Abstract
Microtubules are long, slender polymers of αβ-tubulin found in all eukaryotic cells. Tubulins associate longitudinally to form protofilaments, and adjacent protofilaments associate laterally to form the microtubule. In the textbook view, microtubules are 1) composed of 13 protofilaments, 2) arranged in a radial array by the centrosome, and 3) built into the 9+2 axoneme. Although these canonical structures predominate in eukaryotes, microtubules with divergent protofilament numbers and higher-order microtubule assemblies have been discovered throughout the last century. Here we survey these noncanonical structures, from the 4-protofilament microtubules of Prosthecobacter to the 40-protofilament accessory microtubules of mantidfly sperm. We review the variety of protofilament numbers observed in different species, in different cells within the same species, and in different stages within the same cell. We describe the determinants of protofilament number, namely nucleation factors, tubulin isoforms, and posttranslational modifications. Finally, we speculate on the functional significance of these diverse polymers. Equipped with novel tubulin-purification tools, the field is now prepared to tackle the long-standing question of the evolutionary basis of microtubule structure.
INTRODUCTION
Microtubules are polymers of αβ-tubulin known for their canonical lattice structure. Describing microtubule structure has been a 50-year pursuit, from early examinations of negatively stained specimens by electron microscopy (EM; Ledbetter and Porter, 1963) to near–atomic resolution cryo-EM of purified microtubules (Zhang et al., 2015). The canonical microtubule has 13 protofilaments (Figure 1A; Tilney et al., 1973), and this lattice structure has been found in cells from every supergroup of eukaryotes (reviewed in Unger et al., 1990). The 13-protofilament lattice is not, however, a uniform property of αβ-tubulin polymers. Rather, tubulin polymers have an intrinsic flexibility; purified αβ-tubulin nucleates spontaneously into microtubules ranging from 9 to 16 protofilaments, and 14-protofilament microtubules are the most abundant in vitro (Pierson et al., 1978). Nevertheless, the microtubules found in cells are uniform in their protofilament number. This uniformity in vivo, which contrasts with the variability observed in vitro, indicates that cells specify their protofilament number as 13 during nucleation, for example, with nucleation factors such as the γ-tubulin ring complex (γ-TuRC; Moritz et al., 1995; Zheng et al., 1995). The predominance of 13-protofilament microtubules in protists, fungi, plants, and animals indicates that the last eukaryotic common ancestor (LECA) specified 13-protofilament lattices and that this specification was conserved over 109 years of evolution. What are the pressures, if any, that select for the 13-protofilament lattice? The main explanation for the conservation of 13 protofilaments is what we call the “straight-protofilament hypothesis” (Amos and Schlieper, 2005; Kollman et al., 2010). Because of the helical arrangement of subunits within the lattice, only the 13-protofilament geometry allows protofilaments to run straight relative to the long axis of the microtubule, whereas the protofilaments in other geometries (e.g., 14 protofilaments) supertwist around the microtubule (Figure 1B; Chrétien and Wade, 1991). The straight protofilaments of 13-protofilament microtubules may accommodate kinesin-1, a cargo-bearing motor protein that tracks along single protofilaments (Ray et al., 1993). If kinesin-1 were to walk on a supertwisted microtubule, it would spiral around during long-range transport of organelles, perhaps problematically. According to the straight-protofilament hypothesis, 13-protofilament microtubules offer a selective advantage in the form of effective long-range transport.
FIGURE 1:
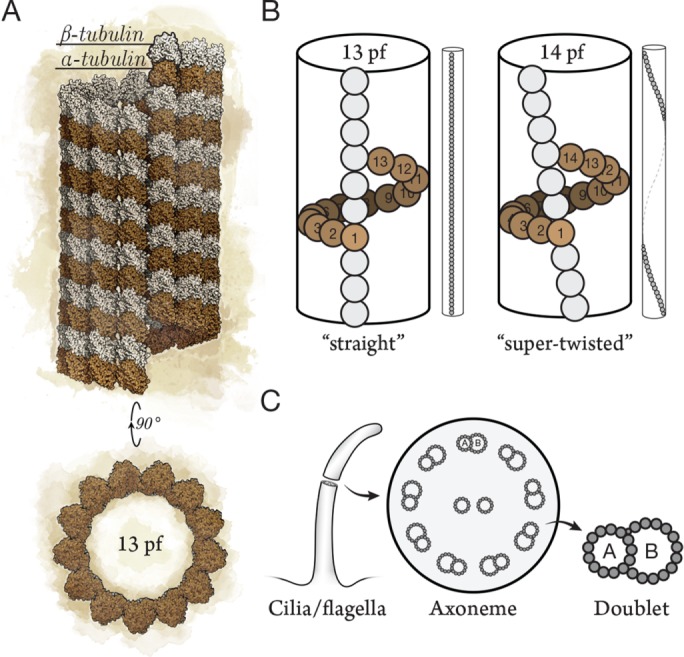
(A) Microtubules are polymers of α/β-tubulin dimers typically composed of 13 protofilaments. (B) Adjacent protofilaments have a longitudinal offset of 9.2 Å between tubulins such that 13-protofilament microtubules have straight protofilaments (left). The lattice of non–13-protofilament microtubules (14 protofilaments shown) must accommodate by imposing a protofilament supertwist (right; not to scale). (C) The functional unit of cilia and flagella (left) is the 9+2 axoneme, with 2 central pair singlets and 9 outer microtubule doublets (middle). Doublets (right) are composed of an incomplete microtubule (10-protofilament B-tubule) clutched to the side of a complete 13-protofilament microtubule (A-tubule). pf, protofilament.
Despite the prevalence of 13 protofilaments, microtubules with other protofilament numbers were discovered in many species during the early heyday of EM. These noncanonical lattices are known to microtubule aficionados primarily as curiosities. At the same time, microtubules in diverse species were found to be arranged in intricate bundles, spirals, rings, and cartwheels, in contrast to the radial arrays and noncentrosomal meshworks typical of tissue culture cells. Here we survey some of these fascinating lattices and morphologies and speculate on their function and evolution. We discuss how cells specify their protofilament numbers using nucleation factors, specific tubulin isoforms, and posttranslational modifications (PTMs) of tubulin. We have named our review after the ancient texts and medieval illuminated manuscripts that described real and mythical creatures. Unlike these historical texts (Bern, 830) and their modern equivalents (Borges, 1957; Gygax, 1977), none of the microtubule structures we describe below are imaginary. We argue that the structural diversity of tubulin polymers raises important questions about microtubule nucleation, about the relationship between microtubule structure and function, and, indeed, about the physiology of the cytoskeleton.
