MTBP, the partner of Treslin, contains a novel DNA-binding domain that associates with double-stranded DNA and G-quadruplex DNA. Mutants of MTBP that lack a functional version of this domain cannot support normal initiation of DNA replication.
Abstract
Treslin, which is essential for incorporation of Cdc45 into the replicative helicase, possesses a partner called MTBP (Mdm2-binding protein). We have analyzed Xenopus and human MTBP to assess its role in DNA replication. Depletion of MTBP from Xenopus egg extracts, which also removes Treslin, abolishes DNA replication. These extracts be can rescued with recombinant Treslin-MTBP but not Treslin or MTBP alone. Thus, Treslin-MTBP is collectively necessary for replication. We have identified a C-terminal region of MTBP (the CTM domain) that binds efficiently to both double-stranded DNA and G-quadruplex (G4) DNA. This domain also exhibits homology with budding yeast Sld7. Mutants of MTBP without a functional CTM domain are defective for DNA replication in Xenopus egg extracts. These mutants display an impaired localization to chromatin and the inability to support loading of Cdc45. Human cells harboring such a mutant also display severe S-phase defects. Thus, the CTM domain of MTBP plays a critical role in localizing Treslin-MTBP to the replication apparatus for initiation.
INTRODUCTION
During the cell cycle, eukaryotic cells must duplicate their genomes faithfully to maintain genomic integrity. The initiation of DNA replication entails the stepwise assembly of numerous proteins onto potential origins of replication (Siddiqui et al., 2013; Tanaka and Araki, 2013; Parker et al., 2017). The origin recognition complex (ORC) and Cdc6 recruit Cdt1 and the mini-chromosome maintenance complex (MCM) to chromatin early in the cell cycle. Prior to S-phase, the MCM complex exists as an inactive precursor of the replicative helicase, the enzyme that will eventually separate the DNA strands for replication. A crucial stage in the process of DNA replication involves the binding of Cdc45 and GINS (go-ichi-ni-san) to MCM to form the CMG (Cdc45-MCM-GINS) complex, which represents the activated version of the helicase. This transformation requires additional structural and regulatory proteins, including two protein kinases. A critical aspect of this process involves phosphorylation of certain key replication proteins by the S-phase cyclin-dependent kinase (S-Cdk).
Studies in the budding yeast system established that S-Cdk phosphorylates proteins known as Sld2 and Sld3 (Tanaka et al., 2007; Zegerman and Diffley, 2007; Labib, 2010; Siddiqui et al., 2013; Tanaka and Araki, 2013). These phosphorylations promote the docking of Sld2 and Sld3 with Dpb11, the yeast homologue of vertebrate TopBP1. These steps are necessary for integration of Cdc45 and GINS with the MCM complex. Furthermore, Sld2 and Sld3 are the two minimally necessary targets of S-Cdk for initiation of S-phase in budding yeast. An important issue has been the extent to which such regulatory mechanisms are conserved in vertebrates, including humans. Vertebrate RecQ4 displays limited sequence homology with Sld2, but the available evidence suggests that it is not a target of S-Cdk for initiation (Sangrithi et al., 2005; Matsuno et al., 2006).
On the other hand, a protein called Treslin (also known as Ticrr) appears to be a genuine functional analogue of yeast Sld3 (Kumagai et al., 2010; Sanchez-Pulido et al., 2010; Sansam et al., 2010). Treslin associates with TopBP1 in a manner that depends on S-Cdk; this regulated binding is essential for the loading of Cdc45 onto MCM and the consequent initiation of S-phase in vertebrates (Boos et al., 2011; Kumagai et al., 2011). Treslin is also subject to negative regulation by the checkpoint-regulatory kinase Chk1 (Guo et al., 2015). Thus, vertebrate cells carefully control the initiation-promoting activity of Treslin.
The issues of how vertebrates, including humans, select and activate origins of replication are not resolved conclusively (Fragkos et al., 2015; Urban et al., 2015; Prioleau and MacAlpine, 2016; Aladjem and Redon, 2017; Parker et al., 2017). These questions are significant because, for example, areas with a paucity of replication origins are more prone to genomic instability (Letessier et al., 2011). There appear not to be highly specific origin sequences in vertebrates like those in budding yeast. Nonetheless, various studies have implicated nucleosome-free regions, CpG islands, G-rich sequences with the potential to form G-quadruplex (G4) DNA, and other features in initiation (Fragkos et al., 2015; Valton and Prioleau, 2016; Hansel-Hertsch et al., 2017; Parker et al., 2017). Interactions with nucleosomes and chromatin-bound proteins also appear to modulate initiation (Fragkos et al., 2015; Parker et al., 2017). These studies have included mapping of nascent DNA strands, localization of ORC, and other methods. However, some technical concerns have been raised about identification of nascent strands (Urban et al., 2015). Moreover, since loaded MCM complexes can move on DNA, the localization of ORC may not necessarily mirror sites of initiation in all cases (Miotto et al., 2016). One avenue to provide further insight into this topic would be to characterize how Treslin associates with prospective sites for initiation. Since this protein acts at the step of helicase activation, which one might expect to be virtually coincident with initiation, the mechanism by which this protein associates with origins may shed light on these problems.
It is largely unknown what parameters regulate the association of Treslin with origins and how this association eventually leads to activation of the replicative helicase. Recent studies have indicated that Treslin has a binding partner in human cells called MTBP (Mdm2-binding protein) that is also important for replication (Boos et al., 2013). However, apart from its association with Treslin, the role of MTBP in replication is unclear. In this report, we have isolated a Xenopus homologue of MTBP and examined its functional relationship with Treslin. We have found that Treslin and MTBP form a stoichiometric complex that is collectively necessary for DNA replication in Xenopus egg extracts. Furthermore, by analyzing the structure of MTBP, we have discovered that it contains a crucial DNA-binding domain that regulates its association with chromatin. This step is important for activation of the replicative helicase in both Xenopus and humans and thus defines a key process in the initiation of DNA replication. Taken together, these findings reveal important insights into the initiation of replication in vertebrates.
RESULTS
Treslin and MTBP form a stoichiometric complex in Xenopus egg extracts
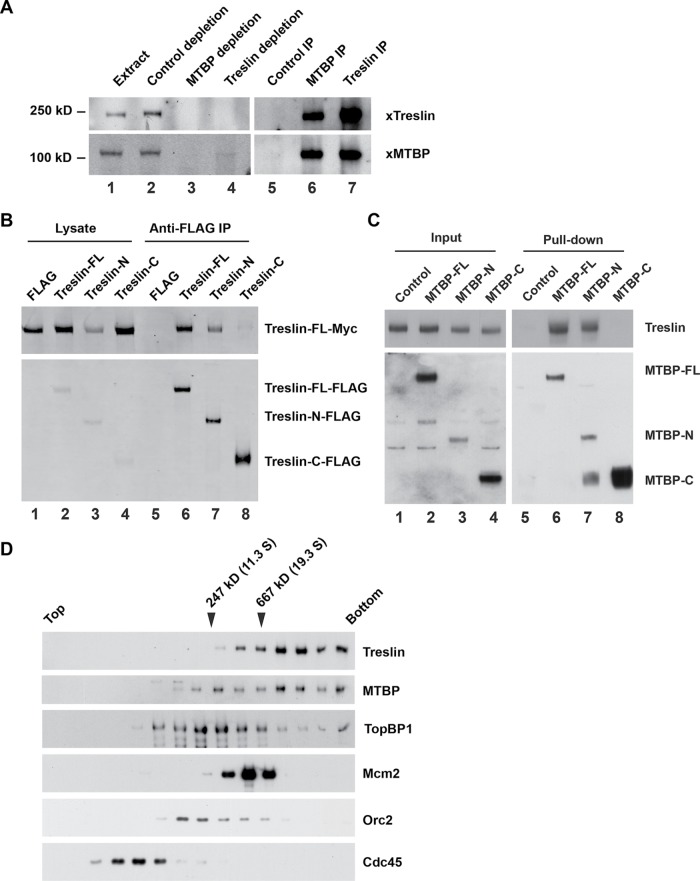
Previous studies identified MTBP as a specific Treslin-interacting protein in human cells (Boos et al., 2013). To investigate further the functional relationship between MTBP and Treslin, we utilized the Xenopus egg extract system, which affords many advantages for the analysis of eukaryotic DNA replication (Arias and Walter, 2004). To evaluate whether the Treslin-MTBP interaction is conserved in this system, we characterized a putative Xenopus homologue of MTBP. We prepared rabbit polyclonal anti-Xenopus MTBP antibodies and found that they recognize an ∼105-kDa polypeptide in Xenopus egg extracts (the calculated molecular mass of Xenopus MTBP is 96 kDa). We performed immunoprecipitation experiments with these anti-MTBP antibodies and could readily observe association of Treslin with MTBP (Figure 1A). Under these immunoprecipitation conditions, MTBP was depleted from the extracts. Significantly, this procedure also resulted in the removal of Treslin from the extracts. In similar experiments with anti-Treslin antibodies, we could efficiently coimmunoprecipitate MTBP with Treslin (Figure 1A). Furthermore, immunodepletion of Treslin resulted in the nearly complete removal of MTBP from the extracts. Therefore, MTBP exists in a stoichiometric complex with Treslin in egg extracts with little or no free MTBP or Treslin.
