B, Kaplan DL. Biomacromolecules. 2012;13:826. doi: 10.1021/bm201731e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai SM, Zhang XL, Lu Q, Sheng WQ, Liu LJ, Dong BJ, Kaplan DL. Biomacromolecules. 2014;15:3044. doi: 10.1021/bm500662z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K, Qian Y, Wang H, Fan L, Huang C, Yin A, Mo X. J. Biomed. Mater. Res A. 2010;95:870. doi: 10.1002/jbm.a.32895. [DOI] [PubMed] [Google Scholar]
- 36.Altman AM, Yan, Matthias Y, Bai X, Bios C, Mathur AB, Song YH, Alt EU. Stem Cells. 2009;27:250. doi: 10.1634/stemcells.2008-0178. [DOI] [PubMed] [Google Scholar]
- 37.Gobin AS, Butler CE, Mathur AB. Tissue Eng. 2006;12:3383. doi: 10.1089/ten.2006.12.3383. [DOI] [PubMed] [Google Scholar]
- 38.Rios CN, Skoracki RJ, Mathur AB. Clin. Orthop. Relat. Res. 2012;470:2541. doi: 10.1007/s11999-012-2475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zang M, Zhang Q, Davis G, Huang G, Jaffari M, Rios CN, Gupta V, Yu P, Mathur AB. Acta Biomater. 2011;7:3422. doi: 10.1016/j.actbio.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 40.Rios CN, Skoracki RJ, Miller MJ, Satterfield WC, Mathur AB. Tissue Eng. Part A. 2009;15:2717. doi: 10.1089/ten.TEA.2008.0360. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Q, Hubenak J, Iyyanki T, Alred E, Turza KC, Davis G, Chang EI, Branch-Brooks CD, Beahm EK, Butler CE. Biomaterials. 2015;73:198. doi: 10.1016/j.biomaterials.2015.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X, Lindsay B, Guido S, Patrick CW. J. Biomed. Mater. Res. A. 2007;81:59. doi: 10.1002/jbm.a.31003. [DOI] [PubMed] [Google Scholar]
- 43.Lu Q, Hu X, Wang XQ, Kluge JA, Lu SZ, Cebe P, Kaplan DL. Acta Biomater. 2010;6:1380. doi: 10.1016/j.actbio.2009.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han FF, Liu SS, Pei YZ, Bai SM, Liu X, Zhao HJ, Ma F, Lu Q, Kaplan DL, Zhu HS. Acta Biomater. 2014;10:921. doi: 10.1016/j.actbio.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 45.Liu SS, Dong CF, Lu Q, Li ZX, Lu GZ, Kaplan DL, Zhu HS. Acta Biomater. 2013;9:8991. doi: 10.1016/j.actbio.2013.06.045. [DOI] [PubMed] [Google Scholar]
- 46.Bai SM, Han HY, Huang XW, Xu WA, Kaplan DL, Zhu HS, Lu Q. Acta Biomater. 2015;20:22. doi: 10.1016/j.actbio.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Zhang CC, Liu LJ, Kaplan DL, Zhu HS, Lu Q. In Vitro. Front. Mater. Sci. 2015;9:382. [Google Scholar]
- 48.Lu GZ, Liu SS, Lin SS, Kaplan DL, Q Lu. Colloid Surf. B. 2014;120:28. doi: 10.1016/j.colsurfb.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 49.Hu X, Park SH, Gil ES, Xia XX, Weiss AS, Kaplan DL. Biomaterials. 2011;32:8979. doi: 10.1016/j.biomaterials.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim UJ, Park J, Kim HJ, Wada M, Kaplan DL. Biomaterials. 2005;26:2775. doi: 10.1016/j.biomaterials.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 51.Pei YZ, Liu X, Liu SS, Lu Q, Liu J, Kaplan DL, Zhu HS. Acta Biomater. 2015;13:168. doi: 10.1016/j.actbio.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wrag LS, Tsioris K, Gi ES, Omenetto FG, Kaplan DL. Adv. Funct. Mater. 2013;23:3403. doi: 10.1002/adfm.201202926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren LL, Kang YQ, Browne C, Bishop J, Yang YZ. Bone. 2014;64:173. doi: 10.1016/j.bone.