ANCESTRAL POLYMERS
We start our survey by considering the evolutionary origins of tubulin polymers. Tubulin-like proteins can be found in bacteria and archaea, indicating that ancestors to αβ-tubulin arose in primitive, unicellular organisms. Like αβ-tubulin, some prokaryotic homologues are part of the cell-division machinery. The most well-studied prokaryotic tubulin, FtsZ, forms the cytokinetic ring of bacteria and archaea (reviewed in Erickson et al., 2010), whereas TubZ and RepX are essential for plasmid partitioning (Larsen et al., 2007; Anand et al., 2008). Unlike αβ-tubulin, these homologues form single-stranded filaments or twisted filament pairs rather than hollow tubes. Lateral interactions between FtsZ filaments can occur, but they lack a defined lattice structure (Nogales et al., 1998; Erickson et al., 2010). The existence of single-stranded filaments in prokaryotes suggests that longitudinal bonds evolved first, whereas the defined lateral interactions that form the microtubule lattice evolved later. Interestingly, defined lateral interactions occur in the polymers of BtubA/BtubB, a tubulin-like heterodimer found in many Prosthecobacter species (Figure 2). BtubA/BtubB polymerizes into 4-protofilament “bacterial microtubules” in vitro (Deng et al., 2017) and perhaps 5-protofilament polymers in cells (Pilhofer et al., 2011). Because BtubA/BtubB appears in a single genus, the genes were probably acquired from a eukaryote by horizontal gene transfer (Martin-Galiano et al., 2011). We can speculate that bacterial microtubules and FtsZ filaments are snapshots of early eukaryotic polymers, before 13 protofilaments became fixed in the LECA.
FIGURE 2:
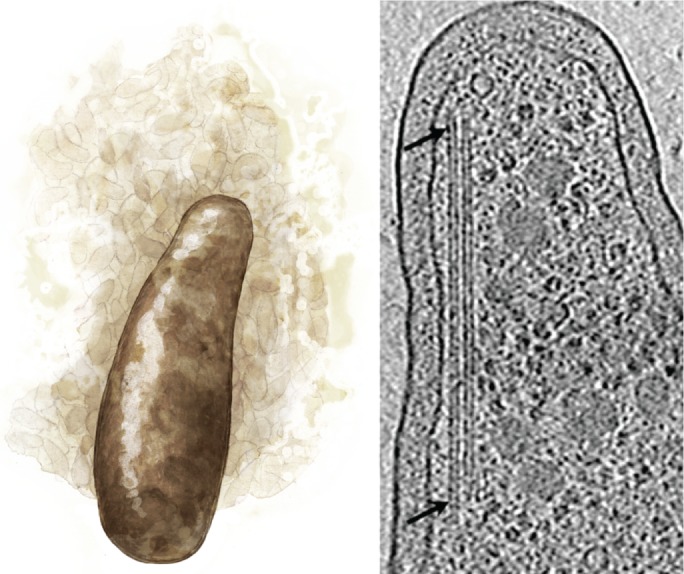
A genus of bacteria, Prosthecobacter (left), is unique in its acquisition of tubulin-like genes that form bacterial microtubules (right, arrows; adapted from Pilhofer et al., 2011).
EUKARYOTIC MICROTUBULE ARCHITECTURES
Although both prokaryotes and eukaryotes require machinery for cell division, eukaryotes also require machinery for internal organization and long-range transport. This requirement is acute in neurons, where synaptic vesicle precursors and mitochondria are carried tens to hundreds of microns down the axon shaft by kinesins. We therefore speculate that eukaryotic microtubule structure evolved straight protofilaments to optimize long-range transport by kinesins, as mentioned above. Many kinesins do not follow a single protofilament during transport, however, but rather drift or wander (Brunnbauer et al., 2012), making it unclear whether straight protofilaments would actually offer a selective advantage. Kinesin-1 may be the exception and not the rule. Furthermore, several eukaryotes, including the model organism Caenorhabditis elegans, do not have 13-protofilament microtubules at all. These examples indicate that 13 protofilaments are not required for the viability of complex organisms. Indeed, in some cases, an atypical lattice may be necessary for complex animal behavior.
Non–13-protofilament microtubules in eukaryotes
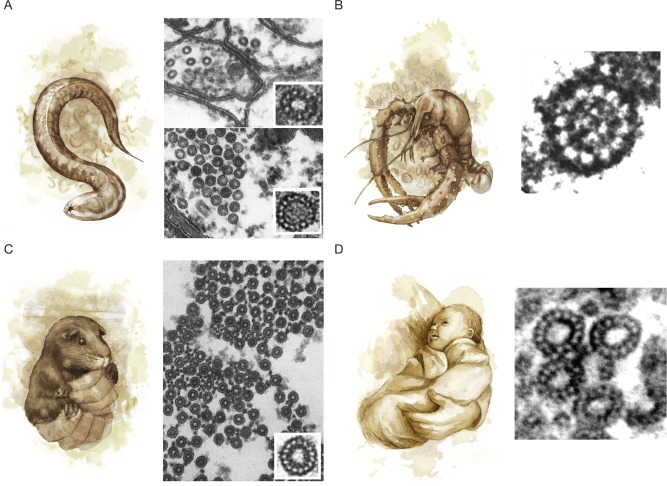
Curious electron microscopists have discovered non–13-protofilament microtubules in plants, animals, and protists. Some of these microtubules have fewer protofilaments and thus smaller diameters. For example, in nematodes, species in both the Rhabditina clade (C. elegans) and the Spirurina clade (Trichostrongylus colubriformis and Ascaridia galli) have 11-protofilament microtubules in the nerve cord as well as in hypodermal, intestinal, and pharyngeal cells (Figure 3A, top right; Chalfie and Thomson, 1982; Davis and Gull, 1983). The occurrence of 11 protofilaments in multiple cell types from multiple clades suggests a shift to 11 protofilaments in the Nematoda phylum. Did the protofilament number in nematodes simply drift or were there pressures that selected for small-diameter microtubules? Nematodes are not alone; 11-protofilament microtubules are also found in the ovary epidermal cells of the grass lily Ornithogalum umbellatum (Kwiatkowska et al., 2006). In some animals, small-diameter microtubules are found in specific cells: lobsters (Nephropidea) and crayfish (Astacoidea) have 12-protofilament microtubules in their nerve cords (Figure 3B), even though neighboring glial cells have 13-protofilament microtubules (Burton et al., 1975). In all the cells described above, the microtubules are uniform in their smaller diameters. We do not know how these cells have changed their nucleation pathways to make uniformly small-diameter microtubules and whether these changes were caused by selective pressures or by genetic drift.