FIGURE 1:
Identification of a Xenopus homologue of MTBP. (A) Xenopus egg extracts (lane 1) were incubated with control, anti-MTBP, or anti-Treslin antibodies coupled to protein A Dynabeads. After removal of the beads by centrifugation, the supernatants (lanes 2–4) and beads (lanes 5–7) were immunoblotted for Treslin and MTBP. Data are representative of at least five independent experiments. (B) Human 293T cells were transfected with a control vector encoding the FLAG tag alone or vectors encoding FLAG-tagged versions of full-length Treslin or the Treslin-N or Treslin-C fragments. Cells were also transfected with a vector encoding full-length Treslin-Myc. Cell lysates were incubated with anti-FLAG beads. Cell lysates (lanes 1–4) and bead-bound proteins (lanes 5–8) were immunoblotted with anti-Myc (top panel) and anti-FLAG antibodies (bottom panel). Data are representative of two independent experiments. (C) Human 293T cells were transfected with a vector encoding the FLAG tag alone or FLAG-tagged versions of full-length MTBP, MTBP-N, or MTBP-C. Cells were also transfected with a vector encoding full-length Treslin-Myc. Cell lysates were incubated with anti-FLAG beads. Cell lysates (lanes 1–4) and bead-bound proteins (lanes 5–8) were immunoblotted with anti-Myc (top panel) and anti-FLAG antibodies (bottom panel). Data are representative of two independent experiments. (D) Native size of Treslin–MTBP complex. Xenopus NPE was subjected to sucrose density gradient centrifugation. Fractions from the gradient were immunoblotted for the indicated proteins. Catalase (247 kDa; 11.3 S) and thyroglobulin (667 kDa; 19.3 S) served as markers. Data are representative of two independent experiments.
Next, we investigated the quaternary structure of this complex. We first asked whether Treslin could form multimers. We co-expressed in human 293T cells different recombinant forms of full-length Treslin that had been tagged at the C-terminal end with FLAG or Myc epitopes (Treslin-FLAG and Treslin-Myc), respectively. In the case of the FLAG constructs, we also prepared N-terminal and C-terminal fragments of Treslin containing residues 1–1257 and 1253–1909 of the protein, denoted Treslin-N and Treslin-C, respectively. As described previously, the Treslin-N fragment is necessary and sufficient for DNA replication in human cells (Kumagai et al., 2011). The Treslin-C fragment contains the regulatory TRCT (Treslin C-terminal) domain, which mediates the inhibition of Treslin by Chk1 (Guo et al., 2015). With anti-FLAG antibodies, we could readily coimmunoprecipitate full-length Treslin-Myc with FLAG-tagged versions of full-length Treslin and the Treslin-N fragment (Figure 1B). However, there was negligible binding of full-length Treslin-Myc to the FLAG-tagged Treslin-C fragment. These observations suggest that Treslin dimerizes (or forms some higher-order multimer) through its N-terminal replication-promoting domain.
Previous studies indicated that two regions in N-terminal domain of Treslin encompassing approximately residues 265–593 are important for binding to MTBP (Boos et al., 2013). However, the putative Treslin-binding region of MTBP has been unknown. To address this issue, we prepared truncation mutants of MTBP that contain roughly the N-terminal 60% (residues 1–532) and C-terminal 40% of the protein (residues 526–904). We designated these constructs as MTBP-N and MTBP-C, respectively. We observed that the MTBP-N fragment bound well to Treslin, but the MTBP-C exhibited no interaction (Figure 1C). Hence, the N-terminal region of MTBP associates with the region of Treslin that is necessary for replication.
To assess the overall size of the Treslin–MTBP complex, we subjected soluble Xenopus nucleoplasmic extract (NPE) fractions to sucrose density gradient centrifugation. We found that Treslin–MTBP migrated as a large complex with an average apparent molecular mass of ∼800 kDa (Figure 1D). TopBP1 mostly did not comigrate with Treslin–MTBP under these conditions. Taken together, these observations suggest that Treslin–MTBP may form at least a tetramer with two copies of each protein.
Treslin-MTBP is collectively essential for DNA replication in Xenopus egg extracts
We proceeded to examine whether MTBP is necessary for DNA replication in Xenopus egg extracts. For this purpose, we immunodepleted MTBP from the extracts with anti-MTBP antibodies (Figure 2A). Consistent with the fact that Treslin-depleted extracts are defective for replication, there was little or no DNA replication in MTBP-depleted extracts (Figure 2, B and C). To assess the specificity of this observation, we attempted to rescue DNA replication in the depleted extracts by the addition of exogenous recombinant proteins. Toward this end, we prepared a recombinant complex of the human forms of Treslin and MTBP in 293T cells. The reason for this choice was twofold. First, we found it difficult to express and purify adequate amounts of the Xenopus Treslin-MTBP complex. Second, this approach afforded us the opportunity to compare mutants of Treslin–MTBP in both the Xenopus and human systems more readily.
FIGURE 2:
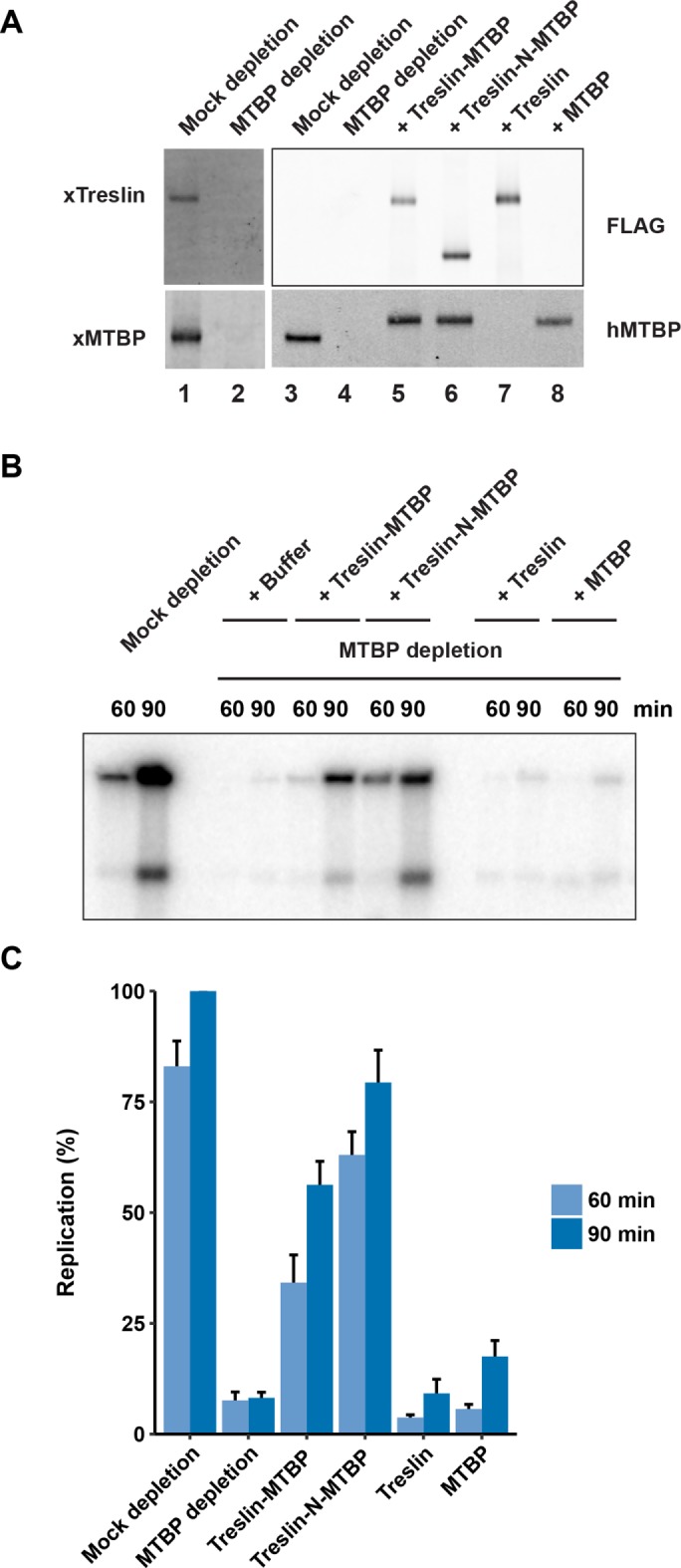
Immunodepletion of MTBP abolishes chromosomal DNA replication in Xenopus egg extracts. (A) Egg extracts were mock-depleted with control antibodies (lanes 1 and 3) or immunodepleted with anti-MTBP antibodies (lanes 2 and 4–8). Following this procedure, extracts were supplemented with buffer alone (lanes 1–4), full-length human Treslin bound to human MTBP (lane 5), human Treslin-N bound to human MTBP (lane 6), full-length human Treslin alone (lane 7), or human MTBP alone (lane 8). Extracts were immunoblotted with antibodies against Xenopus (x) Treslin, Xenopus MTBP, and human (h) MTBP, as well as with anti-FLAG antibodies (as indicated on the sides of the immunoblots). Note that the anti-hMTBP antibodies cross-react with xMTBP. Recombinant Treslin and MTBP proteins were added to a final concentration of ∼10 nM. (B) The indicated extracts were assayed for chromosomal DNA replication at 60 and 90 min. (C) Results of replication assays as in B were compiled from five independent experiments (mean ± SEM). Values are expressed as the percentage of replication at 90 min in mock-depleted extracts.
We could obtain a recombinant Treslin–MTBP complex of good purity by using affinity chromatography to isolate FLAG-tagged Treslin and associated recombinant MTBP from human cells (Supplemental Figure S1). On addition to MTBP-depleted extracts, we observed that this recombinant complex could rescue DNA replication effectively (Figure 2, B and C). By contrast, recombinant Treslin alone could not restore replication. Moreover, a recombinant form of MTBP alone (which we purified separately by virtue of a Twin-Strep tag) was also not significantly active. For these experiments, we also prepared a complex that contains the Treslin-N fragment instead of the full-length protein. In previous studies, we showed that the C-terminal region of Treslin harbors a negative-regulatory, Chk1-binding domain (Guo et al., 2015). Hence, addition of a complex consisting of Treslin-N and MTBP to MTBP-depleted extracts resulted in a higher level of DNA replication in comparison with depleted extracts that were reconstituted with a complex containing full-length Treslin.
In previous studies, we found that recombinant Treslin alone could not rescue DNA replication in Treslin-depleted extracts (Kumagai et al., 2010). We can now rationalize this observation on the basis of the fact that Treslin-depleted extracts also lack MTBP. Furthermore, it seems unlikely that depletion of Treslin-MTBP removes another protein(s) besides Treslin and MTBP that would be important for replication, as addition of recombinant Treslin–MTBP is sufficient to restore DNA synthesis.
