Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a pervasive healthcare-acquired (HA) pathogen with recent emergence as a community-acquired (CA) pathogen. To elucidate whether meat mediates MRSA transmission between animals and humans in Japan, this study examined MRSA isolates from retail meat (n = 8), cows with mastitis (n = 7), and humans (HA-MRSA = 46 and CA-MRSA = 54) by molecular typing, virulence gene analyses, and antimicrobial susceptibility testing. MRSA isolates from retail meat were classified into sequence type (ST) 8/spa type t1767 (n = 4), ST8/t4133 (n = 1), ST59/t3385 (n = 1), ST88/t375 (n = 1), and ST509/t375 (n = 1). All seven MRSA isolates from cows with mastitis were ST8/t1767. 46 HA-MRSA were clonal complex (CC) 5, divided into t002 (n = 30), t045 (n = 12), and t7455 (n = 4). 54 CA-MRSA were classified into 6 different CCs: CC1 (n = 14), CC5 (n = 7), CC8 (n = 29), CC45 (n = 1), CC89 (n = 1), CC509 (n = 1), and into 16 different spa types including newly identified t17177, t17193, and t17194. The majority were CC8/t1767 (n = 16). CC of one CA-MRSA isolate (spa type t1767) was not classified. Among 41 CC8 MRSA (five from meat, seven from cows with mastitis, and 29 CA-MRSA), 14 ST8/SCCmec IVl isolates (three from meat, one from a cow with mastitis, and 10 CA-MRSA) had identical pulsed-field gel electrophoresis patterns and similar spa type (t1767, t4133, and t17177), and were typed as CA-MRSA/J (ST8/SCCmec IVl, positive for sec + sel + tst but negative for Panton–Valentine leukocidin and the arginine catabolic mobile element). These results suggest that there is a transmission cycle of CA-MRSA/J among meat, cows, and humans in Japan, although it is unclear whether the origin is cow.
Introduction
Staphylococcus aureus is an important pathogen that causes infections in humans and is also responsible for disease in animals, such as bovine mastitis [1]. Treatment of infections caused by S. aureus has been further complicated by antimicrobial resistance in the bacteria, particularly methicillin-resistant S. aureus (MRSA) [2]. These infections were initially associated with hospitalization (healthcare-associated MRSA (HA-MRSA)). Recently, MRSA infections have been identified in healthy people living in the community, who had not been in hospital or had any medical treatment [2]. These cases are referred to as community-acquired MRSA (CA-MRSA) and represent a distinct group as compared to HA-MSRA.
The majority of typical HA-MRSA strains have the staphylococcal cassette chromosome mec (SCCmec) I, II, or III, and are classified into sequence type (ST), as defined by multi-locus sequence typing (MLST). ST5/SCCmec II known as the New York/Japan clone, ST22/SCCmec IV known as the epidemic MRSA (EMRSA)-15, and ST36/SCCmec II known as the EMRSA-16, are prevalent HA-MRSA linages [2]. CA-MRSA generally carries SCCmec type IV or V and often produces Panton–Valentine leucocidin (PVL), which causes tissue necrosis and leukocyte destruction [2]. ST8/SCCmec IVa (MRSA USA300), the most common CA-MRSA clone in the United States, is one of the most well-characterized CA-MRSA [3] and is now also a cause of hospital infection [4]. USA300 is positive for the arginine catabolic mobile element (ACME), which enhances colonization and survival in addition to PVL [3]. In the past decade, an increase in CA-MRSA belonging to clonal complex (CC)8, known as the CA-MRSA/J has been reported in Japan [5]; in contrast to USA300, CA-MRSA/J harbors SCCmec IVl and is negative for both PVL and ACME.
More recently, MRSA has been increasingly reported as emerging problem in the veterinary setting; MRSA has been isolated from cows, pigs, horses, and poultry worldwide (livestock-associated MRSA (LA-MRSA) [6]. LA-MRSA generally belongs to ST398 in European countries and ST9 in Asia [6]. It is largely associated with food producing animals but can colonize other host species, include causing infections in humans who are in frequent contact with MRSA-colonized pigs. However, about 20%– 40% of ST398 MRSA cases in humans cannot be epidemiologically linked to contact with livestock animals, indicating an alternative transmission pathway [7]. Additionally, ST398 MRSA also has been detected in retail food (veal, pork, and chicken meat) [8]. Previous studies have suggested that MRSA-positive pigs could contaminate the slaughterhouse environment, and have the potential to contaminate carcasses during the process [9], [10]. These results showed that MRSA are detected at different stages of the meat production chain and persist from farm to folk. The contamination of food products by animal MRSA is a big threat, as it has a potential for wide dissemination in the general population [11].
