Abstract
Treatment with empagliflozin, an inhibitor of the sodium/glucose cotransporter 2 (SGLT2), is associated with slower progression of diabetic kidney disease. In this analysis, we explored the hypothesis that empagliflozin may have an impact on urinary peptides associated with chronic kidney disease (CKD). In this post-hoc, exploratory analysis, we investigated urine samples obtained from 40 patients with uncomplicated type 1 diabetes (T1D) before and after treatment with empagliflozin for 8 weeks to for significant post-therapy changes in urinary peptides. We further assessed the association of these changes with CKD in an independent cohort, and with a previously established urinary proteomic panel, termed CKD273. 107 individual peptides significantly changed after treatment. The majority of the empagliflozin-induced changes were in the direction of “CKD absent” when compare to patients with CKD and controls. A classifier consisting of these 107 peptides scored significantly different in controls, in comparison to CKD patients. However, empagliflozin did not impact the CKD273 classifier. Our data indicate that empagliflozin induces multiple significant changes in the urinary proteomic markers such as mucin and clusterin. The relationship between empagliflozin-induced proteomic changes and clinical outcomes merits further investigation.
Introduction
Diabetes-associated vascular diseases, especially chronic kidney disease (CKD) represent a major burden for developed societies [1]. Today, treatment of CKD in diabetes, also referred to as diabetic kidney disease, DKD, is typically initiated when first symptoms are evident: persistent microalbuminuria or decreased glomerular filtration rate (GFR). Standard treatment is reduction of blood pressure by interfering with the rennin/angiotension/aldosterone systems (RAAS). SGLT2 inhibition may represent an additional renoprotective intervention–beyond the use of renin angiotensin aldosterone system blockade–for DKD. [2,3].
Multiple recent reports have demonstrated the potential of proteomic biomarkers in kidney disease, as reviewed in [4]. While individual biomarkers, like e.g. albuminuria, show considerable variability, this variability can be counteracted by employing high-dimensional classifiers. In such an approach, multiple biomarkers are combined in a predefined model, reducing and combining the biomarkers into one variable expressing the probability of the membership in a specific group (e.g. healthy or CKD) [5]. We have previously developed capillary electrophoresis coupled to mass spectrometry (CE-MS) for the routine application in the assessment of clinically relevant samples, mostly urine [6] and dialysis fluid [7]. The results demonstrated that analysis of the urinary proteome enables discrimination between patients with and without CKD [8], as well as prediction of progression of CKD, irrespective of the underlying disease mechanism [9]. The CE-MS technology is especially well suited to assess small proteins and peptides, without the requirement of tryptic digestion [10].
The aim of this post-hoc, exploratory analysis was to investigate if treatment with empagliflozin impacts the urinary peptidome, with a special emphasis on urinary peptide biomarkers associated with CKD–specifically the CKD273 biomarker, which is associated with CKD progression [11]. Such an impact could serve as indicative of a potential benefit of empagliflozin prior to onset of CKD, and as potential evidence for the use of empagliflozin in the prevention of DKD onset and progression.
Materials and methods
Patients and sample collection
Samples were collected as described in previous work during clamped euglycemia and hyperglycemia in uncomplicated patients with T1D before and after treatment with empagliflozin (25 mg daily) for 8 weeks [12]. All patients consented to the study, which was approved by the University of Toronto University Health Network with approval #11–0213. The available clinical and demographic data were presented previously [12].
Urinary proteome analysis and peptide identification
The urine samples were prepared and analyzed using a P/ACE MDQ capillary electrophoresis system (Beckman Coulter, Fullerton, USA) on line coupled to a MicroTOF MS (Bruker, Bremen, Germany) described previously in detail [13]. Accuracy, precision, selectivity, sensitivity, reproducibility, and stability of the CE-MS method have been previously described [14]. To normalize for variability in urinary output, a set of 29 internal standard peptides were used for calibration, as previously described [15]. Relative abundance of all peptides in a sample is assessed based on the peak area, and normalized to the 29 standard peptides [15]. This procedure has been applied successfully in multiple previous studies, some based on between 1000 and over 10000 individual samples [16,17]. All detected peptides were deposited, matched, and annotated in a MicrosoftSQL database [9], allowing for further analysis and comparison between groups. Sequencing of target peptides was performed as described [18], using Dionex Ultimate 3000 RSLS nano flow system (Dionex, Camberly UK) and a Beckman CE, coupled to an Orbitrap Velos MS instrument (Thermo Scientific). To assess the distribution of peptides in the context of CKD, previously generated datasets [8] were employed. The clinical and demographic data of this cohort are available from the original publication [8].
