Abstract
Protein folding in the cell was originally assumed to be a spontaneous process, based on Anfinsen’s discovery that purified proteins can fold on their own after removal from denaturant. Consequently cell biologists showed little interest in the protein folding process. This changed only in the mid and late 1980s, when the chaperone story began to unfold. As a result, we now know that in vivo, protein folding requires assistance by a complex machinery of molecular chaperones. To ensure efficient folding, members of different chaperone classes receive the nascent protein chain emerging from the ribosome and guide it along an ordered pathway toward the native state. I was fortunate to contribute to these developments early on. In this short essay, I will describe some of the critical steps leading to the current concept of protein folding as a highly organized cellular process.
It is an honor to share the E. B. Wilson Medal with Art Horwich. I have fond memories of our close collaboration and of the excitement we felt when our experiments provided first evidence of protein folding as a chaperone-assisted process. These early findings would have a lasting impact on our scientific careers.
I was introduced to research as a young medical student at Heidelberg University. My doctoral thesis in the biochemistry department under the guidance of Wilhelm Just focused on the functions of peroxisomes in the rat liver. One of our findings was that the peroxisomal membrane system could be induced by thyroid hormones. I was lucky that Walter Neupert at Munich University was invited as an external reviewer of my thesis, and this led to my joining his group in 1985. Walter was famous for his studies on how mitochondria import newly synthesized proteins from the cytosol. His mentorship turned out to be critical, not only professionally but also personally, as he allowed me to attend a molecular biology summer school where I met my future wife, Manajit. Incidentally, I am writing these lines on our 30th wedding anniversary.
Around the time of my arrival in the Munich lab it became clear that proteins had to be unfolded in order to translocate from the cytosol across the mitochondrial membranes. How then would these proteins fold and assemble inside the organelle? I found myself at the right place at the right time to address this fascinating problem.
PROTEIN FOLDING IN MITOCHONDRIA
I was fortunate that Walter put me in touch with Art Horwich (Figure 1), who had conducted a genetic screen in yeast to identify cellular machinery involved in mitochondrial protein import. One of his temperature-sensitive mutants, called mif4 (for mitochondrial import function 4), was particularly puzzling. We embarked on an exciting collaboration and found that mif4 mitochondria remained import-competent, but the imported proteins failed to assemble into their respective oligomeric complexes (Cheng et al., 1989). Intriguingly, the mif4 mutation mapped to the nuclear gene encoding mitochondrial Hsp60, the homologue of Escherichia coli GroEL and the Rubisco subunit-binding protein (RBP) of chloroplasts—large complexes called “chaperonins” (Hemmingsen et al., 1988). GroEL was known as a genetic host factor in phage propagation, and RBP had been observed to bind unassembled subunits of the enzyme Rubisco (Barraclough and Elis, 1980), suggesting a role in mediating protein assembly.
FIGURE 1:

Art Horwich (right) and myself in March 1991 taking a walk in my parents’ village in the northern part of the Black Forest. Photograph by Manajit Hayer-Hartl.
While our initial findings on Hsp60 were consistent with such a role, a second generation of experiments soon revealed the basic function of the chaperonin in polypeptide chain folding. In these experiments, carried out with my student Joachim Ostermann, we targeted the monomeric protein dihydrofolate reductase (DHFR) to mitochondria (Ostermann et al., 1989). Denatured DHFR will refold spontaneously in vitro, but strikingly, this was not what we observed in mitochondria. Instead, the newly imported protein associated with Hsp60 in an unfolded state that was stabilized under ATP limiting conditions. Formation of folded DHFR occurred upon readdition of ATP concomitantly with release from Hsp60. We concluded that Hsp60—and by analogy the other chaperonins—mediated protein folding. Hence the defects in oligomeric assembly observed in the mif4 mitochondria resulted from the failure of protein subunits to fold. These findings in 1989 established the new paradigm of chaperone-assisted protein folding (reviewed in Hartl, 1996).
