NDK5 promotes assembly of motile cilia and flagella with its structure and protein phosphorylation–related reactions instead of the canonical NDK activity. The novel mechanisms and dominant-negative effect of mutated functional NDK5 reveal the remarkable versatility of a molecular platform that is used in diverse cellular processes.
Abstract
Nucleoside diphosphate kinases (NDKs) play a central role in diverse cellular processes using the canonical NDK activity or alternative mechanisms that remain poorly defined. Our study of dimeric NDK5 in a flagellar motility control complex, the radial spoke (RS), has revealed new modalities. The flagella in Chlamydomonas ndk5 mutant were paralyzed, albeit only deficient in three RS subunits. RS morphology appeared severely changed in averaged cryo-electron tomograms, suggesting that NDK5 is crucial for the intact spokehead formation as well as RS structural stability. Intriguingly, ndk5’s flagella were also short, resembling those of an allelic spoke-less mutant. All ndk5’s phenotypes were rescued by expressions of NDK5 or a mutated NDK5 lacking the canonical kinase activity. Importantly, the mutated NDK5 that appeared fully functional in ndk5 cells elicited a dominant-negative effect in wild-type cells, causing paralyzed short flagella with hypophosphorylated, less abundant, but intact RSs, and accumulated hypophosphorylated NDK5 in the cell body. We propose that NDK5 dimer is an RS structural subunit with an additional mechanism that uses cross-talk between the two NDK monomers to accelerate phosphorylation-related assembly of RSs and entire flagella.
INTRODUCTION
It is well established that nucleoside diphosphate kinases (NDKs) transfer a phosphate from NTPs to NDPs (Berg and Joklik, 1953) via a typical ping-pong mechanism. The γ-phosphate of an NTP first forms a high-energy intermediate with a conserved histidine (designated as H121 in this article) located in a narrow catalytic cleft. Following the departure of the resulting NDP, the phosphate is transferred to the recipient NDP containing a different base (reviewed by Lascu and Gonin, 2000). These reactions enable the levels of nucleotide species to be appropriately balanced in diverse cellular compartments. Consistent with the fundamental importance in nucleotide metabolism, NDKs are ubiquitous, expressed in both prokaryotes and eukaryotes, often by multiple NDK genes. Based on sequence similarities, NDKs have been divided into group I (NDK1-4) and group II (NDK5-10) (reviewed by Desvignes et al., 2009). NDKs have many aliases derived from independent research, most notably NM23 or NMEs, because of their elevated transcript levels in non- or low metastatic cancer cells (Steeg et al., 1988).
However, accumulating evidence indicates that NDKs are not merely interconverting NDPs and NTPs. They interact with many molecules (reviewed by Steeg et al., 2011). Direct interactions allow NDKs to stimulate GTP loading of their dynamic GTPase partners that drive constant fission and fusion of diverse membranous compartments (Boissan et al., 2014). Furthermore, NDK2 modulates G proteins and cation channels as a histidine protein kinase (Srivastava et al., 2006; Di et al., 2010; Cai et al., 2014). It is thought that the phosphate transiently linked to H121 in NDKs can be preferentially transferred to specific residues in target proteins instead of to NDPs (reviewed by Attwood and Wieland, 2015). Although histidine phosphorylation is a common prokaryotic regulatory mechanism, NDK2-mediated histidine phosphorylation is a bona fide eukaryotic strategy for regulating critical processes such as lymphocyte activation. A number of NDK-mediated reactions—such as modulations of malignancy of cancer cells (e.g., Steeg et al., 1988; MacDonald et al., 1993, 1996; Chang et al., 1996; Wagner et al., 1997; Carotenuto et al., 2013) and transcriptional regulation of c-myc oncogene (e.g., Postel et al., 1993; Thakur et al., 2009)—do not correlate with the NDK activity or require H121. Similarly, a heterozygous NDK mutation (killer-of-prune or kpn) is lethal to a seemingly healthy Drosophila strain with purple eyes (prune) (Sturtevant, 1956; Rosengard et al., 1989; D’Angelo et al., 2004; Carotenuto et al., 2013; Zhang et al., 2015; reviewed by Takacs-Vellai et al., 2015), although the mutation itself does not affect the NDK activity (Lascu et al., 1992). While metastasis suppression and dominant-negative (DN) lethality naturally have attracted much interest for decades, the molecular underpinnings remain uncertain (Steeg et al., 2011).
Group II NDKs diverged from group I before chordate radiation (Desvignes et al., 2009) and perhaps even before the divergence from the last common eukaryotic ancestor. Most group II NDKs have acquired additional molecular modules and primarily reside in motile cilia and flagella, which likely existed in the eukaryotic ancestor (Mitchell, 2007). Flagellar NDKs are built into the microtubule-based axonemal superstructure in motile cilia and flagella (Ogawa et al., 1996; Padma et al., 2001; Munier et al., 2003; Sadek et al., 2003; Patel-King et al., 2004; Duriez et al., 2007). In humans and other mammals, defects in these NDKs results in Primary cilia dyskinesia (PCD), demonstrating their importance in cilia biology (Duriez et al., 2007; Vogel et al., 2012). However, surprisingly, the H121 residue that is required for phosphate transfer between NDPs and NTPs and conserved in group I NDKs is not strictly conserved in group II NDKs (Munier et al., 2003; Patel-King et al., 2004). Furthermore, respiratory cilia of NDK8 PCD patients lacked the NDK8-containing outer dynein complex (Duriez et al., 2007).
These observations led to a prediction that flagellar NDKs are structural proteins and that the H121 in some NDK domains, and thus the NDK activity, is merely an evolutionary relict (Munier et al., 2003). However, other lines of evidence argue that the NDK activity is important. For example, NDK7 copurifies with the γ-tubulin ring complex from the centrosome that supports ciliogenesis at the interphase, and only recombinant NDK7 with H121 could promote microtubule polymerization from the centrosome in vitro (Liu et al., 2014). NDK5 (alias NME5, NDPK5, NM23-5, and RSP23) is a constitutive subunit of the radial spoke (RS), a molecular complex that exhibits NDK activity (Patel-King et al., 2004) and controls flagellar motility. Mouse NDK5 mutation affects the motility of sperm flagella and of both airway and ependymal cilia but does not disturb left–right asymmetry, a process that requires nodal cilia that lack radial spokes (Vogel et al. 2012). Although ciliary and flagellar dyskinesia is the only known phenotype from RS deficiencies in diverse organisms (e.g., Huang et al., 1981; Jeanson et al., 2015), ndk5 knockout mice are sterile and have sperm that are apoptotic, with short flagella or completely aflagellate (Vogel et al., 2012), similar to mutant mice defective in adenylate kinase 7, which is supposed to produce NTP (Fernandez-Gonzales et al., 2009). These observations raised the possibility that flagellar NDKs are part of a redundant energy metabolic circuitry built into the microtubule-based scaffold to supply NTP for reactions occurring throughout these lengthy organelles (Mitchell et al., 2005; Takei et al., 2014). To elucidate the role of NDK5 in cilia and flagella, we took advantage of in vivo approaches that are uniquely possible in Chlamydomonas (Merchant et al., 2007). The results revealed unexpected actions of NDK5 that shed light on NDKs and flagellar biology.