FIGURE 3:
(A) The nematode C. elegans (left) has diverged from the 13-protofilament microtubules observed in other eukaryotes, with 11-protofilament microtubules in ventral cord neurons (top right) and 15-protofilament microtubules in TRNs (bottom right; adapted from Chalfie and Thomson [1982] and reprinted with permission from Rockefeller University Press). (B) The crayfish Procambarus clarkii (left) has 12-protofilament microtubules in the neurons of the nerve cord (right), whereas supporting glial cells have 13-protofilament microtubules (not shown; adapted from Burton et al. [1975] and reprinted with permission from Rockefeller University Press). (C) The guinea pig Cavia porcellus (left) has bundles of 15-protofilament microtubules in its inner pillar cells (right; adapted from Saito and Hama [1982] and reprinted with permission from Oxford University Press). (D) Divergent protofilament numbers are also found in humans (left). A cross-section through a human blood platelet after treatment with 10 µM ADP (right) shows that microtubules sometimes have 14 protofilaments (top) as opposed to the standard 13 protofilaments (bottom; adapted from Xu and Afzelius [1988] and reprinted with permission from Elsevier).
A clear example of specialized function can be found in the large-diameter microtubules implicated in mechanotransduction. Fifteen-protofilament microtubule bundles are found in mechanosensory cells throughout the animal kingdom, for example, in the pillar cells of the inner ear of guinea pigs and mice (Cavia porcellus and Mus musculus; Figure 3C; Saito and Hama, 1982; Tucker et al., 1992) and the touch receptor neurons (TRNs) of nematodes (C. elegans and T. colubriformis; Figure 3A, bottom right; Chalfie and Thomson, 1982; Davis and Gull, 1983). One hypothesis is that 15-protofilament microtubules provide rigidity that might be necessary for efficient mechanotransduction (Tolomeo and Holley, 1997). Indeed, 15-protofilament microtubules are predicted to be 35% stiffer than those with 13 protofilaments (Gittes et al., 1993). An alternative hypothesis is that 15 protofilaments are better at forming microtubule bundles (Cueva et al., 2012; Topalidou et al., 2012); the cross-links within microtubule bundles can increase rigidity fourfold (Tolomeo and Holley, 1997). Outside of mechanotransduction, the function of large-diameter microtubules is less clear. Fifteen-protofilament microtubules are found in the epidermal cells of insects (Drosophila melanogaster and Blattella germanica; Nagano and Suzuki, 1975; Tucker et al., 1986), but there is no obvious reason to have stiffer microtubules in these cells. Interestingly, some cells increase their protofilament number in response to biochemical signals. For example, microtubules in the marginal band of human blood platelets switch from 13 protofilaments to 14 and 15 protofilaments upon platelet activation (Figure 3D; Xu and Afzelius, 1988). Another striking example occurs in Ciliophora protists (Novisuccinea ovalis and Paramecium tetraurelia), in which 13 protofilaments become 14–16 protofilaments during anaphase (Eichenlaub-Ritter, 1985; Tucker et al., 1985). It may be that these cells express factors that increase protofilament number in response to their change in state. Alternatively, the state change may cause the cells to lose control of their protofilament number; in this case, tubulin’s intrinsic flexibility begins to show through.
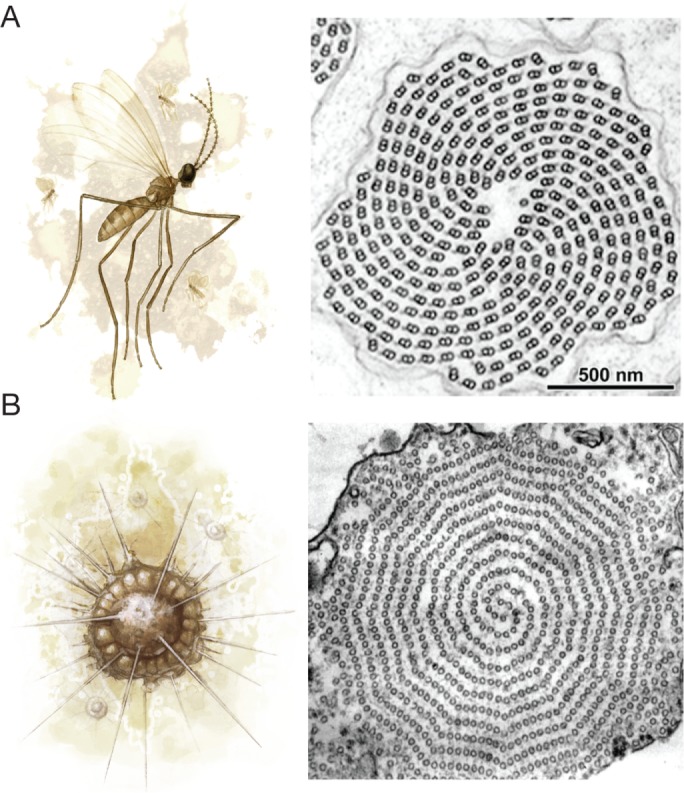
Fifteen- and 16-protofilament microtubules are quite large, but even thicker microtubules are found in the axonemes of insect sperm, where the 9 doublets are surrounded by 9 “accessory microtubules” with typically 13–20 protofilaments (Figure 4A; reviewed in Dallai et al., 2006). In an extreme case, accessory microtubules in the mantidfly Mantispa perla have giant, 40-protofilament “macrotubules” that are filled with polysaccharides (Figure 4B; Dallai et al., 2005). The function of accessory microtubules is not completely understood, but this 9+9+2 configuration might contribute to the sperm’s double-helical waveform (reviewed in Werner and Simmons, 2008). It would be interesting to know whether divergent accessory microtubule protofilament numbers alter the waveform of insect sperm or take on novel roles.
