Loss of the FPC protein Swi1 or Swi3 results in reduced spore viability, delayed replication, changes in recombination, reduced centromeric cohesion stability, and chromosome missegregation in meiosis I and II. These phenotypes reveal a crucial link between meiotic replication fork stability and chromosome segregation.
Abstract
The replication fork protection complex (FPC) coordinates multiple processes that are crucial for unimpeded passage of the replisome through various barriers and difficult to replicate areas of the genome. We examine the function of Swi1 and Swi3, fission yeast’s primary FPC components, to elucidate how replication fork stability contributes to DNA integrity in meiosis. We report that destabilization of the FPC results in reduced spore viability, delayed replication, changes in recombination, and chromosome missegregation in meiosis I and meiosis II. These phenotypes are linked to accumulation and persistence of DNA damage markers in meiosis and to problems with cohesion stability at the centromere. These findings reveal an important connection between meiotic replication fork stability and chromosome segregation, two processes with major implications to human reproductive health.
INTRODUCTION
Meiosis is a conserved cell differentiation process that reduces ploidy and creates genetic diversity through recombination in sexually reproducing organisms. In contrast to mitosis, wherein progenitor and daughter cells carry the same genetic information, meiosis produces daughters with half the number of parental chromosomes. This is accomplished by two consecutive nuclear divisions that follow one round of DNA replication (reviewed in Zickler and Kleckner, 1999; Cavalier-Smith, 2002; Hochwagen, 2008; Ohkura, 2015). Another characteristic that distinguishes meiosis from mitosis is the requirement for programmed double-strand breaks (prDSBs) following DNA synthesis. These are repaired by recombination between homologous chromosomes. The physical connections established in this process are vital for reciprocal exchange of genetic information and for proper chromosome segregation in most organisms (Keeney et al., 1997; Sharif et al., 2002; Davis and Smith, 2003). Lack of prDSBs is associated with chromosome nondisjunction, which contributes to different forms of aneuploidy (Hirose et al., 2011; Sakuno et al., 2011).
Mono-orientation of sister kinetochores in the reductional meiosis I division (MI) is another key feature of meiosis (Kitajima et al., 2003a; Brar et al., 2006; Sakuno et al., 2009, 2011). Separation of homologues to opposite poles of the cell after anaphase I requires that sister kinetochores are mono-oriented, so that they migrate in the same direction on the meiotic spindle. At the onset of anaphase II, as is the case in mitosis, bioriented kinetochores enable sister chromatids to segregate to opposite poles (Yokobayashi and Watanabe, 2005; Hauf et al., 2007; Sakuno et al., 2009; Kim et al., 2015).
Finally, regulation of sister chromatid cohesion is essential for the different chromosome segregation pathways. Gradual loss of cohesin, first on chromosome arms and then at the centromere, facilitates the onset of MI and MII divisions, respectively (Kitajima et al., 2003a; Rabitsch et al., 2004; Brar et al., 2006). Before MI, the meiosis-specific α-kleisin cohesin subunit Rec8 is targeted for degradation by the endopeptidase separase everywhere along the chromosome except the centromere, where Rec8 is protected by shugoshin-PP2A (Kitajima et al., 2004; Marston et al., 2004; Ishiguro et al., 2010). This stepwise process promotes chiasma resolution (necessary for the separation of homologues in MI), while keeping sister chromatids tethered at the centromere until MII (required for reductional division). When cells near MII, Rec8 is entirely removed from the centromeres, allowing the spindle to pull sister chromatids toward opposite poles (Kitajima et al., 2003a; Brar et al., 2006; Katis et al., 2010).
Replication instability, characterized by the dissociation of replisome components that normally ensure efficient resumption of replication following fork stalling or DNA repair (Errico and Costanzo, 2012), is deleterious to genomic integrity and is associated with cancer in humans (Durkin et al., 2008; Allera-Moreau et al., 2012; Coschi et al., 2014; van der Crabben et al., 2016). When it occurs in meiosis, it can reduce fitness of gametes and negatively affect reproductive health (Baudat et al., 2000; Nudell et al., 2000; Baarends et al., 2001; Gu et al., 2007). Although the effects of abnormal replication have been widely studied in mitosis, less is known about the consequences it has in meiosis. Recent work on replication stability genes in fission and budding yeast, however, has shown a link between abnormal replication and meiotic defects (Dolan et al., 2010; Le et al., 2013; Mastro and Forsburg, 2014; Murakami and Keeney, 2014; Wu and Nurse, 2014).
The replication fork protection complex (FPC) is crucial for stabilizing the replisome during replication in vegetative cells and is particularly important in difficult to replicate areas of the genome or during replication stress (reviewed in Leman and Noguchi, 2012). The FPC is also necessary for preventing fork collapse when replication is arrested or when the replisome encounters severe DNA lesions (Noguchi et al., 2003; Shimmoto et al., 2009). The FPC’s primary components are Swi1/Tof1 (human Timeless) and Swi3/Csm3 (human Tipin), which form a heterodimeric complex with interdependent regulation (Noguchi et al., 2004; Chou and Elledge, 2006; Gotter et al., 2007; Ünsal-Kaçmaz et al., 2007).
Although Tof1 (Swi1’s orthologue) was initially identified through its role in fission yeast’s mating-type switching (Egel et al., 1984), work on budding yeast found that it interacts with topoisomerase 1, suggesting an additional function of the FPC in safeguarding genome integrity (Park and Sternglanz 1999). Further examination of the FPC components in various organisms, including humans, revealed the FPC’s involvement in activating the replication and DNA damage checkpoints upon exposure to genotoxic agents (Murakami and Okayama, 1995; Foss, 2001; Noguchi et al., 2003, 2004; Chou and Elledge, 2006; Gotter et al., 2007; Ünsal-Kaçmaz et al., 2007; Yoshizawa-Sugata and Masai, 2007; Smith et al., 2009; Leman et al., 2010).
Destabilization of the FPC by removing either of its components generates substantial DNA damage. Such damage is thought to arise from the uncoupling of the MCM helicase and the DNA polymerases, which generates large ssDNA gaps that are prone to deleterious rearrangements and physical breakage (Noguchi et al., 2003, 2004; Tourrière et al., 2005; Ünsal-Kaçmaz et al., 2007; Dolan et al., 2010). These observations are consistent with a model in which the FPC acts as a sensor of DNA damage (reviewed in Leman and Noguchi, 2012). In this view, Swi1 stabilizes Swi3 at the fork (Chou and Elledge, 2006; Yoshizawa-Sugata and Masai, 2007). Swi3 senses RPA bound to excess ssDNA (Witosch et al., 2014) and communicates with Mrc1 (Claspin), which relays the message to checkpoint effector kinases (Kumagai and Dunphy, 2000; Chini and Chen, 2003). This process allows cells to maintain genome integrity by properly addressing sources of replication instability and DNA damage.
Significantly, the FPC also functions in the regulation of sister chromatid cohesion, a process intimately linked to DNA replication (Ansbach et al., 2008; Leman et al., 2010). In both yeasts, the FPC genetically interacts or indirectly affects Chl1 (the yeast orthologue of human ChlR1), a helicase important for cohesion (Mayer et al., 2001, 2004; Warren et al., 2004; Xu et al., 2007; Ansbach et al., 2008). Loss of FPC components in frog and fission yeast results in dysfunctional chromosome pairing and premature sister chromatid separation, respectively (Errico et al., 2009; Tanaka et al., 2009). Human cell lines depleted of individual FPC components also show cohesion defects, which is consistent with the observation that the FPC physically interacts with Smc1/3 (Leman et al., 2010).
In this work, we examine how replication fork stability facilitates the proper execution of meiosis. We used cells lacking Swi1 and Swi3 (the FPC components) and examined the outcome of their meioses by various means. We observed that fork destabilization did not halt meiotic progression but resulted in defects in recombination. Moreover, lack of each FPC component resulted in abnormal chromosome segregation in both meiotic divisions, which may explain the substantial reduction in spore viability in these strains. These observations support a model in which the FPC contributes to genome stability in meiosis by suppressing DNA damage and promoting proper centromeric cohesion.
RESULTS
FPC mutants have reduced spore viability
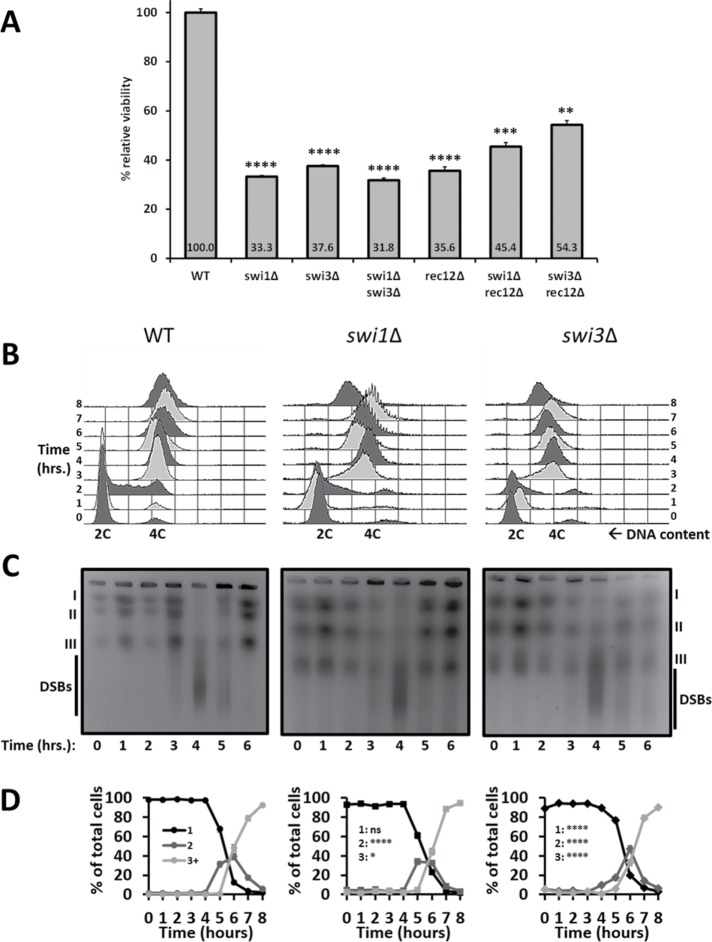
In fission yeast, meiosis is coupled to sporulation (McLeod et al., 1987). Thus defective spores indicate underlying meiotic defects. We crossed heterothallic swi1∆ and swi3∆ strains (FPC mutants) to examine their relative spore viability by random spore analysis. Compared with wild-type cells, the FPC mutants showed a substantial drop in spore viability (Figure 1A). These reductions were similar to those of the swi1∆ swi3∆ and rec12∆ mutants. Intriguingly, loss of Rec12 moderately alleviates the spore viability defects of both swi1∆ and swi3∆ cells. Additionally, plating efficiency revealed no substantial decrease in viability in the FPC and Rec12 mutants compared with wild-type vegetative cells (Figure 1A and Supplemental Figure 1A). Therefore the reduction in spore viability likely reflects meiotic defects.
FIGURE 1:
Spore viability and replication dynamics in FPC mutants. (A) Bulk spore germination of heterothallic cells homozygous for the presence or absence of the FPC and Rec12. Viability was determined by dividing microscope counts of a spore suspension by the resulting colony counts for each strain. Percent figures indicate viability for each genotype and are presented relative to those of the wild-type strain. At least six trials were conducted per genotype (n ≥ 6000). Significance was determined using chi-squared analysis followed by false discovery rate correction for multiple sample comparisons. p values are reported as follows: **, p < 0.0001; ***, p << 0.0001; ****, p <<< 0.0001. Error bars represent 95% confidence intervals. Synchronous meiosis in mat2-102/h- pat1-114/pat1-114 diploids was performed for 8 h. Cells were harvested every hour and examined as indicated in B–D. (B) FACS profiles showing progression of replication through meiotic induction. DNA doubling is denoted as the change from 2C to 4C DNA content. Images are representative of three independent trials. For B–D, hour 0 denotes the time when cells were switched from 25°C to 34°C to elicit meiotic induction. (C) PFGE used to separate whole chromosomes by size and to assess formation and repair of meiotic DSBs. Smears migrating faster than chromosome III represent DSBs. Images of ethidium bromide–stained agarose gels are representative of three different trials. (D) DAPI-stained nuclei were counted for each time point to ascertain progression through meiotic divisions, which are reported as follows: black stands for 1 nucleus, dark gray for 2 nuclei, and light gray for 3 or more nuclei (3+); 2 and 3+ nuclei indicate onset of MI and MII, respectively. n ≥ 900 cells per genotype. Significance was determined using a chi-squared test for trend. p values are reported as follows: ns, not significant; *, p < 0.05; ****, p < 0.0001. Error bars are 95% confidence intervals.