RESULTS
NDK5H121A minigene caused paralyzed short flagella
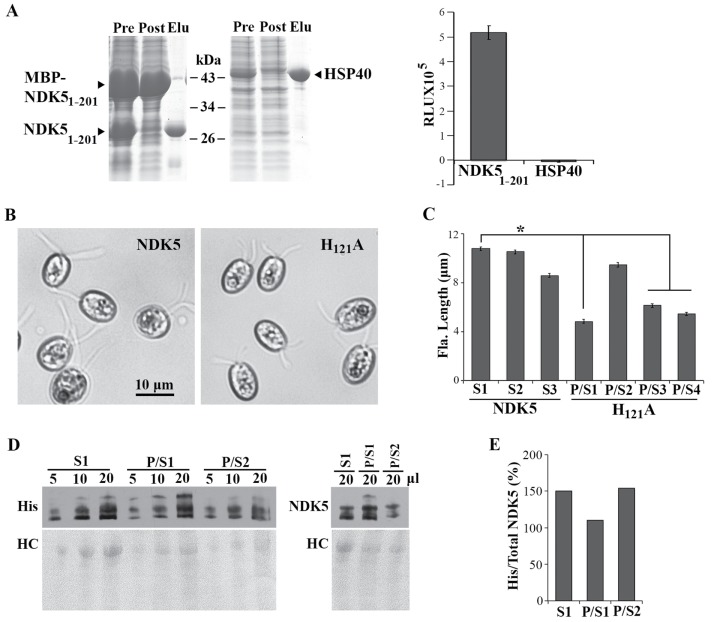
Previous studies demonstrate that isolated RS complexes exhibit NDK activity and that spoke-less mutant axonemes have half of the NDK activity of wild-type (WT) axonemes (Patel-King et al., 2004). To directly test the NDK activity of NDK5 and to overcome the propensity of NDK5 to precipitate (Munier et al., 1998), we expressed in bacteria maltose-binding protein (MBP)-His-NDK51-201 (Donnelly et al., 2006) that contains the conserved NDK domain and the Dpy30 domain but lacks the extended C-terminal tail unique to algal NDK5. After cleavage of the MBP moiety in bacteria, soluble His-NDK51-201 in the extracts was purified by Ni2+ affinity chromatography (Figure 1A, left). The NDK activity was measured using a luciferase-based bioluminometric assay (Karamohamed et al, 1999). As expected, addition of purified His-NDK51-201 to the reaction mixture elicited 5 × 105 relative light units (RLU), compared with −0.004 × 105 RLU for the His-HSP40 control (Figure 1A, right).
FIGURE 1:
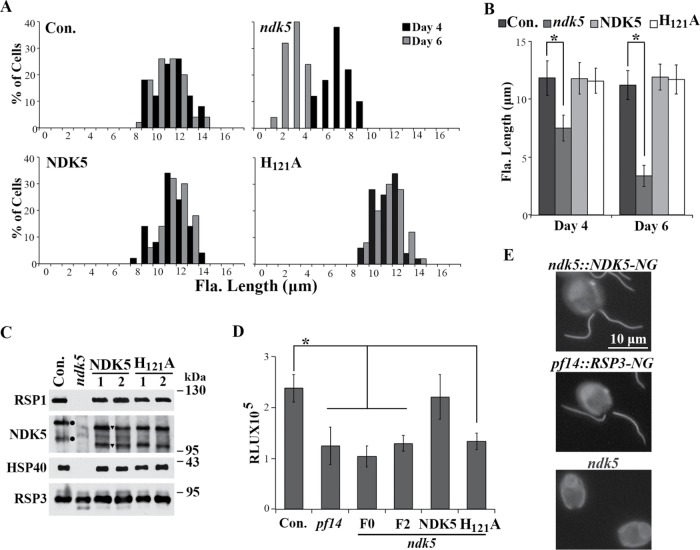
NDK5H121A expressed from a minigene caused paralyzed short flagella in WT cells. (A) Recombinant His-NDK51-201 exhibited NDK activity. Coomassie blue–stained SDS–PAGE showed Ni-NTA purification of His-tagged recombinant proteins (left). Pre, extract from bacteria expressing both MBP-His-NDK51-201 and the TVMV protease that could cleave off MBP; Post, the flow-through; Elu, the eluate. Identical amounts of proteins were analyzed in triplicate for NDK activity using the luciferase-based bioluminescence assay (right). RLU, relative light unit. (B) Representative transgenic strains from WT cells transformed with the minigene expressing full-length His-tagged NDK5 (left) or NDK5H121A (right). NDK5H121A transformants had short paralyzed flagella. The cells were fixed before light microscopy for length measurement. (C) Average flagellar length (Fla. Length) of transgenic strains expressing NDK5 or NDK5H121A. All strains from the NDK5 group, represented by randomly selected S1, S2, and S3 strains, contained only swimmers (S) like the parental strain. Four strains from the NDK5H121A groups contained mostly paralyzed cells but with a fraction of swimmers (P/S). Flagella of three of the four P/S strains had shorter flagella than those of the control strain. Asterisk indicates statistically significant differences (Student’s t test, p < 0.01, n = 50). (D) An anti-His Western blot revealed His-tagged NDK5 polypeptides in the flagellar samples from randomly selected S strains and representative P/S strains (left, top). Different protein loads indicated the signals were not oversaturated. Both His-tagged and untagged-NDK5 proteins were revealed by NDK5 polyclonal antibody (right, top). Total proteins were revealed by Ponceau red (bottom). Dynein, dynein heavy chains. (E) Plot Lanes analysis in the ImageJ software showed the ratio of His-tagged NDK5 vs. total NDK5 in the 20 μl flagellar samples in the Western blots in D.
