Abstract
HIV1-TAT interactive protein (TIP60) is a haploinsufficient tumor suppressor. However, the potential mechanisms endowing its tumor suppressor ability remain incompletely understood. It plays a vital role in virus-induced cancers where TIP60 down-regulates the expression of human papillomavirus (HPV) oncoprotein E6 which in turn destabilizes TIP60. This intrigued us to identify the role of TIP60, in the context of a viral infection, where it is targeted by oncoproteins. Through an array of molecular biology techniques such as Chromatin immunoprecipitation, expression analysis and mass spectrometry, we establish the hitherto unknown role of TIP60 in repressing the expression of the catalytic subunit of the human telomerase complex, TERT, a key driver for immortalization. TIP60 acetylates Sp1 at K639, thus inhibiting Sp1 binding to the TERT promoter. We identified that TIP60-mediated growth suppression of HPV-induced cervical cancer is mediated in part due to TERT repression through Sp1 acetylation. In summary, our study has identified a novel substrate for TIP60 catalytic activity and a unique repressive mechanism acting at the TERT promoter in virus-induced malignancies.
Author summary
A common feature of all malignant cells is their ability of continued proliferation while avoiding replicative senescence. Tumors either activate expression of telomerase reverse transcriptase, TERT (in the case of ~ 85% of tumors) or use a recombination based alternative lengthening of telomeres (remaining 15% of tumors) to maintain their telomere length and thus ensure continued cell replication. Most viruses which cause tumors such as human papillomavirus (HPV) and Hepatitis B virus (HBV) also activate the expression of telomerase and result in malignancy. In this study, we have identified a novel cellular repressor of telomerase, TIP60. TIP60 which is targeted for degradation by HPV oncoprotein E6, acetylates Sp1, a positive regulator of TERT. We show that TIP60-mediated acetylation of Sp1, inhibits Sp1 function at the TERT promoter, thereby reducing TERT expression and thus the growth of HPV-positive cervical cancer cells, HeLa.
Introduction
HIV-1 TAT Interactive protein (TIP60) is a lysine acetyltransferase and a member of the MYST (Moz, Ybf2/Sas3, Sas2 and TIP60) family of evolutionarily related histone acetyltransferases [1]. TIP60 can acetylate histone and non-histone proteins and is involved in a multitude of cellular functions like altering the chromatin structure, transcription and DNA damage repair response [2–4]. TIP60’s role as a bona fide tumor suppressor has also been documented [5]. In return, it is targeted for degradation by many viral oncoproteins like the human papillomavirus (HPV) oncoprotein E6 and adenoviral oncoproteins EIB55K and E4orf6 [6–8]. Infection by the oncogenic HPV is the leading cause of cervical cancer, the second most common cancer in women worldwide. High-risk HPV (HPV 16, HPV 18) is implicated in invasive carcinoma while low-risk HPV (HPV6, HPV8, HPV11) is responsible for genital warts and non-malignant lesions [9]. The mechanism of transformation of normal keratinocytes by HPV is dependent on two oncoproteins, E6 and E7, which interact with and destabilize the tumor suppressors p53 and pRb (Retinoblastoma protein) [10, 11]. E6 also destabilizes TIP60 by cooperating with an E3 cellular ubiquitin ligase, EDD1 [8], and TIP60 in turn represses the E6 promoter, thus maintaining a delicate balance of cellular and viral gene expression [7]. Interestingly, both high and low-risk HPV are capable of destabilizing TIP60 whereas only the high-risk E6 is capable of destabilizing p53. This suggests that TIP60 degradation is an essential and common mechanism in promoting viral gene expression [7, 11].
Telomeres consist of repetitive DNA at the ends of linear chromosomes and their degradation limits the replicative potential of cells [12, 13]. This is a consequence of progressive telomere shortening, occurring with every round of cell division due to the combined contributions of the end replication problem and active telomere processing [14, 15]. Like normal somatic cells, cancer cells would ultimately face telomere-induced replicative senescence but they (re-)activate one of two telomere maintenance pathways to bypass this fate: about 85% of all tumors reactivate telomerase, whereas the remaining 15% employ a recombination based maintenance mechanism, named Alternative Lengthening of Telomeres (ALT) [16, 17]. Telomerase is a reverse transcriptase that can add telomeric repeats de novo, thus counteracting telomere shortening. Its minimal core consists of a catalytic subunit, TERT, and an RNA template for reverse transcription, TERC [18]. TERT expression is regulated at the epigenetic, transcription and protein level by a multitude of cellular factors [19]. Additionally, many oncoviral proteins such as HPV E6 and HBV HBx are known to activate the expression of TERT, thus circumventing senescence and serving their purpose of cellular proliferation [20–23].
E6 collaborates with the transcription factor c-MYC to upregulate TERT in certain cervical cancer cell lines [24]. It also interacts with a cellular ubiquitin ligase E6AP to destabilize a repressor, NFX1-91, on the TERT promoter in a proteasome dependent manner [22, 25, 26]. The collaborative role of c-MYC and another transcription factor, Specificity Protein 1 (Sp1), to activate telomerase has also been reported [27]. This regulation occurs directly at the TERT promoter, which contains E boxes to enable binding of c-MYC and GC boxes that provide binding sites for Sp1 [28]. The dynamic relationship between E6 and TIP60, the reduction in the tumor size upon overexpression of TIP60 as well as reports of TIP60 being present at the TERT promoter [29], have led us to question if TIP60 can regulate TERT expression and if this regulation is dependent on E6. This study indeed identifies the hitherto unknown mechanism of TERT expression regulation by TIP60. We show that TIP60, in contrary to its role in acetylating histones leading to transcriptional activation, serves as a negative regulator of TERT expression by inhibition of Sp1 function. We show that TIP60 acetylates residue K639 on Sp1, which prevents Sp1 binding to the TERT promoter. Moreover, the biological significance of TERT repression by TIP60 is demonstrated by the ability of ectopically expressed TERT to rescue growth defects seen upon TIP60 overexpression in colony forming assays. Thus, this study identifies how TIP60 levels regulate TERT expression by limiting Sp1 binding to the TERT promoter.
Results
TIP60 regulates telomerase activity by altering mRNA expression of TERT
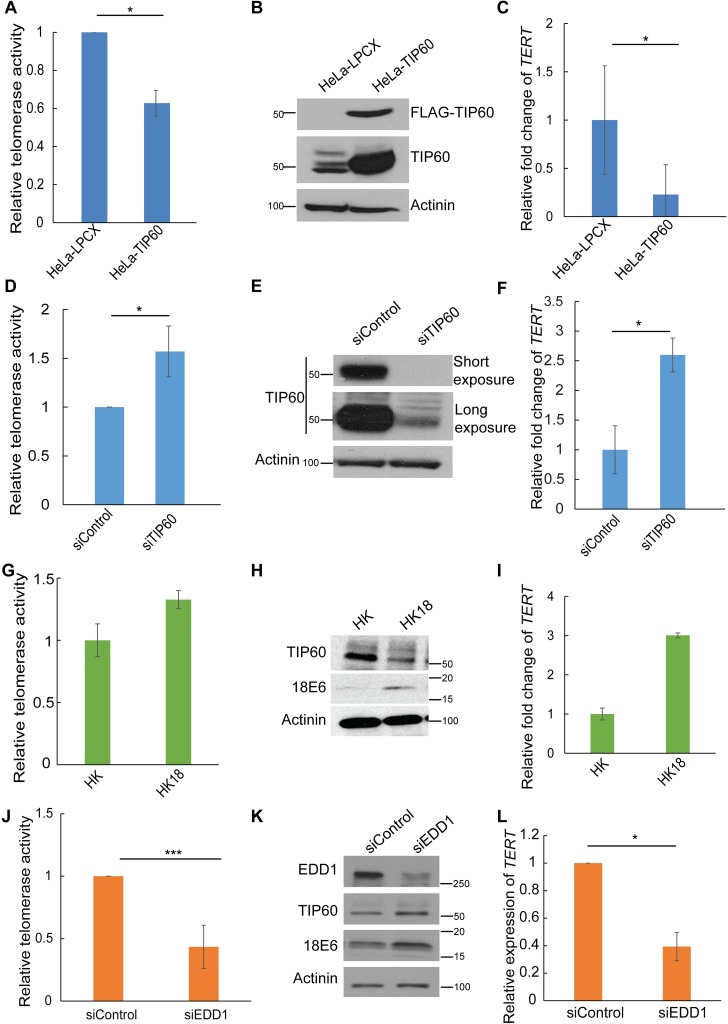
Based on our previous finding of a feedback loop between TIP60 and HPV E6, and the phenotype of reduction in tumor size upon overexpression of TIP60 in vivo [7, 8], here we focused on elucidating the molecular mechanism of this tumor suppressor function of TIP60. Since HPV infection causes immortalization and TIP60 is destabilized by HPV oncogene E6, we investigated if TIP60 regulates TERT expression, a key factor involved in immortalization, in the context of the HeLa cervical cancer cell line that constitutively expresses the viral oncogenes E6 and E7 [30]. In order to investigate TIP60’s role in regulating telomerase activity, we generated HeLa-LPCX (vector) and HeLa-TIP60 overexpressing cell lines [8]. Stable overexpression of Flag-tagged TIP60 results in a decrease in telomerase activity as measured by the quantitative telomere repeat amplification protocol (qTRAP) (Fig 1A and 1B), suggesting that the presence of TIP60 represses telomerase activity. The qTRAP assay serves as a readout for a functional telomerase complex and the associated enzymatic activity. Next, we sought to investigate if this repression was due to a change in the transcript levels of TERT. We measured mRNA levels of TERT in HeLa-LPCX and HeLa-TIP60 cells. In agreement with the changes in telomerase activity, TIP60-overexpressing cells show decreased TERT mRNA expression (Fig 1C). To validate this was specific to TIP60, we depleted TIP60 in HeLa-TIP60 cells by small interfering RNA (siRNA) transfection. Upon transient depletion of TIP60 (Fig 1E), we observed an increase in both telomerase activity and TERT expression (Fig 1D and 1F). The siTIP60 siRNA is a commercially available pool of siRNAs which targets TIP60 and is designed to minimize off-target effects. To further rule out off-target effects of the siRNA, we used another siRNA (siTIP60A) to repeat these experiments in HeLa-TIP60 cells. Transient knockdown of TIP60 using siTIP60A also resulted in an increase in TERT mRNA expression and consequently an increase in relative telomerase activity (S1A, S1B and S1C Fig). Consistent with our observations in HeLa-TIP60 cells, TIP60 depletion in the HeLa parental cells also increased TERT mRNA expression and activity (S1D, S1E and S1F Fig). For all the further experiments, the pool siRNA, siTIP60 has been used.
Fig 1. TIP60 regulates TERT expression and activity.