FIGURE 4:
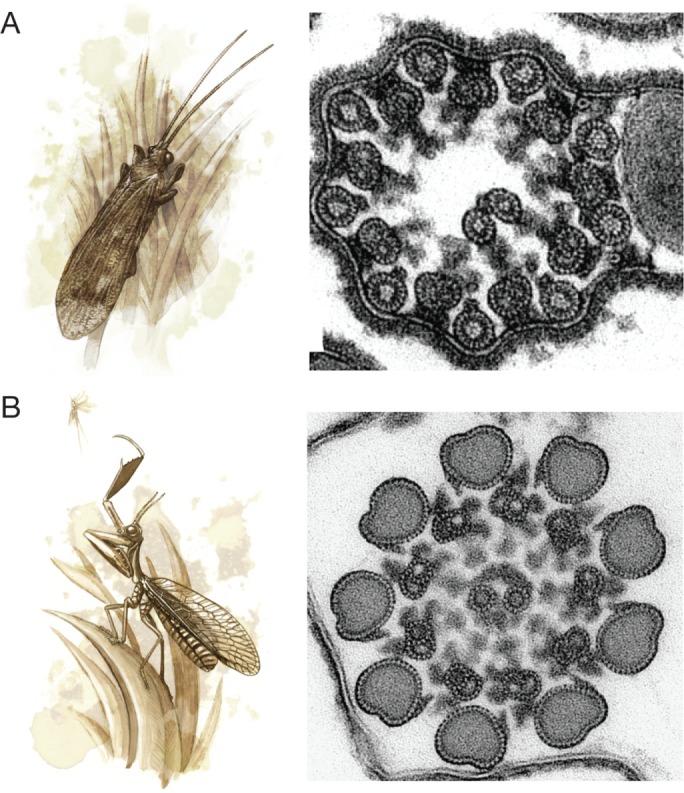
(A) Many insects, such as the caddisfly Stenophylax permistus (left), have unusual sperm axonemes with 9 accessory microtubules surrounding the 9 microtubule doublets in a 9+9+2 configuration (right; adapted from Dallai et al. [2016] and reprinted with permission from the Annual Review of Entomology). (B) The sperm axoneme accessory microtubules of the mantidfly M. perla have 40 protofilaments and are the largest microtubules observed in nature (adapted from Dallai et al. [2005] and reprinted with permission from Elsevier).
DETERMINANTS OF MICROTUBULE ARCHITECTURE
Whatever their roles in cell physiology, non–13-protofilament microtubules provide a series of case studies for the problem of microtubule nucleation. For example, the 11-protofilament microtubules in C. elegans are uniformly 11 protofilaments (Chalfie and Thomson, 1982), despite the fact that C. elegans tubulin forms a range of protofilament numbers when nucleated spontaneously in vitro (Aamodt and Culotti, 1986). How are the structures of noncanonical microtubules specified during nucleation? In a simple view, the protofilament number is established by the angle of the lateral interactions between tubulin subunits; the flexibility of this interprotofilament angle is what produces a range of protofilament numbers when microtubules are nucleated spontaneously. To specify a certain protofilament number, cells need to fix the interprotofilament angle. Cells do so in three ways: 1) with nucleation factors, 2) by expressing specific tubulin isoforms, and 3) through PTM of tubulin.
The simplest way to fix the interprotofilament angle is by providing a nucleation template, such as the 13 γ-tubulins of the γ-TuRC (Figure 5A) or the A-tubules of axonemes, which exclusively nucleate 13-protofilament microtubules (Scheele et al., 1982). The budding yeast (Saccharomyces cerevisiae) γ-TuRC is a conical polymer composed of 7 γ-tubulin small complexes (γ-TuSC), each of which binds 2 γ-tubulins; a partial overlap between one set of γ-TuSCs creates a 13-protofilament template (Zheng et al., 1995; Moritz et al., 2000; Kollman et al., 2010). If the γ-TuRC gained or lost one γ-TuSC, the protofilament number might shift from 13 protofilaments to 15 or 11 protofilaments, respectively, although there is no evidence for such a gain or loss. This idea may explain odd protofilament numbers (e.g., 11 protofilaments in C. elegans), but it cannot explain even protofilament numbers (e.g., 12 protofilaments in lobsters). An important caveat is that the template may not be sufficient to specify the protofilament number. For example, a 14-protofilament GMPCPP microtubule template nucleates a 13-protofilament microtubule lattice (Bechstedt and Brouhard, 2012).
FIGURE 5:
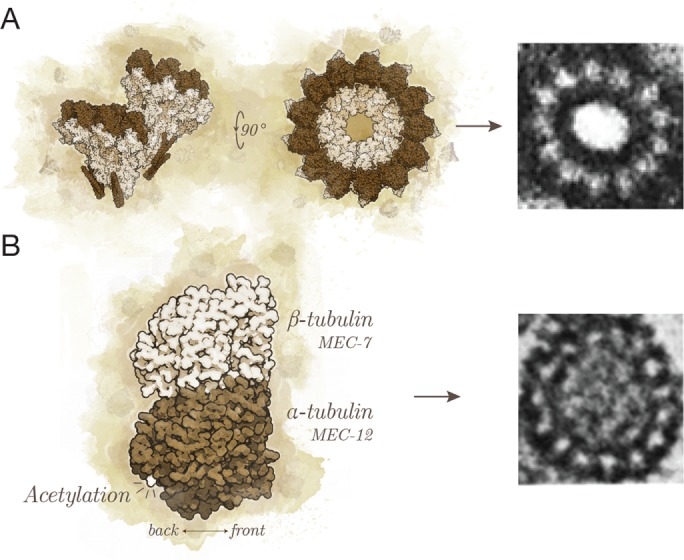
(A) The γ-TuRC provides a template for nucleation with its 13 exposed γ-tubulins (Kollman et al., 2010; left). Shown is a 13-protofilament microtubule nucleated from the centrosome (right; adapted from Evans et al. [1985] and reprinted with permission from Rockefeller University Press). (B) The tubulin dimer (left) can be composed of different isoforms, such as the C. elegans MEC-12/MEC-7 (Savage et al., 1989; Fukushige et al., 1999), and can acquire PTMs, such as acetylation (Cueva et al., 2012; Topalidou et al., 2012). These modifications are able to specify the 15-protofilament microtubules of TRNs in C. elegans (right; adapted from Chalfie and Thomson [1982] and reprinted with permission from Rockefeller University Press).