THE CHAPERONIN FOLDING CAGE
But how did the chaperonins work? George Lorimer made the next advance by reconstituting bacterial Rubisco, a dimeric enzyme, from the denatured state with the help of GroEL and its cofactor GroES (Goloubinoff et al., 1989). To be sure that we measured folding, not assembly, Jörg Martin in my lab chose monomeric proteins (DHFR and rhodanese) as substrates for reconstitution experiments (Martin et al., 1991). Fluorescence spectroscopy revealed that GroEL binds nonnative proteins in a loosely folded, “molten globule”–like conformation, preventing their aggregation. In the presence of GroES, GroEL released the substrate protein in a more folded, less aggregation-prone state.
In 1991 I took on a faculty position at Sloan-Kettering Cancer Center in Manhattan in the new department of Jim Rothman. We next investigated the GroEL system by electron microscopy in collaboration with Wolfgang Baumeister (Langer et al., 1992a). As shown earlier, the ∼800 kDa GroEL complex consists of two stacked, heptameric rings. The new images revealed that unfolded substrate protein binds in the ring center. We also observed that GroES, a heptameric ring of ∼10 kDa subunits, bound like a lid over one end of the GroEL cylinder, concomitant with major conformational changes in the interacting GroEL ring. The GroEL–GroES complex turned out to be highly dynamic, with GroES undergoing cycles of binding and release in a manner regulated by the GroEL ATPase (Martin et al., 1993). Jörg Martin also found that during this reaction, GroES transiently bound to the GroEL ring holding the protein substrate—indirect evidence that GroES encapsulated the protein in the GroEL cavity. Definitive evidence that folding occurs inside the GroEL–GroES cage was obtained 3 years later by Mark Mayhew in my group and Jonathan Weissman in the Horwich lab (Mayhew et al., 1996; Weissman et al., 1996). Both studies concluded that encapsulation in the central GroEL cavity allows a single protein molecule to fold unimpaired by aggregation (Figure 2). Art Horwich’s and Paul Sigler’s crystal structure of the GroEL–GroES complex provided a detailed view of the folding cage (Xu et al., 1997).
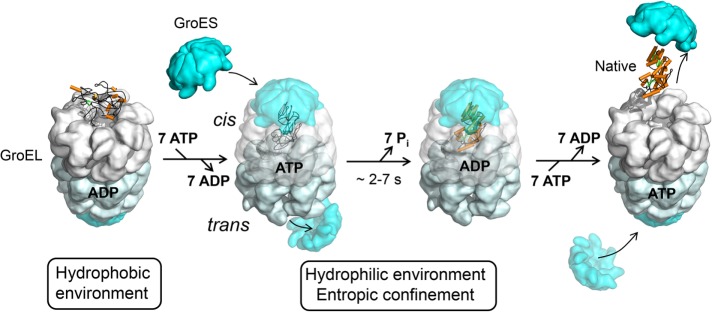
FIGURE 2:
The GroEL/GroES reaction cycle. The current model for protein folding in the chaperonin cage is shown. Substrate binding to GroEL may result in local unfolding (Sharma et al., 2008). ATP binding then triggers a conformational rearrangement of the GroEL apical domains. This is followed by the binding of GroES (forming the cis complex) and substrate encapsulation for folding. At the same time, ADP and GroES dissociate from the opposite (trans) GroEL ring, allowing the release of substrate that had been enclosed in the former cis complex (omitted for simplicity). Substrate remains encapsulated, free to fold, for the time needed to hydrolyze the seven ATP molecules in the newly formed cis complex (∼2–7 s, dependent on temperature). Folding inside the cage may be accelerated due to entropic confinement of dynamic folding intermediates. Binding of ATP and GroES to the trans ring causes the opening of the cis complex. Diagram reproduced from Hartl et al. (2011).
The function of GroEL and GroES is essential, but the natural substrates of the chaperonin system remained unknown. We analyzed the flux of proteins through GroEL (Ewalt et al., 1997) and identified ∼250 GroEL substrates (∼10% of cytosolic proteins) by proteomics (Houry et al., 1999; Kerner et al., 2005). A subset of these proteins proved to be absolutely dependent on GroEL/GroES for folding. These proteins are generally below ∼60 kDa in size and fit into the chaperonin cage. They have complex fold topologies, such as the TIM barrel, and tend to populate aggregation-prone folding intermediates.