Combining peptides into a classifier
To generate a peptide pattern indicative of the impact of empogliflazin, the support vector machine (SVM)-based MosaCluster software [19] was employed. MosaCluster (version 1.7.0) was developed for discrimination between different patient groups in the high-dimensional parameter space by using SVM learning. SVM generates high dimensional models, which rely on features (biomarkers) displaying statistically significant differences between data from patients with a specific disease to controls or other diseases. Each feature allegorizes one dimension in the n-dimensional parameter space [20]. The two classes (here: prior and after empagliflozin treatment) are separated by an n-1 dimensional separating hyperplane. The position of a dataset (a sample) in the n-dimensional dataspace is defined by the amplitude of the features (the peptides used in the classifier). Classification scores provided by this software give a numerical value quantifying the Euclidean distance of the dataset to the maximal margin of the separation hyperplane among cases and controls in multidimensional space, as defined base on the data in the training cohort. A more detailed description has been published recently [14].
Statistical analysis
After testing for normal distribution, continuous data were compared by Wilcoxon rank-sum test, as this test has proven to be of superior statistical power in proteomics datasets [5]. A p-value of <0.05 was considered to be statistically significant. In order to control for the false discovery rate, the p-values were adjusted by the Benjamini and Hochberg method [21].
Results
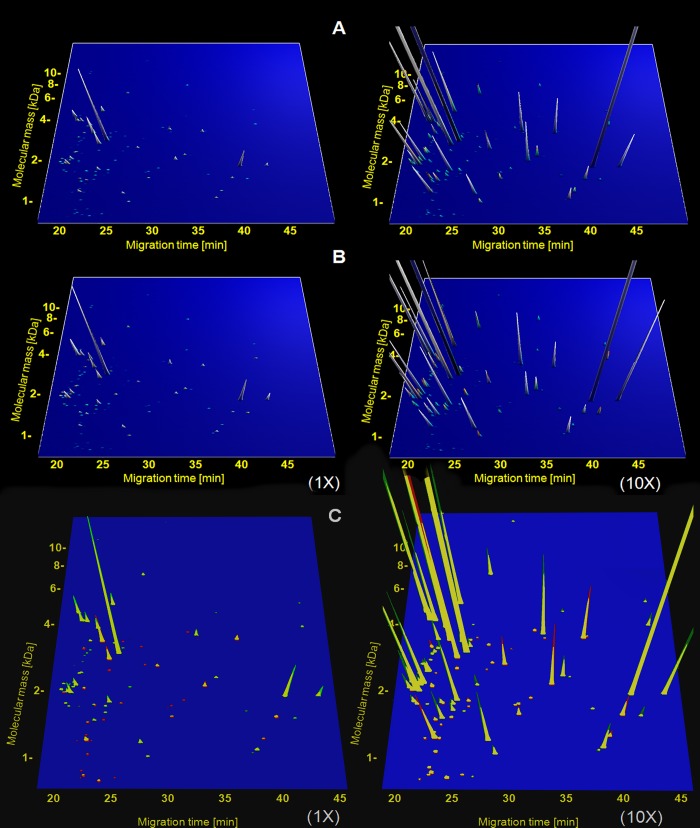
All available samples were analyzed blinded. All data on the individual samples are available in S1 Table. To increase statistical power, data from baseline samples collected during clamped euglycemia and hyperglycemia were combined for the analysis, and the same was done for all follow-up samples obtained after empagliflozin treatment for 8 weeks. The investigation of the proteome of these 160 samples enabled identification of 107 peptides that changed significantly between the baseline and post-treatment samples (S2 Table). Fig 1 illustrates the distribution of the 107 peptides before and after treatment with empagliflozin.
Fig 1. Distribution of the 107 peptides found associated with empagliflozin treatment in the cohort investigated.
Each peptide is displayed based on the mass (in kDa, using a logarithmic scale) and migration time (in minutes). The average relative abundance is represented by the peak intensity. To depict also the lower abundant peptides, the same 107 peptides with abundance magnified 10-fold are shown on the right. Panel A shows the distribution of the 107 peptides before treatment, panel B depicts the distribution after treatment. To ease assessment of changes, the data from patients before and after treatment were superimposed in panel C, peaks were labeled red before EMPA treatment and green after EMPA treatment, the yellow fraction of the peak indicates the overlap in abundance between before and after treatment.
To obtain information on the potential relevance of these changes in the context of CKD, the distribution of these peptides in the cohort of patients with CKD and controls that was used to define CKD273 was investigated. This cohort consisted of 379 healthy controls, and 230 patients with CKD of various etiologies (including 50 patients with diabetic nephropathy) [8]. Of the 107 peptides affected by empagliflozin, 79 showed a greater than 25% change in abundance (either increase or decrease) before and after treatment, and between CKD patients and healthy controls. In the next step we compared the directional change (up- or down-regulation) induced by empagliflozin to the change observed in CKD. The underlying hypothesis was that a change in the same direction in CKD and as a result of empagliflozin treatment may indicate a potential negative effect of empagliflozin, (by inducing a similar response than CKD) and a change in the opposite direction may indicate a potential positive effect (a change towards “healthy”). In 60 of the 79 peptides the change after empagliflozin treatment was opposite to the change observed in CKD, indicating a potential beneficial effect. For 19 peptides, the empagliflozin-induced change was similar to the change observed in CKD.