To further confound the template story, some microtubules—for example, the accessory microtubules in insect sperm—are nucleated without a template. They instead nucleate as outgrowths from a specific location on the B-tubule, one protofilament at a time, before detaching and closing up (Dallai and Afzelius, 1993). It is not known what determines the protofilament number at which the outgrowing microtubule detaches from the B-tubule, but microtubule inner proteins (MIPs) may fix the interprotofilament angles of the outgrowths, as they do in the case of B-tubule outgrowths from the A-tubule (Ichikawa et al., 2017). Template-free microtubule nucleation may also occur in neuronal growth cones, where local nucleation takes place in the absence of γ-TuRCs (Baas and Joshi, 1992; Ma et al., 2004). The neuronal microtubule-associated protein (MAP) doublecortin is sufficient to nucleate 13-protofilament microtubules in vitro (Moores et al., 2004), and we and others have speculated that it might function as a nucleation factor in growth cones (Moores et al., 2004; Bechstedt and Brouhard, 2012). Doublecortin binds the microtubule lattice at the vertex of four tubulin dimers and shares this site with end-binding protein 1 (EB1), which also nucleates 13-protofilament microtubules in vitro (Vitre et al., 2008; Maurer et al., 2012). MAPs, MIPs, and templates are therefore able to specify protofilament numbers by binding different surfaces of the microtubule.
Another way to fix the interprotofilament angle is to change the preferred angle of lateral interactions between tubulins. Cells can change the preferred angle by expressing specific tubulin isoforms or by PTMs, both of which have been shown to modify protofilament numbers. Evidence that the isoform alone can specify protofilament numbers can be found in an experiment in which an endogenous β-tubulin in D. melanogaster was replaced with a testis-specific β-tubulin from the tobacco budworm (Heliothis virescens; Raff et al., 1997). As a result, the 13-protofilament accessory microtubules of the D. melanogaster sperm were transformed to 16 protofilaments. Similarly, the 15-protofilament microtubules in C. elegans TRNs depend on specific α- and β-tubulin isoforms, namely MEC-12 and MEC-7 (Figure 5B; Fukushige et al., 1999; Savage et al., 1989). More recently, acetylation of α-tubulin by acetyltransferases was shown to specify the 15-protofilament microtubules in the TRNs as well (Figure 5B; Cueva et al., 2012; Topalidou et al., 2012). In summary, cells possess numerous tools to specify the protofilament number of their microtubules. Indeed, these tools may have evolved to enable cells to tune the structure of their microtubules in response to selective pressures on complex animal behavior.
HIGHER-ORDER MICROTUBULE ASSEMBLIES
As we have seen, early EM turned up an intriguing diversity at the level of individual tubulin polymers. At the same time, EM also discovered microtubules arranged into a wide range of higher-order assemblies. These assemblies far exceed the commonplace radial arrays and bundles found in most cells. In keeping with the spirit of this review, below we survey a selection of these extraordinary microtubule assemblies.
Specialized assemblies in protists
The greatest diversity of microtubule assemblies is found in the nearly mythical organelles, appendages, and machineries of protists. These unusual microtubule-based organelles are composed of large microtubule bundles with intricate geometries. In ciliates, for example, the order Gymnostomatida (class Nassophorea) feeds through a cytopharyngeal basket that is supported by massive rod structures crammed with hexagonally linked microtubules (Tucker, 1968). Hexagonal packing is a relatively simple geometry, but the bundle geometry can also be complex. Another group of ciliates, the order Suctorida (class Phyllopharyngea), feeds with tentacles that contain a ring of microtubules internally lined with “ribbons” of 7 to 10 microtubules; repeated sliding of the ribbons facilitates ingestion of a prey’s cytoplasm (Bardele, 1972). The number of microtubules within protist organelles can also be remarkably high. Metamonads of the order Oxymonada and Trichomonadida produce an axostyle: a contractile organelle made of thousands of microtubules arranged in spiraling parallel sheets. The undulating axostyle of Saccinobaculus propels the protists through the intestines of termites (McIntosh, 1973). Other metamonads also have microtubule assemblies specialized for invasion. Giardia uses a colossal microtubule-based structure known as the ventral disk to suction onto and colonize human intestinal cells (Elmendorf et al., 2003; Brown et al., 2016). The apicomplexans, on the other hand, invade other cells using the apical complex, a machinery built from numerous highly organized microtubule arrays. One such array in the class Conoidasida is the tightly wound microtubules of the conoid, where each microtubule is not a closed tube but rather a curved sheet of 9 protofilaments (Hu et al., 2002). The intricate microtubule-based machineries of unicellular eukaryotes demonstrate the adaptability of tubulin polymers to the demands of each protist’s niche.
The axonemes of insect sperm and protists
Although multicellular eukaryotes may lack the fantastical microtubule assemblies found in protists, many have elaborations on the familiar axoneme. Insect sperm and protist axonemes frequently deviate from the textbook 9+2 doublet structure (Figure 1C). As few as 3 doublets can suffice, as in the flagella of the apicomplexan Gregarinasina (Prensier et al., 1980). In insect sperm, anywhere from 6 to 16 doublets have been observed (Dallai et al., 1996). Axonemes in the Cecidomyiidea family of insects no longer resemble the canonical form at all and instead have up to 2500 doublets arranged into spirals, rings, and cartwheels (Figure 6A; Dallai et al., 2006). Going further, the Coccoidea superfamily lacks doublets altogether; concentric rings of singlets propel the sperm instead (Baccetti et al., 1982).
FIGURE 6:
(A) A gall midge (left) has cartwheel arrangements of microtubule doublets in the axonemes of its sperm (right; adapted from Dallai et al. [1997] and reprinted with permission from John Wiley & Sons). (B) Heliozoans, such as A. nucleofilum (left), have thin extensions into the environment that contain two interlocking spiral sheets of microtubules that are subdivided into twelve sectors (right; adapted from Tilney and Byers [1969] and reprinted with permission from Rockefeller University Press).
Captivating singlet-based axonemes are also found in Heliozoa protists. Their numerous axopods are thin extensions into the environment that contain two interlocking spiral sheets of microtubules subdivided into 12 sectors (Figure 6B; Tilney and Byers, 1969). Some heliozoans have triangular or hexagonal microtubule arrangements in their axopods (Febvre-Chevalier and Febvre, 1984). These axonemes emanate from a point known as the centroplast or from trilaminar plaques on the nuclear membrane (Tilney, 1971; Cachon et al., 1977). The nucleation factors and cross-linking proteins responsible for these complex arrangements remain a mystery.