(A) Telomerase activity was measured by qTRAP, using the whole cell lysates from HeLa-LPCX and HeLa-TIP60 cells. Heat inactivated samples served as negative control. Stable overexpression of Flag-tagged TIP60 in HeLa cells causes a decrease in the telomerase activity. (B) The stable cell lines generated were validated by western blotting using anti-Flag antibody. α-Actinin serves as a loading control to indicate equal protein amount used for qTRAP assay. (C) RNA was isolated from HeLa-LPCX and HeLa-TIP60 cells and real time PCR was performed to check for TERT expression. (D) TIP60 was transiently depleted in HeLa-TIP60 cells using siRNA and cells were harvested 72 h post transfection. Telomerase activity was measured. (E) Efficiency of TIP60 depletion was confirmed by western blotting. TIP60 was detected using an endogenous TIP60 antibody and α-Actinin serves as loading control. (F) Cellular RNA was isolated and analyzed by real time PCR after knockdown to determine TERT expression. mRNA expression was normalized to GAPDH and plotted as fold change. (G) Relative telomerase activity was measured for HK (human primary keratinocytes) and HK 18 (human keratinocytes expressing integrated HPV 18 genome) cells. (H) Detection of TIP60, 18 E6 and α-Actinin using antibodies for endogenous proteins in HK and HK18 cells. (I) RNA was isolated from HK and HK 18 and TERT expression was detected by qPCR. (J) Transient depletion of EDD1 in HeLa, the ubiquitin ligase known to destabilize TIP60 and measurement of relative telomerase activity. (K) Western blotting of the same samples harvested 72 h after siRNA transfection, with the indicated antibodies to detect endogenous levels of TIP60, EDD1 and E6. (L) qPCR to detect changes in TERT mRNA expression upon EDD1 depletion. Error bars reflect the standard error of mean (SEM) of 3 independent experiments and significance is represented as *, P<0.05.
In order to study if this TERT repression is also observed in primary keratinocytes, which represent a relevant physiological system that bears similarities to early stages of tumorigenesis, we compared TERT expression and TIP60 levels between primary human keratinocytes (HK) and primary keratinocytes which have HPV 18 (HK 18) genome integrated. Consistent with our hypothesis and literature, TIP60 expression was reduced in HK 18 compared to HK cells (Fig 1H). This was also reflected in the increased expression of TERT as well as the increase in relative telomerase activity in HK 18 compared to HK cells (Fig 1G and 1I).
To ensure that these effects were not cell line-specific, we depleted TIP60 in CaSki-TIP60 cells generated using the same retroviral infection as described earlier for HeLa [8]. CaSki is also a cervical cancer line, but these cells constitutively express another high-risk E6 protein, 16E6. Depletion of TIP60 in CaSki-TIP60 cells also resulted in an increase both in telomerase activity (S1G and S1H Fig) and TERT mRNA levels (S1I Fig). This data suggests that TIP60 inhibits telomerase activity by repressing TERT mRNA expression in cervical cancer cell lines and in primary keratinocytes compared to keratinocytes with integrated HPV 18 genome.
HeLa cervical cancer cells are dependent on E6 expression for circumventing senescence and regulating proliferation [30]. Apart from its critical role of degrading tumor suppressor p53 [11], E6 also activates telomerase [22], although the exact mechanism is complex and incompletely characterized. Additionally, the regulatory loop between E6 and TIP60 has already been established [7]. Thus it was imperative to study if the regulation of TERT by TIP60 is dependent on E6. In order to study this intricate feedback, HeLa cells were depleted transiently of both E6 and TIP60 using siRNA transfection and analyzed by western blotting using endogenous TIP60 and E6 antibodies (S2A Fig). In TIP60-depleted samples, the telomerase activity was found to be increased, consistent with our earlier results, and was rescued to control level in the E6 and TIP60 co-depleted samples (S2B Fig). This data suggests that the TIP60 repression of TERT is E6-dependent in cells expressing E6. However, this regulation of TERT by TIP60 is a mechanism that seems to be conserved in cervical cancer cells and is not restricted to only HPV positive cells. C-33A cells, cervical cancer cells without HPV infection, also showed an induction of TERT expression upon TIP60 knockdown (S2C Fig). Furthermore, HPV E6 cooperates with a cellular ligase EDD1 to destabilize TIP60, thus regulating TIP60 levels [8]. In order to study the TIP60-E6-TERT dynamic, and the effect of a physiological increase in TIP60 level in HeLa cells, we transiently depleted EDD1 using siRNA (Fig 1K) and observed a decrease in TERT expression and telomerase activity consistent with the stabilization of TIP60 (Fig 1J and 1L).
TERT promoter is regulated by TIP60 through the transcription factor Sp1
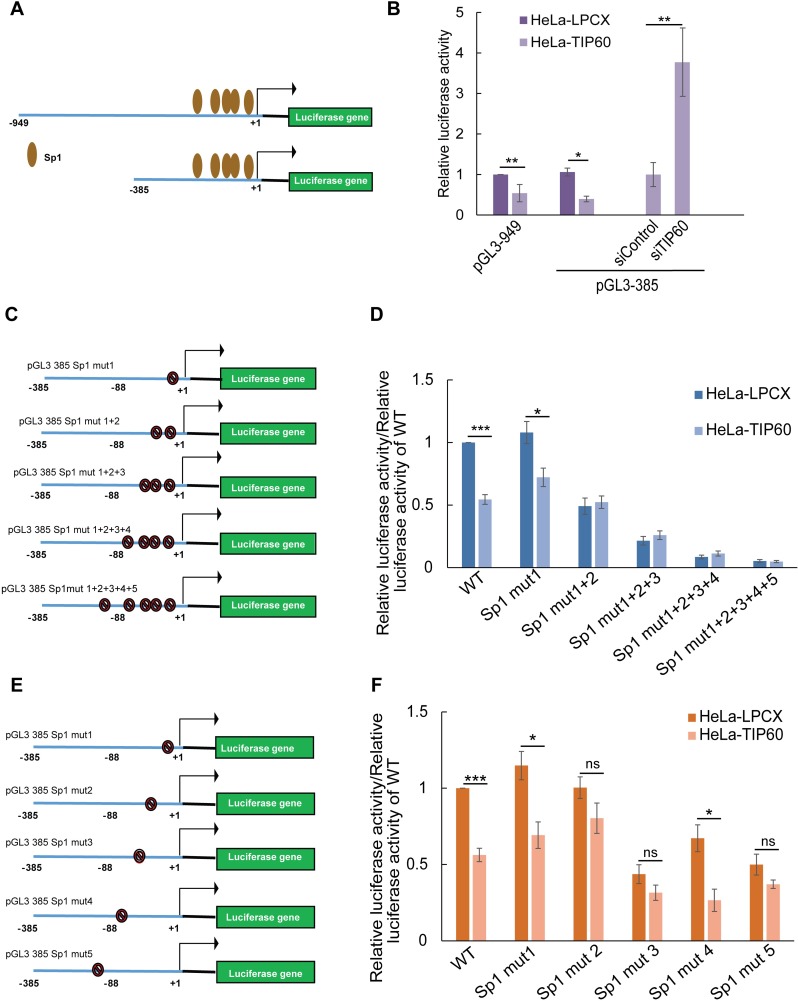
One possible explanation for TERT transcriptional repression is the regulation at the level of the TERT promoter. To test this, we used different constructs of the TERT promoter driving the expression of luciferase in a reporter gene assay (Fig 2A). We used constructs of different length, covering either the full-length promoter sequence (-949 bp to +40 bp) or a shorter version only covering the proximal promoter (-385 bp to +40 bp) [31]. The two constructs were transiently transfected into HeLa-LPCX and HeLa-TIP60 cells and luciferase activity was measured. Compared to HeLa-LPCX cells, the luciferase activity was significantly reduced in HeLa-TIP60 cells, suggesting that TIP60 negatively regulates the TERT promoter (Fig 2B). We observed that the TIP60 repression of luciferase activity occurred in both the promoter constructs. In order to eliminate any cell line specific effect that may arise on comparison of HeLa-LPCX and HeLa-TIP60 cells and also to ascertain specificity of TIP60 to TERT promoter regulation, we transiently depleted TIP60 in HeLa-TIP60 cells. 24 h prior to harvesting, these cells were transfected with pGL3-385 reporter construct and luciferase activity was measured. Consistent with our previous findings, depletion of TIP60 resulted in an increase in TERT expression (S3A and S3B Fig) as well as an increase in the luciferase activity (Fig 2B). As previously reported, the first 181 bp of the TERT promoter contains 5 Sp1 binding sites and a single c-MYC binding site. MAZ, another transcription factor, recognizes and binds GA boxes on promoters of target genes and regulates expression of cellular genes such as c-MYC and Sp1. In order to study if MAZ, Sp1 or MYC play a role in TIP60 mediated TERT repression, first, TIP60 and MAZ were depleted transiently in HeLa cells. There was a decrease in MAZ expression upon TIP60 depletion. However, it was observed that TERT increases both upon depletion of TIP60 alone and also with co-depletion of TIP60 and MAZ (S5E and S5F Fig), thereby suggesting that MAZ may not be involved in this process.
Fig 2. TIP60 regulates the TERT promoter.
(A) Schematic representation of various TERT promoter fragments driving the expression of luciferase gene. Also seen are the known binding sites of the transcription factor, Sp1. (B) HeLa-LPCX (vector), HeLa-TIP60 and HeLa-TIP60 sicontrol/ siTIP60 cells were transiently transfected with luciferase constructs and the cell lysates were processed for luciferase assay after 24 h. (C) Schematic representation of the mutations generated on the TERT promoter of the 5 Sp1 binding sites in the pGL3-385 (WT) luciferase construct. (D) Reporter gene assay with the mutant constructs transiently transfected in HeLa-LPCX and HeLa-TIP60 cells. (E) Schematic of the individual Sp1 site mutations in the pGL3-385 construct. (F) Mutant constructs were transfected into HeLa-LPCX and HeLa-TIP60 cells and luciferase reading was assayed. Relative luciferase activity was plotted as a ratio of firefly to Renilla luciferase activity. For the mutants, luciferase reading was plotted normalized to the pGL3-385 (WT) construct. Error bars reflect the standard error of mean (SEM) of at-least 3 independent experiments and significance is represented as ***, P<0.001, **, P<0.01, *, P<0.05.