FPC mutants do not impede DNA replication
Because the FPC is necessary for proper replication fork progression (Noguchi et al., 2003, 2004), we asked whether the observed meiotic defects in the swi1∆ and swi3∆ strains were related to defects in meiotic S phase (meiS phase). To explore this, we induced synchronous meiosis by trapping cells in G1 through nitrogen starvation and by heat inactivation of Pat1, a negative meiotic regulator. Increasing the temperature to 34°C causes either haploid or diploid cells carrying the pat1-114 allele to enter meiosis. To avoid any confounding effects associated with meiotic induction of haploid cells (Yamamoto and Hiraoka, 2003; Pankratz and Forsburg, 2005), we generated stable h-/mat2-102 pat1-114/pat1-114 diploids in swi1∆ and swi3∆ backgrounds that could enter meiosis only by temperature shift (Pankratz and Forsburg, 2005; Le et al., 2013). We harvested cells over the course of 8 h and examined replication dynamics, DSB formation and repair, and meiotic progression. We monitored completion of the meiS phase by measuring DNA content changes with fluorescence-activated cell sorting (FACS) (reviewed in Sabatinos and Forsburg, 2009).
Wild-type cells finished bulk DNA replication 3 h after meiotic induction, while the FPC mutants exhibited a broadened peak at that time point, which indicates a modest delay in DNA synthesis (Figure 1B). After 4 h, both FPC mutants finished replication similarly to the wild-type diploid. Although rec12∆ cells also showed a slight delay in replication by hour 3, the rec12∆ swi1∆ and rec12∆ swi3∆ mutants did not entirely finish meiS by hour 6 (Supplemental Figure 1B). These observations indicate that the FPC is mostly dispensable for replication progression in an unperturbed meiS phase, except when Rec12 is removed, in which case DNA synthesis is substantially delayed.
Even though fork destabilization did not impede meiS phase progression, it is possible that programmed DSB formation and repair, which follow replication, could be affected. To address this, we carried out pulse-field gel electrophoresis (PFGE) to separate chromosomes and the signature smears of DSB generation by size (Young et al., 2004). In wild-type cells and the FPC mutants, DSBs were initiated 3 h after induction. By hour 4, the majority of DSBs were created, as evidenced by a strong signal beneath chromosome III. DSB resolution occurred in hour 5 with a concomitant return of chromosome signals. By hour 6, all DSBs were repaired, which is consistent with full meiotic entry (Figure 1C and Supplemental Figure 2A). In the absence of Rec12, cells predictably failed to create any DSBs (Cervantes et al., 2000; Ogino et al., 2006). By contrast, rec12∆ swi1∆ and rec12∆ swi3∆ cells exhibited fast-migrating smears at all time points. This agrees with a constitutive level of DNA damage or DNA breaks and corroborates previous observations involving rec12∆ and other replication stability mutants (Dolan et al., 2010; Le et al., 2013) (Supplemental Figures 1C and 2B). These results imply that the FPC is not required for proper induction and repair of prDSBs during meiosis.
Because fork destabilization did not significantly disrupt DSB dynamics (Figure 1C and Supplemental Figure 2A), we asked whether the same was true for meiotic progression. We stained nuclei with 4′,6-diamidino-2-phenylindole (DAPI) and scored for changes in the number of DAPI-stained nuclei over the course of meiosis (Pankratz and Forsburg, 2005; Le et al., 2013). Wild-type cells and the FPC mutants showed a single DAPI mass until hour 4 and began to show two nuclei by hour 5, when cells enter MI. From hour 6 onward, cells continued to divide their nuclei, until most cells showed three or more DAPI masses, indicating completion of MII (Figure 1D). However, relative to the wild-type strain, swi3∆ and swi1∆ cells correspondingly showed delayed-MI and early-MII entry. Thus, although swi1∆, but not swi3∆, cells show similar kinetics to the wild-type strain during metaphase I, both FPC mutants are significantly different from wild-type during both meiotic divisions (Figure 1D and Supplemental Figure 5). Moreover, the rec12∆ and FPC rec12∆ mutants also progressed unimpeded through meiosis. Interestingly, compared with the rec12∆ strain, rec12∆ swi3∆ cells showed delayed MI entry, while both FPC rec12∆ mutants entered MII late (Supplemental Figure 1D). This reduced efficiency in MII dynamics is consistent with similar observations in other replication mutants (Davis and Smith, 2003). These results suggest that fission yeast cells complete meiosis even when fork integrity is compromised, albeit inefficiently in the absence of either FPC component with or without Rec12. Taken together, these observations indicate that destabilizing the fork by eliminating Swi1 or Swi3 is associated with modest defects in meiotic prophase I that may disproportionately affect downstream events.
FPC mutants have persistent DNA damage
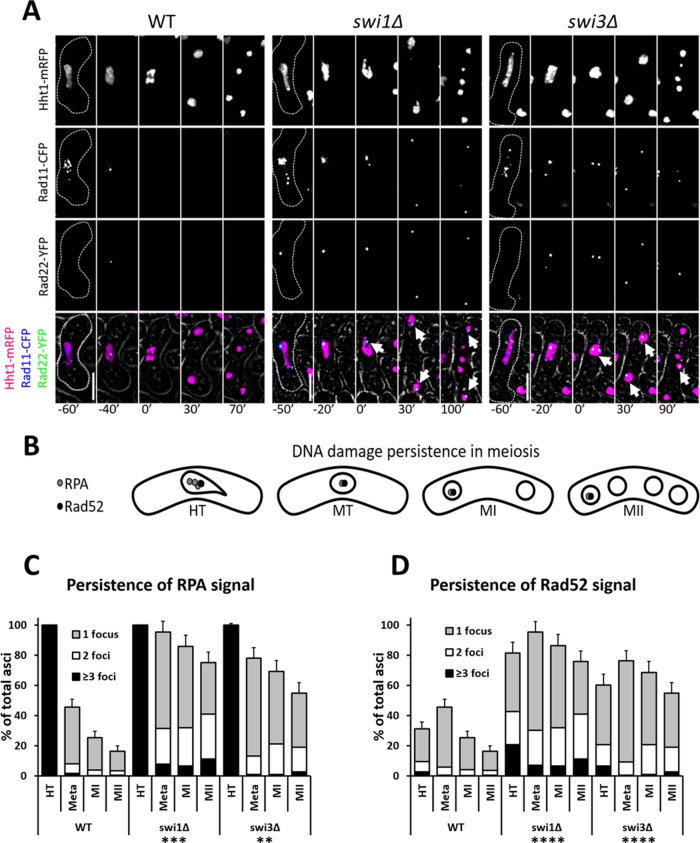
In vegetative cells, absence of the FPC components results in DNA damage accumulation following replication (Noguchi et al., 2004). We asked whether the same was true in meiS phase, and whether such damage persisted through meiosis. We used cells in which histone H3, RPA, and Rad52 (fission yeast’s Hht1, Rad11, and Rad22, respectively) were fluorescently tagged and followed their meioses by live-cell microscopy (Lisby et al., 2004; Sabatinos et al., 2012; Mastro and Forsburg, 2014) (Figure 2, A and B).
FIGURE 2:
Persistence of DNA damage in FPC mutants. (A) Live-cell images of meiotic cells carrying fluorescently tagged histone3 (Hht1-mRFP), RPA (Rad11-CFP), and Rad52 (Rad22-YFP). Panels are shown for individual fluorescent channels. Bright-field panels are omitted due to space limitations. A false-color image is rendered to show merged signals. Dotted cell outlines are overlaid on panels for easier visualization of meiotic cells. Images that show the characteristics of each reported mutant phenotype were used. Selected time frames were chosen for optimal representation of nuclear dynamics. The analyzed meiotic time window encompasses HT, metaphase (MT), MI, and MII. Upright scale bars: 5 µm. (B) Cartoon depicting DNA damage persistence through meiosis in FPC mutants. Gray and black dots represent RPA and Rad52, respectively. (C, D) Quantification of RPA and Rad52 signals in cells from microscopy pictures shown in A. More than 150 cells were scored from at least two independent movies for each genotype. Chi-squared analysis was used to determine significance. p values are reported thus: **, p < 0.01; ***, p < 0.001; ****, p << 0.0001. Error bars represent 95% confidence intervals. RPA and Rad52 signals are binned as follows: light-gray for 1 focus, white for 2 foci, and black for more than 3 foci.
Wild-type cells exhibited RPA signals for most of horse tailing (HT), while a third of cells showed Rad52 signals in late HT, where nuclear oscillation ceases and metaphase I nuclear contraction begins (Figure 2, A–D). This agrees with the timing of chromosome pairing and recombination that ensues after DNA synthesis. When cells reached metaphase I, RPA signals began to decrease, but Rad52 signals showed a transient increase (Figure 2, A–D). This is an expected outcome during the period when most recombination is finalized. As cells entered MI, RPA and Rad52 signals disappeared in most cells and were mainly absent by the time MII was completed (Figure 2, A–D).
By contrast, we observed in the FPC mutants a higher number of cells with DNA damage signals that persisted through meiosis (Figure 2, A–D). In the swi1∆ and swi3∆ strains, the proportion of cells showing RPA foci remained relatively high through the end of MII (Figure 2, A–D). During metaphase I, there was an increase in cells with Rad52 foci, but this fraction gradually decreased in MI and MII. However, compared with the wild-type strain, the FPC mutants showed higher proportions of cells with Rad52 foci at the end of meiosis (Figure 2, A–D). While persistence of DNA damage signals was similar in both FPC mutants, the effect was more pronounced in swi1∆ cells. Three important differences could thus be appreciated between the FPC mutants and wild-type cells: the number of DNA damage signals was greater, more persistent, and at more loci, as evidenced by the larger group of cells showing multiple RPA and Rad52 foci during and after metaphase I. These results reveal that, upon fork destabilization, DNA damage accumulates and remains mostly unrepaired through meiosis, which may negatively affect events important for meiotic segregation.
FPC mutants alter homologous recombination
Rad52 is required to facilitate inter- and intrasister recombination between direct repeats (Muris et al., 1997; van den Bosch et al., 2001; Octobre et al., 2008). Because we observed abnormal Rad52 signals before and during meiosis upon fork destabilization, we asked whether normal recombination dynamics between homologues were affected. To address this question, we made crosses between parents carrying a pair of linked nutritional markers in coupling (++/– –) or in repulsion (+−/−+). We assessed homologous recombination by scoring colony formation on solid media that selected for each tested nutritional outcome. Relative to wild-type cells, the FPC mutants showed 1.2- to 2.8-fold overall decrease in recombination at four distinct intergenic intervals (Table 1). We also assessed intragenic recombination at an ade6 locus containing a recombination hot spot (Schuchert and Kohli, 1988; Szankasi and Smith, 1995; Fleck et al., 1999). In this assay, a similar reduction in recombination was observed, but was especially pronounced in the absence of Swi3.
TABLE 1:
Recombination dynamics in FPC mutants with intergenic and intragenic recombination data given in the top panels and sister chromatid recombination data given in the bottom panel.
| Genetic distance (cM)a | Fold reductionb | p valuec | ||||||
|---|---|---|---|---|---|---|---|---|
| Intergenic interval | Chr | WT | swi1∆ | swi3∆ | swi1∆ | swi3∆ | swi1∆ | swi3∆ |
| his4-lys4d | 2 | 6.81 ± 0.92 | 4.81 ± 0.78 | 2.43 ± 0.83 | 1.42 | 2.80 | 0.0039 | p << 1E-04 |
| leu2-ura2d | 1 | 0.87 ± 0.13 | 0.67 ± 0.20 | 0.40 ± 0.24 | 1.30 | 2.18 | 0.1164 | 0.0051 |
| leu1-his7d | 2 | 2.28 ± 0.41 | 1.08 ± 0.34 | 1.21 ± 0.20 | 2.11 | 1.88 | 0.0004 | 0.0005 |
| his3-leu1e | 2 | 1.13 ± 0.22 | 0.55 ± 0.08 | 0.92 ± 0.08 | 2.05 | 1.23 | 0.0008 | 0.1078 |
| Intragenic intervalf | Chr | Frequency (×10−3) | Fold reductionb | p valueg | ||||
|---|---|---|---|---|---|---|---|---|
| ade6-M26-52 | 3 | 3.34 ± 0.08 (60/17987) | 1.70 ± 0.05 (52/31129) | 0.60 ± 0.04 (7/11838) | 1.96 | 5.57 | 0.0009 | P << 1E-04 |
| Frequency (×10−3) | Fold increaseh | p valueg | ||||||
|---|---|---|---|---|---|---|---|---|
| SCE intervalf | Chr | WT | swi1∆ | swi3∆ | swi1∆ | swi3∆ | swi1∆ | swi3∆ |
| ade6-L469-his3+-ade6-M375/ade6-M375-M210 | 3 | 5.00 ± 0.08 (167/33465) | 16.81 ± 0.24 (186/11064) | 9.13 ± 0.14 (155/16982) | 3.36 | 1.83 | P << 1E-04 | p << 1E-04 |
Significance for genetic distance was determined using a two-tailed Student’s t test. Chi-squared analysis was employed for frequencies. At least five independent trials were carried out per genotype. Error margins represent 95% confidence intervals. Fold change values are relative to wild-type outcomes. At least five independent trials were performed per genotype. Chr, chromosome; WT, wild type.
aHaldane’s formula cM = −50 ln(1−2R) was used to convert recombinant frequencies (R) into centimorgans (cM).
bWild-type cM or frequency values were divided by those of mutants to determine reduction in genetic distance or recombination frequency.
cSignificance calculated by two-tailed student’s t test.
dMarkers in interval were scored in repulsion (+−/−+).
eMarkers in interval were scored in coupling (++/– –).
fFrequency values were calculated by dividing number of Ade+ by total number of colonies screened (shown in parentheses).
gSignificance calculated by chi-squared analysis.
hFrequency values of mutants were divided by those of the wild type to determine increase in recombination frequency.