To elucidate the role of NDK5, we perturbed NDK5 genetically. Although mutations of flagellar genes are mostly recessive (Luck et al., 1977), based on the NDK5’s Dpy30 dimerization domain (Sivadas et al., 2012) and the DN effect observed for Drosophila NDKkpn and heteromeric oligomers (Wilkie, 1994), we predicted that expression of inactive NDK5 might also elicit DN effects. Therefore we first engineered an NDK5-6His minigene with the regulatory elements of the LC8 gene, which encodes an abundant 10-kDa protein present in many molecular complexes, including RSs (Yang et al., 2009). We also built into the plasmid a paromomycin (PMM)-resistance cassette for selecting transformants. To abolish the NDK activity, the codon for H121, which acts as the transient recipient of the NTP γ-phosphate, was mutated to an alanine codon. The NDK5 and NDK5H121A minigenes were transformed into WT cells. All the screened 300 PMM-resistant colonies in the NDK5 control group were indistinguishable from WT cells, indicating that the minigene itself was not harmful. In contrast, four of 300 clones in the NDK5H121A group (P/S1-4) contained Paralyzed cells and Swimmers. In three of the four P/S strains, the flagella were about one-half of the expected 10–12 μm (Figure 1, B and C, p < 0.01). Western blots of flagella did not reveal obvious anomalies but confirmed the presence of His-tagged NDK5 and NDK5H121A in the flagella of the randomly selected S strain and two representative P/S strains (Figure 1D). Quantitative analysis showed similar, if not higher, His-tagged NDK5/Total NDK5 ratios in the S strain flagella (Figure 1E). Thus NDK5H121A, rather than the abundance of His-tagged NDK5 proteins, likely accounted for the motility and length phenotype. While these results were consistent with those in the sperm of ndk5 mice, flagellar lengths in Chlamydomonas varied among the strains; in contrast, none of existing RS mutants was known to exhibit such a prominent length phenotype, except for the LC8 mutant (fla14), whose short flagella were attributed to pleiotropic deficiencies in multiple flagellar complexes (Pazour et al., 1998; Yang et al., 2009). We further investigated an ndk5 recessive mutant and generated genomic constructs to elucidate the role of NDK5 and to test the DN effect of NDK5H121A independently.
Radial spoke defects in paralyzed flagella of an ndk5 insertional mutant
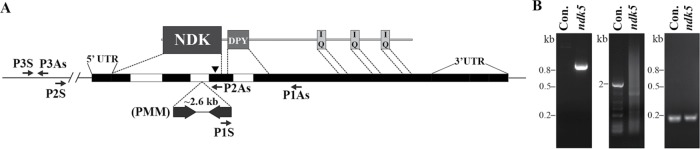
An ndk5 mutant was isolated from a large-scale indexed insertional mutagenesis project (Li et al., 2016). A 21–base pair flanking sequence from high-throughput screening suggested that a PMM-resistance DNA fragment was inserted in the second intron of the NDK5 gene (Figure 2A) in this strain. The strain was paralyzed as expected for RS mutants. The insertion was confirmed by two PCR amplifications of predicted fragments from the genomic DNA using primer pairs that are conducive for either ndk5 or its parental strain control (“Con.” in Figure 2B, left and middle). A nearby region amplified using a primer pair annealing to the middle of the neighboring gene, SPL4, which encodes a putative RNA splicing factor (Figure 2, A and B, right), showed that this region was intact in both strains. The trace amount of full-length NDK5 (Figure 3A and see Discussion) suggests that there is no large deletion at the 5′ end of the NDK5 gene in the mutant.
FIGURE 2:
Characterization of an ndk5 insertional mutant. (A) Schematic illustrating NDK5 protein and gene organization. NDK5 has an NDK domain followed by a Dpy-30 domain and an extended C-terminal tail containing three calmodulin-binding IQ motifs. The ndk5 mutant has an insertion of a 2.6-kb PMM-resistance cassette in the second intron. Black box, exon; white box, intron; black line, flanking sequence; arrowhead, the H121 codon; arrows, PCR primers. (B) PCR diagnosis of the insertion mutagenesis using templates prepared from ndk5 and the parental strain (Con.). A 0.8-kb fragment at the 3′ junction was amplified with the P1S and P1As primer pair from the mutant but not from Con. (left). A nearly 2-kb fragment at the 5′ region, including the second intron, was amplified with the P2S and P2As primer pair from the control but not from ndk5 (middle). An ∼0.2-kb fragment from the neighboring SPL4 gene, ∼5-kb upstream to the NDK5 gene, was amplified with the P3S and P3As primer pair from both templates (right).
FIGURE 3:
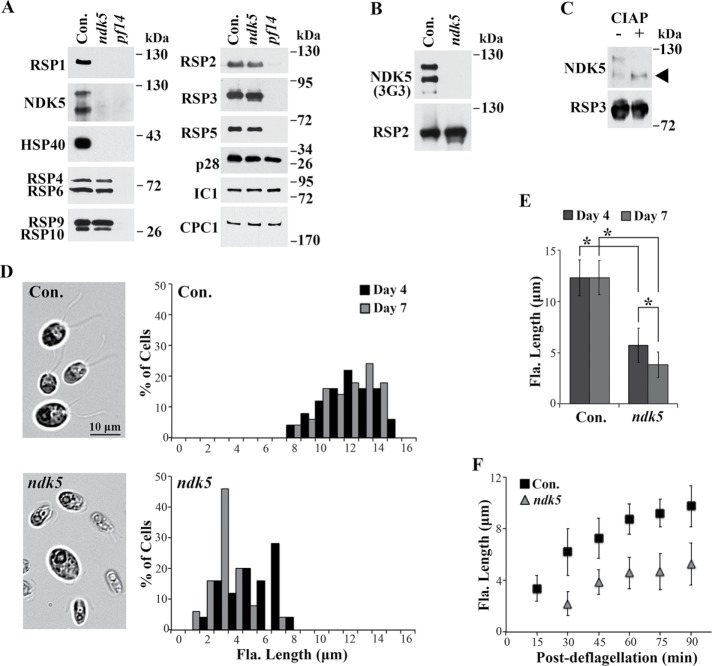
The ndk5 mutant is defective in RSs and flagellar assembly. (A) Western blot analysis of axonemes revealed the absence of RSP1, NDK5, and HSP40 in the paralyzed ndk5 flagella. A fraction of the spoke scaffold protein RSP3 migrated slightly faster than that in WT cells (Con.). The other spoke proteins in the head (RSP4/6 and RSP9/10) and neck (RSP2 and RSP5) appeared normal. The axonemal proteins p28, IC1, and CPC1—subunits of inner dynein arms, outer dynein arms, and the central pair, respectively—were used as controls. The spoke-less pf14 was a negative control. (B) The antigen of mAb 3G3 is NDK5, and not RSP2, which is present in ndk5 axonemes. (C) NDK5 is a phosphoprotein. NDK5 in axonemes receiving the sham CIAP treatment migrated as double bands. In CIAP-treated axonemes, all NDK5 migrate with the lower band (arrowhead). CIAP treatment did not change the migration of the spoke scaffold protein RSP3. (D) Microscopy (left) and histogram (right) revealed short flagella (Fla.) in ndk5 cells cultured in minimal media, in contrast to the full-length flagella of the parental strain (Con.). The length phenotype was more pronounced in late log phase (compare day 4 and day 7, n = 50). (E) Average flagellar length corresponding to D. Asterisks indicate statistically significant differences (p < 0.01, n = 50). (F) ndk5 cells are deficient in flagellar regeneration. Flagella were excised from log-phase cells by pH shock and allowed to regenerate. Aliquots of cells were fixed periodically and imaged. Compared with the control, ndk5 cells regenerated flagella more slowly, and the final lengths were shorter (p < 0.01, n = 20).