Sp1 is known to be involved in the regulation of TERT in collaboration with c-MYC by binding to the GC boxes on the TERT promoter [27] (Fig 2A, S5A Fig), thus making both Sp1 and c-MYC plausible candidates for TIP60 mediated repression. To test if TIP60 regulates TERT promoter through Sp1 or c-MYC, we sequentially mutated the 5 Sp1 binding sites as well as the single c-MYC binding site using site-directed mutagenesis in the pGL3-385 construct of TERT promoter and assessed luciferase activity in HeLa-LPCX and HeLa-TIP60 cells (Fig 2C, S5B Fig) [27]. While mutating the c-MYC binding site did not affect the TIP60-mediated repression, TIP60-overexpressing cells were no longer able to repress the TERT promoter upon mutation of two or more Sp1 binding sites (Fig 2D and S5C Fig). There was a progressive decrease in the luciferase activity with each consecutive mutation of Sp1 binding site consistent with earlier studies (Fig 2D) [27]. To identify the Sp1 binding sites that are affected by TIP60, we also mutated each of the 5 Sp1 binding sites individually using site-directed mutagenesis (Fig 2E). Upon transient transfection of the mutants in HeLa-LPCX and HeLa-TIP60 cells, we observed that mutations of binding sites 1 and 4 did not significantly rescue the TIP60-mediated TERT promoter repression. However, mutation of binding sites 2, 3 and 5 partially abolished the repressive effect of TIP60 (Fig 2F). This data suggests that TIP60 represses the TERT promoter by inhibiting Sp1.
TIP60 inhibits Sp1 occupancy at the TERT promoter
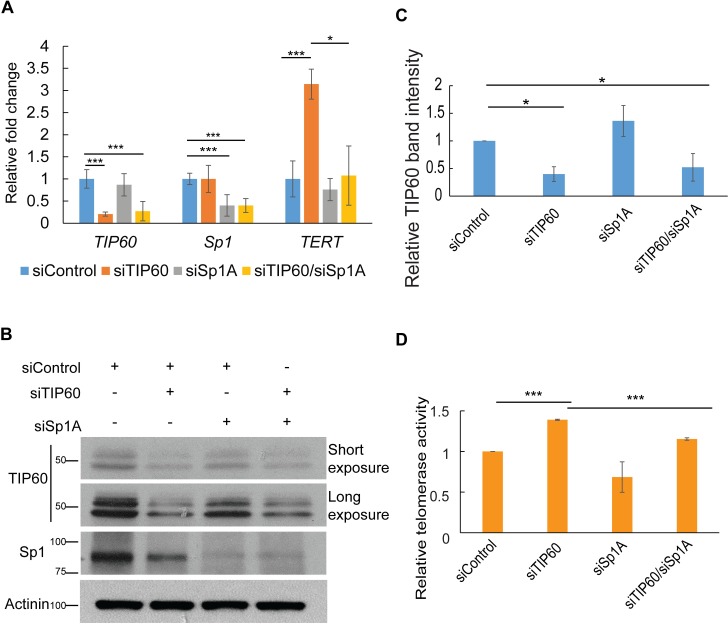
To determine the exact role of Sp1 in TIP60-mediated TERT repression, we first co-depleted TIP60 and Sp1 in HeLa cells using siRNA transfection. The knockdown efficiencies of both Sp1 and TIP60 were greater than 70% both at mRNA and protein levels (Fig 3A, 3B and 3C). The increase in telomerase activity and TERT expression observed upon TIP60 knockdown was partially rescued by co-depletion of TIP60 and Sp1, suggesting that Sp1 acts downstream of TIP60 (Fig 3A and 3D). That TERT expression as well as telomerase activity were reduced upon depletion of Sp1 alone, although to a small extent, is consistent with the findings from other groups (Fig 3A and 3D) [32]. In order to rule out off-target effects of siRNA mediated knockdown, we used another siRNA for depleting Sp1, siSp1 B, and observed a similar rescue in TERT expression upon co-depletion of TIP60 and Sp1 (S4A and S4B Fig).
Fig 3. TIP60 inhibits Sp1 function.
(A, B) TIP60 and Sp1 were depleted in HeLa cells using siRNA transfection. Cells were harvested 72 h post transfection, RNA and protein was isolated to use for real time PCR and western blotting analysis. Real-time PCR analysis was used to study TERT expression upon TIP60 and Sp1 knockdown. All mRNA expression data was normalized to GAPDH and plotted as fold change. Endogenous TIP60 and Sp1 were detected using anti-TIP60 and anti-Sp1 antibodies respectively. All the bands observed when probed with anti-TIP60 correspond to TIP60 protein since they are specifically reduced upon depletion of TIP60. α-Actinin serves as the loading control for the western blot. (C) Quantification of TIP60 bands using Image J software, from three independent experiments has also been depicted. (D) Relative telomerase activity was measured by qTRAP in the same set of samples. Error bars reflect the standard error of mean (SEM) of 3 independent experiments and significance is represented as ***, P<0.001, *, P<0.05.
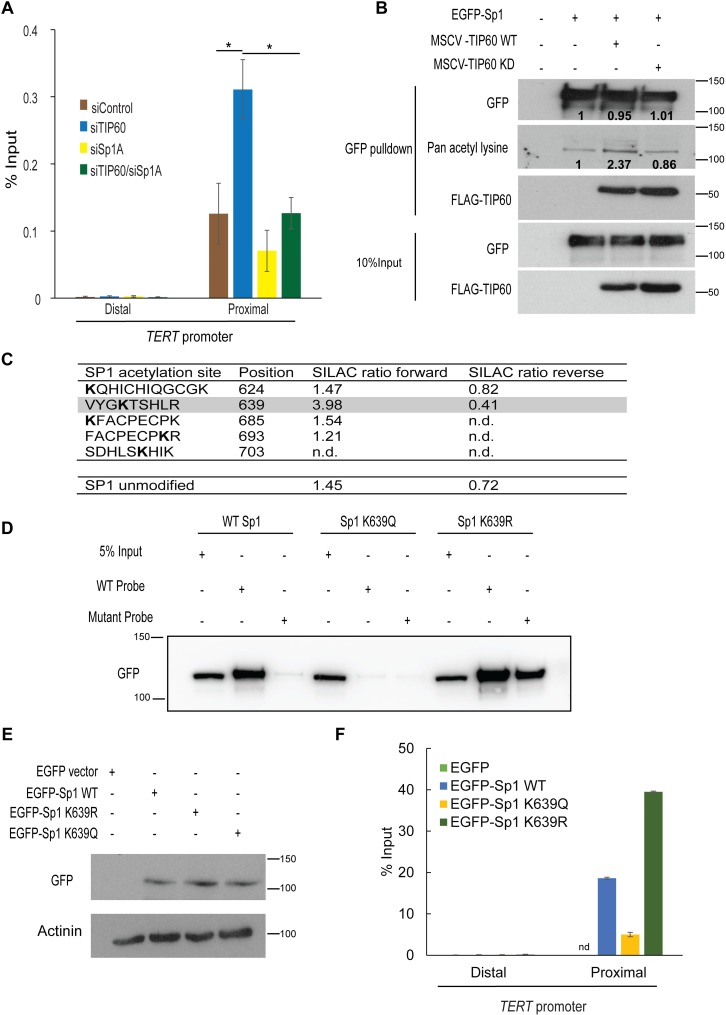
To test whether TIP60 regulates TERT expression by controlling Sp1 binding to the TERT promoter, we performed chromatin immunoprecipitations (ChIP) for Sp1 under TIP60-depleted conditions. Consistent with earlier findings, Sp1 was enriched on the TERT promoter as compared to an upstream element [32] (S5D Fig, Fig 4A). Upon depletion of TIP60, Sp1 occupancy at the TERT promoter was increased, whereas co-depletion of TIP60 and Sp1 showed enrichment similar to control levels (Fig 4A). Together, these data suggest that TIP60 indeed represses TERT by inhibiting Sp1 binding to the TERT promoter and removing its effect as a transcriptional activator. We next sought to identify if TIP60 and Sp1 can associate with each other. Upon pulldown of endogenous TIP60, we identified interaction with endogenous Sp1 (S6A Fig). Since both Sp1 and TIP60 are nuclear proteins, we also repeated the IP in the presence of DNAse, and observed a higher enrichment of both the proteins in this fraction as well as the interaction between the two proteins (S6A Fig), suggesting that indeed TIP60 and Sp1 proteins were found to be associated with each other in HeLa cells.
Fig 4. TIP60 acetylates Sp1 and represses its occupancy at the TERT promoter.
(A) Chromatin immunoprecipitation (ChIP) of Sp1 was performed in HeLa cells depleted of either TIP60 or Sp1 individually or together. Cells were fixed with formaldehyde 72 h post transfection and used for ChIP. An upstream element (distal region) on the TERT promoter served as a negative control for Sp1 binding. Error bars reflect the standard error of mean (SEM) of three independent experiments and significance is represented as *, P<0.05. (B) 1.5 μg of EGFP-Sp1 plasmid was transiently transfected with 1.5 μg of wild-type and catalytically inactive TIP60 (TIP60WT and TIP60KD) into 293T cells and GFP pull-down was performed 48 h post transfection to investigate acetylation of Sp1. The cells were treated with HDAC inhibitors for 5 h prior to harvesting. Sp1 and TIP60 were detected using anti-GFP and anti-Flag antibody respectively. Pan-acetyl lysine antibody was used to detect acetylation on Sp1. Quantification of the bands was performed using Image J software. GFP pulldown was quantified after normalizing to the GFP input in each lane and the pan-acetyl lysine was calculated after normalizing to each individual GFP pulldown. (C) List of acetylated lysines identified on Sp1. SILAC ratios of individual modified peptides are compared to the SILAC ratio of the overall, unmodified Sp1 protein. The VYGKTSHLR peptide modified at position 639 with clearly distinct SILAC ratios is highlighted. (D) WT Sp1 and mutant constructs (Sp1K639R and Sp1K639Q) were transiently transfected in HeLa cells and harvested after 33 h. 100 μg of protein was used for the DNA pull-down and the bound Sp1 was detected with anti-GFP antibody. (E) Sp1 constructs were transfected in HeLa cells and 24 h post transfection, cells were harvested and detection of Sp1 variants was performed using GFP antibody. (F) ChIP of GFP-tagged Sp1 variants in HeLa was performed after transfection of the Sp1 constructs and crosslinking using formaldehyde. n.d refers to the peptides that were identified but not quantified in the experiment.