The C-terminal region of MTBP contains a conserved DNA-binding domain
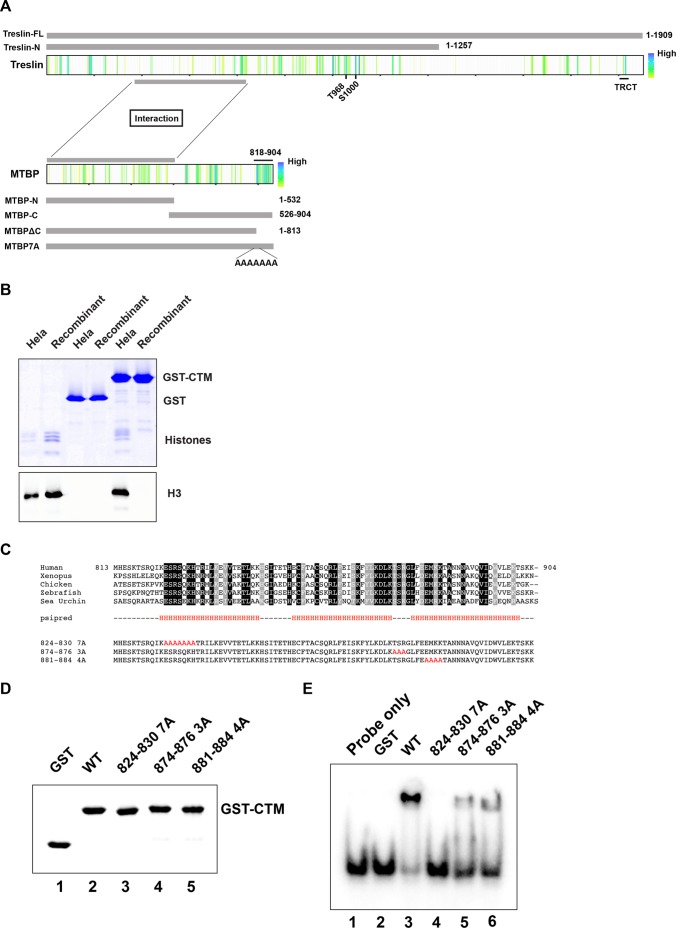
To dissect further the role of MTBP, we sought to define functional domains within this protein. Toward this end, we performed sequence comparisons of MTBP homologues from humans, Xenopus laevis, chicken, zebrafish, and sea urchin (Figure 3A). We utilized a procedure that seeks to highlight functionally important residues (Capra and Singh, 2007). We noticed a highly conserved region of ∼90 amino acids (residues 818–904) near the C-terminal end of MTBP (see also Supplemental Figure S2). Since this region lies outside the Treslin-interacting domain, we speculated that it might have some regulatory role. Therefore, we scrutinized this domain in more detail.
FIGURE 3:
MTBP contains a highly conserved C-terminal domain. (A) Sequences of Treslin (top) and MTBP (bottom) from humans, Xenopus laevis, chicken (Gallus gallus), zebrafish (Danio rerio), and sea urchin (Strongylocentrotus purpuratus) were aligned with Clustal omega. Homology scores were calculated by the method of Capra and Singh (2007). Scores in the top 25% were highlighted with colors ranging from blue to yellow (higher to lower conservation). Regions for interaction between Treslin and MTBP have been denoted. The various constructs of Treslin and MTBP used in this study have been depicted. (B) The CTM domain can associate with nucleosomes. Micrococcal-nuclease digested nucleosomes from HeLa cells (lanes 1, 3, and 5) or recombinant nucleosomes (lanes 2, 4, and 6) were incubated with glutathione-agarose beads containing GST alone (lanes 3 and 4) or GST-CTM (lanes 5 and 6). After washing, the beads were immunoblotted with antibodies against histone H3. Lanes 1 and 2 depict input nucleosomes. Data are representative of at least five independent experiments. (C) Sequences of the CTM domain from the indicated species were aligned with the Clustal program. Predicted helical regions (H) are indicated. Amino acids in the indicated stretches (residues 824–830, 874–876, and 881–884) were mutated to alanine. The sequences of residues in these regions are highlighted in red. (D) Preparations of GST (lane 1) and the indicated forms of GST-CTM (lanes 2–5) were stained with Coomassie blue. (E) A radiolabeled random-sequence double-stranded 30-mer probe (10 nM; lane 1) was tested through EMSA for binding to the indicated proteins (lanes 2–6). Final concentration of proteins was 670 nM. Data are representative of two independent experiments.
We considered the possibility that this region might bind to chromatin or DNA. To address this question, we constructed a fusion protein containing glutathione-S-transferase (GST) and residues 813–904 of MTBP, which we named the C-terminal MTBP (CTM) domain. In some initial experiments, we tested for binding to nucleosomes that had been prepared by digestion of chromatin from human HeLa cells with micrococcal nuclease (MN nucleosomes). We incubated GST-CTM or GST alone (both bound to glutathione agarose beads) with the nucleosomes. After isolating the beads, we monitored binding of MN nucleosomes by immunoblotting for histone H3. We observed that the MN nucleosomes associated efficiently with GST-CTM but not GST alone (Figure 3B). In parallel, we also examined recombinant (R) nucleosomes that consisted of recombinant histones and a 147–base pair DNA sequence with high-affinity for histone octamers (Lowary and Widom, 1998). Significantly, the R nucleosomes exhibited no binding to GST-CTM.
An explanation for these observations is that the CTM domain could be binding to DNA that might be more accessible in at least some MN but not R nucleosomes. Another possibility is that posttranslational modifications on histones could promote binding of Treslin-MTBP to MN nucleosomes. To investigate binding of the CTM domain to DNA, we performed an electrophoretic mobility shift assay (EMSA) with a 32P-labeled DNA substrate. We first utilized a 30-mer of double-stranded DNA (dsDNA) with a randomly generated sequence. We observed that this structure bound quite well to GST-CTM but not to GST alone (Figure 3D; compare lanes 2 and 3 in Figure 3E).
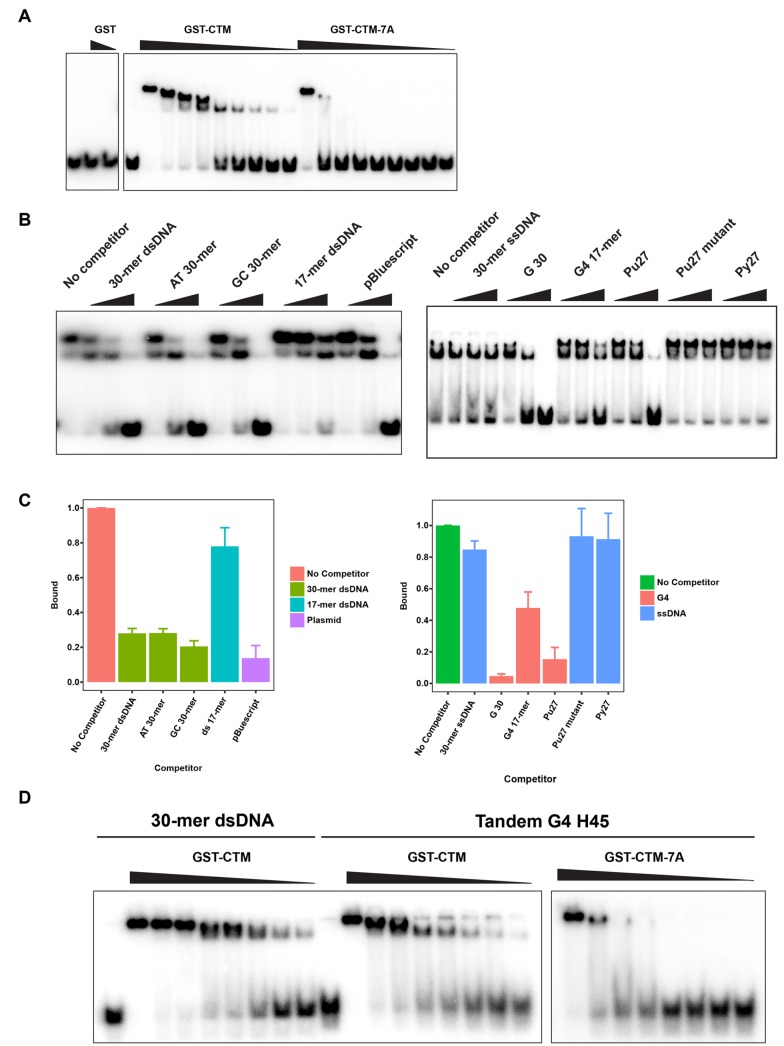
To assess further the specificity of this binding, we sought to develop a mutant of the CTM domain that would be compromised for binding to DNA. We mutated each amino acid in three conserved stretches of the CTM domain to alanine (residues 824–830, 874–876, and 881–884; see Figure 3C). GST fusions of all three mutated domains exhibited reduced binding to the random 30-mer dsDNA, but the 824–830 mutant appeared to be the most deficient (Figure 3D; compare lanes 3–6 in Figure 3E). For the remainder of this study, we chose to utilize the 824–830 mutant, which we refer to as the 7A mutant. In titration experiments with different amounts of wild-type and 7A mutant versions of the GST-CTM protein, we verified that the 7A mutant had substantially lower affinity for the random 30-mer dsDNA (Figure 4A and Supplemental Figure S3A).