In Japan, MRSA have been detected in various food products, including chicken meat [12], [13], duck meat [13], meat products (the details regarding the type of meat product are unclear) [14], and bovine milk [15]. Furthermore, MRSA have been detected in livestock animals, including bovine mastitis [16] and nasal swabs of pigs [17], [18]. Although some MRSA related articles were reported in several origins (animals, meats, and humans), these MRSA isolates were not compared. Therefore, it has remained unsolved the relationship among these MRSA isolated from different origins. To elucidate the relationship among animal, meat, and human isolates, and to assess transmission from animals to humans in Japan, we investigated the characteristics of MRSA from retail meat, cows with mastitis, a common animal disease caused by S. aureus, and humans.
Materials and methods
Sample collection
A total of 5,435 food samples were collected from 2008 to 2009, and eight MRSA were isolated from eight meat samples used in this study. Various types of food (e.g. fish, rice balls) were included in addition to meat in the 5,435 samples, although the number of meat samples was uncertain. Eight MRSA were isolated from retail meat which was purchased in Osaka (n = 5: two ground beef samples and one sample each of pork ribs, ground pork, and Taiwanese frozen duck loin) and Tokyo (n = 3: one sample each of pork ribs, ground beef, and chicken) from 2008 to 2009. All meat, with the exception of the Taiwanese frozen duck loin, was produced domestically. MRSA from meat was isolated using a 1:10 dilution emulsion of the meat sample in sterile phosphate buffer saline. A total of 0.5 ml of the emulsion was added Tryptic to 4.5 ml of Soy Broth (TSB: Becton Dickinson Japan, Tokyo, Japan) with 7.5% NaCl and incubated for 18 to 20 h at 37°C. A loopful of enrichment broth was spread on Mannitol Salt Agar with Egg Yolk (MSEY; Eiken Chemical, Tokyo, Japan) and incubated for 48 h at 37°C. The presumptive colonies of S. aureus (yellow colonies with halo) were streaked and purified onto Trypticase Soy Agar (TSA: Becton Dickinson Japan). Isolates from meat were confirmed to be S. aureus by using PS LATEX (Eiken Chemical). The PCR was performed to confirm of the presence of the mecA gene [19].
Seven MRSA were isolated from seven cows with mastitis in 2011, all bred at the same private farm in Hokkaido. We isolated bacteria from the milk, which were taken from the breast, and identified MRSA to detect the pathogen of the mastitis by request from the owner. The owner of the farm consented to use of the isolates in this study anonymously, including non-disclosure of the city of the farm. We did not perform any animal experiments or field studies in this study. This study also did not involve endangered or protected species. Therefore, the special permission in the authorities for this investigation was not necessary. Milk samples were streaked onto MRSA screening agar (cefoxitin containing Mannitol Salt Agar with Egg Yolk (MS-CFX); Nissui Pharmaceutical, Tokyo, Japan) and overnight at 37°C. The presumptive colonies were further cultured onto TSA and repeatedly sub-cultured to get pure culture. Methicillin resistance was confirmed by testing for the presence of penicillin binding protein 2 (PBP2’) (MRSA-LA; Denka-Seiken, Tokyo, Japan).
A total of 100 human MRSA isolates collected in Kitasato University Hospital from 2014 to 2016 (46 HA-MRSA isolates and 54 CA-MRSA isolates) were obtained from the Infection Control Research Center, Kitasato University, Tokyo, Japan. All HA- and CA-MRSA isolates were recovered from blood samples. Infections were classified as either HA- or CA-MRSA according to origin of MRSA isolates and standard epidemiological definitions established by the U.S. Centers for Disease Control and Prevention [20]. MRSA isolates were classified as HA-MRSA if (i) they were isolated from a culture obtained 48 hours or more after a patient was hospitalized, (ii) the patient had a history of hospitalization, surgery, dialysis, or residence in a long-term care facility within 1 year before the MRSA culture date, (iii) the patient had an indwelling device at the time of culture, or (iv) the patient had a history of MRSA infection or colonization. All other MRSA isolates were considered CA-MRSA. We could not obtain information about the patients (age, symptoms, sex, and places of residence or infection) because of ethical constraints imposed by Kitasato University.
All MRSA isolates were confirmed to be S. aureus by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using the Bruker MALDI Biotyper system (Bruker Daltonics, Bremen, Germany) with the ethanol-formic acid extraction method. All were subsequently confirmed as mecA-positive by PCR [19].