Acetylation of Sp1 on lysine 639 by TIP60 regulates Sp1’s DNA binding ability
As TIP60 is an acetyltransferase that can acetylate histone and non-histone proteins and was found to interact with Sp1, we next investigated whether such a post-translational modification was putatively regulating Sp1 in this context. To test this, we performed pull-down experiments in 293T cells due to their high transient transfection efficiency and robust expression of exogenous protein. 293T cells were transiently transfected with EGFP-Sp1 either alone or with wild-type (MSCV-TIP60WT) or catalytically inactive form of TIP60 (MSCV-TIP60KD). Sp1 was pulled down using GFP TRAP A beads and its acetylation status was determined using a pan-acetyl antibody. We observed that overexpression of wild-type TIP60 increased the acetylation status of Sp1 whereas acetylation levels were unaltered upon overexpression of the catalytically inactive TIP60 (Fig 4B), thereby confirming that Sp1 is indeed a substrate for TIP60’s acetyltransferase activity. In order to identify the lysine residue(s) on Sp1 that are specifically acetylated by TIP60, we performed the GFP pull-down experiments in SILAC (Stable Isotope Labeling with Amino Acids in Cell Culture) labeled cells and analyzed the Sp1 acetylation status between cells overexpressing either TIP60WT or TIP60KD quantitatively by mass spectrometry (S6B Fig). First we ensured that incorporation rate of the labeled amino acids in our input samples was satisfactory (S6C Fig). Here, specific acetylation sites displayed a differential SILAC ratio, whereas non-TIP60 dependent acetylation sites should have SILAC ratios of about 1:1. We identified 5 acetylated lysines on Sp1 (Figs 4C and S6B) of which we also identified a previously known residue K703, a substrate of CBP/p300 acetyltransferase activity [33]. The other four acetylation sites on Sp1 have not been previously reported in other studies. The acetylation spectra for these individual peptides was observed, confirming their existence in our system of study (S7A, S7B and S7C Fig). While most acetylation sites have SILAC ratios similar to the total Sp1 protein, the K639 acetylation site has SILAC ratios indicating specific acetylation by TIP60 (Fig 4C). Interestingly, the K639 residue is part of the first of three zinc finger domains which are involved in Sp1 DNA binding (S6C Fig) [34]. This led us to speculate that acetylation of Sp1 residue K639 by TIP60 might inhibit the occupancy of Sp1 on the TERT promoter by interfering with Sp1’s DNA binding ability. In order to test this hypothesis, using site-directed mutagenesis, we generated two mutations of Sp1, one which was non-acetylable (K639R) and another that mimicked constitutive acetylation (K639Q). We then performed in vitro reconstitution DNA pull-downs with wild-type Sp1 as well as the two mutants. We transfected wild-type Sp1 and the two mutants (Sp1 K639R and Sp1 K639Q) in HeLa cells and performed pull-down experiments using probes with either wild-type Sp1 binding sequence or the mutated Sp1 binding site sequence. We observed that wild-type Sp1 binds to the wild-type DNA probe but not to the probe containing mutated Sp1 binding site, demonstrating the specificity of the experiment. Strikingly, the mutant that mimics constitutive acetylation, Sp1 K639Q, could not bind to either the wild-type or mutant DNA. In contrast, Sp1 K639R, non-acetylable mimetic was found to bind non-specifically to both the wild-type and mutant DNA sequences (Fig 4D). To test if this binding behaviors were recapitulated in vivo, we transiently transfected these GFP tagged Sp1 constructs into HeLa cells (Fig 4E) and performed ChIP with GFP antibody to selectively pulldown only the exogenous Sp1 proteins. Consistent with the DNA pulldown results in Fig 4D, wild-type Sp1 protein was found to be enriched at the TERT promoter compared to the distal region (Fig 4F). Similar results were also obtained with Sp1 K639R mutant whereas the SP1 K639Q mutant was less enriched at the promoter region compared to both Sp1 WT and Sp1 K639R. This suggests that TIP60-mediated acetylation on Sp1 inhibits its DNA binding ability to the Sp1 binding sites found in the TERT promoter. This data shows that Sp1 is an acetylation target of TIP60 and that indeed acetylation at K639 inhibits its DNA binding function, thus providing a likely mechanism for TIP60-dependent transcriptional control of TERT.
TIP60-dependent growth inhibition is due to repression of TERT expression
HeLa-TIP60 cells showed a significant reduction in colony formation ability in vitro and tumor formation in vivo. We questioned if this tumor suppressor role of TIP60 was dependent on repression of the TERT promoter and telomerase activity. Therefore in order to address the biological significance of TIP60-mediated TERT repression, we transiently transfected HeLa-LPCX and HeLa-TIP60 cells with either pBABE (vector) or pBABE-TERT. In this plasmid, TERT expression is not under the control of its endogenous promoter and could thus serve as a useful system to study if TIP60-mediated growth repression, is in part, facilitated by TERT downregulation and if it can be rescued by expression of exogenous TERT. 24 h post transfection, RNA was isolated from the same set of cells which were used in the colony formation assay (CFA) to study TERT expression and we observed a significant overexpression of TERT in both HeLa-LPCX and HeLa-TIP60 (S8A Fig). There was also a decrease in TERT expression in HeLa-TIP60-pBABE as compared to HeLa-LPCX-pBABE, consistent with our earlier findings (S8B Fig).
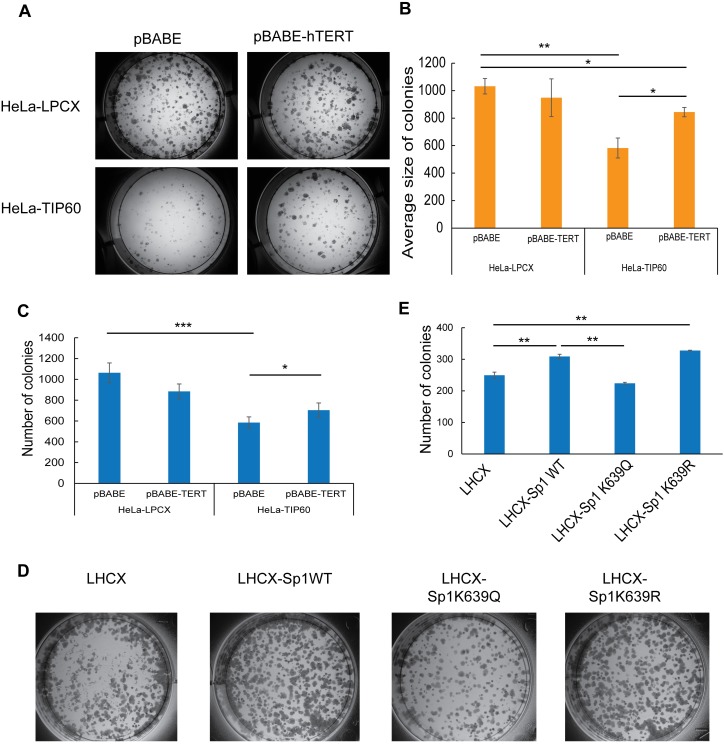
2×103 cells were seeded for CFA. Colonies were fixed and stained with crystal violet after 11 days and their sizes were quantified. We identified a significant reduction in the average size and number of colonies in HeLa-TIP60-pBABE as compared to HeLa-LPCX-pBABE (vector), consistent with our earlier findings [8]. Overexpression of TERT in the HeLa-LPCX background did not affect the colony formation ability. Interestingly, overexpression of TERT in HeLa-TIP60 cells showed a significant increase in the average size and number of colonies as compared to HeLa-TIP60-pBABE, thus rescuing the growth defect caused by TIP60 overexpression (Fig 5A, 5B and 5C).
Fig 5. TIP60-mediated growth defects can be rescued by TERT overexpression and are dependent on Sp1 acetylation.
(A) Representative image from Colony Formation Assay (CFA) where HeLa-LPCX and HeLa-TIP60 cells transiently transfected with pBABE (vector) or pBABE-TERT were seeded at a low density (2×103 cells). The cells were allowed to grow for 11 days before fixing and staining with crystal violet. (B, C) Bar graph shows the quantification performed using Image J software for average size and number of colonies respectively. The overexpression of TIP60 reduces average colony size, which is rescued upon overexpression of TERT in HeLa-TIP60. (D) Sp1 plasmids in LHCX vector backbone were transfected into HeLa cells. Twenty four hours post transfection, 2500 cells were seeded for colony formation assay and representative images are shown. 11 days after, they were fixed and stained with crystal violet and (E) number of colonies was quantified using Image J software. Error bars reflect the standard error of mean (SEM) and significance is represented as *, P<0.05, **, P<0.01, ***, P<0.001.
TIP60-dependent acetylation of Sp1 is involved in mediating the growth inhibition in HeLa cells
Given that Sp1 acetylation could explain TIP60-dependent changes in TERT mRNA levels, we reasoned that this might also be the mechanistic link to explain TIP60-dependent inhibition in colony formation that can be rescued by exogenous expression of TERT. To address this, we examined TERT mRNA levels and colony formation ability upon expression of the Sp1 wild-type and mutant constructs in HeLa (S8C Fig). Overexpression of WT Sp1 as well as Sp1 K639R caused an increase in TERT expression, whereas overexpression of Sp1 K639Q mutant did not result in any change in the TERT expression (S8D Fig). Importantly, overexpression of Sp1 WT and Sp1 K639R resulted in more colonies in CFA in HeLa cells whereas there was no significant difference in the colony formation ability of Sp1 K639Q (Fig 5D and 5E). This data underscores the identified regulatory hierarchy in which TIP60 acetylates Sp1 at K639, which inhibits TERT mRNA expression and in return impacts on the proliferative potential of the affected cancer cells (Fig 6).
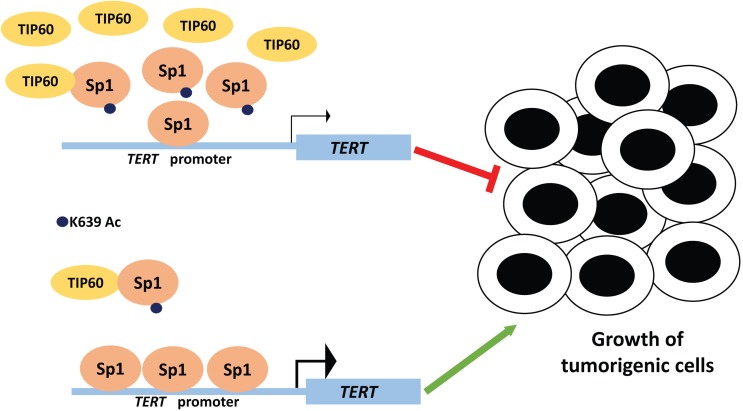
Fig 6. Model of TIP60-mediated TERT repression.
TIP60 inhibits expression of TERT by acetylating Sp1. Decrease in TIP60 levels increases occupancy of Sp1 on TERT promoter resulting in increased expression of TERT and subsequently tumor growth.