FIGURE 4:
The CTM domain associates with dsDNA and G4 DNA. (A) A radiolabeled random-sequence 30-mer dsDNA probe (10 nM; first lane) was incubated with various amounts of GST alone (1 and 2 μM) or with either GST-CTM or GST-CTM-7A (0.0375, 0.075, 0.1, 0.15, 0.2, 0.3, 0.4. 0.6, and 2 μM) in the remaining lanes. Protein concentrations are depicted in descending order. Samples were electrophoresed in polyacrylamide gels and analyzed by phosphorimaging. Data are representative of three independent experiments. (B) The radiolabeled probe from A (10 nM) was incubated with GST-CTM (600 nM) in the absence (first lane) or presence of increasing amounts of the indicated competitor DNAs (remaining lanes). Competitor oligonucleotides were used at concentrations of 10, 50, and 250 nM. The pBluescript plasmid was added at concentrations of 0.2, 1, and 5 ng/μl. Samples were processed for EMSA. Data are representative of three independent experiments. (C) Quantitation of the results from B for samples containing 250 nM competitor DNA or 5 ng/μl pBluescript. Results depict the mean ± SEM for three experiments. (D) Radiolabeled random-sequence 30-mer dsDNA (10 nM) and G4 H45 DNA (10 nM) were incubated in the absence or presence of GST-CTM or GST-CTM-7A, as indicated. The protein concentrations were the same as in A, except that the lowest concentration (0.0375 μM) was omitted. Protein concentrations are depicted in descending order. Samples were processed for EMSA. Data are representative of three independent experiments.
The CTM domain exhibits similarities with the C-terminal domain of yeast Sld7
In budding yeast, Sld7 has been identified as a functionally important partner of Sld3 (Tanaka et al., 2011a, b; Itou et al., 2015). It has been proposed that MTBP may be an analogue of Sld7 (Boos et al., 2013). We obtained structural models of the CTM domain from human MTBP by using the Robetta server (Kim et al., 2004; Song et al., 2013). Interestingly, one model yielded a three-helical structure that bears strong similarity to a three-helical region in the reported crystal structure of the C-terminal region of yeast Sld7 (Itou et al., 2015) (Supplemental Figure S2A). We performed a sequence alignment on the basis of the positions of these three helices and could detect homology between yeast Sld7 and human MTBP in these regions (Pettersen et al., 2004) (Supplemental Figure S2B). Notably, one region with good homology coincided with the 824–830 region of MTBP, which we showed is important for DNA binding. These observations further support the possibility that vertebrate MTBP may be a functional analogue of yeast Sld7.
The C-terminal domain of yeast Sld7 formed a dimer in the crystals used for structural determination (Itou et al., 2015). However, it has not been reported that this domain exists as a dimer in solution. To evaluate whether the CTM domain from MTBP can form dimers, we performed gel filtration experiments (Supplemental Figure S2C). Since GST itself forms dimers, we produced a version of GST-CTM in which tobacco etch virus (TEV) protease could cleave between the GST and CTM portions of the fusion protein. We subjected protease-treated preparations of GST-TEV-CTM to gel filtration in Superdex-200 and found that the isolated CTM fragment clearly migrated with the expected size of a monomer. Therefore, the isolated CTM domain appears not to readily form dimers in solution.
The CTM domain also binds to G-quadruplex (G4) DNA
To evaluate whether other types of DNA could associate with the CTM domain, we first performed competition experiments (Figure 4, B and C). We initially verified that an excess of unlabeled 30-mer dsDNA could compete away binding of GST-CTM to the radioactive 30-mer dsDNA probe. Moreover, pBluescript double-stranded plasmid DNA competed effectively for binding, as did AT-rich or GC-rich 30-mers of dsDNA (see Supplemental Table S1 for oligonucleotide sequences). On the other hand, a single-stranded random-sequence 30-mer DNA could not serve as competitor. Furthermore, the length of the DNA appears to be an important parameter. For example, a double-stranded random-sequence 17-mer of DNA did not compete away binding of GST-CTM to the 30-mer dsDNA.
Next, we investigated the binding of the CTM domain to certain DNA structures that have been implicated in the initiation of replication. Various studies have connected G4 DNA to sites of initiation (Valton and Prioleau, 2016; Hansel-Hertsch et al., 2017). Although it has been reported that human ORC can interact with G4 DNA (Hoshina et al., 2013), a causal relationship between this property and initiation has not been established. To investigate whether the CTM domain might associate with G4 DNA, we first examined various oligonucleotides that can form G4 structures as competitors for binding of GST-CTM to the 30-mer dsDNA (Supplemental Table S1). Significantly, single-stranded G30, which can form G4 structures, could compete very effectively, despite the fact that a random-sequence single-stranded 30-mer was ineffective (Figure 4, B and C).
We also utilized a well-characterized G4 template known as Pu27 that was originally identified in the human c-Myc promoter (Siddiqui-Jain et al., 2002). We observed that Pu27 efficiently competed for binding of GST-CTM to the 30-mer dsDNA (Figure 4, B and C). On the other hand, the complement of this sequence, known as Py27, was inactive. Moreover, a mutant form of Pu27 with nucleotide substitutions that abrogate formation of G4 structures, could not compete away the binding of GST-CTM to the 30-mer dsDNA. Finally, we found that a shorter G4-forming 17-mer oligonucleotide called I100-15 showed reduced activity as a competitor in comparison with Pu27 (Rando et al., 1995; Mukundan et al., 2011). Nonetheless, this 17-mer, which can form a stacked dimeric G4 structure, was more effective than the 17-mer dsDNA (which we designed to contain the I100-15 sequence as one of its two strands).
To monitor more directly the binding of the CTM domain to G4 structures, we performed EMSA experiments. For these assays, we utilized a structure called H45, which contains tandem G4 structures from human telomeric DNA (Bugaut and Alberti, 2015). We observed that the radiolabeled H45 G4 DNA could be quantitatively bound by the GST-CTM protein, as indicated by a retarded electrophoretic mobility (Figure 4D). Thus, the CTM domain associates with this G4 structure quite efficiently.
We proceeded to compare the relative affinity of the CTM domain for this G4 structure versus the 30-mer dsDNA (Figure 4D and Supplemental Figure S3B). By performing titration experiments with different amounts of GST-CTM, we observed that this domain exhibited a comparable affinity for the H45 G4 structure and the 30-mer dsDNA. Finally, we examined binding of the GST-CTM-7A mutant to the H45 G4 structure. As was the case for the 30-mer dsDNA, binding of the 7A mutant to this G4 DNA was substantially reduced in comparison with the wild-type CTM domain (Figure 4D and Supplemental Figure S3B). Taken together, these experiments indicate that the CTM domain is relatively versatile DNA-binding module with high specificity for various DNA structures, including dsDNA and G4 DNA.
The CTM domain of MTBP is essential for DNA replication in egg extracts
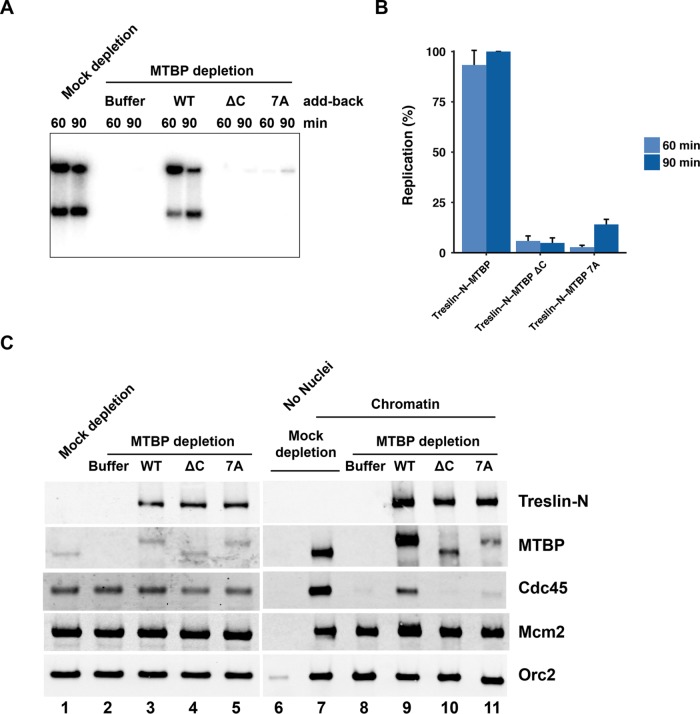
Next, we evaluated whether the CTM domain is important for DNA replication. We first turned to the Xenopus egg extract system. We depleted the endogenous Treslin-MTBP from egg extracts with anti-MTBP antibodies and then tested recombinant complexes containing various mutants of MTBP for their ability to support replication. As expected, depletion of Treslin-MTBP largely abolished DNA replication in the extracts (Figure 5A). In parallel, we added back wild-type or mutant Treslin-MTBP complexes to the depleted extracts. In the case of the mutants, we assayed complexes containing either the 7A mutant of full-length MTBP or a truncation of MTBP that lacks the C-terminal 91 amino acids (∆C). For the Treslin subunit, we utilized the Treslin-N fragment because, as explained above, it yields a higher signal in the replication assays (Supplemental Figure S4). As shown in Figure 5, A and B, a complex containing Treslin-N and wild-type MTBP restored DNA replication. On the other hand, complexes containing the 7A or ∆C mutants of MTBP were essentially inactive for supporting DNA replication. We obtained similar results with complexes containing full-length Treslin and these mutant MTBP proteins (not shown). Overall, the fact that replication is negligible in extracts containing MTBP without a functional CTM domain indicates that this region plays a critical role in DNA replication. Therefore, not only does this region associate with DNA, its presence is also necessary for chromosomal duplication in egg extracts.
FIGURE 5:
The CTM domain of MTBP is essential for DNA replication in egg extracts. (A) Egg extracts were mock-depleted with control antibodies or immunodepleted with anti-MTBP antibodies. Following this procedure, extracts were supplemented with buffer alone or recombinant Treslin-MTBP complexes containing wild-type, ∆C, or 7A versions of MTBP. For the Treslin subunit, we utilized the Treslin-N fragment. Extracts were assayed for chromosomal DNA replication. (B) Compilation of replication assay data from A and additional experiments. Results depict the mean ± SEM for three experiments. Values are expressed as the percentage of replication at 90 min in extracts containing wild-type MTBP. (C) Chromatin-binding assays. Egg extracts were mock-depleted with control antibodies or immunodepleted with anti-MTBP antibodies. Following this procedure, extracts were supplemented with buffer alone (lanes 1, 2, and 6–8) or recombinant Treslin-MTBP complexes containing Treslin-N and wild-type, ∆C, or 7A versions of MTBP (lanes 3–5 and 9–11). Extracts were immunoblotted for the indicated proteins (lanes 1–5). In addition, extracts were incubated in the absence (lane 6) and presence of sperm chromatin (lanes 7–11). After 45 min, chromatin fractions were prepared and immunoblotted for the indicated proteins (lanes 6–11). Data are representative of three independent experiments.