Molecular typing
For all MRSA isolates, SCCmec typing, phage open reading frame (ORF) typing and spa typing were performed. SCCmec typing was performed by multiplex PCR as described previously [21]. SCCmec types I to V were determined based on the mec complex class (mec classes A, B, and C) and the type of ccr (ccr 1, 2, 3, and 5) [21]. For MRSA isolates identified as SCCmecIV, further PCR for detection of the CWASP/J gene (spj) was performed to identify subtype SCCmec IVl [5]. The Clonal complex (CC) of all MRSA isolates were classified by phage ORF typing according to the methods described by Suzuki et al.[22]. Briefly, PCR was performed for the presence of 16 small genomic islets and scored according to islet presence (= 1) or absence (= 0). These scores were then converted to hexadecimal numbers using the internal bin2hex (number, places) function of Microsoft Excel, and the islet pattern (IP) was detected. In many cases, a one-to-one correspondence between the IP and CC identified by multilocus sequence typing (MLST) was observed. The IPs were compared with those previously reported by Suzuki et al.[22], and the CCs of MRSA isolates were classified. spa typing was performed as described previously [23].
Pulsed-field gel electrophoresis (PFGE) was performed for CC8 MRSA isolates with genetic DNA fragments generated using 30 U SmaI (TaKaRa, Otsu, Japan) as previously described [24]. Cluster analysis was performed with the software program BioNumerics v6 (Applied Maths, Sint-Martens-Latem, Belgium) using the Dice coefficient and the unweighted pair group method. MLST for retail meat, cows with mastitis, and CC8 human MRSA isolates was performed as described previously [25]. The founder and CC of each ST were determined using the enhanced version of Based Upon Related Sequence Types (eBURST) [26].
Virulence gene analysis
The presence of genes encoding six staphylococcal enterotoxins, SEA to SEE, which main source of food poisoning [27], in addition to SEL, which CA-MRSA/J carries in Japan frequently [5] (SEs: sea, seb, sec, sed, see, and sel), toxic shock syndrome toxin-1 (TSST-1: tst) [27], which cause of TSS, exfoliative toxin A (ETA: eta) and B (ETB: etb), which are implicated in the cause of staphylococcal scalded-skin syndrome [27], PVL (pvl), which is associated with increased disease severity and found in a high proportion of CA-MRSA strains [19], and ACME (acr) which is a striking feature of USA300 and plays an important role in its growth and survival [3] was determined by PCR using previously reported primers [3], [19][27][28].
Antimicrobial susceptibility testing
Antimicrobial susceptibility was tested by the agar dilution method following Clinical and Laboratory Standards Institute (CLSI) recommendations [29] for the following antibiotics: ampicillin (AMP; Sigma-Aldrich, St. Louis, MO, USA), oxacillin (OXA; Sigma-Aldrich), kanamycin (KAN; Sigma-Aldrich), gentamicin (GEN; Sigma-Aldrich), erythromycin (ERY; Sigma-Aldrich), clindamycin (CLI; Sigma-Aldrich), vancomycin (VAN; Sigma-Aldrich), ciprofloxacin (CIP; Sigma-Aldrich), and tetracycline (TET; Wako Pure Chemical Industries, Osaka, Japan). S. aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 served as quality control strains. The breakpoints of these antimicrobial agents were determined according to CLSI interpretation criteria [29].
Results
Molecular characterization of MRSA isolates from meat, cows with mastitis, and humans
Characteristics of MRSA isolates in this study are summarized in Table 1. Among eight MRSA isolates from meat, two (one from ground pork and one from ground beef) were classified as ST8 (CC8)/t1767/SCCmec IVl, two (one from pork ribs and one from chicken) were ST8 (CC8)/t1767/SCCmec untypable (harbored ccr type 2, but multiplex PCR for mec class was not amplified), one from ground beef was ST8 (CC8)/t4133/SCCmec IVl, one from pork rib was ST88 (CC88)/t1028/SCCmec IV, one from ground beef was ST59 (CC59)/t3385/SCCmec V, and one from Taiwanese frozen duck loin was ST573/t3525/SCCmec IV (Table 1). All seven MRSA isolates from cows with mastitis were classified as ST8 (CC8)/t1767/SCCmec IVl. Among MRSA isolates from humans, all 46 HA-MRSA isolates were classified as CC5/SCCmec II, and were divided into spa type t002 (n = 30), t045 (= 12), and t7455 (n = 4). Fifty-four CA-MRSA isolates yielded 16 different spa types. These 16 spa types belonged to 6 different CCs: 14 of CC1 (t1784: n = 13, t2207: n = 1); 7 of CC5 (t002: n = 5, t045: n = 1, and newly identified t17193: n = 1), 29 of CC8 (t008: n = 2, t986: n = 1, t1476: n = 1, t1767: n = 16, t1852: n = 3, t 4133: n = 1, t12760: n = 1, and newly identified t17177: n = 3 and t17194: n = 1); one of each CC45 (t065), CC89 (t375), and CC509 (t375). The majority of CA-MRSA was CC8/t1767/SCCmec IVl (n = 15), following CC1/t1784/SCCmec IV (n = 12). SCCmec type of CC45 and CC89 were untypable (harbored mec class A, but multiplex PCR for ccr type was not amplified). CC of one CA-MRSA isolate, spa type t1767, was not able to be classified by phage ORF typing because its IP (04C6) was not reported previously. Accordingly, MLST was performed; however, ST was not able to identified because two of seven genes (aroE and glpF) could not amplify using primers described previously [25].