Amino acid sequences were obtained for 46 of the 79 peptides, all listed in Table 1. When investigating these 46 sequenced peptides only, the significant changes in peptides derived from clusterin, alpha-1-antitrypsin, keratin type 2, and mucin were in the direction towards “healthy”, when comparing healthy controls and CKD. The results were less consistent when investigating the change in collagen fragments induced by empagliflozin: larger collagen fragments generally appeared to be increased upon treatment, while smaller fragments appear to be decreased, possibly indicating reduction of specific protease activity involved in the processing of collagen.
Table 1. Lists the 46 peptides significantly different prior to and after treatment with empagliflozin.
| Mass | CE-T | Sequence | Protein Name | Pre EMPA | Post EMPA | EMPA-ind change | BH adj p-value | CKD mean | control mean | CKD-induced |
|---|---|---|---|---|---|---|---|---|---|---|
| 2320 | 20,8 | KNGDDGEAGKpGRPGERGppGPQ | Collagen alpha-1(I) chain | 219,08 | 305,54 | 1,39 | 0,0491 | 43,45 | 148,32 | 0,29 |
| 2328 | 21 | EDPQGDAAQKTDTSHHDQDHP | Short peptide from AAT | 221,04 | 328,15 | 1,48 | 0,0479 | 20,08 | 123,84 | 0,16 |
| 1676 | 29,3 | PGSSGFpGnPGMKGEAGp | Collagen alpha-2(V) chain | 29,56 | 17,38 | 0,59 | 0,0479 | 40,99 | 35,51 | 1,15 |
| 1694 | 23,5 | PpGPpGKNGDDGEAGKpG | Collagen alpha-1(I) chain | 107,8 | 164,62 | 1,53 | 0,0479 | 40,84 | 67,16 | 0,61 |
| 2216 | 33,8 | IGPpGPAGApGDKGESGPSGPAGPTG | Collagen alpha-1(I) chain | 387,62 | 493,17 | 1,27 | 0,0467 | 226,02 | 548,87 | 0,41 |
| 2378 | 23,6 | KNGDDGEAGKpGRpGERGPPGpqG | Collagen alpha-1(I) chain | 15,55 | 4,04 | 0,26 | 0,0450 | 12,39 | 4,11 | 3,02 |
| 2068 | 20,6 | EEDDGEVTEDSDEDFIQP | E3 ubiquitin-protein ligase TRIM33 | 1254,5 | 1751,18 | 1,4 | 0,0432 | 626,98 | 1694,11 | 0,37 |
| 3287 | 25,4 | AGRpGEVGPpGPPGPAGEKGSPGADGPAGAPGTPGPQG | Collagen alpha-1(I) chain | 24,43 | 5,51 | 0,23 | 0,0432 | 4,65 | 16,00 | 0,29 |
| 2007 | 21,4 | GSTGPAGqKGDSGLPGPpGSpGP | Collagen alpha-1(XI) chain | 109,18 | 66,7 | 0,61 | 0,0422 | 15,50 | 51,67 | 0,30 |
| 906,4 | 24,8 | GPpGPpGPpS | Collagen alpha-1(XVIII) chain | 34,59 | 21,06 | 0,61 | 0,0367 | 37,01 | 19,31 | 1,92 |
| 1657 | 25 | GSPGGKGEmGpAGIpGApG | Collagen alpha-1(III) chain | 18,51 | 5,79 | 0,31 | 0,0351 | 9,73 | 7,10 | 1,37 |
| 3281 | 36 | GpAGQDGVGGDKGEDGDPGQPGPpGpSGEAGpPGPPG | Collagen alpha-1(XI) chain | 1077,2 | 817,32 | 0,76 | 0,0333 | 1755,73 | 1915,38 | 0,92 |
| 1401 | 22,6 | DGESGRpGRpGER | Collagen alpha-1(III) chain | 15,51 | 3,84 | 0,25 | 0,0333 | 19,55 | 20,43 | 0,96 |
| 1751 | 23,9 | GpPGpPGKNGDDGEAGKpG | Collagen alpha-1(I) chain | 1215,9 | 1808,18 | 1,49 | 0,0333 | 1022,81 | 1215,80 | 0,84 |
| 3225 | 22,7 | EqGHpGSPGFKGIDGMPGTPGLKGDRGSPGmDG | Collagen alpha-2(IV) chain | 27,84 | 11,96 | 0,43 | 0,0317 | 17,76 | 28,65 | 0,62 |
| 1697 | 29,6 | pGNDGAKGDAGAPGAPGSqG | Collagen alpha-1(I) chain | 41,69 | 21,5 | 0,52 | 0,0286 | 28,06 | 24,27 | 1,16 |