To segregate potential additional mutations that might have been present in ndk5, we backcrossed ndk5 with WT cells. Although ndk5 mated poorly, we managed to restore mating ability by crosses with a high mating strain. The strict cosegregation of PMM resistance and paralyzed progeny confirm that NDK5 is required for flagellar motility but is not required for mating.
RS deficiencies of ndk5 flagella
Western blots probed with NDK5 polyclonal antibody detected two major ∼120-kDa NDK5 bands in control axonemes. They were greatly diminished in both ndk5 and pf14 that lack most RSs (Diener et al., 1993) (Figure 3A). Also diminished were RSP1 located in the spokehead and HSP40 (alias RSP16) that coassembles with NDK5 and RSP2 in the neck region in WT cells. In contrast, RSP2 and the other representative proteins were not significantly affected, including the other four spokehead proteins (RSP4, 6, 9, and 10). Although RSP3, a phosphorylated RS scaffold protein, appeared normal in abundance, a fraction consistently migrated faster. Proteins assayed as additional controls—inner dynein arm p28, outer dynein arm IC1, and CPC1 from the central pair—were present in normal amounts.
The 120-kDa NDK5 doublet bands revealed by the polyclonal antibody and the NDK activity resembled the 120-kDa doublet-spoke protein bands revealed by the monoclonal antibody (mAb) 3G3 and in-gel kinase assays (Williams et al., 1989; Yang et al., 2004). The absence of 3G3-reactive bands in ndk5 axonemes that contained 120-kDa RSP2 (Figure 3B) indicated that NDK5, and not RSP2, is the 3G3 antigen, and that it can phosphorylate proteins, at least in vitro, as expected for NDKs. To determine whether the double bands were due to phosphorylation, we treated control axonemes with calf intestine alkaline phosphatase (CIAP). NDK5 in CIAP-treated axonemes comigrated with the lower band of the NDK5 doublet in the sham-treated control (Figure 3C, top), indicating that the upper band was phosphorylated. In contrast, CIAP treatment did not change RSP3 migration (Figure 3C, bottom).
Short flagella of ndk5
We noticed that ndk5 and paralyzed backcross progeny assembled shorter flagella than controls at log phase (day 3) in Tris-acetate-phosphate (TAP) liquid media; at stationary phase, most cells lack flagella. In minimal media that is commonly used in phenotyping flagellar lengths of mutants (Kubo et al., 2015), most ndk5 cells had flagella suitable for quantitative analysis. Regardless of distribution or averages, ndk5 flagella at steady state were consistently shorter than the controls (Figure 3, D and E). The difference became exacerbated as cultures progressed toward late log phase, when nutrients were becoming depleted (compare day 4 and day 7). For evaluation of flagellar generation rate, flagella were first excised from log-phase cells by pH shock. An aliquot of cells was fixed periodically afterward to assess regenerated flagella. Compared with the control, ndk5 cells regrew flagella more slowly, and new flagella remained shorter even after 90 min (Figure 3F, p < 0.01).
Three-dimensional reconstruction of NDK5 axoneme with cryo-electron tomography
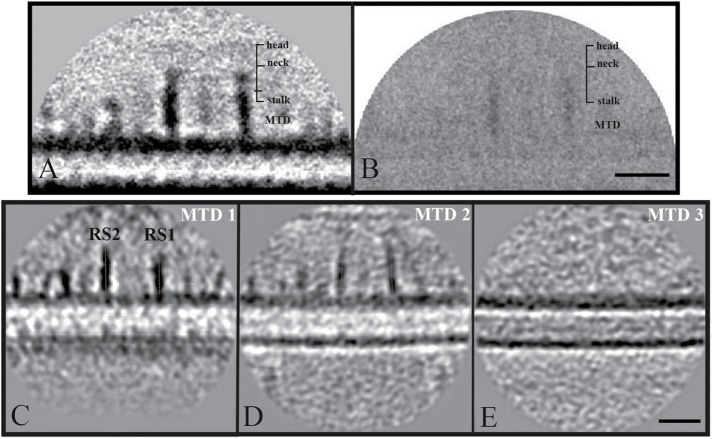
To examine the effect of missing NDK5 (RSP23) on structural integrity of the entire spoke complex, we performed cryo-electron tomography and subsequent subtomogram averaging of ndk5 axoneme (Figure 4). Our initial analysis from the total averages and comparison with the WT and pf24 structures from previously published data (Pigino et al., 2011; Bui et al., 2012) revealed the major structural distortion at the spokeneck and spokehead regions in the radial spokes of ndk5 axoneme. Compared with the WT (Figure 4, A and B), the densities in these regions of ndk5 axoneme (Figure 4, C and D) appeared much more blurred, or nearly missing, similar to the pf24 spoke headless axoneme (Figure 4, E and F). Overall length of the spokes from the average in ndk5 (∼29 nm) mutant flagella is like the headless RSs of the pf24 mutant (Pigino et al., 2011) but considerably shorter than WT spokes (∼43 nm) (Bui et al., 2012).