Table 1. Molecular characterization of MRSA isolates from meat, cow mastitis, and humans (HA-MRSA and CA-MRSA).
| CCa | SCCb mec |
spa | Origin | Resistant isolates | Pattern of virulence genes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Meat (n = 8) |
Cow with mastitis (n = 7) |
CA-MRSA (n = 54) |
HA-MRSA (n = 46) |
AMPf | OXA | KAN | GEN | ERY | CLI | VAN | CIP | TET | ||||
| 1 | IV | t1784 | 13 | 13 | 13 | 1 | 1 | 13 | 0 | 0 | 13 | 0 | sea (n = 9), sea + see (n = 3), negative (n = 1) | |||
| t2207 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | negative | |||||
| 5 | II | t002 | 4 | 30 | 34 | 34 | 32 | 26 | 34 | 33 | 0 | 34 | 27 | seb (n = 13), sec + sel + tst (n = 11), sea + sec + sel + tst (n = 2), sel + tst (n = 1), sel (n = 1), negative (n = 6) | ||
| t045 | 1 | 12 | 13 | 13 | 13 | 7 | 13 | 13 | 0 | 10 | 13 | sec + sel + tst (n = 10), seb + sec + sel + tst (n = 2), sea + sec + see + sel + tst (n = 1) | ||||
| t7455 | 4 | 4 | 4 | 4 | 3 | 4 | 4 | 0 | 4 | 4 | sec + sel + tst (n = 3), seb + sec + sel + tst (n = 1) | |||||
| IV | t002 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | tst (n = 1) | ||||
| t17193 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | negative | |||||
| 8 | IV | t008 | 2 | 2 | 2 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | pvl + acr (n = 2) | |||
| t986 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | negative | |||||
| t1476 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | tst | |||||
| t1767 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | sel + tst | |||||
| t1852 | 3 | 3 | 3 | 3 | 3 | 0 | 0 | 0 | 3 | 0 | negative (n = 3) | |||||
| t17194 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | negative | |||||
| IVl | t1767 | 2 (1 GB, 1 GP)e | 7 | 15 | 24 | 24 | 24 | 19 | 9 | 0 | 0 | 0 | 0 | sec + sel + tst (n = 23), negative (n = 1) | ||
| t4133 | 1 (GB) | 1 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | sec + sel + tst (n = 1), negative (n = 1) | ||||
| t17177 | 3 | 3 | 3 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | sec + sel + tst (n = 3) | |||||
| V | t12760 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | negative (n = 1) | ||||
| UTc | t1767 | 2 (1 PR, 1 C) | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | sec + sel + tst (n = 2) | ||||
| 45 | UTd | t065 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | negative | |||
| 59 | V | t3385 | 1 (GB) | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | sea + seb + sel | |||
| 88 | IV | t1028 | 1 (PR) | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | negative | |||
| 89 | UTd | t375 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | etb | |||
| 509 | II | t375 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | negative | |||
| 573 | IV | t3525 | 1 (TD) | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | sec | |||
| Unclassfied | IV | t1767 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | sed | |||
a; The Clonal complex (CC) of all MRSA isolates were classified by phage ORF typing
b; SCCmec types I to V were determined based on the mec complex class (mec classes A, B, and C) and the type of ccr (ccr 1, 2, 3, and 5)
c; Untypable (ccr type 2 + mec-untypable)
d; Untypable (ccr-untyable + mec class A)
e; GB: ground beef, GP: ground pork, PR: pork ribs, C: chicken, TD: Taiwanese frozen duck loin
f; AMP: ampicillin, OXA: oxacillin, KAN: kanamycin, GEN: gentamicin, ERY: erythromycin, CLI: clindamycin, VAN: vancomycin, CIP: ciprofloxacin, TET: tetracycline
Toxin genes of MRSA isolates from meat, cows with mastitis, and humans
The toxin genes detected in each MRSA isolate are summarized in Table 1. In total, 88% (7/8) from meat, 100% (7/7) from cows with mastitis, 93% (43/46) HA-MRSA isolates, and 70% (38/54) of CA-MRSA isolates carried at least one toxin gene. Among the fourteen CC1/SCCmec IV MRSA isolates, sea (n = 9) was most common, followed by sea + see (n = 3). Among the 51 CC5/SCCmec II MRSA isolates, including 46 HA-MRSA and 5 CA-MRSA isolate, sec + sel + tst (n = 24) was most common, followed by seb (n = 13), seb + sec + sel + tst (n = 3), sea + sec + sel + tst (n = 2), sea + sec + see + sel + tst (n = 1), sel + tst (n = 1), and sel (n = 1). One CC5/SCCmec IV carried tst. Among the nine CC8/SCCmec IV MRSA isolates, pvl + acr (n = 2), sel + tst (n = 1), and tst (n = 1) were observed. Among the twenty-nine CC8/SCCmec IVl MRSA isolates, including three from meat, seven cows with mastitis, and nineteen from humans, 97% (27/29) carried sec + sel + tst. Two CC8 SCCmec untypable isolates from pork ribs and chicken carried sec + sel + tst. One CC59/SCCmec V (isolated from ground beef) carried sea + seb + sel, one CC573/SCCmec IV (isolated from Taiwanese frozen duck) carried sec, and one CC unclassified/SCCmec IV (belonging to CA-MRSA) carried sed.