| 1517 | 41 | FDSDPITVTVPVEV | Clusterin | 49,09 | 279,09 | 5,69 | 0,0286 | 110,21 | 210,33 | 0,52 |
| 3272 | 32,1 | GEVGPAGSPGSNGApGQRGEPGPQGHAGAQGPPGpPG | Collagen alpha-1(III) chain | 29,08 | 11,04 | 0,38 | 0,0279 | 6,39 | 6,10 | 1,05 |
| 1585 | 24,6 | EGSpGHPGQpGPpGPpG | Collagen alpha-1(III) chain | 9,68 | 20,45 | 2,11 | 0,0277 | 6,24 | 14,97 | 0,42 |
| 1928 | 19,5 | DGPRGpTGPIGPPGpAGQPGD | Collagen alpha-1(III) chain | 175,34 | 91,85 | 0,52 | 0,0273 | 29,02 | 67,23 | 0,43 |
| 1142 | 37,3 | GPpGpPGPPGPpA | Collagen alpha-1(VIII) chain | 492,94 | 696,2 | 1,41 | 0,0233 | 134,19 | 404,17 | 0,33 |
| 1239 | 21,2 | GEAGHPGPPGPpGP | Collagen alpha-1(V) chain | 26,03 | 6,73 | 0,26 | 0,0230 | 14,04 | 32,88 | 0,43 |
| 2529 | 27 | GETGPAGRpGEVGPpGPpGPAGEKGSpG | Collagen alpha-1(I) chain | 75,5 | 37,54 | 0,5 | 0,0227 | 34,58 | 48,69 | 0,71 |
| 883,4 | 23,5 | PpGENGKpG | Collagen alpha-1(III) chain | 81,46 | 56,76 | 0,7 | 0,0224 | 52,08 | 72,76 | 0,72 |
| 3417 | 32 | GPpGADGQPGAKGEpGDAGAKGDAGPPGpAGPAGPPGpIG | Collagen alpha-1(I) chain | 1172,9 | 1495,4 | 1,27 | 0,0211 | 692,86 | 1154,42 | 0,60 |
| 1080 | 27,6 | DRGEpGPpGPA | Collagen alpha-1(I) chain | 206,04 | 281,42 | 1,37 | 0,0197 | 45,22 | 145,01 | 0,31 |
| 2487 | 28,2 | GADGQPGAKGEpGDAGAKGDAGPpGPAGP | Collagen alpha-1(I) chain | 646,01 | 460,79 | 0,71 | 0,0197 | 230,53 | 418,62 | 0,55 |
| 2047 | 32,8 | GSNGNpGpPGPSGSpGKDGPpGP | Collagen alpha-1(III) chain | 1282,1 | 827,74 | 0,65 | 0,0192 | 1585,20 | 1925,74 | 0,82 |
| 3686 | 22,2 | GRPEAQPPPLSSEHKEPVAGDAVPGPKDGSAPEVRGA | Neurosecretory protein VGF | 1482 | 2353,74 | 1,59 | 0,0169 | 1426,76 | 1378,98 | 1,03 |
| 1496 | 23,5 | GPpGPpGPpGPpGPPSA | Collagen alpha-1(I) chain | 60,6 | 29,67 | 0,49 | 0,0169 | 98,77 | 72,92 | 1,35 |
| 2825 | 24,5 | ERGEAGIpGVpGAKGEDGKDGSpGEpGANG | Collagen alpha-1(III) chain | 19518 | 22836,6 | 1,17 | 0,0155 | ###### | 16825,06 | 0,92 |
| 2663 | 23,6 | NRGERGSEGSPGHpGQpGPPGpPGApGP | Collagen alpha-1(III) chain | 1707,5 | 1298,97 | 0,76 | 0,0148 | 720,20 | 1021,11 | 0,71 |
| 1262 | 38 | EAVGDGDGDGDADA | ATPase WRNIP1 | 200,94 | 94,99 | 0,47 | 0,0145 | 26,07 | 106,33 | 0,25 |
| 1484 | 22,6 | GPpGKNGDDGEAGKpG | Collagen alpha-1(I) chain | 360,21 | 704,15 | 1,95 | 0,0135 | 135,73 | 318,75 | 0,43 |
| 1792 | 30,8 | RGApGpDGNnGAQGPpGPQ | Collagen alpha-2(I) chain | 83,11 | 24,34 | 0,29 | 0,0094 | 53,75 | 47,05 | 1,14 |
| 1977 | 19,3 | NSESSTVSSGASTATTSESST | Mucin-21 | 186,17 | 379,32 | 2,04 | 0,0092 | 60,40 | 89,04 | 0,68 |
| 2088 | 19,4 | GPPGSPGEDGpAGEpGPPGpEGq | Collagen alpha-1(XV) chain | 208,07 | 368,2 | 1,77 | 0,0092 | 123,45 | 250,72 | 0,49 |
| 1532 | 39,3 | GLpGPpGSNGNpGPpGP | Collagen alpha-1(III) chain | 514,41 | 339,07 | 0,66 | 0,0092 | 258,97 | 322,87 | 0,80 |
| 1935 | 19,9 | GSGGSSYGSGGGSYGSGGGGGGGRG | "Keratin; type II cytoskeletal 1" | 553,01 | 917,44 | 1,66 | 0,0092 | 182,62 | 633,17 | 0,29 |
| 1116 | 36,9 | PpGPpGpPGPpS | Collagen alpha-1(I) chain | 4 | 30,72 | 7,68 | 0,0090 | 68,83 | 78,96 | 0,87 |
| 1539 | 29,7 | PpGEAGKpGEQGVpGD | Collagen alpha-1(I) chain | 236,76 | 168,38 | 0,71 | 0,0082 | 156,03 | 253,95 | 0,61 |