FIGURE 4:
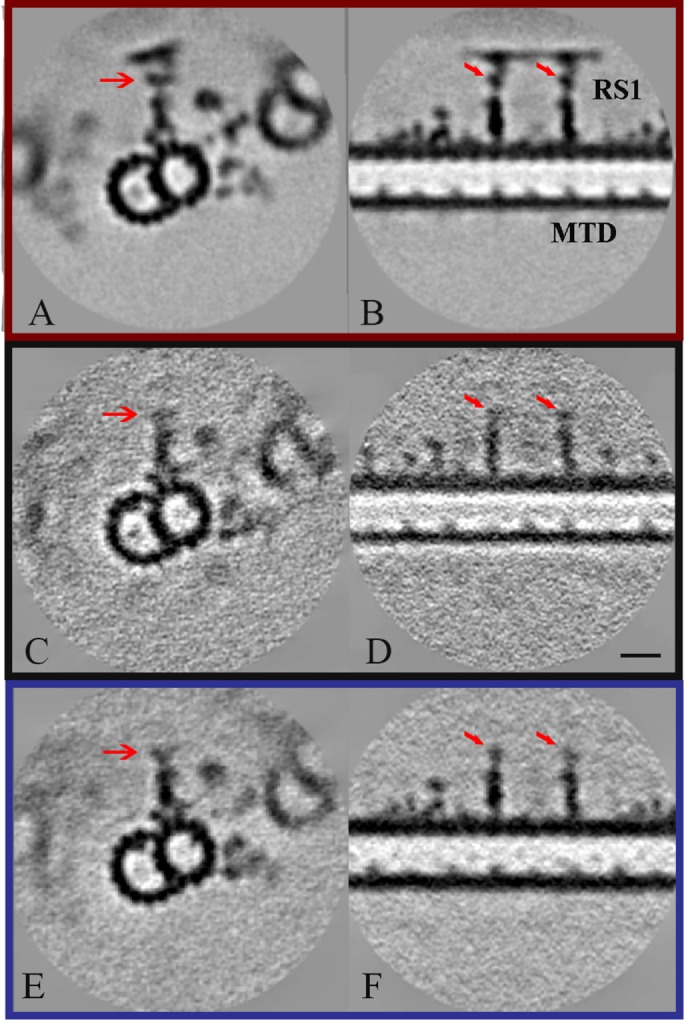
Structure of radial spokes from WT (A, B) (EMD-2131; Bui et al., 2012), ndk5 mutant (C, D), and pf24 mutant (E, F; Pigino et al., 2011) Chlamydomonas. Density maps were obtained by averaging subtomograms involving the 96-nm periodic unit from cryo-electron tomography and shown as transverse (A, C, E) and longitudinal (B, D, F) sections. Red arrows indicate corresponding positions of the RS neck. The density at the position of the spokehead components, RSP1, 4, 6, 9, and 10, are missing in averaged cryo-electron tomograms from the ndk5 strain (C, D) similar to the pf24 headless mutant (E, F). Scale bar: 24 nm.
To assess the subtle differences of the spokes within the ndk5 axoneme and explore the cause of the loss of the spokehead and spokeneck density, we have performed a three-dimensional (3D) variance analysis from aligned subvolumes (Figure 5, A and B), which were included in the final average. The microtubule doublets (MTDs) region displayed no variance as expected and served as a positive control of the success of the density normalization process and the following 3D variance analysis procedures. Interestingly enough, lower variance in the spokeneck and spokehead regions compared with that of the stalk (Figure 5B) excludes the possibility that the RS is fully assembled with bifurcated neck and head but with orientational flexibility with respect to the spoke stalk. Instead, the vast majority of the spokes may have failed assembling the neck and head.
FIGURE 5:
Heterogeneity of radial spokes in ndk5 axonemes. (A) Density map from the total 96 nm–based average shown as longitudinal sections. (B) A corresponding 3D variance map from aligned and normalized particles. Scale bar: 20 nm. The regions with high variance have darker pixel densities. An absence of variance and electron density in the spokehead and spokeneck indicates that the majority of spokes miss the bifurcated neck and head parts. High variance in the spoke stalks infers either orientation or compositional heterogeneity in that region. An absence of variance in individual MTDs serves as a positive control of applied analysis procedures. The representative density map subaveraged from subvolumes reveals typical straight, rigid conformation of spoke stalk with respect to the doublet in MTD 1 (C), in contrast to the subaverage from MTD 2 (D). Red dashed lines are drawn following the spoke-stalk density for the visual representation of the spoke tilt relative to the doublet. The subaverage from MTD 3 (E) shows complete absence of RS assembly. The numbering of the doublets is arbitrary and does not correspond to accepted numbering in the WT axoneme. Scale bar: 24 nm.
The clear signal of the stalk region in the variance map indicates heterogeneity, which was not evident from the final average (Figure 5A). To shed light onto the heterogeneous stalk region, we have looked at the aligned subaverages from a few MTDs (Figure 5, C–E). Indeed, as an example, one of the subaverages showed slightly tilted stalks with respect to MTDs (compare red dashed lines in Figure 5, C and D), and another one showed a complete absence of RSs (Figure 5E). This infers both conformational and compositional heterogeneity in the stalk region.
Full rescue of ndk5 phenotypes by NDK5 and NDK5H121A
To determine whether the disrupted NDK5 gene caused both paralysis and short flagella, we created plasmid pNDK5 that contained the NDK5 genomic DNA and conferred hygromycin (HYG) resistance. To test whether NDK activity is required for NDK5 function, we further mutated this genomic construct to pNDK5H121A. The two constructs were transformed into ndk5 cells. Surprisingly, both plasmids fully rescued ndk5 with efficiencies expected for single plasmid transformations. In one experiment, the rescue rate was 50% for the pNDK5 group (22 swimmer clones out of 44 PMM-resistant transformants) and 33% for the pNDK5H121A group (17 out of 51 screened). The rates varied in replica experiments but were not statistically different between the two groups. The rescued strains from both groups were indistinguishable from the ndk5 parental strain (Con.) in both motility and flagellar length (Figure 6, A and B). Western blot analysis showed that NDK5 polypeptides, HSP40, and RSP1 in the axonemes of rescued strains were restored (Figure 6C). Similar migration of both NDK5 and NDK5H121A expressed from the transgene indicates that the H121A mutation did not affect NDK5 migration or phosphorylation. Therefore H121 is not required for the function or phosphorylation of NDK5 polypeptides. Note that the NDK5 from transgenes (Figure 6C, triangles) migrated faster than the endogenous NDK5 in the control. The distance varied in each electrophoresis, perhaps due to slightly different acrylamide concentration. The variation was also observed in other strains (Yang et al., 2008) and may be due to differences at the evolutionarily divergent C-terminal sequence that interacts with calmodulin. Usefully, this feature allowed us to distinguish exogenous and endogenous NDK5 polypeptides. The NDK5 minigene also rescued ndk5, but none of the rescue clones contained 100% swimmers, suggesting that the minigene was less efficient than the genomic construct.