More recently, in collaboration with Manajit Hayer-Hartl, we discovered that substrate encapsulation in the chaperonin cage not only serves to prevent aggregation but also markedly (up to 100-fold) accelerates the folding of some proteins over their spontaneous folding rate (measured in the absence of aggregation; Brinker et al., 2001; Georgescauld et al., 2014). We attribute this to an effect of confinement that facilitates the conversion of dynamic folding intermediates to the native state by lowering the entropic component of the folding energy barrier (entropic confinement; Tang et al., 2006; Chakraborty et al., 2010). This function of the cage is important in allowing folding to occur on a biologically relevant time scale.
CHAPERONE PATHWAYS
While we studied the chaperonins, significant progress was made by Jim Rothman and others in understanding another chaperone class, the Hsp70s. Evidence emerged that Hsp70s bind hydrophobic peptides (Flynn et al., 1989) and can associate with nascent polypeptide chains during translation (Beckmann et al., 1990), that is, at a stage when the polypeptide is structurally incomplete and not yet able to fold. Taking this into consideration, we envisioned the existence of a coherent pathway in which Hsp70 would interact first with the (growing) polypeptide chain, maintaining it in a nonaggregated, folding competent state, followed by GroEL/GroES-assisted folding of the completed protein. To test this hypothesis, we attempted to reconstitute the proposed chaperone pathway with pure components. The Georgopoulos group had just shown that the ATPase of the E. coli Hsp70, DnaK, was regulated by two additional proteins, DnaJ and GrpE (Liberek et al., 1991). Key to our success in demonstrating the chaperone function of Hsp70 was to include these factors in the reconstitution experiments. Thomas Langer in the lab made several seminal observations (Langer et al., 1992b): Upon dilution of denatured rhodanese into buffer containing DnaK/DnaJ and ATP, rhodanese aggregation was efficiently prevented, but the protein did not fold, even when GroEL and GroES were added. Strikingly, addition of GrpE, the nucleotide exchange factor of DnaK, catalyzed the transfer of the unfolded protein from DnaK/DnaJ to GroEL/GroES for folding. We later confirmed this pathway for other GroEL-dependent proteins, both in vivo and in vitro (Teter et al., 1999; Kerner et al., 2005; Calloni et al., 2012).
Thomas Langer’s reconstitution experiments showed that DnaK (Hsp70) binds unfolded proteins efficiently, but only when combined with DnaJ (Hsp40) in the presence of ATP. We also observed that DnaJ functioned as a chaperone on its own and GrpE was necessary for ATP-dependent cycles of protein binding and release from Hsp70. Experiments with model proteins showed further that Hsp70 binds extended polypeptides, whereas GroEL prefers collapsed molten globule-like states, thus ordering the two chaperone systems along the folding pathway. Collaborative studies with Bernd Bukau revealed that the DnaK system also assists folding through cycles of protein binding and release (Schröder et al., 1993; Szabo et al., 1994). This mechanism is utilized by 20% or more of cytosolic proteins, but fails for the set of proteins that rely on the protected environment of the chaperonin cage for folding.
In 1994, Judith Frydman in the lab extended the principle of a sequential chaperone pathway from bacteria to the eukaryotic cytosol (Frydman et al., 1994; Frydman and Hartl, 1996). Here Hsp70 cooperates with the chaperonin TRiC, which Judith had previously characterized (Frydman et al., 1992). She also discovered that larger multidomain proteins begin to fold during translation in a domain-wise manner, in close association with chaperones. This mechanism serves to avoid nonproductive interactions between folding domains, as shown by Bill Netzer, and thereby solves the problem of folding large proteins (Netzer and Hartl, 1997). The basic organization of the cytosolic chaperone pathway has been highly conserved in evolution, with very similar implementations found in bacteria, archaea, and eukarya (Figure 3).