The CTM domain is necessary for association of MTBP with chromatin and loading of Cdc45 in Xenopus egg extracts
To investigate further the role of the CTM domain, we examined the loading of Cdc45 onto chromatin in the presence of mutant MTBP (Figure 5C). The association of Cdc45 with chromatin correlates closely with activation of the replicative helicase and initiation of replication in egg extracts (Mimura and Takisawa, 1998; Mimura et al., 2000; Walter and Newport, 2000). Furthermore, Treslin is necessary for this process (Kumagai et al., 2010). We prepared mock-depleted and MTBP-depleted extracts and examined the association of Cdc45 with chromatin at 45 min after the addition of sperm chromatin. Typically, at this time, Cdc45 has achieved nearly peak levels on chromatin, but DNA replication is only just beginning. As expected, there was little or no Cdc45 on chromatin in the MTBP-depleted extracts (which also lack Treslin) in comparison with the mock-depleted extracts (Figure 5C). In parallel, we also added back recombinant complexes containing Treslin-N and wild-type, ∆C, or 7A versions of MTBP to the depleted extracts. The complex containing wild-type MTBP restored binding of Cdc45 to chromatin, whereas complexes containing the ∆C or 7A mutants were ineffective in mediating the loading of Cdc45. Finally, we monitored the behavior of Mcm2, a subunit of the MCM complex, and observed that it bound equally well to chromatin from extracts containing the wild-type or mutant forms of MTBP. Therefore, the absence of functional CTM domain in MTBP does not interfere with loading of the MCM complex onto chromatin.
In these experiments, we also followed the binding of MTBP to chromatin. We observed that, in comparison with wild-type MTBP, both the ∆C and 7A proteins showed greatly diminished binding to chromatin (Figure 5C). There was also less binding of Treslin-N to chromatin in extracts containing complexes with the ∆C and 7A MTBP proteins, but the effect was less pronounced than for MTBP. Taken together, these experiments indicate that the C-terminal region of MTBP is critical for the association of MTBP with chromatin. Consequently, Treslin-MTBP complexes that lack a functional CTM domain are unable to recruit Cdc45 to origins.
The CTM domain is necessary for a normal S-phase in human cells
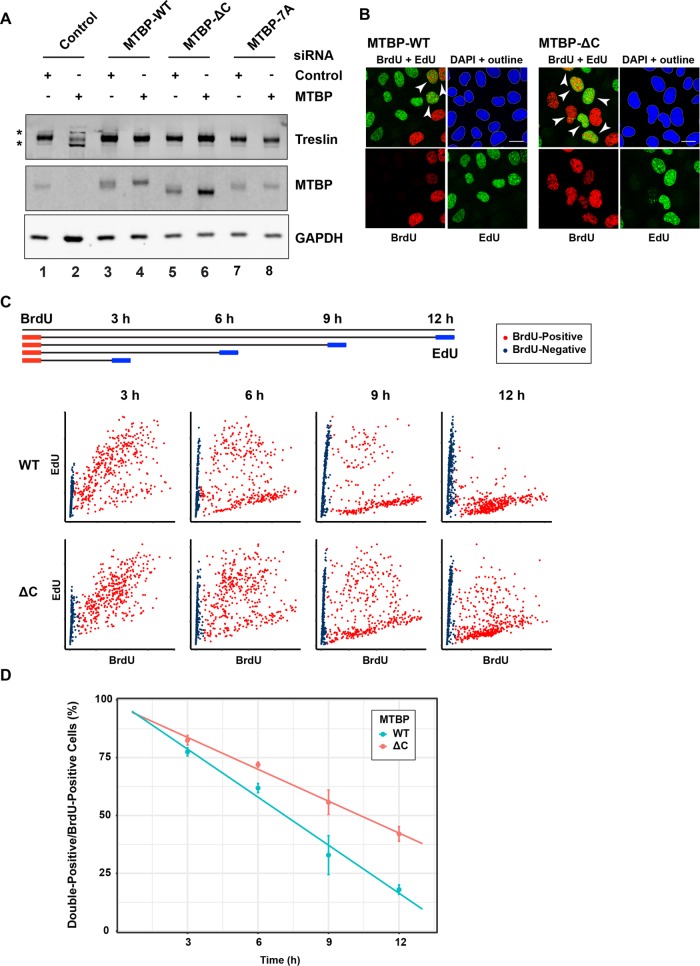
To assess the generality of these observations, we also performed experiments in human cells. For this purpose, we established human U2OS cells lines that could express doxycycline-inducible forms of wild-type, ∆C, or 7A versions of MTBP. We also rendered these forms of MTBP resistant to a combination of two MTBP small interfering RNAs (siRNAs). Next, we ablated the endogenous MTBP from such cells by treatment with these two siRNAs (Figure 6A). This treatment resulted in the very effective removal of MTBP from the cells. We noted that the depletion of MTBP also elicited a substantial reduction in the amount of Treslin in the cells. This observation further supports the existence of a close functional relationship between MTBP and Treslin. Expression of all three forms of recombinant MTBP (i.e., wild type, ∆C, and 7A) restored the expression of Treslin in the siRNA-ablated cells. Thus, binding of MTBP to Treslin is sufficient for the stabilization of Treslin.
FIGURE 6:
Ablation of MTBP from human cells leads to elongation of S-phase. (A) Human U2OS Flp-In T-REx cells were treated with control (lanes 1, 3, 5, and 7) or MTBP siRNA (lanes 2, 4, 6, and 8). As indicated, expression of siRNA-resistant wild-type (WT; lanes 3 and 4), ∆C (lanes 5 and 6), or 7A (lanes 7 and 8) versions of MTBP was induced by addition of doxycycline. Extracts were immunoblotted for human Treslin and MTBP. Loading control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Data are representative of three independent experiments. Asterisks denote background bands in lane 2. (B) The siRNA-treated U2OS cells expressing WT and ∆C versions of siRNA-resistant MTBP (see lanes 4 and 6 in A) were incubated for 30 min with BrdU at t = 0. At 9 h, cells were incubated with EdU for 30 min. Cells were processed for detection of incorporated BrdU and EdU, stained with 4’,6-diamidino-2-phenylindole (DAPI), and examined by confocal microscopy. The images depict staining patterns for BrdU, EdU, BrdU + EdU, and DAPI. Outlines of DAPI-stained nuclei denote areas that were identified for quantitation. Data are representative of three independent experiments. Bar, 10 μm. (C) Design of labeling time-course experiment with BrdU and EdU (top). The various bottom panels depict cells stained for BrdU (x-axis) and EdU (y-axis) at the indicated times. BrdU-positive cells are shown in red, and BrdU-negative cells are shown in blue. Each time point represents 800 cells. (D) Quantitation of the results from panel C. Results were compiled from three independent experiments (mean ± SEM). Paired Student's t test yielded p < 0.01.
We proceeded to monitor DNA replication in these cells by labeling with the nucleoside analogue EdU. We observed that the MTBP-depleted cells showed reduced incorporation of EdU, although it should be noted that these cells also have substantially diminished amounts of Treslin (Supplemental Figure S5). Although these results are generally consistent with those of Boos et al. (2013), these investigators reported that depletion of MTBP led to no decrease or only a minor reduction in the levels of Treslin (see Discussion). Expression of siRNA-resistant, wild-type MTBP effectively restored DNA replication (Supplemental Figure S5). Interestingly, cells expressing the ∆C and 7A mutants of MTBP also displayed good incorporation of EdU. However, these mutant-expressing cells divided more slowly and also appeared unhealthy in comparison with cells expressing wild-type MTBP. Therefore, we performed further analyses to investigate whether these cells were having difficulties with S-phase.
First, we examined whether there might be an elongation of S-phase in cells expressing mutant MTBP (Figure 6, B–D). For this purpose, we incubated cells with BrdU for 30 min. Next, we incubated these cells with EdU for 30 min every 3 h over the course of 12 h. Finally, we stained the cells and performed confocal microscopy to detect incorporation of BrdU and EdU into the DNA. Cells that were positive for BrdU and EdU had been in S-phase since the initial incubation with BrdU. Cells that were positive for BrdU but negative for EdU had left S-phase during the course of the experiment. For siRNA-treated cells expressing wild-type recombinant MTBP, we observed that almost all of the cells had exited S-phase by 12 h. In particular, only 12% of cells were still in S-phase at this point. On the other hand, in the case of cells expressing the ∆C mutant, a markedly greater proportion (42%) were still in S-phase at 12 h. Hence, cells expressing MTBP without a functional CTM domain have a pronounced elongation of S-phase.