Furthermore, because Rad52 antagonizes Dmc1, a RecA-like protein important for homologous recombination (Octobre et al., 2008; Murayama et al., 2013), we examined whether dmc1∆ cells recapitulated the spore viability and recombination outcomes of both FPC mutants. While reduction in spore viability was moderate compared with wild-type cells, the dmc1∆ mutant showed a 1.5-fold decrease in recombination at the his4-lys4 intergenic interval similar to that of the swi1∆ mutant (Supplemental Table 1).
This observation prompted us to ask whether fork destabilization shifted recombination dynamics to favor sister chromatids over homologues as recombination templates. For this, we set up a cross that produced Ade+ offspring only if one of the parent cells performed intra- or intersister recombination during meiosis (Osman et al., 1996; Catlett and Forsburg, 2003; Mastro and Forsburg, 2014) (Table 1). Interestingly, in the absence of Swi1 and Swi3, there was a noticeable two- to threefold increase in sister chromatid exchange (Table 1). These results indicate that fork destabilization alters normal recombination dynamics, potentially reducing chiasmata and thereby affecting meiotic division.
FPC mutants disrupt chromosome segregation
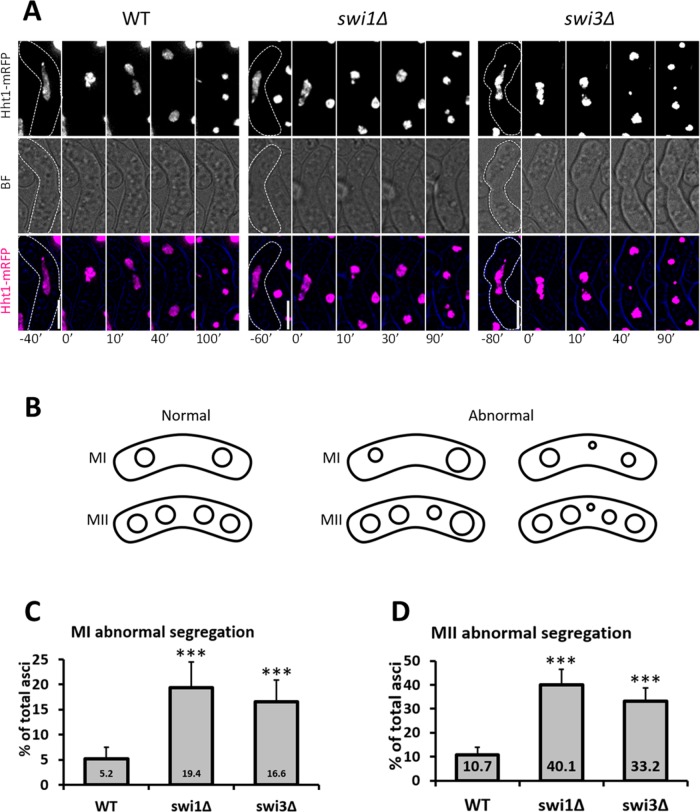
Chromosome segregation is sensitive to changes in recombination (Watanabe and Nurse, 1999; Murakami and Nurse, 2001; Sakuno et al., 2011; Mastro and Forsburg 2014). Thus we asked whether fork destabilization resulted in abnormal nuclear division in MI and MII. We used cells with fluorescently tagged histones (Htt1-mRFP [monomeric red fluorescent protein]) and followed the outcome of meiosis by live-cell microscopy (Figure 3, A and B) (Cooper et al., 1998; Chikashige and Hiraoka, 2001; Klutstein et al., 2015). Whereas few wild-type cells showed abnormal nuclear masses after MI and MII (Figure 3, C and D), the FPC mutants revealed three- to fourfold increases in cells containing asymmetric nuclei and nuclei with extra DNA signal (Figure 3, C and D). Moreover, in the absence of Swi1 and Swi3, cells exhibited two- to fourfold increases in lagging chromosomes following MI and MII (Supplemental Figure 3, A–C). Although lagging chromosomes do not always result in abnormally sized nuclei, increases in this phenotype may indicate underlying division defects (Yokobayashi and Watanabe, 2005). These observations therefore suggest that the FPC is important for proper chromosome segregation in meiosis.
FIGURE 3:
Meiotic segregation in FPC mutants. (A) Live-cell images of meiotic cells carrying fluorescently tagged histone3 (Hht1-mRFP). Panels are shown for a fluorescent channel and bright field. A false-color image is rendered to show merged signals. Dotted cell outlines are overlaid on panels for easier visualization of meiotic cells. Images that show the characteristics of each reported mutant phenotype were used. Displayed time frames were chosen for optimal representation of nuclear dynamics. The meiotic phases shown are HT, metaphase (MT), MI, and MII. Minute 0 (0′) denotes the last nuclear mass contraction in metaphase I before homologous chromosomes separate in anaphase I (10′). Upright scale bars: 5 µm. (B) Cartoon depicting the products of normal and abnormal divisions in MI and MII. (C, D) Quantification of abnormally segregated asci from microscopy images shown in A. More than 150 cells were scored from at least two independent movies for each genotype. Chi-squared analysis was used to determine significance. p values are reported as follows: ***, p < 0.001. Error bars represent 95% confidence intervals.
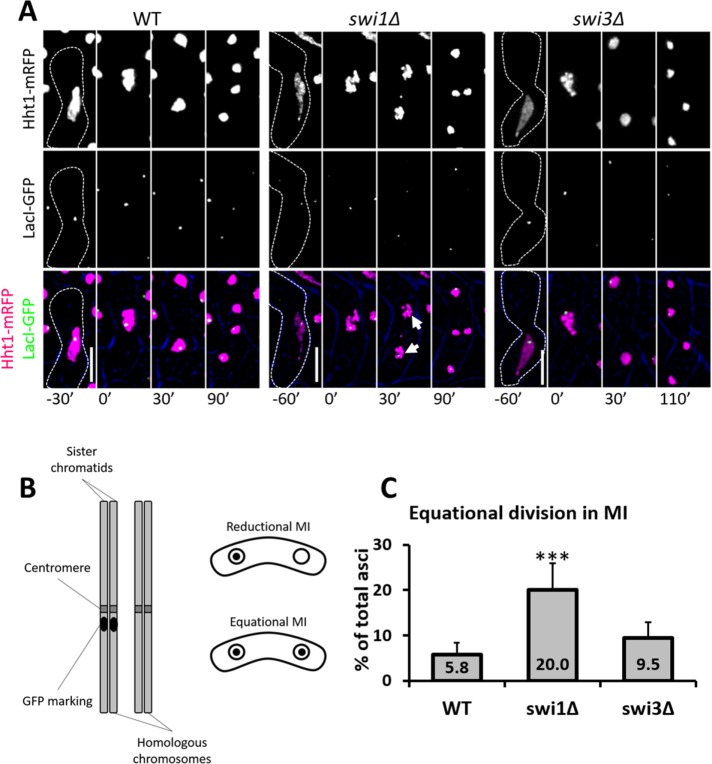
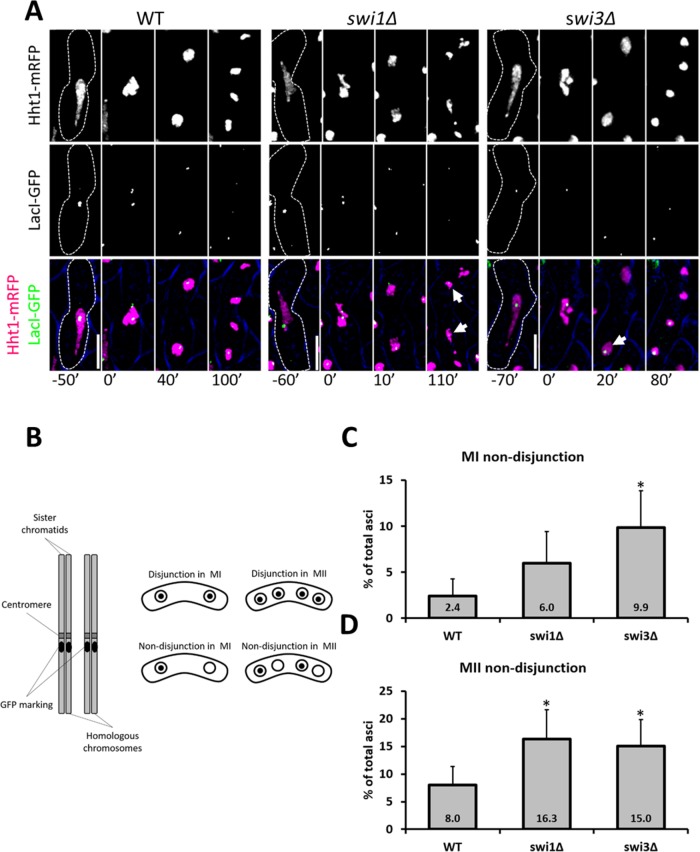
Premature sister chromatid separation or chromosome nondisjunction could explain the missegregation phenotypes observed in the FPC mutants (Yokobayashi and Watanabe, 2005; Hauf et al., 2007; Dudas et al., 2011). To address the first possibility, we crossed cells heterozygous for a green fluorescent protein (GFP) marker integrated at the lys1 locus (near the centromere of chromosome I) and carrying histones fluorescently tagged as indicated earlier. If MI division occurs normally (i.e., reductional division), GFP signal is distributed to one of the two daughter nuclei. However, if equational division occurs (i.e., abnormal MI division), both nuclear spots show GFP signals (Yokobayashi and Watanabe, 2005; Hauf et al., 2007; Hirose et al., 2011) (Figure 4, A and B). Compared with the wild-type strain, swi1∆, but not swi3∆, cells showed a threefold increase in equational division (Figure 4C). This result implies that Swi1 helps to prevent precocious separation of sister chromatids, possibly by contributing to correct monopolar attachment, as was reported previously for cells lacking both Mrc1 (a secondary FPC factor) and Rec12 (Hirose et al., 2011).
FIGURE 4:
MI equational division in FPC mutants. (A) Live-cell images of meiotic cells carrying fluorescently tagged histone3 (Hht1-mRFP) and LacI-GFP on a LacO repeat at lys1 near the centromere of chromosome I. Panels are divided into individual fluorescent channels. Bright-field panels are omitted due to space limitations. A false-color image was generated to show merged signals. Dotted cell outlines are overlaid on panels for easier visualization of meiotic cells. Images that show the characteristics of each reported mutant phenotype were used. Displayed time frames were chosen for optimal representation of nuclear dynamics. The meiotic phases shown are HT, metaphase (MT), MI, and MII. Minute 0 (0′) denotes the last nuclear mass contraction in metaphase I before homologous chromosomes separate in anaphase I. Upright scale bars: 5 µm. (B) Cartoon depicting a pair of homologous chromosomes where one is marked with GFP near the centromeres. Symmetrical distribution of GFP indicates equational division. (C) Quantification of cells showing equational division in MI. More than 150 cells were scored from at least two independent movies for each genotype. Chi-squared analysis was used to determine significance. p values are reported as follows: ***, p < 0.001. Error bars represent 95% confidence intervals.
To examine whether fork destabilization is associated with chromosome nondisjunction, we used a similar strategy, but in a cross homozygous for the GFP marker at lys1. In each meiotic division, if GFP dots are distributed symmetrically, normal division is inferred to have occurred, whereas asymmetric distribution indicates abnormal division (Yokobayashi and Watanabe, 2005; Hauf et al., 2007; Hirose et al., 2011) (Figure 5, A and B). While only swi3∆ cells show elevated nondisjunction in MI (Figure 5C), both FPC mutants show two- to fourfold increases in nondisjunction of sister chromatids in MII (Figure 5D). These results suggest that Swi3 is necessary for proper homologue segregation in MI, while both FPC components contribute to normal sister chromatid separation in MII.
FIGURE 5:
Nondisjunction in MI and MII in FPC mutants. (A) Live-cell images of meiotic cells carrying fluorescently tagged histone3 (Hht1-mRFP) and LacI-GFP on a LacO repeat at lys1 near the centromere of chromosome I. Panels are divided into individual fluorescent channels. Bright-field panels are omitted due to space limitations. A false-color image is provided to show merged signals. Dotted cell outlines are overlaid on panels for easier visualization of meiotic cells. Images that show the characteristics of each reported mutant phenotype were used. Displayed time frames were chosen for optimal representation of nuclear dynamics. The meiotic phases shown are HT, metaphase (MT), MI, and MII. Minute 0 (0′) denotes the last nuclear mass contraction in metaphase I before homologous chromosomes separate in anaphase I (10′). Upright scale bars: 5 µm. (B) Cartoon depicting a pair of homologous chromosomes marked with GFP near the centromeres. Asymmetric distribution of GFP indicates nondisjunction events. (C, D) Quantification of cells showing chromosome nondisjunction in MI and MII. More than 150 cells were scored from at least two independent movies for each genotype. Chi-squared analysis was used to determine significance. p values are reported as follows: *, p < 0.05. Error bars represent 95% confidence intervals.