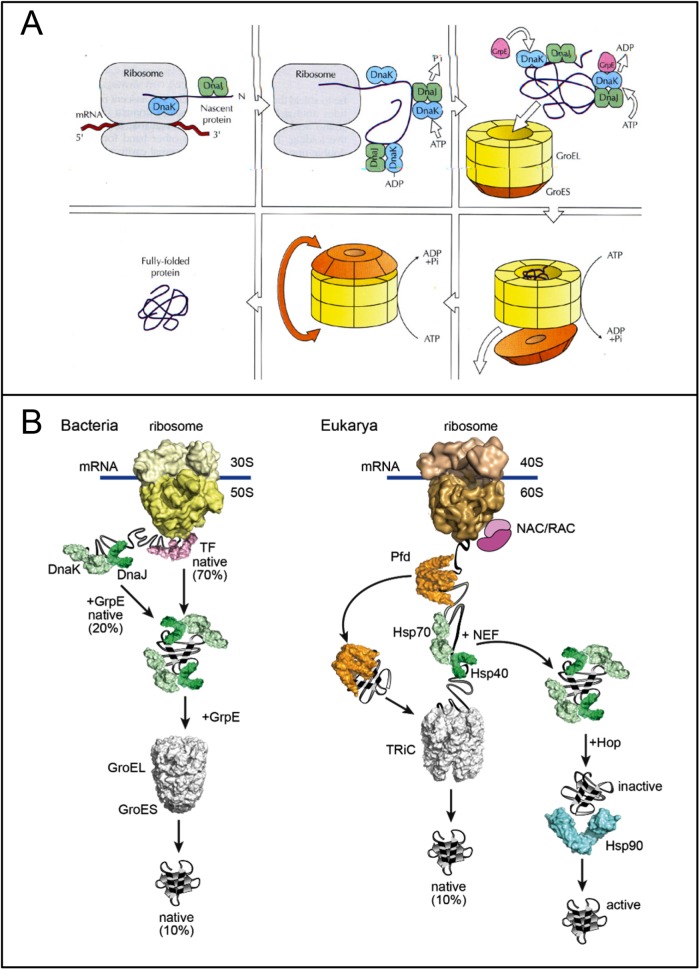
FIGURE 3:
Chaperone pathways in the bacterial and eulkaryotic cytosol. (A) Early model for the E. coli cytosol, shown for a GroEL-dependent protein (reproduced from Martin and Hartl, 1993). In clockwise direction: The nascent chain is stabilized in a folding-competent state during translation by the Hsp70 chaperone system (DnaK/DnaJ). These chaperones bind hydrophobic segments exposed by the extended chain that will later be buried within the folded structure. Upon completion of translation, the protein is unable to fold with DnaK and DnaJ and must be transferred into the central cavity of GroEL. This step requires GrpE, the nucleotide exchange factor of DnaK. Following binding of the protein in a “molten globule”–like conformation into the open ring of GroEL, the protein is encapsulated by GroES in the folding cage. The model was later extended to include the cooperation of DnaK with the ribosome-bound chaperone Trigger factor (Deuerling et al., 1999; Teter et al., 1999) and the finding that the Hsp70 system mediates the folding of proteins that do not require the physical enmvironment of the chaperonin cage (Szabo et al., 1994). (B) Current models for the folding pathways in the bacterial and eukaryotic cytosol reproduced from Balchin et al. (2016). NAC/RAC are eukaryotic ribosome-binding chaperones with a function similar to that of bacterial Trigger factor (TF). Prefoldin (Pfd) recruits the chaperonin TRiC to certain nascent chains. Like TRiC, the Hsp90 chaperone system functions downstream of Hsp70. Hop mediates contacts between Hsp70 and Hsp90.
OUTLOOK
Over the past two decades the chaperone field has developed into a highly active and rapidly expanding branch of molecular life sciences. We are beginning to appreciate the critical role of cooperative chaperone networks in maintaining cellular protein homeostasis and proteome integrity. Understanding these processes at the systems level will be of far-reaching medical relevance, as numerous diseases are linked to protein misfolding and aggregation, including type II diabetes, Alzheimer’s, Parkinson’s, and many others. There is a clear vision that molecular chaperone research will soon enter into the exciting phase of clinical applications.
Acknowledgments
I had the privilege of working with a large number of talented young scientists who deserve my deeply felt gratitude. I apologize to those whose contributions could not be discussed within the format of this essay. I am especially grateful to my wonderful wife and colleague Manajit. I thank my mentors and advisors for continued support and guidance from the beginning of my career, especially Wilhelm Just and Hans Schimassek (Heidelberg University), Walter Neupert (Munich University), Bill Wickner (Dartmouth), and James Rothman (Yale).
Abbreviations used:
- DHFR
dihydrofolate reductase
- Hsp
heat-shock protein.
Footnotes
DOI:10.1091/mbc.E17-07-0480. Mol Biol Cell 28, 2919-2923.