DISCUSSION
It is clear that cells specify the protofilament number of their microtubules during nucleation using nucleation factors, tubulin isoforms, and PTMs. Because cells care about their protofilament numbers, so should we. Although 13 protofilaments are far from universal, their prevalence in every eukaryotic supergroup suggests a persistent selective pressure. But the straight-protofilament hypothesis is hard to reconcile with the 11-protofilament microtubules in C. elegans and the irregular paths of many kinesins. We therefore wondered about other selective pressures for 13 protofilaments. While curating the microtubule structures for this review, we noticed that every axoneme we found had 13-protofilament A-tubules, even in the ciliated neurons of C. elegans (Chalfie and Thomson, 1982). Perhaps the straight protofilaments of the 13-protofilament lattice are the result of selective pressures on cilia and flagella. More specifically, the straight protofilaments of a 13-protofilament A-tubule may be necessary for the B-tubule to attach without twisting around the A-tubule. Similarly, the beating of motile cilia and flagella relies on adjacent doublets sliding parallel to one another; straight protofilaments may therefore be essential for ciliary beating. Indeed, the LECA is thought to have been ciliated, and this ancestral cilium may have originated from cytoplasmic microtubules (reviewed in Carvalho-Santos et al., 2011). If the 13-protofilament lattice arose because of selective pressures on cilia and flagella, it is intriguing that 13 protofilaments nevertheless persist in fungi that lack cilia and flagella. Of course, we cannot rule out the hypothesis that the 13-protofilament lattice was an evolutionary accident. Furthermore, cells may not specify their microtubules in every context; for example, the variable protofilament numbers in human platelets and mitotic ciliates may represent a loss of control rather than a regulated transition. Nevertheless, it is our hypothesis that selective pressures have tuned protofilament numbers in different cell types.
Although microtubule structure is diverse, an overwhelming majority of in vitro studies are performed with tubulin purified from the brains of ungulates, primarily cows and pigs. Although ungulate tubulin has enabled extraordinary progress, recent advances in tubulin expression and purification have allowed us to begin experimenting with other tubulins. For example, yeast tubulin is now obtainable using an inducible expression system (Johnson et al., 2011). Tubulins from other eukaryotes, including C. elegans and Xenopus laevis, have been purified by taking advantage of tubulin’s strong affinity to the TOG (tumor overexpressed gene) domains of XMAP215 family proteins (Widlund et al., 2012). Moreover, human tubulin can be recombinantly expressed and purified in a lepidopteran cell line (Minoura et al., 2013). These techniques have opened the door to comparative studies of tubulin from different species, of tubulin isoforms within the same species, and of tubulin mutations. Indeed, the initial comparisons of different human tubulins or yeast tubulin have already uncovered changes in the parameters of dynamic instability (Geyer et al., 2015; Ti et al., 2016; Vemu et al., 2016).
Solving the structure of microtubules from different species will allow us to determine the structural basis of divergences in lattice structure and dynamic behavior. Advances in EM and three-dimensional reconstruction methods have made near–atomic-resolution models of microtubules possible. These models have revealed the tertiary and quaternary structure of tubulins with extraordinary precision, from residues at the laterally interacting M-loops to the long-range conformational changes that accompany GTP hydrolysis (Zhang et al., 2015; Howes et al., 2017). When near–atomic-resolution EM is combined with novel tubulin-purification methods, we will be able to answer questions about the basis of microtubule quaternary structure and, importantly, about the relationship of microtubule structure to cell physiology.
Acknowledgments
For the gouache and pencil illustrations of organisms, we thank Bruce Worden of Black Market Books, a scientific illustrator, independent comic artist, and good friend. We thank S. Bechstedt, K. H. Bui, and C. Wever for comments on the manuscript. We thank S. Wolfson for the swift kick pertaining to the style of this review. G.J.B. is supported by the Canadian Institutes of Health Research (CIHR) (PJT-148702 and MOP-137055), the Natural Sciences and Engineering Research Council of Canada (RGPIN-2014-03791), and McGill University. S.C. is supported by an Alexander Graham Bell Canada Graduate Scholarship (PGS-D). G.J.B. is a CIHR New Investigator.
Abbreviations used:
- EM
electron microscopy
- LECA
last eukaryotic common ancestor
- PTM
posttranslational modification
- TOG
tumor overexpressed gene
- TRN
touch receptor neuron
- γ-TuRC
γ-tubulin ring complex.
Footnotes
REFERENCES
- Aamodt EJ, Culotti JG. Microtubules and microtubule-associated proteins from the nematode Caenorhabditis elegans: periodic cross-links connect microtubules in vitro. J Cell Biol. 1986;103:23–31. doi: 10.1083/jcb.103.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos LA, Schlieper D. Microtubules and maps. Adv Protein Chem. 2005;71:257–298. doi: 10.1016/S0065-3233(04)71007-4. [DOI] [PubMed] [Google Scholar]
- Anand SP, Akhtar P, Tinsley E, Watkins SC, Khan SA. GTP-dependent polymerization of the tubulin-like RepX replication protein encoded by the pXO1 plasmid of Bacillus anthracis. Mol Microbiol. 2008;67:881–890. doi: 10.1111/j.1365-2958.2007.06100.x. [DOI] [PubMed] [Google Scholar]
- Baas PW, Joshi HC. Gamma-tubulin distribution in the neuron: implications for the origins of neuritic microtubules. J Cell Biol. 1992;119:171–178. doi: 10.1083/jcb.119.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccetti B, Burrini AG, Dallai R, Pallini V. A motile system of singlet microtubules in spermatozoa. Cell Motil. 1982;2:93–101. [Google Scholar]
- Bardele CF. A microtubule model for ingestion and transport in the suctorian tentacle. Z Zellforsch Mikrosk Anat. 1972;126:116–134. doi: 10.1007/BF00306784. [DOI] [PubMed] [Google Scholar]
- Bechstedt S, Brouhard GJ. Doublecortin recognizes the 13-protofilament microtubule cooperatively and tracks microtubule ends. Dev Cell. 2012;23:181–192. doi: 10.1016/j.devcel.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern B. Physiologus Bernensis. Burgerbibliothek Cod. 830;318 [Google Scholar]