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNamara SL, Rnjak-Kovacina J, Schmidt DF, Lo TJ, Kaplan DL. Biomaterials. 2014;35:6941. doi: 10.1016/j.biomaterials.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han HY, Ning HY, Liu SS, Lu Q, Fan ZH, Lu HJ, Lu GZ, Kaplan DL. Adv. Funct. Mater. 2016;26:421. doi: 10.1002/adfm.201504160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Q, Wang XL, Lu SZ, Li MZ, Kaplan DL, Zhu HS. Biomaterials. 2011;32:1059. doi: 10.1016/j.biomaterials.2010.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pei YZ, Liu X, Liu SS, Lu Q, Liu J, Kaplan DL, Zhu HS. Acta Biomater. 2015;13:168. doi: 10.1016/j.actbio.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davidenko N, Schuster CF, Bax BV, Raynal N, Farndale RW, Best SM, Cameron RE. Acta Biomater. 2015;25:131. doi: 10.1016/j.actbio.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kasoju U, Bora N. J. Biomed. Mate.r Res B Appl. Biomater. 2012;100:1854. doi: 10.1002/jbm.b.32753. [DOI] [PubMed] [Google Scholar]
- 60.Wang YY, Wang XY, Shi J, Zhu R, Zhang JH, Zhang ZR, Ma DW, Hou YJ, Lin F, Yang J, Mizuno M. Sci. Rep. 2016;6:39477. doi: 10.1038/srep39477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan CP, Chen Man YB. Phytochem. Anal. 2002;13:129. doi: 10.1002/pca.633. [DOI] [PubMed] [Google Scholar]
- 62.Lin SS, Lu GZ, Liu SS, Bai SM, Liu X, Lu Q, Zuo BQ, Kaplan DL, Zhu HS. J. Mater. Chem. B. 2014;2:2622. doi: 10.1039/C4TB00019F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong XD, Zhao Q, Xiao LY, Lu Q, Kaplan DL. Biomacromolecules. 2016;17:3000. doi: 10.1021/acs.biomac.6b00863. [DOI] [PubMed] [Google Scholar]
- 64.Anghiler A, Lantto R, Kruus K, Arosio C, Freddi C. J. Biotechnol. 2007;127:508. doi: 10.1016/j.jbiotec.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 65.Jones GL, Motta A, Marshall MJ, EI Haj AJ, Cartmell SH. Biomaterials. 2009;30:5376. doi: 10.1016/j.biomaterials.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 66.Chen JP, Chen SH, Lai GJ. Nanoscale Res. Lett. 2012;7:170. doi: 10.1186/1556-276X-7-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baran ET, Tuzlakoğlu K, Mano JF, Reis RL. Mater. Sci. Engineer. C. 2012;32:1314. doi: 10.1016/j.msec.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 68.Bhardwaj N, Kundu SC. Biomaterials. 2012;33:2848. doi: 10.1016/j.biomaterials.2011.12.028. [DOI] [PubMed] [Google Scholar]
- 69.Zeng SG, Liu L, Shi Y, Qiu JQ, Fang W, Rong MD, Guo ZH, Gao WF. PLOS One. 2015;10:e0128658. doi: 10.1371/journal.pone.0128658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dunne LW, Lyyanki T, Hubenak J, Mathur AB. Acta Biomater. 2014;10:3630. doi: 10.1016/j.actbio.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buno KP, Chen XM, Weibel JA, Thiede SN, Garimella SV, Yoder MC, Voytik-Harbin SL. ACS Appl. Mater. Interface. 2016;8:21848. doi: 10.1021/acsami.6b01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lakshmanan R, Kumaraswamy P, Krishnan UM, Sethuraman S. Biomaterials. 2016;97:176. doi: 10.1016/j.biomaterials.2016.02.033. [DOI] [PubMed] [Google Scholar]
- 73.Song HHG, Park KM, Gerecht S. Adv. Drug Deliv. Rev. 2014;79:19. doi: 10.1016/j.addr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.She ZD, Liu WQ, Feng QL. Biomed. Mater. 2009;4:045014. doi: 10.1088/1748-6041/4/4/045014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.