FPC mutants destabilize centromeric cohesion
Some of the missegregation phenotypes associated with fork destabilization can be explained by the effects of reduced homologous recombination (Dudas et al., 2011). However, problems with centromeric cohesion are also associated with abnormal meiotic divisions, as shown previously in Rec8 mutants (Watanabe and Nurse, 1999). We examined dynamics associated with meiotic cohesion using live-cell microscopy. Wild-type and mutant cells carried fluorescently tagged histones and either Rec27-GFP (a component of the linear elements), Sgo1-GFP (shugoshin, a centromeric cohesion protector), or Rec8-GFP (the α-kleisin subunit of meiotic cohesin). Each of these proteins is associated with different aspects of cohesin function and regulation (Watanabe and Nurse, 1999; Kitajima et al., 2004, 2006; Davis et al., 2008).
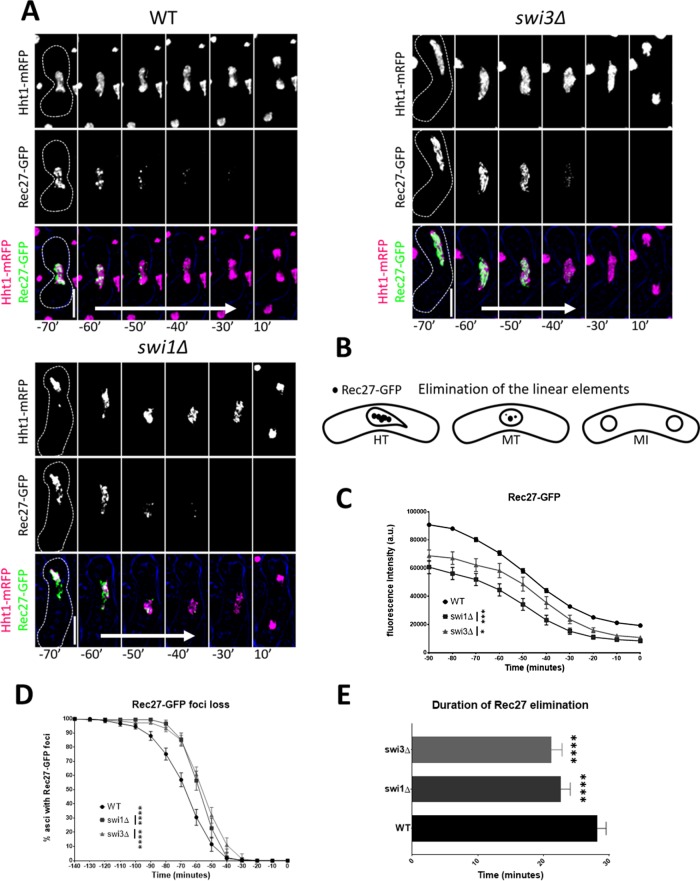
We reasoned that any changes to the temporal dynamics of Rec27, Sgo1, and Rec8 upon fork destabilization would indicate cohesion-related sources of chromosome missegregation. Casein kinase 1 phosphorylation of the cohesin subunit Rec11 (SA3) facilitates loading of the linear element proteins Rec10 and Rec27 along chromosomal axes, which in turn promotes the alignment of homologous chromosomes, Rec12-dependent DNA breakage, and recombination (Ellermeier and Smith, 2005; Davis et al., 2008; Phadnis et al., 2015; Sakuno and Watanabe, 2015). For the examination of Rec27, Sgo1, and Rec8, we measured GFP fluorescence intensity and scored for the disappearance of GFP foci. This complementary approach provides information about GFP signal abundance and GFP organization as cells proceed from late prophase I to just before MI (designated as time point 0) or from late metaphase I to MII in the case of Rec8. The FPC mutants showed 20–30% lower Rec27-GFP intensity before meiosis, and Rec27-GFP foci elimination was 5–7 min faster and occurred 5 min later in metaphase I than in wild-type cells (Figure 6, A–C, and Supplemental Figure 4, A and B). These Rec27 defects are suggestive of changes to the structural integrity of chromosome axes that may affect homologue pairing and recombination. They may also indicate changes to cohesin stability in meiosis.
FIGURE 6:
Elimination of the linear elements in FPC mutants. (A) Live-cell images of meiotic cells carrying fluorescently tagged histone3 (Hht1-mRFP) and Rec27-GFP. Panels are divided into individual fluorescent channels. Bright-field panels are omitted due to space limitations. A false-color image was rendered to show merged signals. Dotted cell outlines are overlaid on panels for easier visualization of meiotic cells. Images that show the characteristics of each reported mutant phenotype were used. Displayed time frames were chosen for optimal representation of nuclear dynamics. The meiotic phases shown are metaphase (MT) and MI. Time preceding the last nuclear mass contraction in metaphase I (0′) before homologous chromosomes separate in anaphase I (10′) is denoted with a negative sign (e.g., −10′). Time 0 is omitted due to space limitations. Upright scale bars: 5 µm. White arrows represent average span of Rec27-GFP elimination. (B) Cartoon depicting elimination of Rec27-GFP during metaphase and before MI. (C) Quantification of Rec27-GFP fluorescence. (D) Quantification Rec27-GFP foci elimination. (E) Duration of Rec27-GFP foci elimination. More than 150 cells were scored from at least two independent movies for each genotype. One-way ANOVA followed by Tukey family-wise comparison was used to determine significance in C and E. Chi-squared analysis was used for D. p values are reported as follows: *, p < 0.05; ***, p < 0.001; ****, p << 0.0001. Error bars represent 95% confidence intervals.
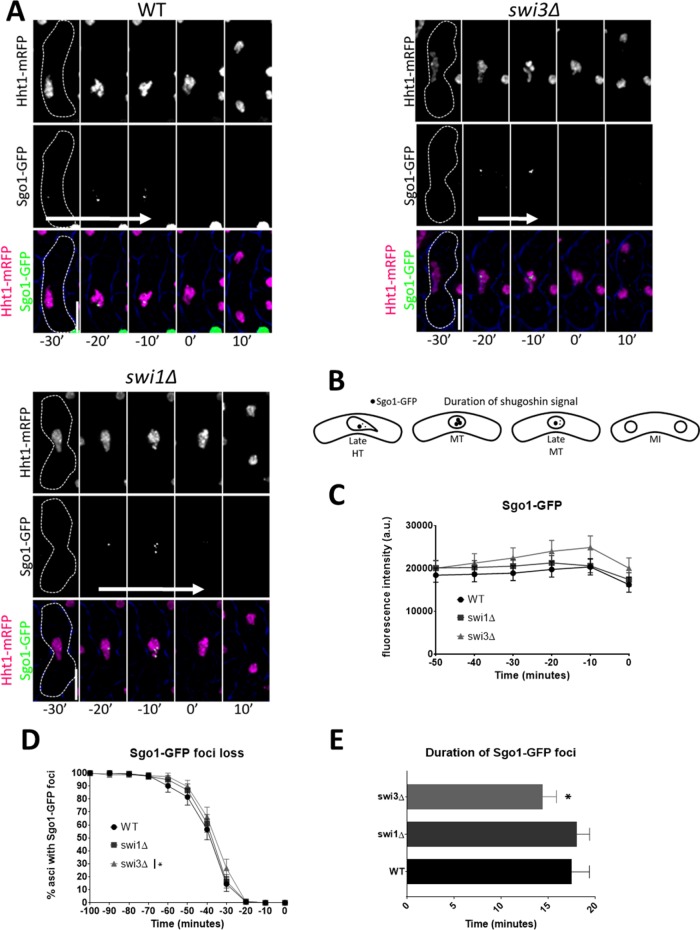
In fission yeast, Sgo1 and PP2A protect centromeric cohesin from separase degradation before anaphase I (Kitajima et al., 2004; Riedel et al., 2006; Ishiguro et al., 2010). Given that swi1∆ cells show increased equational division in MI, which is linked to issues with centromeric cohesin stability (Yokobayashi and Watanabe, 2005; Hauf et al., 2007), we asked whether fork destabilization affects the temporal dynamics of Sgo1 (Figure 7, A–B). Relative Sgo1-GFP signal intensity was similar across genotypes. Intriguingly, we observed that swi3∆, but not the swi1∆ mutant, exhibited a 3-min reduction in the duration of Sgo1-GFP foci compared with wild-type cells (Figure 7, A–D). However, without Swi1, Sgo1-GFP dissipates 2 min later than was observed in the wild-type strain (Supplemental Figure 4, C and D). These results reveal the FPC is necessary for temporal stability of Sgo1 at the centromere and further highlight the separate functions of the FPC components.
FIGURE 7:
Duration of Sgo1 in metaphase in FPC mutants. (A) Live-cell images of meiotic cells carrying fluorescently tagged histone3 (Hht1-mRFP) and Sgo1-GFP. Panels are divided into individual fluorescent channels. Bright-field panels are omitted due to space limitations. A false-color image was generated to show merged signals. Dotted cell outlines are overlaid on panels for easier visualization of meiotic cells. Images that show the characteristics of each reported mutant phenotype were used. Displayed time frames were chosen for optimal representation of nuclear dynamics. The meiotic phases shown are HT, metaphase (MT), and MI. Time preceding the last nuclear mass contraction in metaphase I (0′) before homologous chromosomes separate in anaphase I (10′) is denoted with a negative sign (e.g., −10′). Upright scale bars: 5 µm. White arrows indicate average span of Sgo1-GFP signal. (B) Cartoon depicting duration of Sgo1-GFP signal during metaphase and before MI. (C) Quantification of Sgo1-GFP fluorescence. (D) Quantification of Sgo1-GFP foci in metaphase I. (E) Duration of Sgo1-GFP foci. More than 150 cells were scored from at least two independent movies for each genotype. One-way ANOVA followed by Tukey family-wise comparison was used to determine significance in C and E. Chi-squared analysis was used for D. p values are reported as follows: *, p < 0.05. Error bars represent 95% confidence intervals.
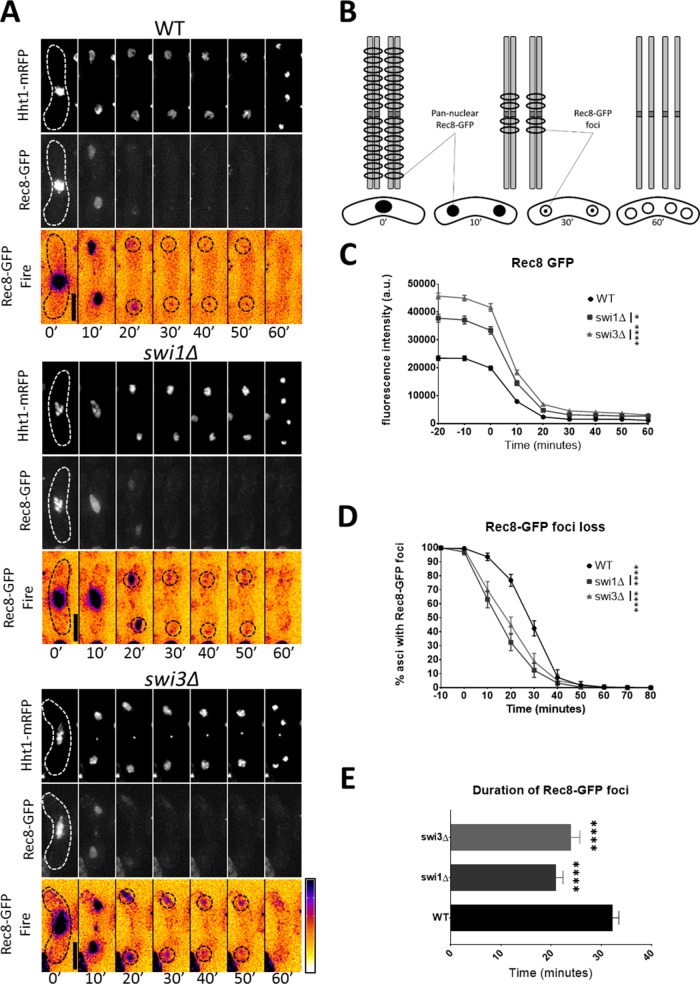
Rec8 is a subunit of cohesin that helps keep sister chromatids together following meiS phase (Watanabe and Nurse, 1999; Watanabe et al., 2001). During anaphase I, Rec8 is removed from chromosome arms, which facilitates chiasma resolution and segregation of homologous chromosomes (Buonomo et al., 2000; Kitajima et al., 2003b; Brar et al., 2006). Nevertheless, Rec8 is retained at the centromere until right before anaphase II to ensure correct segregation of sister chromatids (Kitajima et al., 2003b, 2004; Riedel et al., 2006). In addition, Rec8 and Moa1 facilitate mono-orientation of kinetochores in MI, thus promoting segregation of homologous chromosomes to opposite poles of the cell (Yokobayashi and Watanabe, 2005; Hauf et al., 2007). Because lack of Swi1 and Swi3 is associated with abnormal MI division and anomalous Sgo1 timing before anaphase I, we asked whether fork destabilization would disrupt the temporal dynamics of centromeric cohesion. For this, we used live-cell imaging to follow Rec8-GFP from late metaphase I to MII (Figure 8, A and B, and Supplemental Figure 4G).