Discussion
Telomerase has been an attractive therapeutic target for cancer therapy because it is selectively activated in cancers and stem cells but not expressed in most differentiated tissues [35]. Although multiple factors regulating TERT have been identified in the past, there is a dearth of knowledge regarding mechanisms that repress the endogenous TERT levels in normal somatic cells. Human papillomavirus protein, E2, which is known to repress expression of HPV E6 and E7 and is inactivated upon integration of HPV genome into the host, is also known to repress the TERT promoter and associated telomerase activity. Furthermore, this repressive function of E2 is mediated by its recruitment to the TERT promoter and through Sp1 binding sites [9, 36]. This supports our findings on the significance of Sp1 in regulating TERT expression and that negative regulators of Sp1 function are potent repressors of telomerase expression. Tumor suppressors have been shown to negatively regulate telomerase expression [37]and our study identifies for the first time, TIP60, to be a negative regulator of TERT. In the past, the unconventional repressive activity associated with the histone acetyl transferase, TIP60, has been studied and it is known to repress viral oncogenes like HPV E6, adenovirus E1A as well as some cellular genes [6, 7, 38]. Since the HPV Long control region (LCR), which regulates transcription of the viral oncogenes, is known to harbor binding sites for Sp1 and we have identified TIP60-mediated acetylation to inhibit Sp1 function at the TERT promoter, we were curious to study if modulation of TIP60 levels altered Sp1 occupancy at the HPV LCR. We observed no significant increase in enrichment of Sp1 at HPV LCR regions upon TIP60 depletion (S9 Fig) as compared to siControl treated cells. On the contrary, Sp1 occupancy was decreased significantly in one region of HPV LCR upon TIP60 depletion. The specificity of Sp1 ChIP was established by decreased occupancy upon both Sp1 knockdown as well as co-depletion of TIP60 and Sp1 (S9 Fig). This hints at the role of perhaps other members of the Sp family of transcription factors or other transcription factors at the HPV LCR region.
Although mechanisms of post-translational and post-transcriptional control of TERT have been elucidated, the predominant form of regulation across many cancer types is the transcriptional control [39, 40]. Previous studies have shown a good correlation of TERT expression with the promoter activity using luciferase reporter assays [28, 40] thus reiterating the importance of transcriptional control of TERT and hence telomerase activity. In our study, HeLa cells overexpressing TIP60 showed a decreased telomerase activity (Fig 1A) that is consistent with the decrease in TERT expression (Fig 1C) and depletion of TIP60 increases telomerase activity (Fig 1D) as well as TERT expression (Fig 1F).
This data indicates that TIP60 represses TERT expression in HeLa cells that constitutively express the viral oncoprotein E6, which, together with cellular transcription factors like c-MYC and Sp1 up-regulates TERT expression [24, 41] and destabilizes endogenous TERT repressors like NFX1-91 to activate the promoter [25]. Our data also shows the dependence on E6 for TERT regulation by TIP60 (S2 Fig), indicating that this mechanism could be a feature unique to virus-induced tumors. Several other onco-viruses like HBV, Epstein Barr Virus (EBV), Human T-lymphotropic virus 1 (HTLV-1) and adenovirus are also known to up-regulate TERT transcriptionally [21, 42, 43]. It is interesting to note that some of these are also known to destabilize TIP60 in a fashion similar to HPV E6 and adenovirus [6, 7]. Thus, we speculate that destabilization of TIP60 by viral onco-proteins, which in turn leads to increased telomerase activity, could be a general mechanism that allows virus infected cells to evade senescence. It will be interesting to validate the results in other virus-induced cancers since this would provide an opportunity for therapeutic intervention specific only to the tumor cells. This idea is further supported by the notion that many viral oncogenes also rely on Sp1 to regulate TERT transcriptionally [20, 44]. Our study has identified a novel role of TIP60 in regulating the function of Sp1 at the TERT promoter by acetylating Sp1 at K639 (Fig 4) and we speculate that this may be a feature common to all virus-induced cancers.
Sp1 plays an important regulatory role in TERT activation, however, it has been shown that levels of Sp1 binding to the TERT promoter region are similar in TERT expressing and negative cells [45], which suggests that there must be a differential regulation of Sp1 activity in tumorigenic cells expressing TERT. From our data, TIP60 seems to be a likely candidate that could functionally regulate Sp1 (Fig 4). TIP60-dependent acetylation of Sp1 provides a window for therapeutic intervention in cancers driven by Sp1 overexpression. Identification of the lysine 639 on Sp1 as a specific target of TIP60 offers the possibility of a diagnostic tool for TERT-driven cancers. Earlier reports on post-translational modifications of Sp1 indicate that the acetylation of Sp1 occurs at its DNA binding domain, and that acetylated Sp1 loses its DNA binding ability at different promoters such p21 and BAK [33, 46]. Identification of TIP60 as the acetyltransferase responsible for acetylating Sp1 (Fig 4C) leads us to speculate if expression of other downstream targets of Sp1 also changes upon modulating TIP60 levels. Sp1 is also known to play an anti-senescence role in the context of telomere uncapping induced senescence. In cell culture models that use a TRF2 dominant negative mutant, Sp1 expression is down-regulated, leading to senescence phenotype [47], thereby highlighting the importance of this transcription factor in telomere biology and reiterating the possibility to target it in cancers that activate telomerase.
In conclusion, this study identifies a novel transcriptional repressor of the catalytic subunit of telomerase and suggests a mechanism of action mediated at the TERT promoter through Sp1 (Fig 6). TIP60-mediated TERT repression might be one of the mechanisms by which virus-infected cells escape replicative senescence.
Materials and methods
Cell culture
HeLa cells (ATCC CCL-2), 293T cells (ATCC CRL-3216) and C-33A cells (ATCC HTB-31) were cultured in Dulbecco’s modified Eagle’s media (DMEM) high glucose (Sigma Cat. No. D-5796) with 10% fetal bovine serum (Sigma Cat. No. F-7524), 1% penicillin streptomycin (Gibco Cat. No. 15140–122), Human keratinocytes (ATCC PCS-200-011) were grown in Dermal Cell Basal Medium (ATCC PCS-200-030) supplemented with Bovine Pituitary Extract (BPE), rh TGF-α, L-glutamine, hydrocortisone hemisuccinate, insulin, epinephrine and apo-transferrin at 37°C and 5% CO2. HPV18-transformed keratinocytes (HK18) were generated and grown as described elsewhere [48]. For SILAC labeling, HEK293T cells were incubated in DMEM (-Arg, -Lys) medium containing 10% dialyzed fetal bovine serum (Gibco) supplemented with 42 mg/l 13C615N4 L-arginine and 73 mg/l 13C615N2 L-lysine (Cambridge Isotope Laboratories) or the corresponding non-labeled amino acids, respectively. Stable cell lines used in the study, HeLa-LPCX, HeLa-TIP60 and CaSki-TIP60 were generated as described elsewhere [8].
siRNA transfection
Sp1 and E6 were depleted by transiently transfecting siRNA targeting the open reading frame of these proteins [2, 7, 32]. A commercially available siRNA for TIP60 (siTIP60) was purchased (Sigma Aldrich, SASI_Hs01_00073301) and used in experiments. Sequence of siTIP60A used was as follows- (Sense 5’ UGAUCGAGUUCAGCUAUGA 3’; antisense 5’ UCAUAGCUGAACUCGAUCA 3’). siSP1B siRNA was purchased from SantaCruz (Cat. No. sc-29487). Briefly, 1 million cells were seeded into a 10-cm plate and 20 nM siRNA (siSp1A, siSP1 B, siTIP60A) or 10 nM siRNA (siTIP60, siE6) was used for transfection along with 15 μl Lipofectamine RNAiMax (Invitrogen, Cat. No. 56532) following the manufacturer’s protocol. Six hours post transfection, the transfection mixture was replaced by growth medium and the cells were harvested 72 h after transfection for further analysis.
Quantitative telomerase repeat amplification protocol (qTRAP)
qTRAP is a real-time PCR based technique that measures the ability of telomerase to add telomeric repeat sequences to the ends of its substrates. We modified the conventional TRAP assay using the TRAPeze telomerase detection kit (Millipore, Cat. No. S7700) to facilitate accurate detection. Briefly, TIP60 was depleted by siRNA transfection and the cells were harvested after 72 h. The cell pellet was lysed using 1 CHAPS lysis buffer provided with the kit (10 mM Tris-HCl, pH 7.5; 1 mM MgCl2; 1 mM EGTA; 0.1 mM benzamidine; 5 mM β-mercaptoethanol; 0.5% CHAPS; 10% glycerol), protein was quantified and then the TRAP reaction was set up with 200 nM of telomerase primer TS (5’ AATCCGTCGAGCAGAGTT 3’) and anchored return primer ACX (5’ GCGCGGCTTACCCTTACCTTACCCTAACC 3’). The reaction was carried out in the ABI Prism 7500 (Applied Biosystems) thermal cycler with the samples incubated for 20 min at 25°C and amplified in 32 PCR cycles with 30 sec at 95°C and 90 sec at 60°C followed by 72°C for 1 min. The threshold Ct values were determined and data was analyzed by comparing to a standard curve generated by serial dilution of the siControl/LPCX vector samples. Heat inactivated lysates serve as negative control.
RNA isolation
Total RNA was isolated using TRIZOL reagent (Life Technologies, Cat. No. 15596–026) according to manufacturer’s protocol.
Real-time PCR (RT-PCR) analysis
The total RNA was converted into complementary DNA (cDNA) using iSCRIPT cDNA synthesis kit (Bio-Rad Cat. No. 170–8891). This served as a template for RT-PCR analysis using iTaq Universal SYBR Green Supermix (Bio-Rad Cat. No. 172-5124) on an Applied Biosystems 7500 Fast Real Time PCR system using the following conditions for the run—50°C for 2 min, 95°C for 10 min and amplified by 40 cycles of PCR, 95°C for 15 sec and 60°C for 1 min. The melt curves for all qPCR reactions were also obtained. Results were analyzed and represented as fold change. The primers used are specified in the primer list (Table 1).
Table 1. Quantitative PCR primers.
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| TIP60 | AATGTGGCCTGCATCCTAAC | TGTTTTCCCTTCCACTTTGG |
| E6 | CCAGAAACCGTTGAATCCAG | GTTGGAGTCGTTCCTGTCGT |
| TERT | CTACTCCTCAGGCGACAAGG | TGGAACCCAGAAAGATGGTC |
| Sp1 | CTATAGCAAAATGCCCCAGGT | TCCACCTGCTGTGTCATCAT |
Western blot analysis
Proteins were separated on 8% SDS-PAGE gel, transferred onto a nitrocellulose membrane, and detected with primary antibody (in 3% milk in TBST or 4% PBST) against α-Actinin (B-12, SantaCruz, Cat. No. sc-166524, 1:1000), E6 (SantaCruz, Cat. No. sc-365089, 1:500), FLAG (SantaCruz, Cat. No. sc-807, 1:1000), Sp1 (SantaCruz, Cat. No. sc-59, 1:500), TIP60 (generated in the lab 1:500), Pan-acetyl lysine (SantaCruz, Cat. No. sc-8663R, 1:500), GFP-B2 (SantaCruz, Cat. No. sc-9996, 1:1000) and GFP (ROCHE, no. 11814460001 1:4000). Quantification of blots was done using Image J software. For the immunoprecipitation experiments, 4 μg of cell lysate from HeLa cells was used and immunoprecipitated using endogenous TIP60 antibody overnight. The interacting Sp1 was detected using the endogenous antibody described earlier by western blotting.