- Borges JL. Manual de Zoología Fantástica. Mexico City, Mexico: Fondo de Cultura Económica; 1957. [Google Scholar]
- Brown JR, Schwartz CL, Heumann JM, Dawson SC, Hoenger A. A detailed look at the cytoskeletal architecture of the Giardia lamblia ventral disc. J Struct Biol. 2016;194:38–48. doi: 10.1016/j.jsb.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunnbauer M, Dombi R, Ho T-H, Schliwa M, Rief M, Ökten Z. Torque generation of kinesin motors is governed by the stability of the neck domain. Mol Cell. 2012;46:147–158. doi: 10.1016/j.molcel.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Burton PR, Hinkley RE, Pierson GB. Tannic acid-stained microtubules with 12, 13, and 15 protofilaments. J Cell Biol. 1975;65:227–233. doi: 10.1083/jcb.65.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachon J, Cachon M, Tilney LG, Tilney MS. Movement generated by interactions between the dense material at the ends of microtubles and non-actin-containing microfilaments in Sticholonche zanclea. J Cell Biol. 1977;72:314–338. doi: 10.1083/jcb.72.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Tracing the origins of centrioles, cilia, and flagella. J Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Thomson JN. Structural and functional diversity in the neuronal microtubules of Caenorhabditis elegans. J Cell Biol. 1982;93:15–23. doi: 10.1083/jcb.93.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D, Wade RH. New data on the microtubule surface lattice. Biol Cell. 1991;71:161–174. doi: 10.1016/0248-4900(91)90062-r. [DOI] [PubMed] [Google Scholar]
- Cueva JG, Hsin J, Huang KC, Goodman MB. Post-translational acetylation of α tubulin constrains protofilament number in native microtubules. Curr Biol. 2012;22:1066–1074. doi: 10.1016/j.cub.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallai R, Afzelius BA. Development of the accessory tubules of insect sperm flagella. J Submicrosc Cytol Pathol. 1993;25:499–504. [Google Scholar]
- Dallai R, Afzelius BA, Mamaev B. Flagellar axonemes with 10 microtubular doublets in spermatozoa from gall-midges (Diptera, Cecidomyiidae) Acta Zool. 1996;77:153–160. [Google Scholar]
- Dallai R, Gottardo M, Beutel RG. Structure and evolution of insect sperm: new interpretations in the age of phylogenomics. Annu Rev Entomol. 2016;61:1–23. doi: 10.1146/annurev-ento-010715-023555. [DOI] [PubMed] [Google Scholar]
- Dallai R, Lupetti P, Frati F, Mamaev BM, Afzelius BA. Characteristics of spermatozoa from five gall-midge species (Diptera, Cecidomyiidae) Acta Zool. 1997;78:33–37. [Google Scholar]
- Dallai R, Lupetti P, Mencarelli C. Unusual axonemes of hexapod spermatozoa. Int Rev Cytol 254, 45–99. 2006. [DOI] [PubMed]
- Dallai R, Lupetti P, Osella G, Afzelius BA. Giant sperm cells with accessory macrotubules in a neuropteran insect. Tissue Cell. 2005;37:359–366. doi: 10.1016/j.tice.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Davis C, Gull K. Protofilament number in microtubules in cells of two parasitic nematodes. J Parasitol. 1983;69:1094–1099. [PubMed] [Google Scholar]
- Deng X, Fink G, Bharat TAM, He S, Kureisaite-Ciziene D, Löwe J. Four-stranded mini microtubules formed by Prosthecobacter BtubAB show dynamic instability. Proc Natl Acad Sci USA. 2017;114:201705062. doi: 10.1073/pnas.1705062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U. Spatiotemporal control of functional specification and distribution of spindle microtubules with 13, 14 and 15 protofilaments during mitosis in the ciliate Nyctotherus. J Cell Sci. 1985;76:337–355. doi: 10.1242/jcs.76.1.337. [DOI] [PubMed] [Google Scholar]
- Elmendorf HG, Dawson SC, McCaffery JM. The cytoskeleton of Giardia lamblia. Int J Parasitol. 2003;33:3–28. doi: 10.1016/s0020-7519(02)00228-x. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L, Mitchison T, Kirschner M. Influence of the centrosome on the structure of nucleated microtubules. J Cell Biol. 1985;100:1185–1191. doi: 10.1083/jcb.100.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febvre-Chevalier C, Febvre J. Axonemal microtubule pattern of Cienkowskya mereschkovskyi and a revision of heliozoan taxonomy. Orig Life. 1984;13:315–338. [Google Scholar]
- Fukushige T, Siddiqui ZK, Chou M, Culotti JG, Gogonea CB, Siddiqui SS, Hamelin M. MEC-12, an alpha-tubulin required for touch sensitivity in C. elegans. J Cell Sci. 1999;112:395–403. doi: 10.1242/jcs.112.3.395. [DOI] [PubMed] [Google Scholar]
- Geyer EA, Burns A, Lalonde BA, Ye X, Piedra F-A, Huffaker TC, Rice LM. A mutation uncouples the tubulin conformational and GTPase cycles, revealing allosteric control of microtubule dynamics. eLife. 2015;4:e10113. doi: 10.7554/eLife.10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygax G. Monster Manual. Lake Geneva, WI: TSR; 1977. [Google Scholar]
- Howes SC, Geyer EA, LaFrance B, Zhang R, Kellogg EH, Westermann S, Rice LM, Nogales E. Structural differences between yeast and mammalian microtubules revealed by cryo-EM. J Cell Biol. 2017:jcb.201612195. doi: 10.1083/jcb.201612195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Roos DS, Murray JM. A novel polymer of tubulin forms the conoid of Toxoplasma gondii. J Cell Biol. 2002;156:1039–1050. doi: 10.1083/jcb.200112086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M, Liu D, Kastritis PL, Basu K, Hsu TC, Yang S, Bui KH. Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins. Nat Commun. 2017;8:15035. doi: 10.1038/ncomms15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V, Ayaz P, Huddleston P, Rice LM. Design, overexpression, and purification of polymerization-blocked yeast αβ-tubulin mutants. Biochemistry. 2011;50:8636–8644. doi: 10.1021/bi2005174. [DOI] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska M, Popłońska K, Stepiński D, Hejnowicz Z. Microtubules with different diameter, protofilament number and protofilament spacing in Ornithogalum umbellatum ovary epidermis cells. Folia Histochem Cytobiol Pol Acad Sci Pol Histochem Cytochem Soc. 2006;44:133–138. [PubMed] [Google Scholar]
- Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter MC, Porter KR. A “microtubule” in plant cell fine structure. J Cell Biol. 1963;19:239–250. doi: 10.1083/jcb.19.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Shakiryanova D, Vardya I, Popov SV. Quantitative analysis of microtubule transport in growing nerve processes. Curr Biol. 2004;14:725–730. doi: 10.1016/j.cub.2004.03.061. [DOI] [PubMed] [Google Scholar]