FIGURE 8:
Persistence of centromeric cohesion in FPC mutants. (A) Live-cell images of meiotic cells carrying fluorescently tagged histone3 (Hht1-mRFP) and Rec8-GFP. Panels are divided into individual fluorescent channels. Bright-field panels are omitted due to space limitations. A false-color image was generated using ImageJ’s Fire LUT to better show intensity of GFP signals. Dotted cell outlines are overlaid on panels for easier visualization of meiotic cells. Images that show the characteristics of each reported mutant phenotype were used. Displayed time frames were chosen for optimal representation of nuclear dynamics. The meiotic phases shown are metaphase (MT), MI, and MII. Minute 0 (0′) denotes the last nuclear mass contraction in metaphase I before homologous chromosomes separate in anaphase I (10′). Upright scale bars: 5 µm. (B) Cartoon depicting gradual elimination of Rec8, first at chromosome arms and then at the centromere. Rec8 dynamics are shown in the context of MI and MII division. (C) Quantification of Rec8-GFP fluorescence from metaphase I to MII. (D) Quantification of Rec8-GFP foci loss before MII. (E) Duration of Rec8-GFP foci loss. More than 150 cells were scored from at least two independent movies for each genotype. One-way ANOVA followed by Tukey family-wise comparison was used to determine significance in C and E. Chi-squared analysis was used for D. p values are reported as follows: *, p < 0.05; ****, p << 0.0001. Error bars represent 95% confidence intervals.
In wild-type cells, Rec8-GFP signal goes from pan-nuclear (observed in late metaphase I to early anaphase) to a pair of foci that localize to the centromeres and generally persist for more than half an hour (Figure 8, A, B, and E). This signal pattern following MI corresponds to Rec8 elimination at chromosome arms, persistence at the centromere, and degradation before MII (Watanabe and Nurse, 1999; Kitajima et al., 2003b; Le et al., 2013) (Figure 8B). In the FPC mutants, Rec8-GFP signal intensity was higher, but persistence of centromeric cohesion was 8–10 min shorter, and Rec8-GFP foci loss occurred 12–15 min earlier than in wild-type cells (Figure 8, C–E, and Supplemental Figure 4, F and G). This result agrees with the general observation 10 min after MI that Rec8-GFP signal remains as centromeric foci in the wild-type strain but dissociates throughout the nucleus in the FPC mutants (Figure 8, A–E, and Supplemental Figure 4, F and G).
We next investigated whether Rec8 localization at the centromere is affected over the period that encompasses the MI to MII transition, when centromeric Rec8 is protected to keep sister chromatids together until anaphase II. Cells were synchronized in G1 phase of the cell cycle by nitrogen starvation, and meiosis was induced by Pat1 inactivation. We then performed chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR). Although the FPC mutants and wild-type cells were statistically similar, fork destabilization consistently led to modest reductions in centromeric Rec8 before and after MI (Supplemental Figure 4E). These results, along with the observation that sister chromatids in the FPC mutants are left untethered at the centromeres longer than in wild-type cells (Supplemental Figure 4, F and G), indicate that fork destabilization accelerates elimination of centromeric cohesion, which not only precipitates early separation of sister chromatids in MI but also contributes to their abnormal segregation in MII.
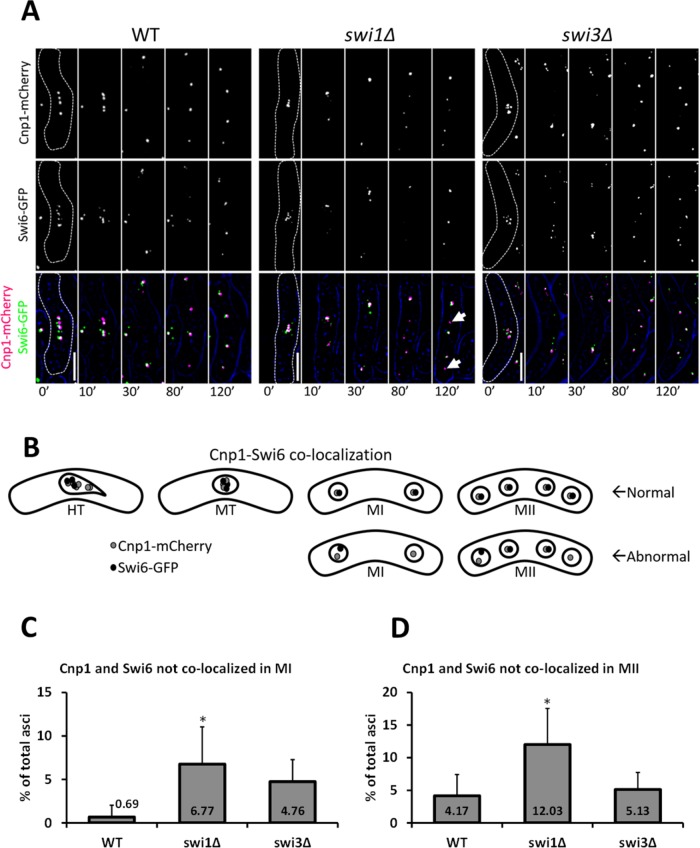
Swi6 (the fission yeast homologue of human HP1) is essential for heterochromatin formation and for the recruitment of cohesin and Sgo1 to the centromere (Ekwall et al., 1995; Nonaka et al., 2002; Kitajima et al., 2003b; Kawashima et al., 2007; Yamagishi et al., 2008). To further corroborate the centromeric cohesion defects thus far observed, we asked whether fork destabilization impacts centromeric localization of Swi6. We used live-cell microscopy to monitor colocalization of Cnp1-mCherry (the fission yeast homologue of mammalian CENP-A, a centromere-specific histone variant; Takahashi et al., 2000) and Swi6-GFP following MI and MII (Figure 9, A and B). We reasoned that colocalization dynamics of these proteins after each division should reflect the overall ability of cells to mobilize heterochromatin back to the centromere. We observed that swi1∆, but not swi3∆ or wild-type cells, had a significant three- to 10-fold increase in cells where Cnp1 and Swi6 did not colocalize after each meiotic division (Figure 9, C and D). Although the effect is statistically modest, it nonetheless suggests Swi1 is necessary for correct recruitment of heterochromatin to the centromere.
FIGURE 9:
Recruitment of heterochromatin to the centromere in FPC mutants. (A) Live-cell images of meiotic cells carrying fluorescently tagged Cnp1 and Swi6. Panels are divided into individual fluorescent channels. Bright-field panels are omitted due to space limitations. A false-color image was generated to show merged signals. Dotted cell outlines are overlaid on panels for easier visualization of meiotic cells. Images that show the characteristics of each reported mutant phenotype were used. Displayed time frames were chosen for optimal representation of nuclear dynamics. The meiotic phases shown are metaphase (MT), MI, and MII. Minute 0 (0′) denotes the last nuclear mass contraction in metaphase I before homologous chromosomes separate in anaphase I (10′). Upright scale bars: 5 µm. (B) Cartoon depicting localization of Cnp1-mCherry and Swi6-GFP in metaphase, MI, and MII. (C, D) Quantification of cells showing Cnp1-mCherry and Swi6-GFP dots not colocalized in both meiotic divisions. More than 130 cells were scored from at least two independent movies for each genotype. Chi-squared analysis was used to determine significance. p values are reported as follows: *, p < 0.05. Error bars represent 95% confidence intervals.
DISCUSSION
The FPC travels with the replisome and is important for coupling the MCM helicase with the DNA polymerases (Tourrière et al., 2005; Ünsal-Kaçmaz et al., 2007; Leman et al., 2010; Leman and Noguchi, 2012). This function helps prevent excessive unwinding and ssDNA accumulation during replication stress. Areas with extended ssDNA segments are prone to fork reversal and unregulated genomic recombination (Noguchi et al., 2003, 2004). In fission yeast, the FPC’s primary components are Swi1 and Swi3 (Noguchi et al., 2004). Swi1 associates with chromatin through its DDT domain, found in DNA-binding homeobox-containing proteins and different transcription and chromatin-remodeling factors (Noguchi et al., 2012). This domain is also important for recruiting Swi3 (Noguchi et al., 2012). Stable association of Swi1 and Swi3 facilitates binding to the fork of Mrc1, a secondary FPC component necessary for replication processivity and activation of the replication checkpoint (Shimmoto et al., 2009; Matsumoto et al., 2010). Together these proteins act as sensors of replication stress that protect genomic integrity during DNA duplication. Furthermore, Swi1 and Swi3 are also involved in restarting the fork after exposure to S-phase stressors and play a role in establishing and maintaining proper sister chromatid cohesion in vegetative cells (Noguchi et al., 2004; Matsumoto et al., 2005; Ansbach et al., 2008; Rapp et al., 2010).
In fission yeast, meiS phase takes longer to complete than S phase in vegetative conditions (Wu and Nurse, 2014). DNA damage induced at this stage does not elicit checkpoint arrest and can be repaired through meiotic recombination. In checkpoint mutants, however, excess damage may persist until at least the first meiotic division (Pankratz and Forsburg, 2005). Perturbations to meiS phase are associated with recombination and segregation problems downstream (Watanabe et al., 2001; Wu and Nurse, 2014). Because the FPC contributes to replication processivity and the response to DNA damage, we asked whether absence of its components would disrupt any aspect of the meiotic program.
Loss of spore viability
We observed that the FPC mutants showed decreased spore viability (Figure 1A), which is consistent with previous reports of similar outcomes in other replication-stability mutants (Dolan et al., 2010; Le et al., 2013; Mastro and Forsburg, 2014; Murakami and Keeney, 2014; Wu and Nurse, 2014). The reduction seen in the swi1∆ swi3∆ strain is similar to that of swi1∆ cells and agrees with Swi1’s function of stabilizing Swi3 at the fork (Leman et al., 2010; Leman and Noguchi, 2012; Noguchi et al., 2012). Interestingly, combining rec12∆ with the swi1∆ or swi3∆ deletions partially suppressed 33–45% spore unviability observed in the single mutants (Figure 1A). This observation suggests that the damage-induced recombination intermediates previously seen during vegetative growth in the FPC mutants (Noguchi et al., 2003, 2004) may also be present during meiotic prophase I and may serve as recombination sources, albeit suboptimal ones, when prDSBs are abolished (Cervantes et al., 2000; Pankratz and Forsburg, 2005).
Meiotic replication and DSB formation
Absence of the FPC components did not halt meiotic progression but was associated with moderately delayed replication (Figure 1, B–D). It is possible that replisome progression was affected by unstable DNA structures like ssDNA gaps or rearranged forks that result from the uncoupling of DNA helicase and the DNA polymerases (Noguchi et al., 2003, 2004; Leman et al., 2010). In that context, physical impediments to fork progression would be alleviated by homologous recombination, which relies on Rec12 for DSB formation. Therefore Rec12 may assist fork progression by stimulating the removal of DNA barriers generated by unstable forks. This scenario is consistent with the observations that swi1∆ and swi3∆ cells repair DSBs as well as the wild-type strain (Figure 1C and Supplemental Figure 2A). Also, when lacking Rec12, the FPC mutants exhibit substantial delay in replication and generate low-migrating PFGE smears indicative of constitutive DNA damage and incomplete DSB repair (Supplemental Figures 1C and 2B). However, because these meiotic induction experiments were carried out in pat1-114 genetic backgrounds, in which chromosome segregation is not entirely normal (Cipak et al., 2012), we cannot eliminate the possibility that the observed results are pat1-114 specific and thus may be different from wild-type meiosis.
Persistent DNA damage
Deletion of swi1 and swi3 generates increased RPA and Rad52 foci in vegetative cells (Noguchi et al., 2004). In the FPC mutants, we observed elevated signals for both markers that intensified in late HT and persisted until MII. This result is consistent with an FPC role in surveilling excess ssDNA levels and in maintaining stability at the fork, and with failure of meiotic cells to respond to meiS phase damage (Pankratz and Forsburg, 2005). A secondary consequence of accumulating large pools of Rad52 is the impact this may have on recombination, because its dissociation is crucial for Rad51 binding (New et al., 1998; Shinohara and Ogawa, 1998). We observed normal formation of prDSBs by PFGE, suggesting that the fork protection complex is not associated with activation of Rec12. We analyzed four distinct intergenic loci and one intragenic locus to assess recombination activity in the FPC mutants (Table 1). We observed a general approximately twofold decrease in homologous recombination similar to that of dmc1∆ cells at the his4-lys4 intergenic locus (Supplemental Table 1), in which exchange between homologues is reduced. This prompted us to ask whether a decrease was also true for sister chromatid exchange. Deletion of the FPC components in meiosis led to increased recombination between sister chromatids (Table 1). This alteration to recombination dynamics, while potentially ensuring resolution of surplus recombination substrates, may compromise downstream meiotic activity, which depends on chiasmata formation and resolution for proper chromosome segregation (Dudas et al., 2011; Hirose et al., 2011).
Chromosome missegregation
We used live-cell imaging to follow the FPC mutants through meiosis (Figure 3, A–D, and Supplemental Figure 3, A–C). We observed that, without Swi1 and Swi3, cells have increased lagging chromosomes and chromosome missegregation in both meiotic divisions. We asked whether the observed division anomalies resulted from premature sister separation or chromosome nondisjunction (Figure 4, A–C). Interestingly, we found separate functions of Swi1 and Swi3 during MI. Swi1 appears to facilitate accurate monopolar attachment, because its absence is associated with an increase in equational division. By contrast, Swi3 seems to mediate correct disjunction of homologous chromosomes in MI. In MII, both FPC mutants exhibited increased nondisjunction of sister chromatids (Figure 5, A–D). Thus the missegregation phenotypes of the FPC mutants may be partially related to their reduced homologous recombination.