Transient transfection and luciferase assay
For the luciferase assay, 500 ng of pGL3, pGL3-949 and pGL3-385 were transfected along with 50 ng of Renilla luciferase construct into HeLa-LPCX and HeLa-TIP60 stable cells in a 12-well plate (1×105 cells per well) using 1 μl of Lipofectamine 2000 (Invitrogen Cat. No. 11668019) for each well following the manufacturer’s instructions. The cells were harvested 24 h after transfection and whole cell lysates were used to determine luciferase activity using the Dual-luciferase reporter 1000 assay system (Promega Cat. No. E1980). All firefly luciferase readings were normalized with Renilla luciferase readings.
Mutagenesis
To generate mutants of Sp1 binding sites on the TERT promoter, the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent, Cat. No. 200522) was used according to the manufacturer’s instructions. The primers used for mutagenesis are provided in Table 2.
Table 2. Site-directed mutagenesis primers.
| Forward primer (5'-3') | Reverse primer (5'-3') | WT sequence (5’-3’) | |
|---|---|---|---|
| Sp1mut1 | GCCGCGAGGAGAGGTATGGGCCGCGGAAAGG | CCTTTCCGCGGCCCATACCTCTCCTCGCGGC | TCCGCGGCCCCGCCCTCTCCT |
| Sp1mut2 | AGGGGAGGGTTTGGGAGGGCCCGGAGGG | CCCTCCGGGCCCTCCCAAACCCTCCCCT | CCTCCCAGCCCCTCCCCTTC |
| Sp1mut3 | CCCGGAGGGGTTTGGGCCGGGGACCC | GGGTCCCCGGCCCAAACCCCTCCGGG | CGGGTCCCCGGCCCAGCCCCC |
| Sp1mut4 | GGGTCGGGACGGGTATGGGTCCGCGCGGA | TCCGCGCGGACCCATACCCGTCCCGACCC | CCTCCGCGCGGACCCCGCCCCG |
| Sp1mut5 | CGCGCGGAGGAGGTTGAGCTGGAAGGTG | CACCTTCCAGCTCAACCTCCTCCGCGCG | CCCTTCACCTTCCAGCTCCGCCTCCT |
| Myc mutant | CAGTCCCTCCGCCACAAAGGAAGCGCGGTCCTG | CAGGACCGCGCTTCCTTTGTGGCGGAGGGACTG | ACCGCGCTTCCCACGTGGC |
Chromatin immunoprecipitation (ChIP)
ChIP was performed following the protocol as described elsewhere (7). Briefly, cells were cross-linked with 1% formaldehyde for 10 min at room temperature. The cells were lysed using SDS Lysis buffer for ChIP (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8). The lysate was then sonicated for 15 cycles at 40% amplitude (15 sec off ON and 45 sec OFF). The sonicated samples were then diluted in ChIP dilution buffer (0.01% SDS, 1% Triton X100, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8, 167 mM NaCl) and used for the immunoprecipitation with anti-Sp1 or anti-GFP antibody. After an over-night incubation with 10 μg of anti-Sp1 antibody or 4 μg of GFP antibody, the bound DNA was washed sequentially with low salt wash buffer (0.1% SDS, 1% Triton X 100, 2 mM EDTA, 20 mM Tris-HCl pH8, 150 mM NaCl), high salt wash buffer (0.1% SDS, 1% Triton X 100, 2 mM EDTA, 20 mM Tris-HCl pH 8, 500 mM NaCl), LiCl wash buffer (0.25 M LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8) and TE wash buffer (10 mM Tris-HCl pH 8, 1 mM EDTA) to remove non-specific sequences and eluted in the elution buffer (84 mg NaHCO3, 1 ml 10% SDS, 9 ml H2O). Then the samples were reverse cross-linked using NaCl at 65°C for 4 h. The eluted DNA was purified and used for qPCR. qPCR was performed in 40 cycles with 95°C for 15 sec and 53°C for 1 min for TERT promoter primers and 95°C for 15 sec and 60°C for 1 min for TERT upstream primers with the products of ChIP, using primers described in Table 3.
Table 3. Chromatin immunoprecipitation primers.
| Forward primer (5'-3') | Reverse primer (5'-3') | |
|---|---|---|
| TERT promoter | TGCCCCTTCACCTTCCA | CTGAAACTCGCGCCG |
| TERT upstream element | GGCTACTGCACGCACCTTTTA | CAAAGATGGCACAGCCTCCG |
GFP pull-down
293T cells were transiently transfected with plasmid expressing EGFP-Sp1 (2 μg) in the presence of TIP60 (2 μg) or a catalytically dead form of TIP60 (2 μg). Cells were harvested 48 h post transfection, after treatment with Histone deacetylase (HDAC) inhibitors (5 μM Sodium butyrate, 1 μM Trichostatin A and 1 μM Nicotinamide) for 5 h prior to harvest. Cells were lysed using lysis buffer (10 mM Tris Cl pH 7.5, 150 mM NaCl, 0.5 M EDTA and 0.2% NP-40) and incubated with GFP-TRAP_A beads (Chromotek, Cat. No. gta-20) following the manufacturer’s protocol. The pull-down samples were analyzed by western blotting and the acetylation on Sp1 was detected using a pan-acetyl lysine antibody.
Colony formation Assay
2×103 cells were seeded and allowed to grow in complete medium supplemented with antibiotics for 11 days. The colonies were fixed and stained using 0.1% Crystal violet in 20% methanol. The images were quantified using Image J software (http://imagej.nih.gov/ij/). The parameters for particle size on Image J were set from 0-infinity and the circularity as 0–1 and all particles in this dimension were included to calculate number and size of colonies.
Mass spectrometry analysis
In-gel digestion and MS analysis was performed essentially as previously described by Butter et al [49]. In brief, samples were reduced in 10 mM DTT 1 h at 56°C followed by alkylation with 55 mM iodoacetamide (Sigma) for 45 min in the dark. Tryptic digest was performed in 50 mM ammonium bicarbonate buffer with 2 μg trypsin (Promega) at 37°C overnight. Peptides were desalted on StageTips and analyzed by nanoflow liquid chromatography on an EASY-nLC 1200 system coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Peptides were separated on a C18-reversed phase column (25 cm long, 75 μm inner diameter) packed in-house with ReproSil-Pur C18-QAQ 1.9 μm resin (Dr Maisch). The column was mounted on an Easy Flex Nano Source and temperature controlled by a column oven (Sonation) at 40°C. A 105-min gradient from 2 to 40% acetonitrile in 0.5% formic acid at a flow of 225 nl/min was used. Spray voltage was set to 2.4 kV. The Q Exactive HF was operated with a TOP20 MS/MS spectra acquisition method per MS full scan. MS scans were conducted with 60,000 and MS/MS scans with 15,000 resolution. The raw files were processed with MaxQuant [50] version 1.5.2.8 with preset standard settings for SILAC-labeled samples digested with trypsin and the re-quantify option was activated. Carbamidomethylation was set as fixed modification while methionine oxidation, protein N-acetylation and lysine-acetylation were considered as variable modifications. Search results were processed with MaxQuant filtered with a false discovery rate of 0.01.
In vitro DNA reconstitution pull-down
Biotinylated DNA baits consisting of the 2nd Sp1 binding site in the TERT promoter or a mutated sequence were prepared as previously described [51]. For primer sequences see Table 4. Sp1 WT or Sp1 mut DNA was immobilized on 250 μg paramagnetic streptavidin beads (Dynabeads MyOne C1, Life Technologies) on a rotation wheel for 30 min at room temperature. Subsequently, baits were incubated with 100 μg of nuclear extract in PBB buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 0.5% Igepal CA-630 (Sigma)) while rotating for 2 h at 4°C. 20 μg sheared salmon sperm DNA (Ambion) was added as a competitor for DNA binding. After three washes with PBB buffer, bound proteins were eluted in 2× Laemmli buffer (Sigma-Aldrich), boiled for 5 min at 95°C and separated on a 4–12% NuPAGE Novex Bis-Tris precast gel (Life Technologies).
Table 4. DNA pull-down primers.
| Forward primer (5'-3') | Reverse primer (5'-3') | |
|---|---|---|
| WT Probe | AGAGGAAGGGGAGGGGCTGGGAGGGCCCGG | CTCCGGGCCCTCCCAGCCCCTCCCCTTCCT |
| Mutant Probe | AGAGGAAGGGGAGGGTTTGGGAGGGCCCGG | CTCCGGGCCCTCCCAAACCCTCCCCTTCCT |
Statistical analysis
All statistical analyses were performed using unpaired two-tailed student’s t-test. Error bars indicate standard error of mean (SEM) for the indicated number of biological repeats. Significance is represented as *P<0.05, **P<0.01, ***P<0.001.
Supporting information
(A) TIP60 was transiently depleted in HeLa-TIP60 cells using siTIP60A and cells were harvested 72 h post transfection. Telomerase activity was measured. (B) Efficiency of TIP60 depletion was confirmed by western blotting. TIP60 was detected using an endogenous TIP60 antibody and Actin serves as loading control. (C) Cellular RNA was isolated and analyzed by real time PCR after knockdown to determine TERT expression. mRNA expression was normalized to GAPDH and plotted as fold change. (D) Transient depletion of TIP60 in HeLa cells using siRNA transfection and measurement of telomerase activity by qTRAP. (E) Western blotting analysis on the same set of cells to verify the knockdown of TIP60. Endogenous TIP60 was detected using anti-TIP60 antibody. (F) Real-time PCR analysis to study the expression of TERT upon TIP60 knockdown. mRNA data was normalized to GAPDH and plotted as fold change. (G) Relative telomerase activity was measured after transient knockdown of TIP60 using siTIP60 in CaSki-TIP60 cells. (H) Depletion of TIP60 was verified by western blotting analysis using anti-TIP60 antibody to detect TIP60. All the bands observed when probed with anti-TIP60 antibody correspond to TIP60 protein since they are specifically reduced upon depletion of TIP60. (I) TERT expression was also checked after TIP60 depletion using qPCR. mRNA data was normalized to GAPDH and plotted as fold change. Error bars reflect the standard error of mean (SEM) of 3 independent experiments and significance is represented as *, P<0.05, ***, P<0.001.
(TIF)
(A) Co-depletion of TIP60 and E6 using siRNA was performed in HeLa cells. Seventy two hours post transfection of TIP60 and E6 siRNA, cells were harvested and analyzed by western blotting with antibodies to detect endogenous TIP60 and E6. All the bands observed when probed with anti-TIP60 antibody correspond to TIP60 protein since they are specifically reduced upon depletion of TIP60. α-Actinin serves as an internal control as well as to indicate equal protein amount used for qTRAP assay. (B) Telomerase activity was measured for the same set of samples using qTRAP. (C) C-33A cells are cervical cancer cells which are not infected by HPV. In these cells, TIP60 was transiently depleted using siRNA and cells were harvested 72 h post transfection. Real-time PCR analysis was used to study the expression of TERT upon TIP60 depletion. Error bars reflect the standard error of mean (SEM) of 3 independent experiments and significance is represented as *, P<0.05, **, P<0.01
(TIF)
(A, B) Transient depletion of TIP60 using siRNA and increase in TERT expression was verified in the same set of cells used for luciferase assay in Fig 2B.