| 3209 | 22,7 | PpGESGREGApGAEGSpGRDGSpGAKGDRGETGP | Collagen alpha-1(I) chain | 5616,4 | 4002,63 | 0,71 | 0,0077 | 3257,70 | 6895,61 | 0,47 |
| 2048 | 19,9 | KGEDGDPGqpGPPGpSGEAGpP | Collagen alpha-1(XI) chain | 25,37 | 68,37 | 2,69 | 0,0070 | 0,00 | 20,53 | 0,00 |
| 2679 | 23,6 | PGMPGADGpPGHPGKEGppGEKGGQGpPG | Collagen alpha-1(V) chain | 2842,7 | 2158,62 | 0,76 | 0,0068 | 2330,40 | 2398,19 | 0,97 |
| 2695 | 23,5 | AGSPGSNGApGQRGEpGpQGHAGAqGPPGP | Collagen alpha-1(III) chain | 2652,7 | 2079,6 | 0,78 | 0,0068 | 3134,21 | 2728,53 | 1,15 |
| 3554 | 31,1 | GPAGpAGERGEQGPAGSpGFqGLPGPAGPPGEAGKPGEq | Collagen alpha-1(I) chain | 72,83 | 24,19 | 0,33 | 0,0068 | 27,93 | 54,70 | 0,51 |
Mass, migration, and sequence of the individual peptides are given as identifiers. The distribution before and after empagliflozin treatment, expressed as relative abundance is given, the induced fold-change, and the p-value after adjustment for multiple testing. In addition, the distribution of these peptides in a cohort of patients with CKD and controls [8], as well as the observed fold-change in this cohort is listed.
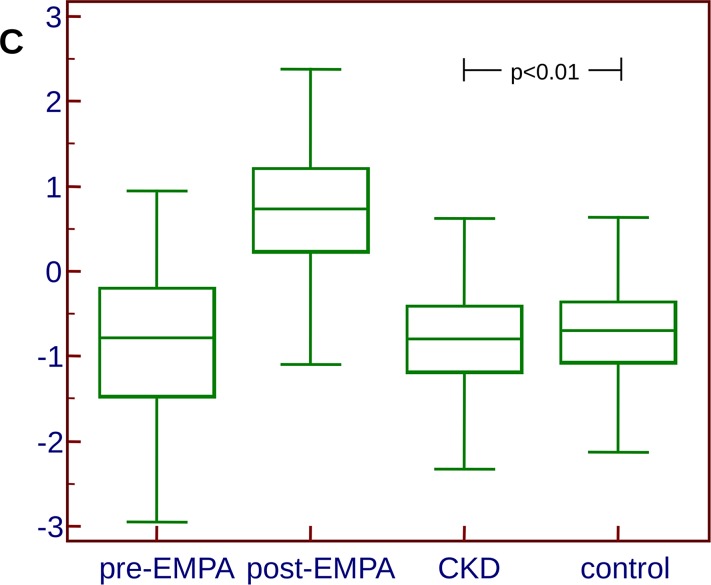
To investigate how the changes in these 107 peptides relate to CKD, we combined all 107 peptides into an SVM-based classifier to receive a composite score, using the data obtained from all samples investigated in the context of empagliflozin treatment. The classifier was trained using all 160 samples, and allowed clear differentiation between the two groups (treated and untreated) in the training set. This classifier was subsequently applied onto the 609 samples that were also employed in the discovery of CKD273 in the past [8] to investigate if a significant difference in the scoring in the two groups can be detected, which would indicate a potential impact of empagliflozin treatment on CKD (in both directions, either promoting or suppressing its development). In this independent dataset from patients with CKD of different etiologies and controls, the mean scoring of the CKD cases was -0.835, the mean scoring of the controls was -0.711 (p = 0.0075 for the between-group difference, Fig 2). These data further support the initial findings, that the changes induced with empagliflozin are directional pointing away from CKD and towards the healthy controls (of the 107 peptides significantly changing in abundance after empagliflozin treatment, 60 peptides pointing towards healthy, 19 pointing towards CKD, 28 appear unchanged, as outlined above), indicating a potential benefit of empagliflozin.
Fig 2. Box-Whisker blots of the scoring representing the proteome changes induced as a result of empagliflozin treatment.
The clear separation of the groups prior and after treatment is evident. However, when applying this classifier to a cohort of CKD patients and controls, the controls score slightly, yet significantly (p<0.01) higher (indicative of empagliflozin treatment) than the CKD patients.