Antimicrobial susceptibility
All MRSA isolates in this study were resistant to β-lactams (AMP and OXA); however, they were susceptible to VAN. In addition, CC8/SCCmec IVl was resistant to KAN (29/29: 100%) and GEN (24/29: 83%). CC5/SCCmec II was resistant to all tested antimicrobial agents except for VAN.
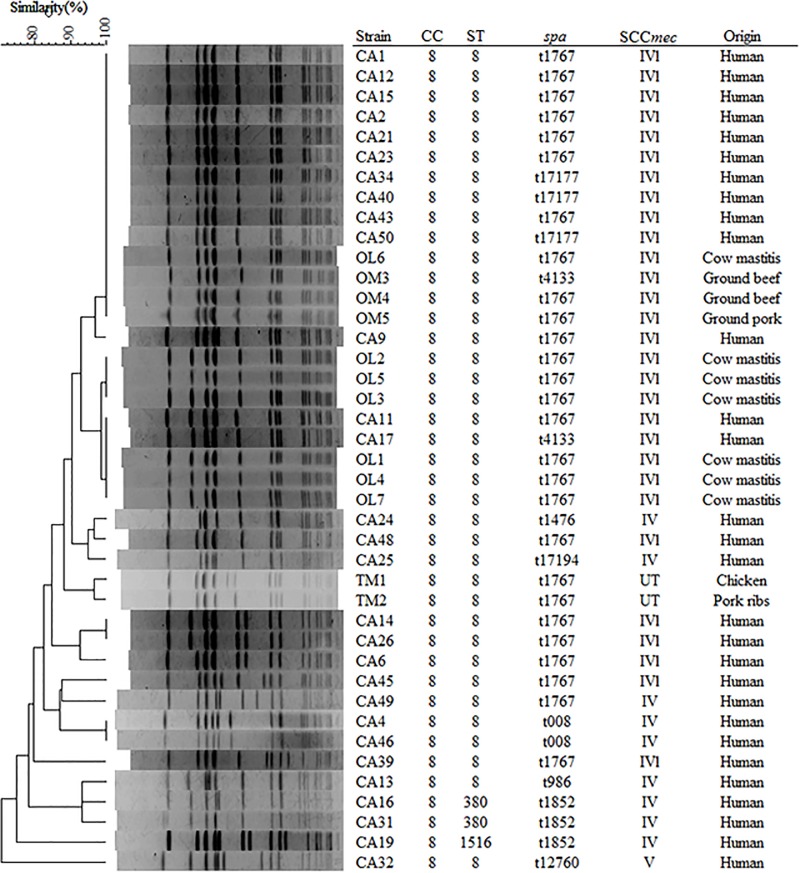
PFGE analysis and MLST of CC8 MRSA isolates
PFGE analysis and MLST were performed to classify MRSA isolates according to CC8 (n = 41, determined by phage ORF typing), which was the common clone among retail meat (n = 5), cows with mastitis (n = 7), and CA-MRSA from humans (n = 29) (Fig 1). CC8 isolates were classified into a total of three STs: ST8 (allelic profile 3-3-1-1-4-4-3), ST380 (3-3-61-42-4-4-3), and ST1516 (3-3-1-42-4-4-3). Three CC8/SCCmec IVl MRSA isolates from meat (two from beef and one from pork), one from a cow with mastitis, and ten human CA-MRSA isolates showed 100% PFGE similarity. Similarly, two human CA-MRSA isolates and three from cow with mastitis showed 100% similarity with different origin. These were ST8/SCCmec IVl containing three spa types with similar repeat profiles (t1767: 11-19-12-21-17-34-24-24-34-22-25, t4133: 11-12-21-17-34-24-24-34-22-25, and t17177: 11-19-12-21-17-34-24-24-24-24-34-22-25).