(TIF)
(A, B) TIP60 and Sp1 were depleted in HeLa cells using siRNA transfection. Sp1 was depleted using a second siRNA, siSp1B. Cells were harvested 72 h post transfection, RNA and protein was isolated to use for real time PCR and western blotting analysis. All mRNA expression data was normalized to GAPDH and plotted as fold change. Endogenous TIP60 and Sp1 were detected using anti-TIP60 and anti-Sp1 antibodies respectively.
(TIF)
(A, B) Representation of MYC binding site and the mutant generated on the TERT promoter (385 construct) in the pGL3 vector. (C) Luciferase reporter gene assay with the MYC binding site mutation. Relative luciferase activity was calculated as a ratio of the firefly luciferase signal to the Renilla luciferase signal. (D) Diagrammatic representation of the TERT promoter region to describe the location of primers designed for ChIP. The TERT promoter primers amplify a region 200 bp upstream of the transcription start site (TSS). The upstream element primers amplify a region of the promoter 3500 bp upstream of the TSS. (E) Transient co-depletion of TIP60 and MAZ in HeLa cells and detection of TERT expression by qPCR.
(TIF)
(A) Endogenous TIP60 protein was pulled down from HeLa cell lysates and associated Sp1 protein was detected by endogenous Sp1 antibody, both in the presence and absence of DNAse. (B) Schematic representation of the steps in the SILAC based mass spectrometry analysis. (C) Quantification of the incorporation rate of ‘heavy’ arginine and lysine in the cells used for the SILAC experiment. (D) Representation of the different domains of Sp1 including the three Zinc finger motifs as obtained from UniProt (http://www.uniprot.org/).
(TIF)
(A) Representative MS1 spectra of the Sp1 peptide VYGK(ac)TSHLR2+. In the forward experiment (upper panel) the heavy peptide is more abundant than the light peptide, whereas in the reverse experiment (lower panel) the opposite trend is visible. (B) Representative MS2 spectra for the VYGK(ac)TSHLR2+ peptide both with heavy (upper panel) and light labeling (lower panel) providing sequence evidence for the existence of the Sp1 modification. (C) Representative MS2 spectra providing sequence evidence for 4 additional Sp1 acetylation sites that according to the SILAC quantification are not dependent on TIP60.
(TIF)
(A, B) mRNA expression data of TERT normalized to GAPDH and plotted as fold change from cells seeded for CFA in Fig 5A. (C, D) Sp1 constructs were transiently transfected into HeLa cells and 24 h post transfection, cells were seeded for CFA. The remaining cells were used to isolate RNA and study Sp1 and TERT expression respectively, by qPCR.
(TIF)
(A) Graphical representation of the primers designed along HPV LCR. (B) Sp1 ChIP was performed in HeLa cells transiently depleted of either TIP60 or Sp1 alone or together. Three regions along the HPV LCR were used to detect Sp1 occupancy upon modulating TIP60 levels. Error bars represent standard error of mean (SEM) of three independent experiments.
(TIF)
List of all the peptides identified in our experimental set up.
(XLSX)
Acknowledgments
We thank the members of the Jha laboratory for helpful discussion and comments. We thank Dr Vanitha Krishna Subbaiah for generation of the stable cell lines used in this study. We thank Prof Francoise Thierry for the generous gift of HK18 cells used in this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
SJ was supported by grants from National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centers of Excellence initiative to the Cancer Science Institute of Singapore (R-713-006-014-271), National Medical Research Council (NMRC CBRG-NIG BNIG11nov001) and Ministry of Education Academic Research Fund (MOE AcRF Tier 1 T1-2012 Oct -04 and T1-2016- Apr-01). SJ, DK and DGT were supported by the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education’s Tier 3 grants (MOE2014-T3-1-006). DGT was supported by a STaR Investigator Award and an RCE Core grant from the NRF, Singapore, and NIH grants CA66996, CA197697, and HL131477. DR and SH were supported by a post-graduate fellowship awarded by Yong Loo Lin School of Medicine, National University of Singapore. KKL, YZ, SSB, and BHC were supported by a postgraduate fellowship awarded by the Cancer Science Institute of Singapore, National University of Singapore. HSK was supported by a post-graduate fellowship awarded by NGS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14(2):147–54. doi: 10.1016/j.gde.2004.02.009 . [DOI] [PubMed] [Google Scholar]
- 2.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28(8):2690–700. doi: 10.1128/MCB.01983-07 ; PubMed Central PMCID: PMC2293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102(37):13182–7. doi: 10.1073/pnas.0504211102 ; PubMed Central PMCID: PMC1197271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24(6):841–51. doi: 10.1016/j.molcel.2006.11.026 ; PubMed Central PMCID: PMC1766330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448(7157):1063–7. doi: 10.1038/nature06055 . [DOI] [PubMed] [Google Scholar]
- 6.Gupta A, Jha S, Engel DA, Ornelles DA, Dutta A. Tip60 degradation by adenovirus relieves transcriptional repression of viral transcriptional activator EIA. Oncogene. 2013;32(42):5017–25. doi: 10.1038/onc.2012.534 ; PubMed Central PMCID: PMC3955737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jha S, Vande Pol S, Banerjee NS, Dutta AB, Chow LT, Dutta A. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol Cell. 2010;38(5):700–11. doi: 10.1016/j.molcel.2010.05.020 ; PubMed Central PMCID: PMC2886028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subbaiah VK, Zhang Y, Rajagopalan D, Abdullah LN, Yeo-Teh NS, Tomaic V, et al. E3 ligase EDD1/UBR5 is utilized by the HPV E6 oncogene to destabilize tumor suppressor TIP60. Oncogene. 2016;35(16):2062–74. doi: 10.1038/onc.2015.268 . [DOI] [PubMed] [Google Scholar]
- 9.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2(5):342–50. doi: 10.1038/nrc798 . [DOI] [PubMed] [Google Scholar]
- 10.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56(20):4620–4. . [PubMed] [Google Scholar]
- 11.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–36. . [DOI] [PubMed] [Google Scholar]
- 12.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85(18):6622–6. ; PubMed Central PMCID: PMCPMC282029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, et al. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci U S A. 1992;89(21):10114–8. ; PubMed Central PMCID: PMCPMC50288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu P, Takai H, de Lange T. Telomeric 3' overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell. 2012;150(1):39–52. doi: 10.1016/j.cell.2012.05.026 ; PubMed Central PMCID: PMCPMC3392515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olovnikov AM. [Principle of marginotomy in template synthesis of polynucleotides]. Dokl Akad Nauk SSSR. 1971;201(6):1496–9. . [PubMed] [Google Scholar]
- 16.Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Semin Cancer Biol. 2011;21(6):349–53. doi: 10.1016/j.semcancer.2011.10.001 ; PubMed Central PMCID: PMCPMC3370415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11(5):319–30. doi: 10.1038/nrg2763 . [DOI] [PubMed] [Google Scholar]
- 18.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337(6205):331–7. doi: 10.1038/337331a0 . [DOI] [PubMed] [Google Scholar]
- 19.Aisner DL, Wright WE, Shay JW. Telomerase regulation: not just flipping the switch. Curr Opin Genet Dev. 2002;12(1):80–5. . [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Shi W, Luan F, Xu S, Yang F, Sun W, et al. Hepatitis B virus X protein upregulates transcriptional activation of human telomerase reverse transcriptase. Virus Genes. 2010;40(2):174–82. doi: 10.1007/s11262-009-0441-3 . [DOI] [PubMed] [Google Scholar]
- 21.Zou SQ, Qu ZL, Li ZF, Wang X. Hepatitis B virus X gene induces human telomerase reverse transcriptase mRNA expression in cultured normal human cholangiocytes. World J Gastroenterol. 2004;10(15):2259–62. doi: 10.3748/wjg.v10.i15.2259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Dakic A, Zhang Y, Dai Y, Chen R, Schlegel R. HPV E6 protein interacts physically and functionally with the cellular telomerase complex. Proc Natl Acad Sci U S A. 2009;106(44):18780–5. doi: 10.1073/pnas.0906357106 ; PubMed Central PMCID: PMC2773972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380(6569):79–82. doi: 10.1038/380079a0 . [DOI] [PubMed] [Google Scholar]
- 24.McMurray HR, McCance DJ. Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J Virol. 2003;77(18):9852–61. doi: 10.1128/JVI.77.18.9852-9861.2003 ; PubMed Central PMCID: PMC224601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gewin L, Myers H, Kiyono T, Galloway DA. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004;18(18):2269–82. doi: 10.1101/gad.1214704 ; PubMed Central PMCID: PMC517520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gewin L, Galloway DA. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J Virol. 2001;75(15):7198–201. doi: 10.1128/JVI.75.15.7198-7201.2001 ; PubMed Central PMCID: PMCPMC114450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyo S, Takakura M, Taira T, Kanaya T, Itoh H, Yutsudo M, et al. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT). Nucleic Acids Res. 2000;28(3):669–77. ; PubMed Central PMCID: PMC102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, et al. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59(3):551–7. . [PubMed] [Google Scholar]
- 29.Xu M, Katzenellenbogen RA, Grandori C, Galloway DA. An unbiased in vivo screen reveals multiple transcription factors that control HPV E6-regulated hTERT in keratinocytes. Virology. 2013;446(1–2):17–24. doi: 10.1016/j.virol.2013.07.014 ; PubMed Central PMCID: PMC3787310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeFilippis RA, Goodwin EC, Wu L, DiMaio D. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J Virol. 2003;77(2):1551–63. doi: 10.1128/JVI.77.2.1551-1563.2003 ; PubMed Central PMCID: PMC140828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Toh L, Lau P, Wang X. Human telomerase reverse transcriptase (hTERT) is a novel target of the Wnt/beta-catenin pathway in human cancer. J Biol Chem. 2012;287(39):32494–511. doi: 10.1074/jbc.M112.368282 ; PubMed Central PMCID: PMCPMC3463325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L, Ishihara K, Ichimura T, Fujita N, Hino S, Tomita S, et al. MCAF1/AM is involved in Sp1-mediated maintenance of cancer-associated telomerase activity. J Biol Chem. 2009;284(8):5165–74. doi: 10.1074/jbc.M807098200 . [DOI] [PubMed] [Google Scholar]