REFERENCES
- Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- Barraclough R, Ellis RJ. Protein synthesis in chloroplasts. IX. Assembly of newly-synthesized large subunits into ribulose bisphosphate carboxylase in isolated intact pea chloroplasts. Biochim Biophys Acta. 1980;608:18–31. doi: 10.1016/0005-2787(80)90129-x. [DOI] [PubMed] [Google Scholar]
- Beckmann RP, Mizzen LE, Welch WJ. Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M. Dual function of protein confinement in chaperonin-assisted protein folding. Cell. 2001;107:223–233. doi: 10.1016/s0092-8674(01)00517-7. [DOI] [PubMed] [Google Scholar]
- Calloni G, Chen T, Schermann SM, Chang H-C, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Chakraborty K, Chatila M, Sinha J, Shi Q, Poschner BC, Sikor M, Jiang G, Lamb DC, Hartl FU, Hayer-Hartl M. Chaperonin-catalyzed rescue of entropically trapped states in protein folding. Cell. 2010;142:112–122. doi: 10.1016/j.cell.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Cheng MY, Hartl FU, Martin J, Pollock RA, Kalousek F, Neupert W, Hallberg EM, Hallberg RL, Horwich AL. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989;337:620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- Ewalt KL, Hendrick JP, Houry WA, Hartl FU. In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell. 1997;90:491–500. doi: 10.1016/s0092-8674(00)80509-7. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Chappell TG, Rothman JE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- Frydman J, Hartl FU. Principles of chaperone-assisted folding: differences between in vitro and in vivo mechanisms. Science. 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Georgescauld F, Popova K, Gupta AJ, Bracher A, Engen JR, Hayer-Hartl M, Hartl FU. GroEL/ES chaperonin modulates the mechanism and accelerates the rate of TIM-barrel domain folding. Cell. 2014;157:922–934. doi: 10.1016/j.cell.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and MgATP. Nature. 1989;342:884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hemmingsen SM, Woolford C, van der Vies SM, Tilly K, Dennis DT, Georgopoulos CP, Hendrix RW, Ellis RJ. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang H-C, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, Hartl FU. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992b;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Langer T, Pfeifer G, Martin J, Baumeister W, Hartl FU. Chaperonin-mediated protein folding: GroES binds to one end of the GroEL cylinder, which accommodates the protein substrate within its central cavity. EMBO J. 1992a;11:4757–4765. doi: 10.1002/j.1460-2075.1992.tb05581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Hartl FU. Protein folding in the cell: Molecular chaperones pave the way. Structure. 1993;1:161–164. doi: 10.1016/0969-2126(93)90017-b. [DOI] [PubMed] [Google Scholar]
- Martin J, Langer T, Boteva R, Schramel A, Horwich AL, Hartl FU. Chaperonin-mediated protein folding at the surface of GroEL through a “molten globule”-like intermediate. Nature. 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- Martin J, Mayhew M, Langer T, Hartl FU. The reaction cycle of GroEL and GroES in chaperonin-assisted protein folding. Nature. 1993;366:228–233. doi: 10.1038/366228a0. [DOI] [PubMed] [Google Scholar]
- Mayhew M, da Silva ACR, Martin J, Erdjument-Bromage H, Tempst P, Hartl FU. Protein folding in the central cavity of the GroEL-GroES chaperonin complex. Nature. 1996;379:420–426. doi: 10.1038/379420a0. [DOI] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- Ostermann J, Horwich AL, Neupert W, Hartl FU. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature. 1989;341:125–130. doi: 10.1038/341125a0. [DOI] [PubMed] [Google Scholar]
- Schröder H, Langer T, Hartl FU, Bukau B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Chakraborty K, Müller BK, Astola N, Tang Y-C, Lamb DC, Hayer-Hartl M, Hartl FU. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell. 2008;133:142–153. doi: 10.1016/j.cell.2008.01.048. [DOI] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schröder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y-C, Chang H-C, Roeben A, Wischnewski D, Wischnewksi N, Kerner MJ, Hartl FU, Hayer-Hartl M. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125:903–914. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang DA, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell. 1999;97:755–765. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- Weissman JS, Rye HS, Fenton WA, Beechem JM, Horwich AL. Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- Xu ZH, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–749. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]