FIGURE 6:
Genomic DNA expressing NDK5 or NDK5H121A rescued both the paralysis and length phenotypes of ndk5. (A) Flagellar-length (Fla. length) distribution showed that the flagella of ndk5 transgenic strains expressing NDK5 or NDK5H121A were longer than that of ndk5 but similar to that of the ndk5 parental strain. Cells cultured in minimal media were fixed at two indicated time points, and the lengths were measured from micrographs; n = 50. (B) Average flagellar length of corresponding data from Figure 4A. Asterisk indicates statistically significant differences (p < 0.01). (C) Western blot analysis of axonemes from representative transformants. RSP1, HSP40, and NDK5 polypeptides were restored. Note that NDK5 and NDK5H121A expressed by the BAC-derived transgene (triangle) migrated identically but faster than endogenous NDK5 (dot). (D) H121A mutation abolished NDK activity. The NDK activity of axonemes from ndk5 transformants expressing NDK5 was restored to the activity level of WT axonemes (Con.), whereas the activity of axonemes from NDK5H121A transformants, spoke-less mutant pf14, and ndk5 strains (either F0 or F2 from backcross) was ∼50% lower. The relative NDK activity was the RLU value normalized to the control. Quadruple aliquots from each sample were measured. Asterisk indicates statistically significant differences (Tukey HSD test, p < 0.01). (E) Live-cell imaging of transgenic ndk5 and pf14, respectively, expressing fluorescent NDK5 (top) and RSP3 (middle). ndk5 (bottom) was a negative control. NG-tagged NDK5 and RSP3 appeared identical in distribution and intensity throughout flagella.
We then compared NDK activities of various axonemes of equal weights. The spoke-less pf14, F0 ndk5, F2 ndk5, and, importantly, ndk5 rescued with NDK5H121A exhibited similar NDK activities that were roughly one-half lower than the positive control and the transgenic ndk5 rescued with NDK5 (Figure 6D). Therefore H121A mutation indeed abolished the NDK activity of NDK5. Thus the deficiencies in RS composition, motility, and flagellar lengths of the ndk5 strain are caused by diminished NDK5; and NDK activity of NDK5 requires H121 as do other NDKs, but the enzymatic activity is dispensable for its function and phosphorylation. The phenotypes of strains generated in this project are summarized in Table 1. We further performed site-directed mutagenesis on pNDK5H121A to replace residues predicted to bind NTP or catalyze phosphotransfer, including K12A, N118A, R108A, and H55A (Tepper et al., 1994; Tiwari et al., 2004). The four constructs with double mutations still fully rescued ndk5, indicating that the mechanism of NDK5 action has diverged substantially from the primordial NDK catalytic mechanism.
TABLE 1:
Strains described in this study and their corresponding motility, flagellar generation, and RS assembly phenotypes.
| WT | ndk5 | ndk5::NDK5 | ndk5::H121A | WT::H121A(P/S) | WT::H121A(P) | |
|---|---|---|---|---|---|---|
| Motility | S | P | S | S | P/S | P |
| Flagellar generation | ++ | + | ++ | ++ | ++ | + |
| HSP40; RSP1 | ++ | − | ++ | ++ | ++ | + |
| NDK5 | ++ | − | ++ | ++ | ++ | + |
| NDK5 Pho. | ++ | N/A | ++ | ++ | ++ | + |
| RSP3 | ++ | ++ | ++ | ++ | ++ | + |
| RSP3 Pho. | ++ | ++ | ++ | ++ | ++ | + |
+, Reduction in flagellar lengths, protein abundance, or phosphorylation (Pho.); ++, similar to the WT strain;
−, absence of proteins; N/A, not applicable; P, paralyzed cells; S, swimmers.
We also converted the genomic construct to express fluorescent NDK5 with a tag containing NeonGreen (NG), a monomeric fluorescent protein with spectral properties similar to EGFP but 2.7-fold brighter (Shaner et al., 2013). Fluorescent NDK5 expressed in ndk5 transgenic cells decorated the entire flagella, as did NG-tagged RSP3 expressed in pf14 transgenic cells (Figure 6E). The similar location and intensity of NDK5 and RSP3 suggest that each RS contains two NDK5 molecules, as was found for RSP3 and some other RSPs (Kohno et al., 2011; Sivadas et al., 2012; Oda et al., 2014), and suggests that NDK5 in the RS promotes full-length flagella.
Rescue of the short flagella of a spoke-less pf14 allelic mutant
We have noticed that flagellar lengths of pf14 allelic mutants that lack most RSs vary considerably with growth conditions and from strain to strain. We hypothesized that the putative short flagella phenotype of pf14 might be obscured by growth conditions and by backcrosses that led to preferential selection of paralyzed pf14 progeny with long flagella conferred by other genes. To test this, we cultured three pf14 strains—cc1032, cc2496 and cc613—in minimal media. The latter two strains were derived from a single pf14 isolate, whereas cc1032 was derived independently (Luck et al., 1977; Diener et al., 1993; see annotation in the Chlamydomonas Resource Center [www.chlamycollection.org/]). As expected, all three strains were paralyzed, and flagellar lengths of the cc1032 and cc2496 strains were within the normal range, around 10–11 μm throughout the culture. However, as in ndk5, flagella of cc613 were short, 7.32 ± 0.95 μm on day 3 and barely 5 μm on day 5 (Figure 7). Transformation of an RSP3 genomic construct into cc613 cells rescued paralysis. Importantly, flagellar lengths (as shown in a representative rescued strain cc613::RSP3) nearly doubled and were no longer sensitive to culture durations. Therefore, like ndk5, spoke-less mutants are deficient in flagellar generation, but this prominent phenotype has been overlooked due to variations in the genetic background.