Fig 1. Genetic relationships among CC8 MRSA isolates.
UPGMA dendrogram showing genetic relatedness among representative CC8 MRSA isolates as determined by PFGE with SmaI. UT; ccr type 2 + mec-untypable.
Discussion
This study showed that three MRSA isolates from retail meat, one MRSA from a cow with mastitis, and ten CA-MRSA isolates were closely related according to spa type, and identical according to PFGE pattern, ST, and SCCmec, they all had the characteristics of ST8/SCCmec IVl. Our study is the first to detect the closel molecular epidemiological relationship of MRSA among retail meat, cows with mastitis, and CA-MRSA from humans in Japan.
ST8 MRSA isolated in this study showed a similar genotype and antimicrobial susceptibility pattern to ST8 CA-MRSA/J in Japan, which has some different characteristics from foreign countries. In Japan, a ST8 CA-MRSA/J strain was described; ST8 CA-MRSA/J which can be characterized as carrying SCCmec IVl, spa type t1767, negative for PVL and ACME, positive for sec, sel, and tst, and resistant to gentamicin [4]. It is reported that 37.5% (18/48) of CA-MRSA from human were typed as ST8 CA-MRSA/J in Japan [5]. Gentamicin is used in an outpatient for the treatment of skin infections in Japan [30]. Therefore, antibiotic therapy based on antimicrobial susceptibility test is needed for skin infections caused by MRSA. ST8/SCCmec IVa, positive for PVL and ACME MRSA (USA300), is a predominant CA-MRSA genotype in the US and worldwide [31]. USA300 clones show resistance to many non-β-lactams (macrolides, fluoroquinolones, and tetracycline) in addition to β-lactams [31]. ST8 MRSA isolates used in this study showed the same genotype and antimicrobial profile, aminoglycosides resistance, as those of CA-MRSA/J. Although the geographical area where human MRSA was derived, as well as the sample size of meat and animals were all limited, this study revealed that ST8 CA-MRSA/J spreads not only to the human community setting, but also among meat and living livestock (cow), but not yet to the healthcare setting.
Four STs (ST8, ST59, ST88, and ST573) were identified in isolates from retail meat in this study. All of these STs are human-associated types: ST8 in the United States and worldwide [31], ST59 in Taiwan [31], ST88 in Africa and Asia [32], ST573 which is a rare clone found previously in Taiwan [33] and Australia [34]. Three of four STs (ST8, ST59, and ST88) were found primarily in human CA-MRSA in Japan [2]. MRSA isolates from retail chicken and duck meat in Japan were ST8/SCCmec IV, and regarded as CA-MRSA [13]. These observations suggest a relationship of MRSA between retail meats and humans. However, these four STs have not been reported in Japanese livestock animals, and the prevalence of MRSA is low [0.9%–8% [17][18]] in Japanese livestock. Considering these reports, there is a strong possibility that MRSA isolates from meat in this study are contaminated from humans, although we cannot draw any definitive conclusions regarding the source of contaminated retail meat with MRSA.
Among STs from retail meat, ST573 MRSA from Taiwanese duck loin was first isolated in Japan. ST573 is a rare clone, found previously in Taiwanese children [SCCmec V, 0.3% (1/294)] [35], healthcare settings [SCCmec IV, 2.4% (5/206)] in Taiwan [33], and in community settings [SCCmec V, 0.05% (2/4,099)] in Australia [34]. There are several reports of pathogens associated with the import of food; an oxacillin-susceptible mecA-positive S. aureus (OS-MRSA), has never been isolated in Europe, was isolated from imported cheese. It might indicate that OS-MRSA may enter the EU via the import of food [36]. Although the source of ST573/SCCmec IV MRSA from Taiwanese frozen duck in this study is unclear, it might have been brought to Japan via imported meat from Taiwan, the only region where ST573 carrying SCCmec IV MRSA has been detected [33].
Since S. aureus can produce enterotoxins, it also poses a threat to humans who ingest food contaminated with these toxins [37]. Staphylococcal food poisoning, characterized by vomiting and diarrhea, is a leading cause of food-borne illness in Japan [38]. Food sources of S. aureus have expanded to include livestock animal products and low-fat milk [38]. Toxic shock syndrome (TSS), which can be life-threatening, is defined by clinical and laboratory evidence of fever, rash, desquamation, hypotension, and multiple organ failure caused not only by toxic shock syndrome toxin-1 (TSST-1), but also by enterotoxins [39]. In this study, 88% (7/8) of MRSA isolates from retail meat were positive for enterotoxin genes and tst, higher than previous study [28.6% [13]]. In Japan, the contamination rate of MRSA in meat is low [0.45% to 1.5% [12][13]], but MRSA isolates from retail meat frequently carry virulence genes, and the spread of MRSA can cause human disease via the handling of contaminated retail meat.