- 33.Hung JJ, Wang YT, Chang WC. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol Cell Biol. 2006;26(5):1770–85. doi: 10.1128/MCB.26.5.1770-1785.2006 ; PubMed Central PMCID: PMCPMC1430254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suske G. The Sp-family of transcription factors. Gene. 1999;238(2):291–300. . [DOI] [PubMed] [Google Scholar]
- 35.Ducrest AL, Szutorisz H, Lingner J, Nabholz M. Regulation of the human telomerase reverse transcriptase gene. Oncogene. 2002;21(4):541–52. doi: 10.1038/sj.onc.1205081 . [DOI] [PubMed] [Google Scholar]
- 36.Lee D, Kim HZ, Jeong KW, Shim YS, Horikawa I, Barrett JC, et al. Human papillomavirus E2 down-regulates the human telomerase reverse transcriptase promoter. J Biol Chem. 2002;277(31):27748–56. doi: 10.1074/jbc.M203706200 . [DOI] [PubMed] [Google Scholar]
- 37.Lin SY, Elledge SJ. Multiple tumor suppressor pathways negatively regulate telomerase. Cell. 2003;113(7):881–9. . [DOI] [PubMed] [Google Scholar]
- 38.Kim JW, Song PI, Jeong MH, An JH, Lee SY, Jang SM, et al. TIP60 represses transcriptional activity of p73beta via an MDM2-bridged ternary complex. J Biol Chem. 2008;283(29):20077–86. doi: 10.1074/jbc.M800161200 . [DOI] [PubMed] [Google Scholar]
- 39.Horikawa I, Barrett JC. Transcriptional regulation of the telomerase hTERT gene as a target for cellular and viral oncogenic mechanisms. Carcinogenesis. 2003;24(7):1167–76. doi: 10.1093/carcin/bgg085 . [DOI] [PubMed] [Google Scholar]
- 40.Horikawa I, Cable PL, Afshari C, Barrett JC. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 1999;59(4):826–30. . [PubMed] [Google Scholar]
- 41.Oh ST, Kyo S, Laimins LA. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J Virol. 2001;75(12):5559–66. doi: 10.1128/JVI.75.12.5559-5566.2001 ; PubMed Central PMCID: PMC114268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinha-Datta U, Horikawa I, Michishita E, Datta A, Sigler-Nicot JC, Brown M, et al. Transcriptional activation of hTERT through the NF-kappaB pathway in HTLV-I-transformed cells. Blood. 2004;104(8):2523–31. doi: 10.1182/blood-2003-12-4251 . [DOI] [PubMed] [Google Scholar]
- 43.Terrin L, Dal Col J, Rampazzo E, Zancai P, Pedrotti M, Ammirabile G, et al. Latent membrane protein 1 of Epstein-Barr virus activates the hTERT promoter and enhances telomerase activity in B lymphocytes. J Virol. 2008;82(20):10175–87. doi: 10.1128/JVI.00321-08 ; PubMed Central PMCID: PMC2566252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glasspool RM, Burns S, Hoare SF, Svensson C, Keith WN. The hTERT and hTERC telomerase gene promoters are activated by the second exon of the adenoviral protein, E1A, identifying the transcriptional corepressor CtBP as a potential repressor of both genes. Neoplasia. 2005;7(6):614–22. ; PubMed Central PMCID: PMC1501281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horikawa I, Cable PL, Mazur SJ, Appella E, Afshari CA, Barrett JC. Downstream E-box-mediated regulation of the human telomerase reverse transcriptase (hTERT) gene transcription: evidence for an endogenous mechanism of transcriptional repression. Mol Biol Cell. 2002;13(8):2585–97. doi: 10.1091/mbc.E01-11-0107 ; PubMed Central PMCID: PMC117927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waby JS, Chirakkal H, Yu C, Griffiths GJ, Benson RS, Bingle CD, et al. Sp1 acetylation is associated with loss of DNA binding at promoters associated with cell cycle arrest and cell death in a colon cell line. Mol Cancer. 2010;9:275 doi: 10.1186/1476-4598-9-275 ; PubMed Central PMCID: PMCPMC2972244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An HJ, Lee HJ, Jang S, Jung YJ, Choi SS, Park SC, et al. Transcription factor Sp1 prevents TRF2(DeltaBDeltaM)-induced premature senescence in human diploid fibroblasts. Mol Cell Biochem. 2016;414(1–2):201–8. doi: 10.1007/s11010-016-2672-7 . [DOI] [PubMed] [Google Scholar]
- 48.Demeret C, Garcia-Carranca A, Thierry F. Transcription-independent triggering of the extrinsic pathway of apoptosis by human papillomavirus 18 E2 protein. Oncogene. 2003;22(2):168–75. doi: 10.1038/sj.onc.1206108 . [DOI] [PubMed] [Google Scholar]
- 49.Butter F, Kappei D, Buchholz F, Vermeulen M, Mann M. A domesticated transposon mediates the effects of a single-nucleotide polymorphism responsible for enhanced muscle growth. EMBO Rep. 2010;11(4):305–11. doi: 10.1038/embor.2010.6 ; PubMed Central PMCID: PMCPMC2854606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. doi: 10.1038/nbt.1511 . [DOI] [PubMed] [Google Scholar]
- 51.Kappei D, Butter F, Benda C, Scheibe M, Draskovic I, Stevense M, et al. HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment. EMBO J. 2013;32(12):1681–701. doi: 10.1038/emboj.2013.105 ; PubMed Central PMCID: PMCPMC3680732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) TIP60 was transiently depleted in HeLa-TIP60 cells using siTIP60A and cells were harvested 72 h post transfection. Telomerase activity was measured. (B) Efficiency of TIP60 depletion was confirmed by western blotting. TIP60 was detected using an endogenous TIP60 antibody and Actin serves as loading control. (C) Cellular RNA was isolated and analyzed by real time PCR after knockdown to determine TERT expression. mRNA expression was normalized to GAPDH and plotted as fold change. (D) Transient depletion of TIP60 in HeLa cells using siRNA transfection and measurement of telomerase activity by qTRAP. (E) Western blotting analysis on the same set of cells to verify the knockdown of TIP60. Endogenous TIP60 was detected using anti-TIP60 antibody. (F) Real-time PCR analysis to study the expression of TERT upon TIP60 knockdown. mRNA data was normalized to GAPDH and plotted as fold change. (G) Relative telomerase activity was measured after transient knockdown of TIP60 using siTIP60 in CaSki-TIP60 cells. (H) Depletion of TIP60 was verified by western blotting analysis using anti-TIP60 antibody to detect TIP60. All the bands observed when probed with anti-TIP60 antibody correspond to TIP60 protein since they are specifically reduced upon depletion of TIP60. (I) TERT expression was also checked after TIP60 depletion using qPCR. mRNA data was normalized to GAPDH and plotted as fold change. Error bars reflect the standard error of mean (SEM) of 3 independent experiments and significance is represented as *, P<0.05, ***, P<0.001.
(TIF)
(A) Co-depletion of TIP60 and E6 using siRNA was performed in HeLa cells. Seventy two hours post transfection of TIP60 and E6 siRNA, cells were harvested and analyzed by western blotting with antibodies to detect endogenous TIP60 and E6. All the bands observed when probed with anti-TIP60 antibody correspond to TIP60 protein since they are specifically reduced upon depletion of TIP60. α-Actinin serves as an internal control as well as to indicate equal protein amount used for qTRAP assay. (B) Telomerase activity was measured for the same set of samples using qTRAP. (C) C-33A cells are cervical cancer cells which are not infected by HPV. In these cells, TIP60 was transiently depleted using siRNA and cells were harvested 72 h post transfection. Real-time PCR analysis was used to study the expression of TERT upon TIP60 depletion. Error bars reflect the standard error of mean (SEM) of 3 independent experiments and significance is represented as *, P<0.05, **, P<0.01
(TIF)
(A, B) Transient depletion of TIP60 using siRNA and increase in TERT expression was verified in the same set of cells used for luciferase assay in Fig 2B.
(TIF)
(A, B) TIP60 and Sp1 were depleted in HeLa cells using siRNA transfection. Sp1 was depleted using a second siRNA, siSp1B. Cells were harvested 72 h post transfection, RNA and protein was isolated to use for real time PCR and western blotting analysis. All mRNA expression data was normalized to GAPDH and plotted as fold change. Endogenous TIP60 and Sp1 were detected using anti-TIP60 and anti-Sp1 antibodies respectively.
(TIF)
(A, B) Representation of MYC binding site and the mutant generated on the TERT promoter (385 construct) in the pGL3 vector. (C) Luciferase reporter gene assay with the MYC binding site mutation. Relative luciferase activity was calculated as a ratio of the firefly luciferase signal to the Renilla luciferase signal. (D) Diagrammatic representation of the TERT promoter region to describe the location of primers designed for ChIP. The TERT promoter primers amplify a region 200 bp upstream of the transcription start site (TSS). The upstream element primers amplify a region of the promoter 3500 bp upstream of the TSS. (E) Transient co-depletion of TIP60 and MAZ in HeLa cells and detection of TERT expression by qPCR.
(TIF)
(A) Endogenous TIP60 protein was pulled down from HeLa cell lysates and associated Sp1 protein was detected by endogenous Sp1 antibody, both in the presence and absence of DNAse. (B) Schematic representation of the steps in the SILAC based mass spectrometry analysis. (C) Quantification of the incorporation rate of ‘heavy’ arginine and lysine in the cells used for the SILAC experiment. (D) Representation of the different domains of Sp1 including the three Zinc finger motifs as obtained from UniProt (http://www.uniprot.org/).
(TIF)
(A) Representative MS1 spectra of the Sp1 peptide VYGK(ac)TSHLR2+. In the forward experiment (upper panel) the heavy peptide is more abundant than the light peptide, whereas in the reverse experiment (lower panel) the opposite trend is visible. (B) Representative MS2 spectra for the VYGK(ac)TSHLR2+ peptide both with heavy (upper panel) and light labeling (lower panel) providing sequence evidence for the existence of the Sp1 modification. (C) Representative MS2 spectra providing sequence evidence for 4 additional Sp1 acetylation sites that according to the SILAC quantification are not dependent on TIP60.
(TIF)
(A, B) mRNA expression data of TERT normalized to GAPDH and plotted as fold change from cells seeded for CFA in Fig 5A. (C, D) Sp1 constructs were transiently transfected into HeLa cells and 24 h post transfection, cells were seeded for CFA. The remaining cells were used to isolate RNA and study Sp1 and TERT expression respectively, by qPCR.
(TIF)
(A) Graphical representation of the primers designed along HPV LCR. (B) Sp1 ChIP was performed in HeLa cells transiently depleted of either TIP60 or Sp1 alone or together. Three regions along the HPV LCR were used to detect Sp1 occupancy upon modulating TIP60 levels. Error bars represent standard error of mean (SEM) of three independent experiments.
(TIF)
List of all the peptides identified in our experimental set up.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.