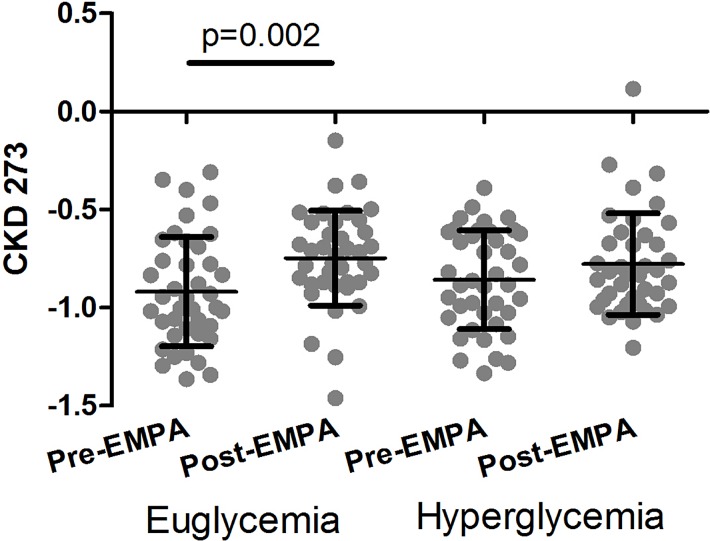
To further assess the effect of empagliflozin, we applied CKD273, a high-dimensional classifier based on 273 urinary peptides that were found significantly changed in CKD [8,22], onto the 160 datasets obtained in this study. All baseline samples scored negative, indicating absence of CKD, in line with the clinical findings. As shown in Fig 3, levels of CKD273 increased within the negative (CKD absent) range during clamped euglycemia, but not during clamped hyperglycemia. Overall, no consistent impact of the empagliflozin treatment on CKD273 could be observed in this cohort.
Fig 3. Distribution of the CKD273 scoring in patients before and after treatment with empagliflozin.
Discussion
In this analysis we investigated the impact of empagliflozin treatment on the urine proteome in type I diabetes patients with preserved kidney function. In several recent studies urinary proteomic changes were reported as a consequence of therapeutic intervention, both based on drugs [13,23], but also on diet or lifestyle [24,25]. Our aim was to determine if a) empagliflozin treatment has an impact on urinary peptides, and b) if such an impact of empagliflozin treatment would be in the direction of CKD (indicating a negative effect) or towards “healthy” (indicating a positive effect). We have in several recent studies identified collagen type I fragments as being decreased in the urine in the initial phase of onset of CKD [9,11]. Though this process was not consistently affected by empagliflozin treatment, among the collagen fragments, larger fragments generally showed changes in the same direction observed in CKD, while the smaller fragments generally showed changes towards healthy. Further, we detected an increase in a specific clusterin peptide after empagliflozin treatment. Peptides derived from clusterin were found decreased in CKD [8], indicating that the observed change presented here represents an improvement with respect to CKD development.
Of special interest is the upregulation of a mucin fragment as a result of empagliflozin treatment. In a recent manuscript we have identified the decrease of mucin fragments in urine as a major component in the CKD-induced changes [26]. The data presented here indicate that empagliflozin treatment may reverse these CKD-induced changes. To assess the impact of the combined changes in urinary peptides observed as a result of empagliflozin treatment, we generated a composite classifier integrating all 107 peptides. When applying this classifier onto an independent cohort of healthy controls and patients with CKD, we detected a significantly higher scoring in the group of healthy individuals, indicating a positive impact of empagliflozin.
Empagliflozin is the first compound that to our knowledge demonstrates a rapid and significant impact on urinary peptides in the context of diabetes and CKD. In our previous investigations, while short term treatment with irbesartan [23] did not show a significant impact on urine peptides, longer-term treatment for 2 years did show an impact. No overlap exists between the changes induced by irbesartan and the changes observed here as a result of empagliflozin, suggesting that empagliflozin has a different, direct impact on urinary peptides, although the mechanisms responsible for these changes are not yet known.
We also investigated the impact of empagliflozin treatment on CKD273, a urinary peptide-based classifier associated with CKD onset and progression [9,11,17]. All patients in the study scored negative for CKD. While this observation was in agreement with the clinical characteristics, we did expect that at least some patients would present borderline or positive scoring for CKD. In these patients, we hypothesized to see an impact of empagliflozin treatment on the classification, a change towards “healthy”. However, all samples scored “healthy” in this primary prevention cohort, and significant improvement of the “healthy” scoring as a result of empagliflozin treatment was not to be expected. The CKD273 score increased within the “CKD absent” range during clamped euglycemia, and we were unable to detect an impact of empagliflozin on CKD273 during clamped hyperglycemic conditions. Since levels of CKD273 were all in the CKD absent range at all studied time points in this cohort, it is difficult to draw any conclusions about prognostic implications in patients with baseline CKD.
This exploratory, post-hoc study has limitations: The sample size was small and the treatment period was only 8 weeks, perhaps not sufficiently long to observe significant changes in CKD273. The results may not be generalizable to patients with either type 2 diabetes or patients with evidence of renal disease. Whether SGLT2 inhibition impacts on CKD273 in patients with a background of renal disease is unknown, and will be examined in future work.
In conclusion, empagliflozin has a significant impact on specific urine peptides, and the proteomic changes induced by empagliflozin suggest a potential nephroprotective effect. However, this potential nephroprotective effect needs to be investigated and confirmed in dedicated clinical trials that are designed to assess the impact of empagliflozin in patients with CKD, and investigate how these proteomic changes correlate with clinical outcomes.