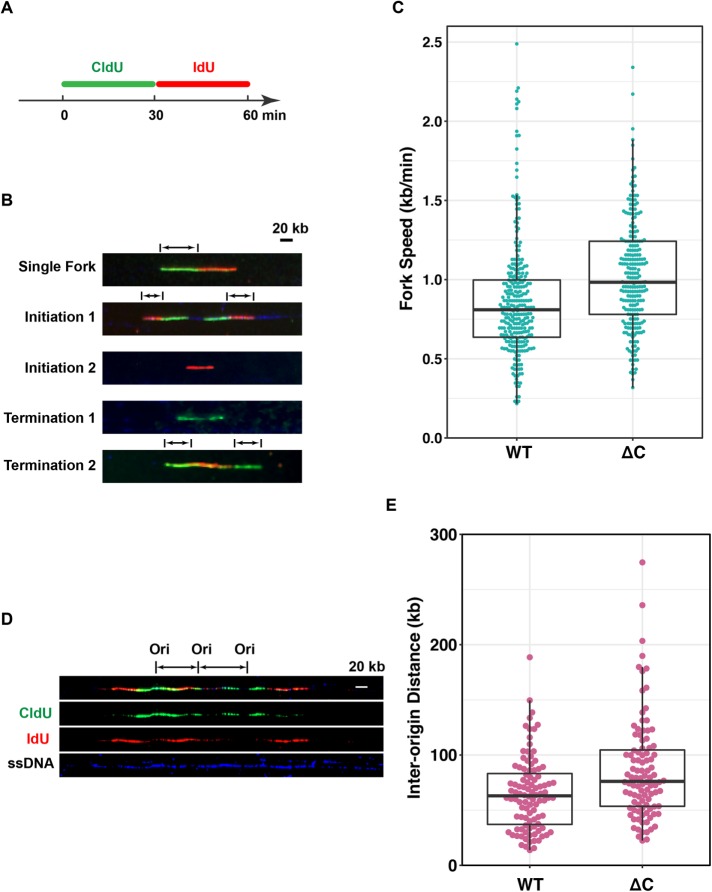
This protracted S-phase could be due to slower DNA elongation or diminished origin firing or both. To examine these possibilities, we performed DNA fiber studies to observe the spatial distribution of replication in siRNA-treated cells expressing wild-type or ∆C versions of MTBP (Figure 7). We sequentially incubated the cells with CldU and IdU for 30 min each and then prepared DNA fiber spreads. We first quantitated rates of fork elongation. Interestingly, the rates of fork elongation in cells expressing the MTBP ∆C mutant were actually significantly faster than in cells expressing wild-type MTBP, even though S-phase was notably slower in the mutant-expressing cells (Figure 7, A–C).
FIGURE 7:
Human cells expressing MTBP without a functional CTM domain exhibit diminished origin firing. (A) Schematic of procedure for labeling of DNA fibers. Human cells expressing wild-type (WT) and ∆C versions of MTBP were treated with siRNA, labeled sequentially with CldU and IdU, and processed for DNA combing as described under Materials and Methods. (B) Representative images used for determination of replication fork velocities. Arrows indicate the lengths of tracks used for measurement of fork velocities. (C) Quantitation of replication fork velocities in cells expressing MTBP-WT or MTBP-∆C. Three independent experiments were performed with measurement of more than 150 fibers each. A Mann-Whitney U test yielded p < 0.001. Box plot depicts second quartile, median, and third quartile. (D) Representative images used for determination of inter-origin distances. (E) Quantitation of inter-origin distances in cells expressing MTBP-WT or MTBP-∆C. Results are compilation of two independent experiments with 50 random samples measured for each condition in each experiment. A Mann-Whitney U test yielded p < 0.001. Box plot depicts the second quartile, median, and third quartile.
Various studies have indicated that the extent of origin firing can influence fork elongation (Petermann et al., 2010; Zhong et al., 2013). In particular, these studies established an inverse correlation between the frequency of origin firing and the rate of fork elongation. This phenomenon may be due to the availability of replication factors and DNA precursors. Accordingly, we directly examined the extent of origin firing in the siRNA-treated cells by measuring interorigin distances. We observed that there was a significant increase in inter-origin distances in cells expressing the ∆C version of MTBP in comparison with cells expressing wild-type MTBP (Figure 7, D and E). Therefore, origin firing occurs less frequently in cells containing a form of MTBP that lacks a functional CTM domain. Although these cells compensate by accelerating fork elongation, there is nonetheless still a significant lengthening of S-phase. Overall, these observations indicate that the CTM domain is important for origin firing in both Xenopus egg extracts and human cells.
DISCUSSION
In this report, we have investigated the mechanisms that underlie the initiation of chromosomal DNA replication in vertebrate cells. In particular, we have focused on the Cdk-mediated activation of the replicative helicase, which is a key process in the initiation of S-phase. As a handle on this problem, we have examined the role of Treslin and its more recently identified MTBP subunit. We have found that Treslin-MTBP functions as an integral unit in promoting replication. Furthermore, analysis of MTBP has revealed that this protein possesses a specific DNA-binding domain that is necessary for normal DNA replication.
In principle, Treslin-MTBP could utilize a DNA-binding domain in a number of ways. For example, this domain could recognize DNA at a prospective origin. Alternatively, this domain could act during activation of the replicative helicase. This process would involve melting of the DNA helix and movement of single-stranded DNA (ssDNA) to achieve an arrangement in the CMG complex for processive unwinding of the DNA. Our observations suggest that MTBP acts at a quite early step, such as origin selection. One reason is that mutants of MTBP without a functional CTM domain cannot interact efficiently with chromatin in egg extracts. Consequently, these mutants cannot mediate the recruitment of Cdc45 onto chromatin. Thus, the CTM domain appears to function before Cdc45 has achieved stable association with the MCM complex. Moreover, we have also observed that the CTM domain does not interact effectively with ssDNA.
It has been thought that MTBP might correspond to the vertebrate analogue of budding yeast Sld7, which interacts with Sld3 in the yeast system. However, the supporting evidence has largely been that MTBP associates with Treslin, the vertebrate form of Sld3. In this report, we have identified a functionally critical region of MTBP, the CTM domain, that displays some predicted structural similarity as well as actual sequence homology with the C-terminal region of Sld7. It will be interesting to assess whether this region is important for the function of Sld7 in budding yeast.
During our siRNA experiments, we found that ablation of MTBP from human U2OS cells also results in a substantial reduction in the intracellular concentration of Treslin. These results differ somewhat from those of Boos et al. (2013), who stated that ablation of MTBP from HeLa cells did not affect the levels of Treslin. However, these investigators mentioned that depletion of MTBP had a minor effect on the levels of Treslin in a subset of experiments with HeLa and U2OS cells. At least in our experiments, the fact that MTBP-depleted human cells have significantly reduced amounts of Treslin raises the issue of whether the decrease in replication in these cells is actually due to the absence of MTBP. Our findings in this paper have helped to clarify this issue. In particular, we have found that antibody-treated Xenopus egg extracts that lack both Treslin and MTBP can be rescued with recombinant Treslin-MTBP but not Treslin or MTBP alone. In addition, extracts containing mutant forms of MTBP without a functional CTM domain are defective for DNA replication even though such extracts contain a sufficient supply of Treslin. Furthermore, human cells harboring a version of MTBP without a functional CTM domain display S-phase defects even though such cells have normal amounts of Treslin. Notably, the absence of Sld7 in budding yeast also leads to a reduction in the amount of Sld3 (Tanaka et al., 2011b). Although Sld7 is not essential for cell growth in yeast, its absence does slow S-phase progression.
An intriguing aspect of this work is that Xenopus egg extracts and human cells appear to differ in the extent to which they require MTBP for initiation. There is essentially no replication in egg extracts containing versions of MTBP that lack a functional CTM domain. Therefore, the implication is that most if not all origins in this system have a stringent dependency on this region of MTBP. On the other hand, human cells harboring MTBP without a functional domain do successfully enter S-phase. However, these cells encounter difficulties during the progression of S-phase because of diminished origin firing.
The chromatin in egg extracts typically assembles within 30–45 min after the addition of demembranated sperm nuclei to the extracts. Therefore, this chromatin has not yet acquired some features of the mature chromatin in adult somatic cells (Fisher and Fisher, 2011). One possibility is that MTBP is strictly necessary for replication of chromatin with characteristics of an embryonic state. A potential implication is that adult somatic cells contain a mixture of different classes of origins. Those origins with a character more similar to an embryonic state would require MTBP, while others would have a more relaxed dependency on MTBP. Another possibility is that cancer cells like U2OS cells might be more permissive for origin firing. It will be interesting to compare the origins in human cells that depend on the CTM domain more stringently with those can fire without this domain.
The features that define replication origins in vertebrates, including humans, have not been defined conclusively (Aladjem and Redon, 2017). Although it is generally agreed that there is not a simple consensus sequence for origins as is the case in budding yeast, various studies have indicated roles for certain types of DNA sequences, states of chromatin, or nuclear organization in initiation (Fragkos et al., 2015; Valton and Prioleau, 2016; Hansel-Hertsch et al., 2017; Parker et al., 2017). For example, nucleosome-free regions, CpG islands, G-rich regions capable of forming G4 DNA, and other characteristics have been implicated as having some role in initiation. Nonetheless, these features do not appear to be an absolute necessity for all origins. Our findings have indicated that the CTM region of MTBP is a relatively versatile domain that can interact with various DNA structures including dsDNA, G4 DNA, and possibly others. Furthermore, since there would be at least two copies of MTBP in the native Treslin-MTBP complex, there would be two CTM domains present in this complex. This architecture might allow Treslin-MTBP to interact with multiple DNA sequence motifs in a combinatorial manner. In this scenario, Treslin-MTBP may be able to recognize an array of potential origins.
In this report, we have identified G4 DNA as a potential contributor to recognition of origins by MTBP. Various mapping studies have indicated that G4 DNA is enriched at certain origins (Valton and Prioleau, 2016; Hansel-Hertsch et al., 2017). However, not all G4-containing regions function as origins, and some origins appear not to contain G4 DNA. Our findings indicate that, although MTBP can associate with G4 DNA, it can interact with dsDNA as well. Therefore, MTBP would not be restricted to G4-containing regions, but such regions may nonetheless facilitate recruitment of MTBP when present. If Treslin-MTBP were to act in a combinatorial manner, then G4 DNA may function in concert with other origin determinants in some cases.
In closing, we have provided a substantial insight into how a key regulator of replication initiation interacts with the DNA in vertebrate cells. This information may help in the elucidation of further details of this critical process in vertebrates.
MATERIALS AND METHODS
Xenopus MTBP
Xenopus MTBP (GenBank: BC070600.1) was cloned from a Xenopus oocyte cDNA library with Phusion High Fidelity DNA polymerase (NEB). For production of a His10-tagged version of amino acids 437-860 from Xenopus MTBP, the appropriate DNA sequence was cloned into pH10UE (Hu et al., 2008).