Cohesin regulators
Another possibility is that these phenotypes result from defects in centromeric cohesion (Watanabe and Nurse, 1999; Kitajima et al., 2003a; Yokobayashi and Watanabe, 2005). We addressed this by examining dynamics of cohesion-associated markers. We looked at the temporal dynamics of Rec27 because it is important for homologue pairing and its elimination coincides with DSB repair and chiasmata resolution (Figure 6, A–C, and Supplemental Figure 4, A and B). We found that fork destabilization is associated with reduced Rec27-GFP signal and with accelerated Rec27-GFP foci elimination, which may contribute to recombination anomalies and suggests possible cohesion issues (Phadnis et al., 2015; Sakuno and Watanabe, 2015). Because Sgo1 protects centromeric cohesion before the onset of anaphase I (Kitajima et al., 2004; Ishiguro et al., 2010), we also examined its temporal dynamics in metaphase I (Figure 7, A–C, and Supplemental Figure 3, C and D). For this, we used Sgo1-GFP strains that did not exhibit centromere localization issues, as was previously reported for a Sgo1-GFP construct lacking a 3′ untranslated region sequence (Rabitsch et al., 2004). We observed that deletion of Swi3, but not Swi1, decreases the timing of Sgo1 signal. This result implies that Swi3 is important for normal protection of centromeric cohesion and that this function may be independent of its association with Swi1 at the fork. Interestingly, however, absence of Swi1 shifts the timing of Sgo1 centromeric dissociation to a later stage in metaphase I (Supplemental Figure 4, C and D), thus also affecting Sgo1 regulation of Rec8.
Cohesin instability at the centromere
Rec8 is the cohesin subunit whose separase-dependent degradation ensures proper segregation of sister chromatids in MII (Kitajima et al., 2003a; Ishiguro et al., 2010). Because both FPC mutants showed abnormal MII division, we used live-cell imaging to examine the duration of centromeric Rec8 from MI to MII (Figure 8, A–D, and Supplemental Figure 4, F and G) (Le et al., 2013). In the FPC mutants, relative Rec8-GFP intensity was higher than in wild-type cells before and during meiosis. On the basis of this result, we surmise that lack of the FPC components may interfere with Rec8-GFP dissociation dynamics, respectively leading to delayed and early Rec8-GFP elimination at chromosome arms and the centromere. Indeed, we observed that, in swi1∆ and swi3∆ cells, most Rec8-GFP foci significantly disappeared within the first 30 min of meiosis. This meant that for the remaining 20–30 min before MII, cells lacking either FPC component were mostly untethered at the centromere (Figure 8, A–D, and Supplemental Figure 4, F and G). This may explain the high proportion of cells that exhibited nondisjunction issues in MII.
To corroborate this finding, we examined the localization of Rec8 at the centromere during the period when it is actively protected. Relative to wild-type cells, the FPC mutants consistently showed modest reductions of centromeric Rec8 (Supplemental Figure 4E). Although this result is insufficient to entirely explain the FPC mutants’ missegregation phenotype, it suggest that subtle changes in centromere dynamics may impair the function of other cohesin regulators. Indeed, because the heterochromatin protein Swi6 helps to recruit both cohesin and Sgo1 to the centromere (Yamagishi et al., 2008), we asked whether Swi6 recruitment to centromeres is affected in the FPC mutants during MI and MII (Figure 9, A–D). We found that, without Swi1, cells miscolocalize Swi6 relative to Cnp1 (a centromeric histone). This observation confirms that the FPC is necessary to maintain cohesion stability at the centromere, a result that has been previously reported for vegetative cells (Ansbach et al., 2008).
FPC contribution to genome stability in meiosis
In light of these results, we propose a model in which the FPC contributes to proper meiotic segregation by suppressing unregulated generation of ssDNA and by ensuring cohesion stability at the centromere (Figure 10 and Supplemental Figure 5). These two possibilities agree with the apparent separate functions of the FPC components. Because Swi1 regulates Swi3’s binding to the fork (Noguchi et al., 2012), we reasoned that phenotypes shared by both mutants can be attributed to the absence of Swi3. However, Swi1 may retain independent functions that can, if disrupted, compound those of a Swi3 protein unbound to the fork. Consistent with observations previously made of Tipin (reviewed in Leman and Noguchi, 2012), Swi3 likely monitors ssDNA content by sensing RPA binding. In the absence of Swi3, the number of RPA and Rad52 foci increases. This suggests that decreased recombination may lie behind some of the homologue nondisjunction issues observed in both FPC mutants, but particularly in swi3∆ cells.
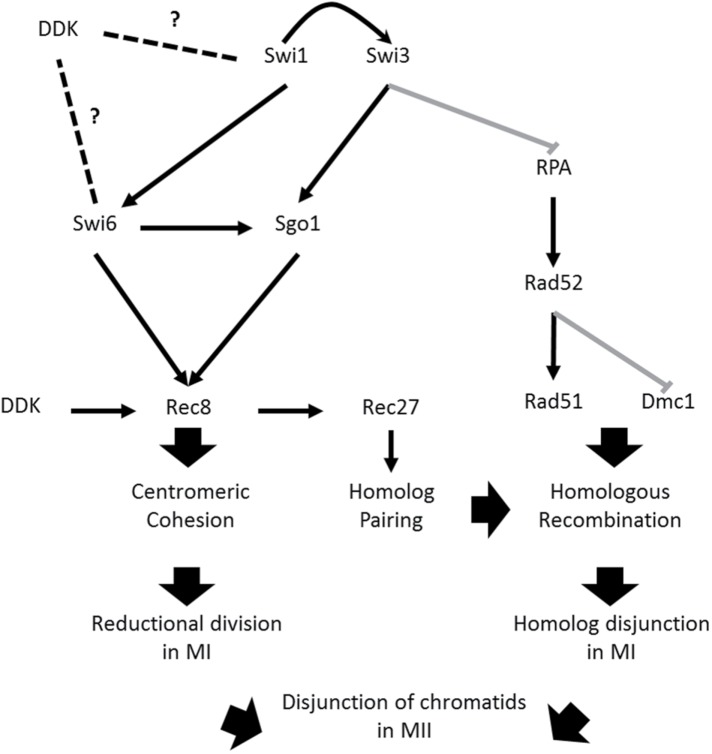
FIGURE 10:
Model of FPC contribution to meiotic segregation. Proposed model explaining the missegregation phenotypes of FPC mutants. Swi1 stabilizes Swi3 at the fork. Both Swi1 and Swi3 help to suppress generation of large ssDNA gaps bound by excess RPA. Normal RPA levels facilitate proper binding of Rad52 to nucleoprotein filament, which in turn promotes Rad51 binding. Both Rad51 and Dmc1 promote normal homologous recombination, which guarantees proper disjunction of homologous chromosomes in MI. Swi1 is necessary for normal recruitment of Swi6 to the centromere. Centromeric Swi6 helps recruit Sgo1, which stabilizes Rec8 at the centromere. Stable centromeric cohesion ensures monopolar attachment of sister kinetochores, which is necessary for correct reductional segregation in MI. Suppression of DNA damage and maintenance of cohesion stability at the centromere ensures accurate separation of sister chromatids. DDK physically interacts with the FPC components and Swi6. It is also involved in the regulation of Rec8. Thus DDK is shown as having a potential role in modulating cohesion stability at the centromere by a yet unknown mechanism (hence the dashed lines on the side).
In the absence of Swi1, we observed problems with Swi6 recruitment to the centromere. As shown previously, this effect may disrupt centromeric Rec8 by preventing efficient cohesin and Sgo1 association (Yamagishi et al., 2008). Moreover, swi1∆ and swi3∆ cells showed defects in the timing of Sgo1 association, an outcome that may further compromise cohesin stability (Figure 10 and Supplemental Figure 5). Thus we propose that Swi1 is important for monopolar attachment and reductional division in MI by maintaining cohesion stability at the centromere. This connection with Rec8 dynamics may also confirm the FPC mutants’ problems with recombination via disruption of Rec27-GFP foci elimination. Given that the FPC physically interacts with Hsk1-Dfp1, fission yeast’s Dbf4-dependent kinase (DDK), and DDK has a role in regulating meiotic replication, DSB formation, and cohesion (Matsumoto et al., 2005, 2010; Matos et al., 2008; Katis et al., 2010; Le et al., 2013), our model predicts DDK or a yet unknown DDK substrate as the link that mechanistically connects replication with meiotic segregation, which would confirm similar observations made in budding yeast (Murakami and Keeney, 2014).
Significance to human reproductive health
Mammalian meiosis is characterized by a long-lasting prophase I (Reichman et al., 2017). Insults to recombination and segregation can severely compromise reproductive outcomes (Herbert et al., 2015). Increasingly, it has become apparent that factors involved in replication during cell proliferation can also regulate important aspects of the meiotic program (Mastro and Forsburg, 2014; Murakami and Keeney, 2014; Wu and Nurse, 2014; Nguyen et al., 2017). In this work, we observed that the FPC components are required for proper cohesion dynamics. When the FPC is destabilized, cells exhibit increased missegregation in both meiotic divisions with concomitant persistence of DNA damage markers. In fission yeast, thus lack of either FPC component significantly reduces the fitness of meiotic products. Given that FPC functions are conserved from yeast to human (Leman and Noguchi, 2012), our findings have major implications for reproductive health. It is possible that many cohesinopathies (van der Lelij et al., 2010) and aneuploidies known to result in embryonic lethality and arrested development may arise from replication-related processes. Further examination of additional prophase I–related events will reveal commonalities and differences that define the functional trajectories of the mitotic and meiotic cell division programs.
MATERIALS AND METHODS
Cell growth and culture
Supplemental Table 2 shows the strains used in this work. Detailed descriptions of general fission yeast culture conditions, media, and standard techniques are found in Sabatinos and Forsburg (2010). Liquid and solid yeast extract plus supplements (YES) was used for the construction and maintenance of all strains, except for when pombe minimal glutamate (PMG) with the appropriate supplements was required. For plating efficiency, cells were picked from single colonies and grown in 10-ml cultures at 32°C to mid–log phase. Cells were counted with a hemocytometer, and 500 cells were spread with glass beads on each plate. Plates were incubated at 32°C for 3–5 d, after which colonies were counted.
Spore viability and recombination
Spore viability and recombination assays were carried out by picking single colonies from heterothallic strains, which were subsequently mixed in 5 µl sterile water on malt extract (ME) plates. Mated strains were incubated at 25°C for 2–3 d. Colony matter from individual mating patches was swiped and resuspended in a 1-ml 0.5% glusulase solution (PerkinElmer, Boston, MA). The glusulase spore suspension was incubated with constant rotation at 25°C for 12–16 h. Spores were counted with a hemocytometer, and 500–1000 spores were dispersed per YES plate. Spores were allowed to germinate and proliferate at 32°C for 3–5 d. Subsequently colonies were counted and, in recombination assays, replica plated onto solid PMG media with appropriate supplements.
To distinguish and eliminate diploid cells from recombination data, phloxin B was added to YES replica plates in which diploids look red, while haploids look pale pink. Recombination dynamics were examined by crossing parent strains carrying linked nutritional markers in coupling ++/– or in repulsion +−/−+. Thus only colonies that were (+−) or (++, – –) arose from actual recombination events, not from diploids that survived glusulase treatment. Genetic distance was calculated using Haldane’s formula as described in Smith (2009): cM = −50 ln (1 − 2R), where genetic distance is in cM and R is the recombinant fraction. Restoration of the Ade+ phenotype was used as a measure of intragenic recombination between parent strains respectively carrying the ade6-M26 and ade6-52 alleles. Experiments were repeated at least six times, with at least 1000 spores plated per trial. Sister chromatid recombination was calculated as previously described by Catlett and Forsburg (2003). To account for postmeiotic recombination between sister chromatids, spores were plated on YE plates. Any white colonies that emerged resulted from true meiotic events, while sectored colonies arose only after the first cell division.
For assays involving tetrad dissection (e.g., strain construction involving similar markers), cell matter was taken from individual mating patches and spread on YES plates. Asci were dissected using a microscope with a micromanipulator, after which spores were treated as described earlier. A two-tailed t test was employed to determine significance for genetic distances. A chi-squared test followed by false discovery rate correction for multiple sample comparisons, which controls for the proportion of significant results that are actually false positives, was used in all other instances yielding frequency and proportion data, except for when sample size merited use of Fisher’s exact test.