Supporting information
Original ID of the samples is given on the top. For all peptides or proteins, internal Peptide ID, mass and migration is given. In addition, sequence, original protein, as well as start and stop amino acid position is given, where applicable.
(XLSX)
The table lists the mass (in Da) and migration time (CE-T, in minutes), the relative abundance before (Pre EMPA) and after (Post EMPA) treatment, and the fold-change induced by empagliflozin (EMPA-ind change). Further, the average abundance in patients with CKD (CKD mean) and controls (control mean) as well as the fold-change observed in CKD (CKD-induced) is listed. Where applicable, the Sequence, and protein Name and Symbol are given. For all peptides, the unadjusted p-value (for the difference prior and after empagliflozin treatment), the AUC, and the Benjamini-Hochberg (BH) adjusted p-value is listed.
(XLSX)
Acknowledgments
We are indebted and extend our gratitude to all study participants who took part in this research study for their time and commitment to this study protocol. We thank the student volunteers, nursing, and administrative staff who were essential in carrying out this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
DC has received consulting fees or speaking honorarium or both from Janssen, Boehringer Ingelheim-Eli Lilly, AstraZeneca, Merck, Sanofi, and his institution has received operating funding from Janssen, Boehringer Ingelheim-Eli Lilly, AstraZeneca, Merck. HM is the co-founder and co-owner of Mosaiques Diagnostics. BAP has received fees for continuing medical education events from Janssen and Boehringer Ingelheim, has served as an advisor for Boehringer Ingelheim, and his research institute has received research grants on his behalf from Boehringer Ingelheim. The funder provided support in the form of salaries for author HM, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Jha V, Garcia-Garcia G, Iseki K, Li Z, Naicker S, Plattner B et al. (2013) Chronic kidney disease: global dimension and perspectives. Lancet 382: 260–272. doi: 10.1016/S0140-6736(13)60687-X [DOI] [PubMed] [Google Scholar]
- 2.Wanner C, Inzucchi SE, Lachin JM, Fitchett D, von Eynatten M, Mattheus M et al. (2016) Empagliflozin and Progression of Kidney Disease in Type 2 Diabetes. New England Journal of Medicine 375: 323–334. doi: 10.1056/NEJMoa1515920 [DOI] [PubMed] [Google Scholar]
- 3.Heerspink HJL, Perkins BA, Fitchett DH, Husain M, Cherney DZI (2016) Sodium Glucose Cotransporter 2 Inhibitors in the Treatment of Diabetes Mellitus: Cardiovascular and Kidney Effects, Potential Mechanisms, and Clinical Applications. Circulation 134: 752–772. doi: 10.1161/CIRCULATIONAHA.116.021887 [DOI] [PubMed] [Google Scholar]
- 4.Mischak H, Delles C, Vlahou A, Vanholder R (2015) Proteomic biomarkers in kidney disease: issues in development and implementation. Nat Rev Nephrol 11: 221–232. doi: 10.1038/nrneph.2014.247 [DOI] [PubMed] [Google Scholar]
- 5.Dakna M, Harris K, Kalousis A, Carpentier S, Kolch W, Schanstra JP et al. (2010) Addressing the challenge of defining valid proteomic biomarkers and classifiers. BMC Bioinformatics 11: 594 doi: 10.1186/1471-2105-11-594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mischak H, Schiffer E, Zürbig P, Dakna M, Metzger J (2009) Urinary proteome analysis using capillary electrophoresis coupled mass spectrometry: a powerful tool in clinical diagnosis, prognosis and therapy evaluation. Journal of Medical Biochemistry 28: 223–234. [Google Scholar]
- 7.Kaiser T, Hermann A, Kielstein J.T., Wittke S, Bartel S, Krebs R et al. (2003) Capillary electrophoresis coupled to mass spectrometry to establish polypeptide patterns in dialysis fluids. J Chromatogr A 1013: 157–171. [DOI] [PubMed] [Google Scholar]
- 8.Good DM, Zürbig P, Argiles A, Bauer HW, Behrens G, Coon JJ et al. (2010) Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteomics 9: 2424–2437. doi: 10.1074/mcp.M110.001917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schanstra JP, Zurbig P, Alkhalaf A, Argiles A, Bakker SJ, Beige J et al. (2015) Diagnosis and prediction of CKD progression by assessment of urinary peptides. J Am Soc Nephrol 26: 1999–2010. doi: 10.1681/ASN.2014050423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stalmach A, Albalat A, Mullen W, Mischak H (2013) Recent advances in capillary electrophoresis coupled to mass spectrometry for clinical proteomic applications. Electrophoresis 34: 1452–1464. doi: 10.1002/elps.201200708 [DOI] [PubMed] [Google Scholar]
- 11.Pontillo C, Jacobs L, Staessen JA, Schanstra JP, Rossing P, Heerspink HJ et al. (2016) A urinary proteome-based classifier for the early detection of decline in glomerular filtration. Nephrol Dial Transplant [DOI] [PubMed] [Google Scholar]