Antibodies
Affinity purified antibodies against Xenopus and human Treslin were described previously (Kumagai et al., 2010). For antibodies against Xenopus MTBP, residues 437–860 were used as the antigen. A His10-tagged version of this polypeptide was produced in Escherichia coli Rosetta(DE3)pLysS cells and purified as described previously except that the elution buffer contained 0.5 M imidazole (Kumagai et al., 2011). These antibodies were produced in a commercial facility (Pocono Rabbit Farm and Laboratory). Other antibodies are described in Supplemental Table S2.
Xenopus egg extracts
Whole cytoplasmic extracts from Xenopus laevis eggs were prepared as before (Kumagai et al., 2010). NPE fractions from Xenopus eggs was prepared as described (Lebofsky et al., 2009; Guo et al., 2015). Immunodepletions, DNA replication assays, and preparation of chromatin fractions were also described previously (Kumagai et al., 1998, 2010; Guo et al., 2015). We used whole cytoplasmic egg extracts for all of the experiments in this paper except for the sucrose density gradient analyses, for which we used the NPE fraction. All procedures involving Xenopus laevis frogs were approved by the Caltech Institutional Animal Care and Use Committee.
Recombinant GST fusion proteins
To produce GST fusion proteins containing versions of the CTM domain from human MTBP (residues 813–904), appropriate DNA was amplified by PCR and cloned into pGEX4T-3 using NEBuilder HiFi DNA assembly master mix (NEB). Proteins were produced and purified as described previously (Guo et al., 2015).
Plasmids for expression in human cells
The plasmid pcDNA5/TO-Treslin-SF (which also encodes the SV40 NLS, an S-peptide tag, and a 3X-FLAG tag at the C-terminal end of human Treslin) was created as described (Guo et al., 2015). The production of pcDNA5/TO-Treslin-Myc was described previously (Kumagai et al., 2011). pcDNA5/TO-MTBP-TS (which encodes the SV40 NLS and the Twin-Strep tag at the C-terminal end of human MTBP) was created in a similar manner. Point and deletion mutants were created by PCR-based methods using Q5 High Fidelity DNA polymerase and HiFi DNA assembly master mix (NEB). All mutants were validated by DNA sequencing.
Human tissue culture cells
Human 293T and U2OS cells were cultured in DMEM supplemented with 10% fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin. Cells were tested at least monthly for mycoplasma by DAPI staining. For protein–protein interaction studies in 293T cells, cultures in six-well dishes (2 ml) were transfected with 1 µg total of the appropriate plasmids in the presence of 3 µg polyethyleneimine. After 24 h, cells were harvested and resuspended in 200 µl 20 mM HEPES-KOH (pH 7.5) containing 500 mM NaCl, 0.5% NP-40, 5 mM EDTA, 1 mM dithiothreitol (DTT), 10 mM β-glycerolphosphate, 1 mM NaF, 0.1 mM sodium vanadate, 0.6 µM tautomycin, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 10 µg/ml each of pepstatin, chymostatin, and leupeptin. The lysate was centrifuged at 16,000 × g for 10 min. The supernatant was diluted with 460 µl 20 mM HEPES-KOH (pH 7.5) containing 5 mM MgCl2 and incubated with 0.25 U/µl Benzonase for 30 min on ice. The lysate was clarified by centrifugation for 10 min. Proteins in the supernatant were immunoprecipitated with 1 µg anti-FLAG antibody or 1 µg Strep-MAB antibody bound to protein G Dynabeads for 1 h. Beads were washed three times with HEPES-buffered saline (HBS; 10 mM HEPES-KOH, pH 7.5, and 150 mM NaCl) containing 0.5% NP-40.
Purification of human Treslin–MTBP complex
Human 293T cells on a 150-mm plate (30-ml culture) were transfected with 2.3 µg pcDNA5/TO-Treslin-SF and 18.4 µg pcDNA5/TO-MTBP-TS in the presence of 63 µl of 1 mg/ml polyethyleneimine. After 48 h, cells were washed in phosphate-buffered saline (PBS) and lysed in 3 ml lysis buffer (10 mM HEPES-KOH, pH 7.5, 0.3 M NDSB-201 [3-(1-pyridinio)-1-propanesulfonate], 150 mM NaCl, 0.1% NP-40, 1 mM DTT, 0.5 mM PMSF, and 10 µg/ml each of pepstatin, chymostatin, and leupeptin). The lysate was centrifuged at 11,700 × g for 10 min. The supernatant was incubated for 2 h at 4°C with 40 µl anti-FLAG M2 agarose beads that had been prewashed in 0.1 M glycine (pH 3.5). Beads were washed three times with lysis buffer, three times with lysis buffer lacking NP-40, and once with elution buffer (10 mM HEPES-KOH, pH 7.5, 0.5 M NDSB-201, 150 mM NaCl, and 1 mM DTT). Beads were suspended in a total of 100 µl of elution buffer containing 0.25 mg/ml 3X-FLAG peptide and 10 µg/ml each of pepstatin, chymostatin, and leupeptin, and eluted for 1 h at 4°C. The eluate was dialyzed in egg lysis buffer (ELB; 10 mM HEPES-KOH, pH 7.7, 250 mM sucrose, 50 mM KCl, and 2.5 mM MgCl2) containing 1 mM DTT.
Purification of MTBP
Human 293T cells on 150 mm plate (30 ml) were transfected with 21 µg pcDNA5/TO-MTBP-TS in the presence of 63 µl of 1 mg/ml polyethyleneimine. After 48 h, cells were lysed in 3 ml of 10 mM HEPES-KOH (pH 7.5) containing 500 mM NaCl, 0.5% NP-40, 5 mM EDTA, 1 mM DTT, 1 mM PMSF, and 10 µg/ml each of pepstatin, chymostatin, and leupeptin. MTBP-TS was purified with Streptactin XT beads (IBA Life Sciences) and eluted in 10 mM HEPES-KOH (pH 7.5) containing 500 mM NaCl, 1 mM DTT, and 10 mM biotin. Bovine serum albumin (BSA) was added to 0.1 mg/ml and the protein was dialyzed in ELB overnight.
EMSA experiments
High-performance liquid chromatography–purified oligonucleotides purchased from IDT were used for producing 32P-labeled probes. Double-stranded oligonucleotides (1 pmol/µl) were denatured in 10 mM Tris-HCl (pH 7.4) containing 1 mM EDTA at 95°C for 5 min and annealed by slow cooling to room temperature. The oligonucleotides were phosphorylated in the presence of γ-[32P]ATP with T4 polynucleotide kinase. Various concentrations of proteins and 10 nM probe were incubated in 10 mM Tris-HCl (pH 7.5) containing 100 mM NaCl, 5% glycerol, 1 mM DTT, and 0.1 mg/ml BSA for 15 min at room temperature. The samples were electrophoresed in 10% polyacrylamide gels in 0.5× Tris-borate-EDTA (TBE) buffer at room temperature.
Oligonucleotides known to form G4 structures were denatured in 10 mM Tris-HCl (pH 7.4) containing 60 mM KCl and 1 mM EDTA at 95°C for 5 min and annealed by slow cooling to room temperature. The oligonucleotides were purified in 15% polyacrylamide gels and labeled with 32P as above. The EMSA reaction was performed as above except that the buffer contained 62.5 mM KCl and 37.5 mM NaCl. The gels were run in 0.5× TBE at 4°C.
Sucrose gradient centrifugation
A sucrose gradient (1.4 ml) was formed by layering 200 µl each of 10%, 15%, 20%, 25%, 30%, 35%, and 40% sucrose in ELB and incubating for 2 h at room temperature and 1 h at 4°C. NPE (25 µl) was diluted with 1 volume of ELB containing no sucrose and loaded onto the gradient. The samples were centrifuged at 35,000 rpm for 16 h at 4°C in a TLS55 rotor in a Beckman TL100 ultracentrifuge.
Gel filtration chromatography
A TEV protease site was created between GST and the CTM coding sequence in pGEX-CTM. The GST-TEV-CTM protein was expressed and purified as for GST-CTM. GST-TEV-CTM was digested with AcTEV protease (Thermo Fisher) in TEV protease buffer at 4°C overnight. The digested protein was chromatographed in a Superdex-200 column equilibrated with HBS containing 1 mM DTT.
Structural modeling
The Robetta full-chain protein structure prediction server was used to model the structure of the CTM domain (Kim et al., 2004; Song et al., 2013). Amino acids 813–904 from human MTBP were used as the input. One of five models generated was highly similar to the structure of the C-terminal domain of yeast Sld7 (Itou et al., 2015). A sequence alignment was created by superimposing the modeled structure of the CTM domain on the published Sld7 C-terminal structure and using the UCSF Chimera package (Pettersen et al., 2004).
Production of U2OS cell lines capable of expressing MTBP and its mutants
U2OS Flp-In T-REx cells (kindly provided by Stephen Blacklow, Harvard Medical School, MA) were maintained in the presence of 15 μg/ml Blasticidin S (Malecki et al., 2006). Cells were transfected with pcDNA5/FRT/TO encoding siRNA-resistant forms of full-length human MTBP and its mutants (7A and ∆C) with the Fugene 6 transfection reagent (Promega). These constructs also contained the SV40 NLS and the Twin-Strep tag at the C-terminal end (MTBP-TS). Cells were selected with 200 μg/ml Hygromycin B and 15 μg/ml Blasticidin S. Single colonies that expressed the desired protein on addition of 1 µg/ml doxycycline were isolated and used for analysis.
siRNA treatment
Expression of siRNA-resistant MTBP was induced in the U2OS-MTBP cell lines with 1 µg/ml doxycycline for 24 h before the transfection. Cells were transfected with 100 nM control siRNA (Dharmacon ON-TARGET plus nontargeting siRNA #1) or MTBP siRNA (a 1:1 mixture of Dharmacon ON-TARGET plus MTBP siRNA #1 and #2) in the presence of Lipofectamine RNAiMAX (Thermo Fisher Scientific). See Supplemental Table S1 for oligonucleotide sequences. After 24 h, cells were transfected again with 50 nM control or MTBP siRNA. Cells were maintained in 1 µg/ml doxycycline except for several hours during and after the transfection. After 96 h, cells were processed for characterization of functional effects.