FIGURE 7:
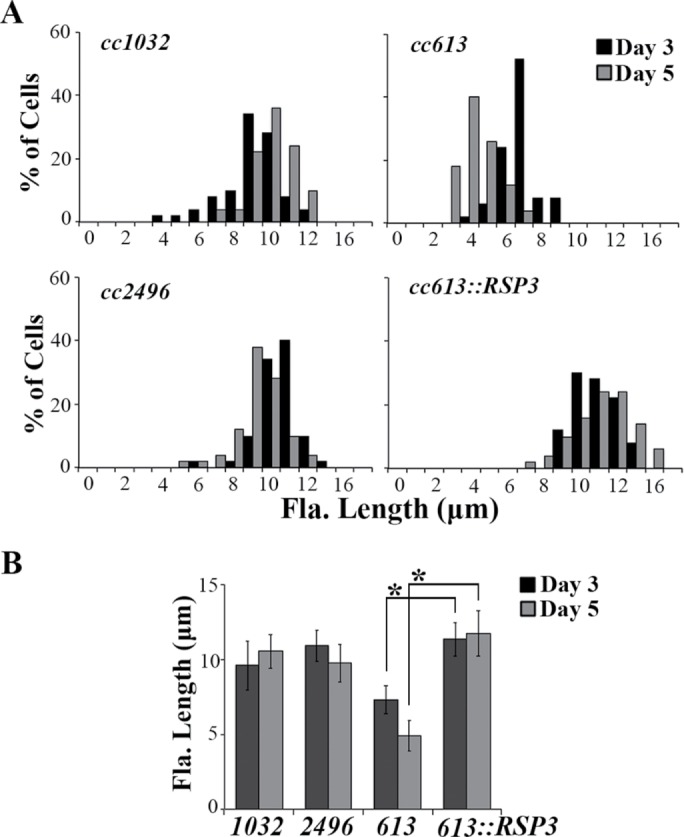
Diminished RSs result in short flagella. (A) Flagellar-length (Fla. length) distribution of three pf14 strains cultured in minimal media. The defect in the RSP3 gene of cc613 and cc2496 that were derived from one common isolate was identical. Only flagella of cc613 were particularly short, and this was exacerbated at late log phase (day 5). The length phenotype and paralysis were rescued by a RSP3 transgene. (B) Average flagellar lengths of pf14 cells. Asterisks indicate statistically significant differences (p < 0.01, n = 50).
DN effect of NDK5H121A genomic DNA
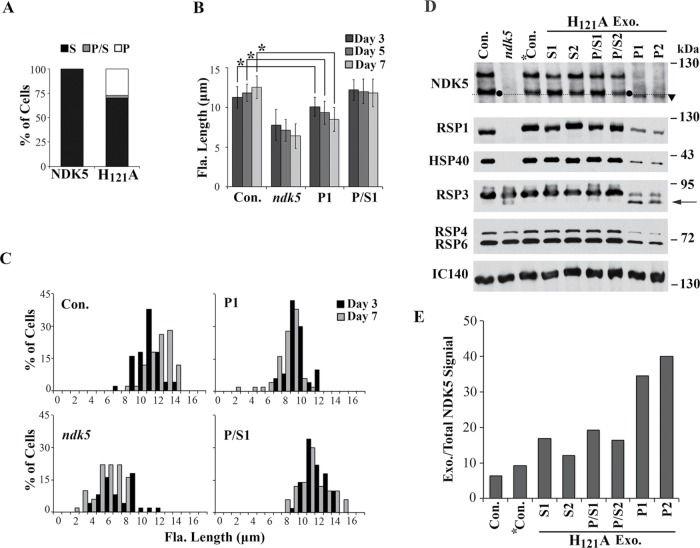
Figure 1 shows that NDK5H121A-His expressed from a minigene in Chlamydomonas had a DN effect as Drosophila NDKkpn. To test this independently, we transformed WT cells with the untagged pNDK5H121A genomic construct. All transgenic clones in the pNDK5 control group were swimmers (S), whereas some clones for the pNDK5H121A group contained paralyzed (P) cells. The percentages of clones with motility phenotypes varied among repeated transformations. In one experiment, 12 out of 44 PMM-resistant clones (27%) contained entirely P cells, whereas 1 clone (2%) had both P and S cells (Figure 8A). The flagella of P strains were also short, albeit not as short as ndk5 flagella, and the length phenotype was exacerbated as the cells approached stationary phase (Figure 8, B and C, compare day 3 and day 7). Therefore NDK5H121A—although fully functional when expressed in ndk5—has a DN effect in the presence of NDK5, regardless of whether the mutant protein is expressed from the minigene or genomic DNA, or with or without a His tag.
FIGURE 8:
NDK5H121A expression from a genomic transgene in WT cells impaired flagellar motility, length, RS assembly, and RS phosphorylation. (A) The percentages of clones with swimmers (S), paralyzed cells (P), or a mixture (P/S) expressing NDK5 (first bar) or NDK5H121A (second bar) from one experiment. Average flagellar length (B) and flagellar-length (Fla. length) distribution (C) of representative strains cultured in minimal media showed that flagellar lengths of P strains (P1) were between the control and ndk5, whereas P/S strains (P/S1) had flagella of normal length. Asterisks indicate statistically significant differences (p < 0.01, n = 50). (D) Western blot analysis of axonemes from transformants with different motility levels. RSPs in the flagella of P/S strains (P/S1 and P/S2) appeared normal but reduced in P strains (P1 and P2) (top). In addition, NDK5 and the scaffold protein RSP3 were hypophosphorylated (arrow), especially NDK5H121A (triangle). IC140, a subunit of the inner dynein arm, was the loading control. *Con., a WT strain transformed with pNDK5. Dashed line, the arbitrary line to distinguish exogenous proteins expressed from the transgenes. (E) The ratio of rapidly migrating NDK5 from the transgenes relative to total NDK5 in D. Ratio of exogenous protein in Con. strain represents the background. Exo., Exogenous.
Unexpectedly, Western blots of randomly selected strains showed that, unlike ndk5 axonemes, DN axonemes from the two P strains (P1 and P2) actually contained all RSPs tested, including RSP1, HSP40, and fast-migrating NDK5H121A from the transgene (Figure 8D, arrowhead). However, all RSPs were evidently less abundant than in controls, while NDK5 and RSP3 appeared less phosphorylated. The loading control was IC140 of the inner dynein I1. To compare the abundance of endogenous and exogenous NDK5, we analyzed the Western blot with the Plot Lanes program after several attempts that failed to fully separate the two populations. The fast-migrating NDK5H121A (below the dashed line in Figure 8D) constituted 34% and 40% of total NDK5 in the flagellar samples from the two P strains (Figure 8E). This fraction was substantially lower for the other strains: 17%, 12%, 19%, and 16% for S1, S2, P/S1, and P/S2 strains, respectively, and 6% and 9% for the control strains. The similar ratios in S strain and P/S strain samples suggest that a threshold level of NDK5H121A is needed to cause paralysis, whereas the level of NDK5H121A is higher to cause paralysis in all cells. The pNDK5H121A-NG that should have been useful to distinguish the two populations cannot elicit DN effects, perhaps due to the NG moiety.