- 12.Cherney DZ, Perkins BA, Soleymanlou N, Maione M, Lai V, Lee A et al. (2014) Renal hemodynamic effect of sodium-glucose cotransporter 2 inhibition in patients with type 1 diabetes mellitus. Circulation 129: 587–597. doi: 10.1161/CIRCULATIONAHA.113.005081 [DOI] [PubMed] [Google Scholar]
- 13.Haubitz M, Good DM, Woywodt A, Haller H, Rupprecht H, Theodorescu D et al. (2009) Identification and validation of urinary biomarkers for differential diagnosis and dvaluation of therapeutic intervention in ANCA associated vasculitis. Mol Cell Proteomics 8: 2296–2307. doi: 10.1074/mcp.M800529-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mischak H, Vlahou A, Ioannidis JP (2013) Technical aspects and inter-laboratory variability in native peptide profiling: The CE-MS experience. Clin Biochem 46: 432–443. doi: 10.1016/j.clinbiochem.2012.09.025 [DOI] [PubMed] [Google Scholar]
- 15.Jantos-Siwy J, Schiffer E, Brand K, Schumann G, Rossing K, Delles C et al. (2009) Quantitative Urinary Proteome Analysis for Biomarker Evaluation in Chronic Kidney Disease. J Proteome Res 8: 268–281. doi: 10.1021/pr800401m [DOI] [PubMed] [Google Scholar]
- 16.Nkuipou-Kenfack E, Bhat A, Klein J, Jankowski V, Mullen W, Vlahou A et al. (2015) Identification of ageing-associated naturally occurring peptides in human urine. Oncotarget 6: 34106–34117. doi: 10.18632/oncotarget.5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pontillo C, Zhang Z, Schanstra J, Jacobs L, Zuerbig P, Thijs L et al. (2017) Prediction of chronic kidney disease stage 3 by CKD273, a urinary proteomic biomarker. Kidney International Reports in press: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein J, Papadopoulos T, Mischak H, Mullen W (2014) Comparison of CE-MS/MS and LC-MS/MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis 35: 1060–1064. doi: 10.1002/elps.201300327 [DOI] [PubMed] [Google Scholar]
- 19.Lankisch TO, Metzger J, Negm AA, Vobetakuhl K, Schiffer E, Siwy J et al. (2011) Bile proteomic profiles differentiate cholangiocarcinoma from primary sclerosing cholangitis and choledocholithiasis. Hepatology 53: 875–884. doi: 10.1002/hep.24103 [DOI] [PubMed] [Google Scholar]
- 20.Girolami M, Mischak H, Krebs R (2006) Analysis of complex, multidimensional datasets. Drug Discov Today: Technologies 3: 13–19. doi: 10.1016/j.ddtec.2006.03.010 [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B (Methodological) 57: 125–133. [Google Scholar]
- 22.Pontillo C, Mischak H (2017) Urinary peptide-based classifier CKD273: towards clinical application in chronic kidney disease. Clinical Kidney Journal 10: 192–201. doi: 10.1093/ckj/sfx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen S, Mischak H, Zürbig P, Parving HH, Rossing P (2010) Urinary proteome analysis enables assessment of renoprotective treatment in type 2 diabetic patients with microalbuminuria. BMC Nephrol 11: 29 doi: 10.1186/1471-2369-11-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmerli LU, Schiffer E, Zürbig P, Kellmann M, Mouls L, Pitt A et al. (2008) Urinary proteomics biomarkers in coronary artery disease. Mol Cell Proteomics 7: 290–298. doi: 10.1074/mcp.M700394-MCP200 [DOI] [PubMed] [Google Scholar]
- 25.Silva S, Bronze MR, Figueira ME, Siwy J, Mischak H, Combet E et al. (2015) Impact of a 6-wk olive oil supplementation in healthy adults on urinary proteomic biomarkers of coronary artery disease, chronic kidney disease, and diabetes (types 1 and 2): a randomized, parallel, controlled, double-blind study. Am J Clin Nutr 101: 44–54. doi: 10.3945/ajcn.114.094219 [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Ravassa S, Pejchinovski M, Yang W, Zurbig P, Lopez B et al. (2017) A urinary fragment of mucin1 subunit alpha is a novel biomarker associated with renal dysfunction in the general population. Kidney International Reports in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Original ID of the samples is given on the top. For all peptides or proteins, internal Peptide ID, mass and migration is given. In addition, sequence, original protein, as well as start and stop amino acid position is given, where applicable.
(XLSX)
The table lists the mass (in Da) and migration time (CE-T, in minutes), the relative abundance before (Pre EMPA) and after (Post EMPA) treatment, and the fold-change induced by empagliflozin (EMPA-ind change). Further, the average abundance in patients with CKD (CKD mean) and controls (control mean) as well as the fold-change observed in CKD (CKD-induced) is listed. Where applicable, the Sequence, and protein Name and Symbol are given. For all peptides, the unadjusted p-value (for the difference prior and after empagliflozin treatment), the AUC, and the Benjamini-Hochberg (BH) adjusted p-value is listed.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.