Conclusion
This study showed that ST8 CA-MRSA/J is detected in the community setting, including retail meat and cows, and suggested that there is the transmission route of ST8 CA-MRSA/J among these sources. However, the direction of transfer of MRSA could not be established, and the results might not be reflective of Japan overall because the number of MRSA isolates from meat and animals was very low. Additional studies are needed to determine the origin of MRSA from retail meat, confirm the distribution of ST8 CA-MRSA/J in living animals, and assess the risk of the spread of MRSA to consumers and others who handle meat.
Supporting information
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Juhász-kaszanyitzky É, Jánosi S. MRSA Transmission between Cows and Humans. Emerg Infect Dis 2007;13:3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto T, Nishiyama A, Takano T, Yabe S, Higuchi W, Razvina O, et al. Community-acquired methicillin-resistant Staphylococcus aureus: community transmission, pathogenesis, and drug resistance. J Infect Chemother 2010;16:225–254. doi: 10.1007/s10156-010-0045-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 4.Uhlemann A-C, Otto M, Lowy FD, DeLeo FR. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect Genet Evol 2014;21:563–574. doi: 10.1016/j.meegid.2013.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwao Y, Takano T, Higuchi W, Yamamoto T. A new staphylococcal cassette chromosome mec IV encoding a novel cell-wall-anchored surface protein in a major ST8 community-acquired methicillin-resistant Staphylococcus aureus clone in Japan. J Infect Chemother 2012;18:96–104. doi: 10.1007/s10156-011-0348-5 [DOI] [PubMed] [Google Scholar]
- 6.Chuang Y-Y, Huang Y-C. Livestock-associated meticillin-resistant Staphylococcus aureus in Asia: an emerging issue? Int J Antimicrob Agents 2015;45:334–340. doi: 10.1016/j.ijantimicag.2014.12.007 [DOI] [PubMed] [Google Scholar]
- 7.Deiters C, Günnewig V, Friedrich AW, Mellmann A, Köck R. Are cases of methicillin-resistant Staphylococcus aureus clonal complex (CC) 398 among humans still livestock-associated? Int J Med Microbiol 2015;305:110–113. doi: 10.1016/j.ijmm.2014.11.007 [DOI] [PubMed] [Google Scholar]
- 8.Pantosti A. Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front Microbiol 2012;3:127 doi: 10.3389/fmicb.2012.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molla B, Byrne M, Abley M, Mathews J, Jackson CR, Fedorka-Cray P, et al. Epidemiology and genotypic characteristics of methicillin-resistant Staphylococcus aureus strains of porcine origin. J Clin Microbiol 2012;50:3687–3693. doi: 10.1128/JCM.01971-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhegghe M, Herman L, Haesebrouck F, Butaye P, Heyndrickx M, Rasschaert G. Preliminary evaluation of good sampling locations on a pig carcass for livestock-associated MRSA isolation. Int J Food Contam 2015;2:5 doi: 10.1186/s40550-015-0013-3 [Google Scholar]
- 11.Kluytmans JAJW. Methicillin-resistant Staphylococcus aureus in food products: cause for concern or case for complacency? Clin Microbiol Infect 2010;16:11–15. doi: 10.1111/j.1469-0691.2009.03110.x [DOI] [PubMed] [Google Scholar]
- 12.Kitai S, Shimizu A, Kawano J, Sato E, Nakano C, Uji T, et al. Characterization of methicillin-resistant Staphylococcus aureus isolated from retail raw chicken meat in Japan. J Vet Med Sci 2005;67:107–110. [DOI] [PubMed] [Google Scholar]
- 13.Ogata K, Narimatsu H, Suzuki M, Higuchi W, Yamamoto T, Taniguchi H. Commercially distributed meat as a potential vehicle for community-acquired methicillin-resistant Staphylococcus aureus. Appl Environ Microbiol 2012;78:2797–2802. doi: 10.1128/AEM.07470-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazuyasu M, Wataru I, Takaomi W. Detection of methicillin-resistant Staphylococcus aureus (MRSA) from foodstuffs, foods and cooking facilities. Japanese J Food Microbiol 2002;19:127–131. [Google Scholar]
- 15.Hata E, Katsuda K, Kobayashi H, Uchida I, Tanaka K, Eguchi M. Genetic variation among Staphylococcus aureus strains from bovine milk and their relevance to methicillin-resistant isolates from humans. J Clin Microbiol 2010;48:2130–2139. doi: 10.1128/JCM.01940-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hata E. Bovine mastitis outbreak in Japan caused by methicillin-resistant Staphylococcus aureus New York/Japan clone. J Vet Diagn Invest 2016;28:291–298. doi: 10.1177/1040638716643126 [DOI] [PubMed] [Google Scholar]