- Martin-Galiano AJ, Oliva MA, Sanz L, Bhattacharyya A, Serna M, Yebenes H, Valpuesta JM, Andreu JM. Bacterial tubulin distinct loop sequences and primitive assembly properties support its origin from a eukaryotic tubulin ancestor. J Biol Chem. 2011;286:19789–19803. doi: 10.1074/jbc.M111.230094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR. The axostyle of Saccinobaculus: II. Motion of the microtubule bundle and a structural comparison of straight and bent axostyles. J Cell Biol. 1973;56:324–339. doi: 10.1083/jcb.56.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoura I, Hachikubo Y, Yamakita Y, Takazaki H, Ayukawa R, Uchimura S, Muto E. Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Lett. 2013;587:3450–3455. doi: 10.1016/j.febslet.2013.08.032. [DOI] [PubMed] [Google Scholar]
- Moores CA, Perderiset M, Francis F, Chelly J, Houdusse A, Milligan RA. Mechanism of microtubule stabilization by doublecortin. Mol Cell. 2004;14:833–839. doi: 10.1016/j.molcel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guénebaut V, Heuser J, Agard DA. Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Nagano T, Suzuki F. Microtubules with 15 subunits in cockroach epidermal cells. J Cell Biol. 1975;64:242–245. doi: 10.1083/jcb.64.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Downing KH, Amos LA, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Mol Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- Pierson GB, Burton PR, Himes RH. Alterations in number of protofilaments in microtubules assembled in vitro. J Cell Biol. 1978;76:223–228. doi: 10.1083/jcb.76.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. Microtubules in bacteria: ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 2011;9:e1001213. doi: 10.1371/journal.pbio.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensier G, Vivier E, Goldstein S, Schrevel J. Motile flagellum with a “3 + 0” ultrastructure. Science. 1980;207:1493–1494. doi: 10.1126/science.7189065. [DOI] [PubMed] [Google Scholar]
- Raff EC, Fackenthal JD, Hutchens JA, Hoyle HD, Turner FR. Microtubule architecture specified by a β-tubulin isoform. Science. 1997;275:70–73. doi: 10.1126/science.275.5296.70. [DOI] [PubMed] [Google Scholar]
- Ray S, Meyhöfer E, Milligan RA, Howard J. Kinesin follows the microtubule’s protofilament axis. J Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Hama K. Structural diversity of microtubules in the supporting cells of the sensory epithelium of guinea pig organ of corti. J Electron Microsc (Tokyo) 1982;31:278–281. [PubMed] [Google Scholar]
- Savage C, Hamelin M, Culotti JG, Coulson A, Albertson DG, Chalfie M. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989;3:870–881. doi: 10.1101/gad.3.6.870. [DOI] [PubMed] [Google Scholar]
- Scheele RB, Bergen LG, Borisy GG. Control of the structural fidelity of microtubules by initiation sites. J Mol Biol. 1982;154:485–500. doi: 10.1016/s0022-2836(82)80008-9. [DOI] [PubMed] [Google Scholar]
- Ti S-C, Pamula MC, Howes SC, Duellberg C, Cade NI, Kleiner RE, Forth S, Surrey T, Nogales E, Kapoor TM. Mutations in human tubulin proximal to the kinesin-binding site alter dynamic instability at microtubule plus- and minus-ends. Dev Cell. 2016;37:72–84. doi: 10.1016/j.devcel.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG. How microtubule patterns are generated. J Cell Biol. 1971;51:837–854. doi: 10.1083/jcb.51.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Bryan J, Bush DJ, Fujiwara K, Mooseker MS, Murphy DB, Snyder DH. Microtubules: evidence for 13 protofilaments. J Cell Biol. 1973;59:267–275. doi: 10.1083/jcb.59.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Byers B. Studies on the microtubules in heliozoa. J Cell Biol. 1969;43:148–165. doi: 10.1083/jcb.43.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolomeo JA, Holley MC. Mechanics of microtubule bundles in pillar cells from the inner ear. Biophys J. 1997;73:2241–2247. doi: 10.1016/S0006-3495(97)78255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I, Keller C, Kalebic N, Nguyen KCQ, Somhegyi H, Politi KA, Heppenstall P, Hall DH, Chalfie M. Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr Biol. 2012;22:1057–1065. doi: 10.1016/j.cub.2012.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JB. Fine structure and function of the cytopharyngeal basket in the ciliate nassula. J Cell Sci. 1968;3:493–514. doi: 10.1242/jcs.3.4.493. [DOI] [PubMed] [Google Scholar]
- Tucker JB, Mathews SA, Hendry KA, Mackie JB, Roche DL. Spindle microtubule differentiation and deployment during micronuclear mitosis in Paramecium. J Cell Biol. 1985;101:1966–1976. doi: 10.1083/jcb.101.5.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JB, Milner MJ, Currie DA, Muir JW, Forrest DA, Spencer M-J. Centrosomal microtubule-organizing centres and a switch in the control of protofilament number for cell surface-associated microtubules during Drosophila wing morphogenesis. Eur J Cell Biol. 1986;41:279–289. [Google Scholar]
- Tucker JB, Paton CC, Richardson GP, Mogensen MM, Russell IJ. A cell surface-associated centrosomal layer of microtubule-organizing material in the inner pillar cell of the mouse cochlea. J Cell Sci. 1992;102:215–226. doi: 10.1242/jcs.102.2.215. [DOI] [PubMed] [Google Scholar]
- Unger E, Böhm KJ, Vater W. Structural diversity and dynamics of microtubules and polymorphic tubulin assemblies. Electron Microsc Rev. 1990;3:355–395. doi: 10.1016/0892-0354(90)90007-f. [DOI] [PubMed] [Google Scholar]
- Vemu A, Atherton J, Spector JO, Szyk A, Moores CA, Roll-Mecak A. Structure and dynamics of single-isoform recombinant neuronal human tubulin. J Biol Chem. 2016;291:12907–12915. doi: 10.1074/jbc.C116.731133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B, Coquelle FM, Heichette C, Garnier C, Chrétien D, Arnal I. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat Cell Biol. 2008;10:415–421. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- Werner M, Simmons LW. Insect sperm motility. Biol Rev. 2008;83:191–208. doi: 10.1111/j.1469-185X.2008.00039.x. [DOI] [PubMed] [Google Scholar]
- Widlund PO, Podolski M, Reber S, Alper J, Storch M, Hyman AA, Howard J, Drechsel DN. One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol Biol Cell. 2012;23:4393–4401. doi: 10.1091/mbc.E12-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Afzelius BA. Early changes in the substructure of the marginal bundle in human blood platelets responding to adenosine diphosphate. J Ultrastruct Mol Struct Res. 1988;99:254–260. doi: 10.1016/0889-1605(88)90069-9. [DOI] [PubMed] [Google Scholar]
- Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]