PFGE
Stable diploids were created using ade6-M210/M216 complementation in parent strains carrying the mat2-102 and pat1-114 alleles to perform synchronous meiosis as described by Catlett and Forsburg (2003) with a few adjustments. Briefly, cultures were grown in Edinburgh minimal medium (EMM) plus supplements, except adenine, to an OD595 = 0.7–1.0, after which cells were starved for 16.5–17 h in EMM minus nitrogen. Starved cultures were refed EMM containing a 1:2 dilution of supplements (minus adenine) plus NH4Cl and shifted from 25°C to 34°C for meiotic induction. Subsequently cells were harvested every hour (FACS samples were also taken), treated with sodium azide, and washed in phosphate-buffered saline (PBS) and citrate-phosphate sorbitol EDTA. A lysing enzymes solution containing 0.2 mg/ml 100T Zymolyase (Nacalai Tesque, Kyoto, Japan) and 0.45 mg/ml Trichoderma harzianum–lysing enzymes (Sigma, St. Louis, MO) was used to digest the cell wall. Following the first time point, the lysing solution was titrated to 50% and 25% in the second and all subsequent time points, respectively, as performed by Cervantes et al. (2000). Lysate from cells was mixed with 2% CleanCut Agarose (Bio-Rad, Hercules, CA) to generate pulse-field gel plugs, which were then incubated at 55°C in a proteinase K (Bioline Reagents, Taunton, MA)/sarkosyl-EDTA solution for 48 h, with an intermediate change of protease solution after 24 h. The DNA plugs were then washed in Tris-EDTA (TE) and equilibrated in Tris-acetate-EDTA (TAE). A Bio-Rad Chef II Pulse Field Machine was employed to separate DNA by size using a 1% Mega Base Agarose (Bio-Rad, Hercules, CA)/TAE gel. The pulse-field apparatus was run for 48 h at 2 V/cm, with an 1800-s switch time and a 106° angle. The gel was stained in ethidium bromide and visualized in a ChemiDoc Imager (Bio-Rad, Hercules, CA). ImageJ (National Institutes of Health, Bethesda, MD) was used to quantify DSB formation and repair as performed previously (Borde et al., 2000; Mastro and Forsburg, 2014). Briefly, the ethidium bromide signal beneath chromosome III was measured and divided by the summed chromosome signal values of each time point. Significance was computed using one-way analysis of variance (ANOVA) followed by Tukey’s family-wise comparisons.
Live-cell imaging
For live-cell imaging, heterothallic strains were grown in PMG with appropriate supplements at 25°C or 32°C to late log phase (OD595 = ∼0.8). Cells were concentrated and washed in liquid ME and incubated at 25°C for 12–20 h in an air shaker. Starved cells were pelleted in a microfuge and spread on pads made with 2% agarose in liquid sporulation media on top of glass microscope slides. Cover lids were carefully placed on prepared pads and moved clockwise with index finger for two rotations to generate a cell monolayer. Once cell matter was properly distributed, pads were sealed with 1:1:1 Vaseline/lanolin/paraffin (at indicated ratios by weight).
Imaging was carried out as described in Mastro and Forsburg (2014) and Sabatinos et al. (2015) with a few modifications. Cells were staged at 25°C using a DeltaVision microscope with softWorRx version 4.1 (GE Healthcare; Issaquah, WA) equipped with a 60×/ 1.4 NA Plan-Apo lens, solid-state illuminator, and 12-bit charge-coupled device (CCD) camera. Fluorescent proteins were excited and detected with the following filter sets and exposure times: GFP: excitation (ex)475/28, emission (em)525/50, 0.15 s; DsRed or mCherry or red fluorescent protein (RFP): (ex)575/25, (em)632/60, 0.08 s; cerulean or cyan fluorescent protein (CFP): (ex)438/24, (em)470/24, 0.15 s; yellow fluorescent protein (YFP): (ex)513/17, (em)559/38, 0.15 s. The following polychroic mirrors were used: GFP/mCherry Chroma ET C125705 approximately: 520/50–630/80; Semrock CFP/YFP/DsRed 61008 bs approximately: 415/20–462/32–535/5–635/74. Long-term time-lapse videos used 13 0.5-µm z-sections. Images were deconvolved and maximum-intensity projected (softWoRx). Projected fluorescence images were combined with transmitted light images. Images were contrast adjusted using an equivalent histogram stretch on all samples.
GFP focus discrimination was performed by applying a signal threshold twice as large as the average nuclear background. When examining the spatial and temporal dynamics of fluorescently tagged proteins, an event was determined to have ceased if it failed to occur again after three consecutive frames. Frames were taken every 10 min. Measurement of GFP intensity was carried out using ImageJ. Circles of constant size were overlaid on undivided or divided nuclei to measure mean GFP intensity. Background signal was further removed to calculate net GFP signal intensity using the formula: netintensity = grossintensity−[mean backgroundintensity × nuclear area]. At least two independent experiments and five fields per experiment were assessed for each sample. Cell counts were pooled and presented as proportions ±95% confidence interval, which was calculated using sum-squared error rules. Significance was evaluated using a chi-squared test or Fisher’s exact test, depending on sample size. For continuous GFP-intensity data in Figures 6 – 8, significance was determined by calculating the area under the curve of each line and then using one-way ANOVA with subsequent Tukey’s family-wise comparisons for all genotypes. Similarly, but for GFP-foci loss data in Figures 6 – 8, a chi-squared test for trend was employed.
Flow cytometry and microscopy
Whole-cell FACS was carried out as described in detail in Sabatinos and Forsburg (2009, 2015) and Sabatinos et al. (2015) with minor changes. To perform cell cycle analysis or microscopy, cells were fixed in 70% ethanol. Following rehydration in 50 mM sodium citrate, cells were treated with 1 µM SytoxGreen (Invitrogen, Carlsbad, CA) plus 10 µg/ml RNase A and incubated at 36°C for 1–2 h. Samples were then sonicated before being analyzed on a FACScan machine (BD Biosciences, San Jose, CA). DAPI staining was done by first rehydrating fixed cells in PBS and placing them to dry on positively charged glass slides. Mount solution (50% glycerol and 1 µg/ml DAPI) was then applied before cells were photographed on a Leica DMR wide-field epifluorescence microscope equipped with a 63×/1.62 NA Plan-Apo objective lens, 100-W Hg arc lamp for excitation, and a 12-bit Hamamatsu ORCA-100 CCD camera. Images were acquired using OpenLab version 3.1.7 (ImproVision, Lexington, MA) software and analyzed with ImageJ.
Chromatin immunoprecipitation
ChIP was performed as described in Rougemaille et al. (2008) with minor changes. Briefly, synchronous meiosis was achieved as mentioned above. Cells were harvested every hour, treated with 37% formaldehyde (1% final volume) followed by quenching with 0.25 M glycine, and washed in Tris-buffered saline (TBS) before being stored at −80°C. Pelleted cells were lysed by bead-beating in a FastPrep machine (MP Biomedicals, Santa Ana, CA), and the resulting lysate was sonicated four times for 15 s (15% duty cycle) with a Branson Sonifier 450 microtip sonicator. Protein concentration was determined using the Bradford method. Some of the lysate was stored as input, while the rest was treated with 1:200 mouse anti-GFP antibody (Abcam 290) (Abcam, Cambridge, MA) and later combined with protein A Dynabeads (Invitrogen, Carlsbad, CA). After several washes (in lysis buffer, wash buffer, and Tris-EDTA), the protein–DNA portion was separated from the Dynabeads using TE and 1% SDS at 65°C for 30 min, followed by centrifugation. To reverse the formaldehyde cross-link, the resulting elution was then incubated at 65°C for at least 12 h (input included). Afterward, 2 mg/ml proteinase K (Bioline Reagents, Taunton, MA) was used for 2 h at 37°C to eliminate any remnant protein from the samples. DNA was isolated with a Qiagen PCR Purification Kit and analyzed by Q-PCR using a CFX Connect Real-Time System (Biorad, Hercules, CA). Fold values were calculated using the 2-∆∆Ct (Livak) method (Livak and Shmittgen, 2001). To determine fold change in cohesion, the input and the immunoprecipitated fractions were used as the calibrator and test parameters, respectively, while act1 was used as the reference gene (testing for Rec8-GFP localization at the euchromatin) and the centromeric dg locus as the target locus (testing for Rec8-GFP localization at the heterochromatin-rich centromere). Significance was determined using a Wilcoxon signed-rank test.
Supplementary Material
Acknowledgments
We thank all current members of the Forsburg lab, Amanda Jensen, Ruben Petreaca, Nimna Ranatunga, Oscar Aparicio, Sean Curran, and Matthew Michael for technical support and early review of this article. Our appreciation also goes to Tara Mastro, Marc Green, and Sarah Sabatinos for their assistance with PFGE, ChIP, live-cell imaging, and statistical analysis. We also extend our gratitude to Gerald Smith and Yoshinori Watanabe for providing us with strains used to analyze cohesin regulation. This work was made possible by National Institutes of Health grants R35 GM118109 and R01 GM081418 to S.L.F.
Abbreviations used:
- CCD
charge-coupled device
- DAPI
4′,6-diamidino-2-phenylindole
- DDK
Dbf4-dependent kinase
- EMM
Edinburgh minimal medium
- FACS
fluorescence-activated cell sorting
- FPC
fork protection complex
- GFP
green fluorescent protein
- HT
horse tailing
- ME
malt extract
- meiS
meiotic S phase
- mCherry
monomeric Cherry
- MI
meiosis I
- MII
meiosis II
- mRFP
monomeric red fluorescent protein
- PBS
phosphate-buffered saline
- PFGE
pulse-field gel electrophoresis
- PMG
pombe minimal glutamate
- prDSB
programmed double-strand break
- TAE
Tris-acetate-EDTA
- TE
Tris-EDTA
- YES
yeast extract plus supplements.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-02-0101) on August 30, 2017.
REFERENCES
- Allera-Moreau C, Rouquette I, Lepage BA, Oumouhou N, Walschaerts M, Leconte E, Schilling V, Gordien K, Brouchet L, Delisle MB, et al. DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis. 2012;1:e30. doi: 10.1038/oncsis.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansbach AB, Noguchi C, Klansek IW, Heidlebaugh M, Nakamura TM, Noguchi E. RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol Biol Cell. 2008;19:595–607. doi: 10.1091/mbc.E07-06-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarends WM, van der Laan R, Grootegoed JA. DNA repair mechanisms and gametogenesis. Reproduction. 2001;121:31–39. doi: 10.1530/rep.0.1210031. [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- Borde V, Goldman AS, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- Brar GA, Kiburz BM, Zhang Y, Kim JE, White F, Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Catlett MG, Forsburg SL. Schizosaccharomyces pombe Rdh54 (TID1) acts with Rhp54 (RAD54) to repair meiotic double-strand breaks. Mol Biol Cell. 2003;14:4707–4720. doi: 10.1091/mbc.E03-05-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Origins of the machinery of recombination and sex. Heredity. 2002;88:125–141. doi: 10.1038/sj.hdy.6800034. [DOI] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Hiraoka Y. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol. 2001;11:1618–1623. doi: 10.1016/s0960-9822(01)00457-2. [DOI] [PubMed] [Google Scholar]
- Chini CCS, Chen J. Human claspin is required for replication checkpoint control. J Biol Chem. 2003;278:30057–30062. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci USA. 2006;103:18143–18147. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipak L, Hyppa RW, Smith GR, Gregan J. ATP analog-sensitive Pat1 protein kinase for synchronous fission yeast meiosis at physiological temperature. Cell Cycle. 2012;11:1626–1633. doi: 10.4161/cc.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- Coschi CH, Ishak CA, Gallo D, Marshall A, Talluri S, Wang J, Cecchini MJ, Martens AL, Percy V, Welch I, et al. Haploinsufficiency of an RB–E2F1–Condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov. 2014;4:840–853. doi: 10.1158/2159-8290.CD-14-0215. [DOI] [PubMed] [Google Scholar]
- Davis L, Rozalén AE, Moreno S, Smith GR, Martín-Castellanos C. Rec25 and Rec27, novel linear-element components, link cohesin to meiotic DNA breakage and recombination. Curr Biol. 2008;18:849–854. doi: 10.1016/j.cub.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Smith GR. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics. 2003;163:857–874. doi: 10.1093/genetics/163.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan WP, Le AH, Schmidt H, Yuan JP, Green M, Forsburg SL. Fission yeast Hsk1 (Cdc7) kinase is required after replication initiation for induced mutagenesis and proper response to DNA alkylation damage. Genetics. 2010;185:39–53. doi: 10.1534/genetics.109.112284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas A, Ahmad S, Gregan J. Sgo1 is required for co-segregation of sister chromatids during achiasmate meiosis I. Cell Cycle. 2011;10:951–955. doi: 10.4161/cc.10.6.15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Ragland RL, Arlt MF, Mulle JG, Warren ST, Glover TW. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc Natl Acad Sci USA. 2008;105:246–251. doi: 10.1073/pnas.0708097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R, Beach DH, Klar AJ. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc Natl Acad Sci USA. 1984;81:3481–3485. doi: 10.1073/pnas.81.11.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- Ellermeier C, Smith GR. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Cosentino C, Rivera T, Losada A, Schwob E, Hunt T, Costanzo V. Tipin/Tim1/And1 protein complex promotes Polα chromatin binding and sister chromatid cohesion. EMBO J. 2009;28:3681–3692. doi: 10.1038/emboj.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Costanzo V. Mechanisms of replication fork protection: a safeguard for genome stability. Crit Rev Biochem Mol Biol. 2012;47:222–235. doi: 10.3109/10409238.2012.655374. [DOI] [PubMed] [Google Scholar]
- Fleck O, Lehmann E, Schär P, Kohli J. Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat Genet. 1999;21:314–317. doi: 10.1038/6838. [DOI] [PubMed] [Google Scholar]
- Foss EJ. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics. 2001;157:567–577. doi: 10.1093/genetics/157.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter AL, Suppa C, Emanuel BS. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol. 2007;366:36–52. doi: 10.1016/j.jmb.2006.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu AH, Liang J, Lu NX, Wu B, Xia YK, Lu CC, Song L, Wang SL, Want XR. Association of XRCC1 gene polymorphisms with idiopathic azoospermia in a Chinese population. Asian J Androl. 2007;9:781. doi: 10.1111/j.1745-7262.2007.00325.x. [DOI] [PubMed] [Google Scholar]