- 17.Baba K, Ishihara K, Ozawa M, Tamura Y, Asai T. Isolation of meticillin-resistant Staphylococcus aureus (MRSA) from swine in Japan. Int J Antimicrob Agents 2010;36:352–354. doi: 10.1016/j.ijantimicag.2010.06.040 [DOI] [PubMed] [Google Scholar]
- 18.Sato T, Usui M, Motoya T, Sugiyama T, Tamura Y. Characterisation of meticillin-resistant Staphylococcus aureus ST97 and ST5 isolated from pigs in Japan. J Glob Antimicrob Resist 2015;3:283–285. doi: 10.1016/j.jgar.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 19.Al-Talib H, Yean CY, Al-Khateeb A, Hassan H, Singh K-KB, Al-Jashamy K, et al. A pentaplex PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus and Panton-Valentine Leucocidin. BMC Microbiol 2009;9:113 doi: 10.1186/1471-2180-9-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- 21.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 2007;51:264–274. doi: 10.1128/AAC.00165-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki M, Matsumoto M, Takahashi M, Hayakawa Y, Minagawa H. Identification of the clonal complexes of Staphylococcus aureus strains by determination of the conservation patterns of small genomic islets. J Appl Microbiol 2009;107:1367–1374. doi: 10.1111/j.1365-2672.2009.04321.x [DOI] [PubMed] [Google Scholar]
- 23.Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol 1999;37:3556–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulvey MR, Chui L, Ismail J, Louie L, Murphy C, Chang N, et al. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J Clin Microbiol 2001;39:3481–3485. doi: 10.1128/JCM.39.10.3481-3485.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 2000;38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST : Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol 2000;38:1032–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cremonesi P, Luzzana M, Brasca M, Morandi S, Lodi R, Vimercati C, et al. Development of a multiplex PCR assay for the identification of Staphylococcus aureus enterotoxigenic strains isolated from milk and dairy products. Mol Cell Probes 2005;19:299–305. doi: 10.1016/j.mcp.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 29.Wayne P. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement (M100-S24). Clin Lab Stand Inst 2014.
- 30.Iwaki M, Noguchi N, Nakaminami H, Sasatsu M, Ito M. Antimicrobial activity and frequency of spontaneous gentamicin-resistant mutants in bacteria related skin infections. Yakugaku Zasshi 2011;131:1653–1659. [DOI] [PubMed] [Google Scholar]
- 31.David MZ, Daum RS. Community-Associated Methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 2010;23:616–687. doi: 10.1128/CMR.00081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr Opin Microbiol 2012;15:588–595. doi: 10.1016/j.mib.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 33.Sheng W-H, Wang J-T, Lauderdale T-L, Weng C-M, Chen D, Chang S-C. Epidemiology and susceptibilities of methicillin-resistant Staphylococcus aureus in Taiwan: emphasis on chlorhexidine susceptibility. Diagn Microbiol Infect Dis 2009;63:309–313. doi: 10.1016/j.diagmicrobio.2008.11.014 [DOI] [PubMed] [Google Scholar]
- 34.Nimmo GR, Coombs GW. Community-associated methicillin-resistant Staphylococcus aureus (MRSA) in Australia. Int J Antimicrob Agents 2008;31:401–410. doi: 10.1016/j.ijantimicag.2007.08.011 [DOI] [PubMed] [Google Scholar]
- 35.Chen C-J, Hsu K-H, Lin T-Y, Hwang K-P, Chen P-Y, Huang Y-C. Factors associated with nasal colonization of methicillin-resistant Staphylococcus aureus among healthy children in Taiwan. J Clin Microbiol 2011;49:131–137. doi: 10.1128/JCM.01774-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez-Lázaro D, Oniciuc E-A, García PG, Gallego D, Fernández-Natal I, Dominguez-Gil M, et al. Detection and characterization of Staphylococcus aureus and methicillin-resistant S. aureus in foods confiscated in EU borders. Front Microbiol 2017;8 doi: 10.3389/fmicb.2017.01344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hennekinne J-A, De Buyser M-L, Dragacci S. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol Rev 2012;36:815–836. doi: 10.1111/j.1574-6976.2011.00311.x [DOI] [PubMed] [Google Scholar]
- 38.Asao T, Kumeda Y, Kawai T, Shibata T, Oda H, Haruki K, et al. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol Infect 2003;130:S0950268802007951 doi: 10.1017/S0950268802007951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 2000;13:16–34, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.