Fluorescence-activated cell sorting (FACS) analysis
Cells were incubated with 20 µM EdU for 30 min before harvesting. After collection, cells were fixed in 70% ethanol. The click reaction was performed for 30 min in Tris-buffered saline (TBS) containing 4 mM CuSO4, 2 µM Alexa 488 azide, and 10 mM sodium ascorbate (Salic and Mitchison, 2008). Cells were treated in 0.2 mg/ml RNase A and stained with 2 µM 7-AAD. FACS analysis was performed with a FACSCalibur flow cytometer (BD Biosciences).
Dual labeling of cells with BrdU and EdU
Cells were incubated with 20 µM BrdU for 30 min. After various time intervals, cells were incubated with 20 µM EdU for 30 min. Following fixation with 2% formaldehyde for 10 min and permeabilization with 0.5% Triton X-100 for 10 min, cells were treated with 2 N HCl for 30 min. Subsequently, the click reaction was performed for 30 min in TBS containing 4 mM CuSO4, 4 µM Alexa 488 azide, and 10 mM sodium ascorbate. Cells were then stained with 1 µg/ml mouse anti-BrdU antibodies (MoBU-1; Thermo Fisher Scientific) followed by incubation with goat anti-mouse Alexa 594 antibodies (Thermo Fisher Scientific). Nuclei were stained with 0.5 µg/ml DAPI for 10 min. Images were obtained with a Zeiss LSM 710 laser scanning confocal microscope with a 20× Plan-Apochromat objective. Image analysis was performed on Sum Slices Projection Images generated from confocal stacks.
DNA combing assays
Cells expressing wild-type and ∆C versions of MTBP were treated with MTBP siRNA for 96 h. Cells were labeled with 100 µM CldU for 30 min, washed sequentially with PBS and DMEM, labeled with 100 µM IdU for 30 min, and finally washed in cold PBS before harvesting by treatment with trypsin. Cells were embedded in agarose plugs (75,000 cells per 90 µl). DNA was extracted using the FiberPrep kit (Genomic Vision) and combed onto vinylsilane-coated coverslips (Combicoverslips) at 300 µm/s using a combing machine assembled according to a published procedure (Kaykov et al., 2016). Combed DNA was denatured in 0.5 NaOH for 25 min and washed in PBS. The coverslips were incubated with mouse anti-BrdU (B44) for detection of IdU, rat anti-BrdU (BU1/75 ICR1) for detection of CldU, and rabbit anti-ssDNA for 1 h at 37°C, and washed with PBS. Finally, coverslips were incubated with Alexa 594 goat anti-mouse immunoglobulin G (IgG), Alexa 488 goat anti-rat IgG, and Alexa 647 goat anti-rabbit IgG, and washed with PBS. Images were obtained by wide-field microscopy using Zeiss AxioObserver.Z1 and LSM 800 microscopes with 40× Plan-Apochromat objectives.
Quantification and statistical analysis
EMSA gels were scanned with a Typhoon FLA 9500 phosphorimager and the images were quantified with Fiji (ImageJ). Microscope images were loaded into CellProfiler (Lamprecht et al., 2007). Nuclei were identified by DAPI staining and outlined. Integrated intensities for the EdU signal and BrdU signal for each nucleus were quantified. For each experimental condition, 500–2000 nuclei were quantified with CellProfiler. Error bars depict the mean ± SEM. “n” values are indicated in the figure legends. Statistical analyses (Student's t test and Mann-Whitney U test) were performed in RStudio using the t.test and wilcox.test functions, respectively.
Supplementary Material
Acknowledgments
We are grateful to Kanomi Sasaki-Capela and Bashar Alhoch for technical assistance. We also thank Rochelle Diamond for assistance with the FACS analyses. Imaging studies were performed in the Caltech Biological Imaging Center, which is supported by the Beckman Institute and the Arnold and Mabel Beckman Foundation. We are also grateful to Lea Goentoro for access to the Zeiss AxioObserver.Z1 microscope. This work was supported by National Institutes of Health grants GM-043974 and GM-070891 to W.G.D.
Abbreviations used:
- CTM
C-terminal MTBP
- DAPI
4’,6-diamidino-2-phenylindole
- dsDNA
double-stranded DNA
- DTT
dithiothreitol
- FACS
fluorescence-activated cell sorting
- G4
G-quadruplex
- GINS
go-ichi-ni-san
- GST
glutathione-S-transferase
- MCM
mini-chromosome maintenance
- MTBP
Mdm2-binding protein
- NPE
nucleoplasmic extract
- ORC
origin recognition complex
- PMSF
phenylmethylsulfonyl fluoride
- TBE
Tris-borate-EDTA
- TEV
tobacco etch virus
- TRCT
Treslin C-terminal.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-07-0448) on September 6, 2017.
REFERENCES
- Aladjem MI, Redon CE. Order from clutter: selective interactions at mammalian replication origins. Nat Rev Genet. 2017;18:101–116. doi: 10.1038/nrg.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. Initiation of DNA replication in Xenopus egg extracts. Front Biosci. 2004;9:3029–3045. doi: 10.2741/1457. [DOI] [PubMed] [Google Scholar]
- Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, Ponting CP, Diffley JF. Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr Biol. 2011;21:1152–1157. doi: 10.1016/j.cub.2011.05.057. [DOI] [PubMed] [Google Scholar]
- Boos D, Yekezare M, Diffley JF. Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science. 2013;340:981–984. doi: 10.1126/science.1237448. [DOI] [PubMed] [Google Scholar]
- Bugaut A, Alberti P. Understanding the stability of DNA G-quadruplex units in long human telomeric strands. Biochimie. 2015;113:125–133. doi: 10.1016/j.biochi.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Capra JA, Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev. 2011;21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Fragkos M, Ganier O, Coulombe P, Mechali M. DNA replication origin activation in space and time. Nat Rev Mol Cell Biol. 2015;16:360–374. doi: 10.1038/nrm4002. [DOI] [PubMed] [Google Scholar]
- Guo C, Kumagai A, Schlacher K, Shevchenko A, Shevchenko A, Dunphy WG. Interaction of Chk1 with Treslin negatively regulates the initiation of chromosomal DNA replication. Mol Cell. 2015;57:492–505. doi: 10.1016/j.molcel.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol. 2017;18:279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- Hoshina S, Yura K, Teranishi H, Kiyasu N, Tominaga A, Kadoma H, Nakatsuka A, Kunichika T, Obuse C, Waga S. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J Biol Chem. 2013;288:30161–30171. doi: 10.1074/jbc.M113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu RG, Wang H, Xia Z, Varshavsky A. The N-end rule pathway is a sensor of heme. Proc Natl Acad Sci USA. 2008;105:76–81. doi: 10.1073/pnas.0710568105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itou H, Shirakihara Y, Araki H. The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3. Acta Crystallogr D Biol Crystallogr. 2015;71:1649–1656. doi: 10.1107/S1399004715010457. [DOI] [PubMed] [Google Scholar]
- Kaykov A, Taillefumier T, Bensimon A, Nurse P. Molecular combing of single DNA molecules on the 10 megabase scale. Sci Rep. 2016;6:19636. doi: 10.1038/srep19636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J Cell Biol. 2011;193:995–1007. doi: 10.1083/jcb.201102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, Walter JC. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol Biol. 2009;521:229–252. doi: 10.1007/978-1-60327-815-7_13. [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Masuda T, Matsui T, Takisawa H. Central role for Cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells. 2000;5:439–452. doi: 10.1046/j.1365-2443.2000.00340.x. [DOI] [PubMed] [Google Scholar]
- Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Ji Z, Struhl K. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc Natl Acad Sci USA. 2016;113:E4810–E4819. doi: 10.1073/pnas.1609060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukundan VT, Do NQ, Phan AT. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011;39:8984–8991. doi: 10.1093/nar/gkr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MW, Botchan MR, Berger JM. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit Rev Biochem Mol Biol. 2017;52:107–144. doi: 10.1080/10409238.2016.1274717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci USA. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Prioleau MN, MacAlpine DM. DNA replication origins—where do we begin? Genes Dev. 2016;30:1683–1697. doi: 10.1101/gad.285114.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando RF, Ojwang J, Elbaggari A, Reyes GR, Tinder R, McGrath MS, Hogan ME. Suppression of human immunodeficiency virus type 1 activity in vitro by oligonucleotides which form intramolecular tetrads. J Biol Chem. 1995;270:1754–1760. doi: 10.1074/jbc.270.4.1754. [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Diffley JF, Ponting CP. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr Biol. 2010;20:R509–510. doi: 10.1016/j.cub.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 2010;24:183–194. doi: 10.1101/gad.1860310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui K, On KF, Diffley JF. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol. 2013;5:a012930. doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol. 2013;5:a010371. doi: 10.1101/cshperspect.a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol. 2011a;21:2055–2063. doi: 10.1016/j.cub.2011.11.038. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Umemori T, Endo S, Muramatsu S, Kanemaki M, Kamimura Y, Obuse C, Araki H. Sld7, an Sld3-associated protein required for efficient chromosomal DNA replication in budding yeast. EMBO J. 2011b;30:2019–2030. doi: 10.1038/emboj.2011.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JM, Foulk MS, Casella C, Gerbi SA. The hunt for origins of DNA replication in multicellular eukaryotes. F1000Prime Rep. 2015;7:30. doi: 10.12703/P7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton AL, Prioleau MN. G-Quadruplexes in DNA replication: a problem or a necessity? Trends Genet. 2016;32:697–706. doi: 10.1016/j.tig.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Nellimoottil T, Peace JM, Knott SR, Villwock SK, Yee JM, Jancuska JM, Rege S, Tecklenburg M, Sclafani RA. The level of origin firing inversely affects the rate of replication fork progression. J Cell Biol. 2013;201:373–383. doi: 10.1083/jcb.201208060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.