- Hauf S, Biswas A, Langegger M, Kawashima SA, Tsukahara T, Watanabe Y. Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I. EMBO J. 2007;26:4475–4486. doi: 10.1038/sj.emboj.7601880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert M, Kalleas D, Cooney D, Lamb M, Lister L. Meiosis and maternal aging: insights from aneuploid oocytes and trisomy births. Cold Spring Harb Perspect Biol. 2015;7:a017970. doi: 10.1101/cshperspect.a017970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Suzuki R, Ohba T, Hinohara Y, Matsuhara H, Yoshida M, Itabashi Y, Murakami H, Yamamoto A. Chiasmata promote monopolar attachment of sister chromatids and their co-segregation toward the proper pole during meiosis I. PLoS Genet. 2011;7:e1001329. doi: 10.1371/journal.pgen.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A. Meiosis. Curr Biol. 2008;18:R641–R645. doi: 10.1016/j.cub.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin–PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol. 2010;12:500–506. doi: 10.1038/ncb2052. [DOI] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell. 2010;18:397–409. doi: 10.1016/j.devcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 2007;21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kim J, Ishiguro KI, Nambu A, Akiyoshi B, Yokobayashi S, Kagami A, Ishiguro T, Pendas A, Takeda N, Sakakibara Y, et al. Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature. 2015;517:466–471. doi: 10.1038/nature14097. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Miyazaki Y, Yamamoto M, Watanabe Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J. 2003a;22:5643–5653. doi: 10.1093/emboj/cdg527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro KI, Iemura SI, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Yokobayashi S, Yamamoto M, Watanabe Y. Distinct cohesin complexes organize meiotic chromosome domains. Science. 2003b;300:1152–1155. doi: 10.1126/science.1083634. [DOI] [PubMed] [Google Scholar]
- Klutstein M, Fennell A, Fernández-Álvarez A, Cooper JP. The telomere bouquet regulates meiotic centromere assembly. Nat Cell Biol. 2015;17:458–469. doi: 10.1038/ncb3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Le AH, Mastro TL, Forsburg SL. The C-terminus of S. pombe DDK subunit Dfp1 is required for meiosis-specific transcription and cohesin cleavage. Biol Open. 2013;2:728–738. doi: 10.1242/bio.20135173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman AR, Noguchi E. Local and global functions of Timeless and Tipin in replication fork protection. Cell Cycle. 2012;11:3945–3955. doi: 10.4161/cc.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman AR, Noguchi C, Lee CY, Noguchi E. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci. 2010;123:660–670. doi: 10.1242/jcs.057984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- Mastro TL, Forsburg SL. Increased meiotic crossovers and reduced genome stability in absence of Schizosaccharomyces pombe Rad16 (XPF) Genetics. 2014;198:1457–1472. doi: 10.1534/genetics.114.171355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell. 2008;135:662–678. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Ogino K, Noguchi E, Russell P, Masai H. Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J Biol Chem. 2005;280:42536–42542. doi: 10.1074/jbc.M510575200. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Shimmoto M, Kakusho N, Yokoyama M, Kanoh Y, Hayano M, Russell P, Masai H. Hsk1 kinase and Cdc45 regulate replication stress-induced checkpoint responses in fission yeast. Cell Cycle. 2010;9:4627–4637. doi: 10.4161/cc.9.23.13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC (Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, Newitt R, Aebersold R, Boone C, Brown G, et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod M, Stein M, Beach D. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. EMBO J. 1987;6:729. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Keeney S. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell. 2014;158:861–873. doi: 10.1016/j.cell.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Nurse P. Regulation of premeiotic S phase and recombination-related double-strand DNA breaks during meiosis in fission yeast. Nat Genet. 2001;28:290–293. doi: 10.1038/90142. [DOI] [PubMed] [Google Scholar]
- Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- Murayama Y, Kurokawa Y, Tsutsui Y, Iwasaki H. Dual regulation of Dmc1-driven DNA strand exchange by Swi5–Sfr1 activation and Rad22 inhibition. Genes Dev. 2013;27:2299–2304. doi: 10.1101/gad.218693.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris DF, Vreeken K, Schmidt H, Ostermann K, Clever B, Lohman PH, Pastink A. Homologous recombination in the fission yeast Schizosaccharomyces pombe: different requirements for the rhp51+, rhp54+ and rad22+ genes. Curr Genet. 1997;31:248–254. doi: 10.1007/s002940050202. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Nguyen H, James NG, Nguyen L, Nguyen TP, Vuong C, Ortega MA, Jameson DM, Ward WS. Higher order oligomerization of the licensing ORC4 protein is required for polar body extrusion in murine meiosis. J Cell Biochem. 2017 doi: 10.1002/jcb.25949. doi:10.1002/jcb.25949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi C, Rapp JB, Skorobogatko YV, Bailey LD, Noguchi E. Swi1 associates with chromatin through the DDT domain and recruits Swi3 to preserve genomic integrity. PLoS ONE. 2012;7:e43988. doi: 10.1371/journal.pone.0043988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, Du LL, Russell P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol. 2003;23:7861–7874. doi: 10.1128/MCB.23.21.7861-7874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, McDonald WH, Yates JR, Russell P. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol. 2004;24:8342–8355. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Nudell D, Castillo M, Turek PJ, Pera RR. Increased frequency of mutations in DNA from infertile men with meiotic arrest. Hum Reprod. 2000;15:1289–1294. doi: 10.1093/humrep/15.6.1289. [DOI] [PubMed] [Google Scholar]
- Octobre G, Lorenz A, Loidl J, Kohli J. The Rad52 homologs Rad22 and Rti1 of Schizosaccharomyces pombe are not essential for meiotic interhomolog recombination, but are required for meiotic intrachromosomal recombination and mating-type-related DNA repair. Genetics. 2008;178:2399–2412. doi: 10.1534/genetics.107.085696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K, Hirota K, Matsumoto S, Takeda T, Ohta K, Arai KI, Masai H. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc Natl Acad Sci USA. 2006;103:8131–8136. doi: 10.1073/pnas.0602498103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H. Meiosis: an overview of key differences from mitosis. Cold Spring Harb Perspect Biol. 2015;7:a015859. doi: 10.1101/cshperspect.a015859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Fortunato EA, Subramani S. Double-strand break-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Genetics. 1996;142:341–357. doi: 10.1093/genetics/142.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz DG, Forsburg SL. Meiotic S-phase damage activates recombination without checkpoint arrest. Mol Biol Cell. 2005;16:1651–1660. doi: 10.1091/mbc.E04-10-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Sternglanz R. Identification and characterization of the genes for two topoisomerase I-interacting proteins from Saccharomyces cerevisiae. Yeast. 1999;15:35–41. doi: 10.1002/(SICI)1097-0061(19990115)15:1<35::AID-YEA340>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Phadnis N, Cipak L, Polakova S, Hyppa RW, Cipakova I, Anrather D, Karvaiova L, Mechtler K, Smith GR, Gregan J. Casein kinase 1 and phosphorylation of cohesin subunit Rec11 (SA3) promote meiotic recombination through linear element formation. PLoS Genet. 2015;11:e1005225. doi: 10.1371/journal.pgen.1005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenhaber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Rapp JB, Noguchi C, Das MM, Wong LK, Ansbach AB, Holmes AM, Arcangioli B, Noguchi E. Checkpoint-dependent and-independent roles of Swi3 in replication fork recovery and sister chromatid cohesion in fission yeast. PLoS ONE. 2010;5:e13379. doi: 10.1371/journal.pone.0013379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichman R, Alleva B, Smolikove S. Prophase I: preparing chromosomes for segregation in the developing oocyte. In: Arur S, editor. Signaling-Mediated Control of Cell Division: From Oogenesis to Oocyte-to-Embryo Development. Springer International; 2017. pp. 125–173. [DOI] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- Rougemaille M, Shankar S, Braun S, Rowley M, Madhani HD. Ers1, a rapidly diverging protein essential for RNA interference-dependent heterochromatic silencing in Schizosaccharomyces pombe. J Biol Chem. 2008;283:25770–25773. doi: 10.1074/jbc.C800140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatinos SA, Forsburg SL. Measuring DNA content by flow cytometry in fission yeast. In: Vengrova S, Dalgaard J, editors. DNA Replication: Methods and Protocols. Humana Press: 2009. pp. 449–461. [DOI] [PubMed] [Google Scholar]
- Sabatinos SA, Forsburg SL. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 2010;470:759–795. doi: 10.1016/S0076-6879(10)70032-X. [DOI] [PubMed] [Google Scholar]
- Sabatinos SA, Forsburg SL. Measuring DNA content by flow cytometry in fission yeast. In: Vengrova S, Dalgaard J, editors. DNA Replication: Methods and Protocols. Humana Press: 2015. pp. 79–97. [DOI] [PubMed] [Google Scholar]
- Sabatinos SA, Green MD, Forsburg SL. Continued DNA synthesis in replication checkpoint mutants leads to fork collapse. Mol Cell Biol. 2012;32:4986–4997. doi: 10.1128/MCB.01060-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatinos SA, Ranatunga NS, Yuan JP, Green MD, Forsburg SL. Replication stress in early S phase generates apparent micronuclei and chromosome rearrangement in fission yeast. Mol Biol Cell. 2015;26:3439–3450. doi: 10.1091/mbc.E15-05-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- Sakuno T, Tanaka K, Hauf S, Watanabe Y. Repositioning of aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I. Dev Cell. 2011;21:534–545. doi: 10.1016/j.devcel.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Sakuno T, Watanabe Y. Phosphorylation of cohesin Rec11/SA3 by casein kinase 1 promotes homologous recombination by assembling the meiotic chromosome axis. Dev Cell. 2015;32:220–230. doi: 10.1016/j.devcel.2014.11.033. [DOI] [PubMed] [Google Scholar]
- Schuchert P, Kohli J. The ade6-M26 mutation of Schizosaccharomyces pombe increases the frequency of crossing over. Genetics. 1988;119:507–515. doi: 10.1093/genetics/119.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif WD, Glick GG, Davidson MK, Wahls WP. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome. 2002;1:1. doi: 10.1186/1475-9268-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmoto M, Matsumoto S, Odagiri Y, Noguchi E, Russell P, Masai H. Interactions between Swi1-Swi3, Mrc1 and S phase kinase, Hsk1 may regulate cellular responses to stalled replication forks in fission yeast. Genes Cells. 2009;14:669–682. doi: 10.1111/j.1365-2443.2009.01300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Smith GR. Genetic analysis of meiotic recombination in Schizosaccharomyces pombe. Methods Mol Biol. 2009;557:65–76. doi: 10.1007/978-1-59745-527-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Fu MA, Brown EJ. Tim–Tipin dysfunction creates an indispensible reliance on the ATR–Chk1 pathway for continued DNA synthesis. J Cell Biol. 2009;187:15–23. doi: 10.1083/jcb.200905006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Kubota Y, Tsujimura T, Kumano M, Masai H, Takisawa H. Replisome progression complex links DNA replication to sister chromatid cohesion in Xenopus egg extracts. Genes Cells. 2009;14:949–963. doi: 10.1111/j.1365-2443.2009.01322.x. [DOI] [PubMed] [Google Scholar]
- Tourrière H, Versini G, Cordón-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Ünsal-Kaçmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch M, Vreeken K, Zonneveld JB, Brandsma JA, Lombaerts M, Murray JM, Lohman PHM, Pastink A. Characterization of RAD52 homologs in the fission yeast Schizosaccharomyces pombe. Mutat Res. 2001;461:311–323. doi: 10.1016/s0921-8777(00)00060-4. [DOI] [PubMed] [Google Scholar]
- van der Crabben SN, Hennus MP, McGregor GA, Ritter DI, Nagamani SC, Wells OS, Harakalova M, Chinn I, Alt A, Vondrova L, et al. Destabilized SMC5/6 complex leads to chromosome breakage syndrome with severe lung disease. J Clin Invest. 2016;126:2881–2892. doi: 10.1172/JCI82890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lelij P, Chrzanowska KH, Godthelp BC, Rooimans MA, Oostra AB, Stumm M, Zdzienicka MZ, Joenje H, de Winter JP. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am J Hum Genet. 2010;86:262–266. doi: 10.1016/j.ajhg.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CD, Eckley DM, Lee MS, Hanna JS, Hughes A, Peyser B, Jie C, Irizarry R, Spencer FA. S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol Biol Cell. 2004;15:1724–1735. doi: 10.1091/mbc.E03-09-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature. 2001;409:359–363. doi: 10.1038/35053103. [DOI] [PubMed] [Google Scholar]
- Witosch J, Wolf E, Mizuno N. Architecture and ssDNA interaction of the Timeless-Tipin-RPA complex. Nucleic Acids Res. 2014;42:12912–12927. doi: 10.1093/nar/gku960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PYJ, Nurse P. Replication origin selection regulates the distribution of meiotic recombination. Mol Cell. 2014;53:655–662. doi: 10.1016/j.molcel.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Boone C, Brown GW. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics. 2007;176:1417–1429. doi: 10.1534/genetics.107.072876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 2003;22:2284–2296. doi: 10.1093/emboj/cdg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005;123:803–817. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem. 2007;282:2729–2740. doi: 10.1074/jbc.M605596200. [DOI] [PubMed] [Google Scholar]
- Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.