Hypophosphorylated NDK5 in the cell body
RSP3 is hypophosphorylated in preassembled RSs in extracts from WT cell bodies (Qin et al., 2004) and in mutant flagella with reduced amounts of RSs (Gupta et al., 2012). To elucidate the underpinning of the DN effects, we analyzed NDK5 in cell body extracts from various strains by Western blot analysis (Figure 9). Axonemes (lanes 1 and 2) and cell body extracts (lanes 5 and 6) of ndk5 and its parental strain were controls. The lanes loaded with both axonemes and cell body extracts (lanes 3, 4, and 9) tested whether sample preparations affected polypeptide migration. Given reduced RSs in the DN flagella, we also included a pf14 cell body extract (lane 8) as a control for impaired RS assembly. As cell bodies typically harbor enough axonemal proteins for the regeneration of two half-length flagella (Rosenbaum and Child, 1967), we reasoned that RSs that failed to assemble in flagella may accumulate in the cell body (Diener et al., 2011).
FIGURE 9:
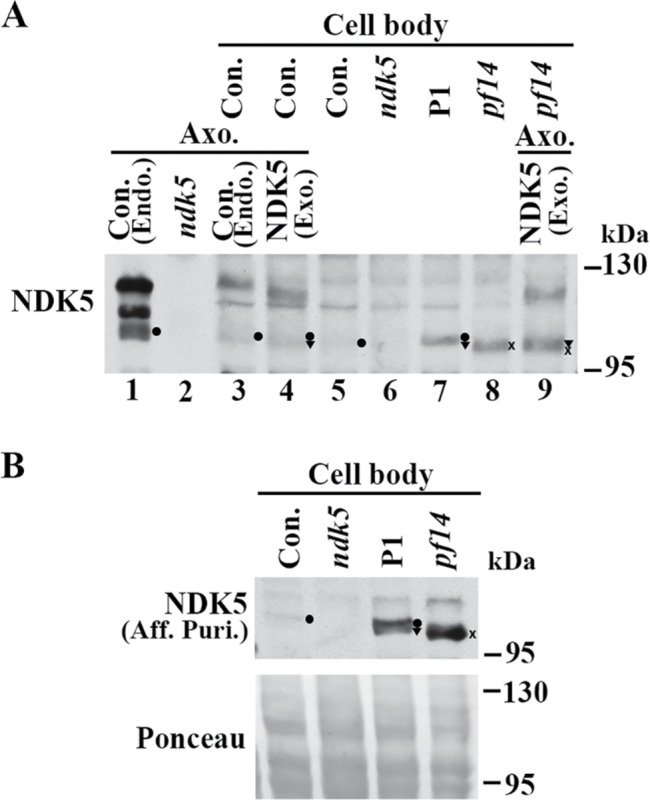
Accumulated hypophosphorylated NDK5 polypeptides in the cell body of the DN strain. (A) A representative Western blot of cell body extracts probed with anti-NDK51-201 serum. NDK5 in the DN strain P1 (lane 7) was more abundant than that in the control (lane 5) but similar to that in pf14 (x in lane 8), a control for reduced RS abundance in flagella. Negative control was ndk5 (lane 6). NDK5 bands in axonemes, by themselves or added to cell body extracts (lanes 1–4 and 9), served as markers for phosphorylated NDK5 and hypophosphorylated NDK5 that are endogenous (dot), compared with that from the transgene (triangle) or in pf14 (x). The axoneme sample loaded in lane 3 was one-sixth of the sample loaded in lane 1. (B) A Western blot probed with affinity-purified (Aff. Puri.) anti-NDK58-586 polyclonal antibody independently confirmed the hypophosphorylated state of endogenous NDK5 (dot) in the control. The band was undetectable in ndk5. The protein load was shown by Ponceau S stain (bottom).
Anti-NDK51-201 polyclonal antibody revealed a faint putative NDK5 band in the parental control cell body extracts, which largely comigrated with the hypophosphorylated NDK5 band in control axonemes (Figure 9A, compare dots in lanes 1, 3, and 5). Similar bands were more prominent in cell body extracts of the P1 DN strain that contained both endogenous (dot in lane 7) and exogenous NDK5 (triangle) or of pf14 that migrated slightly faster (x in lane 8). These bands in the cell body of all strains except ndk5 were confirmed by probing the duplicated blot with affinity-purified polyclonal anti-NDK58-586 (Patel-King et al., 2004) (dot, triangle, and x in Figure 9B, top). Protein loads are shown in the Ponceau S–stained blot (bottom). The 3G3 mAb was too weak to reveal NDK5 in cell body samples. Duplicated control blots probed for other RSPs contained multiple background bands and were not informative.
NDK5 is primarily hypophosphorylated in the cell body extracts, suggesting that it becomes phosphorylated after entering flagella. The P1 cell body extract contained more NDK5 polypeptides than the control but was similar to pf14 (Figure 9A, compare lanes 5, 7, and 8), indicating that NDK5H121A, which causes DN effects, is not excessively overexpressed, and is not degraded either, unlike the presumptive effect caused by the Drosophila kpn mutation (Biggs et al., 1988). The varied abundance of NDK5H121A expressed by minigene or genomic constructs in individual transformants may account for variations in phenotype severity, a scenario similar to kpn mutants (Biggs et al., 1988).
DISCUSSION
NDK plays an important role in multiple biological processes. Only some of them involve the nucleoside phosphorylase activity for which the proteins were named. More uncertain are mechanisms in the events where the NDK activity is dispensable. The ability to manipulate the gene in transgenic strains of Chlamydomonas provides a unique opportunity to investigate the NDK activity–independent mechanism and the DN effects of NDK.
The structural roles of NDK5 for proper assembly of the RS head–neck region
While both Western blot and cryo-electron tomographic analysis reveal RS deficiencies in the ndk5 flagella, the severities differ drastically. Except for RSP1, HSP40, and NDK5, all the other components in the head–neck region, appear in normal abundance in Western blots (Figure 3). However, entire spokeheads (Figures 4 and 5) are invisible in the comparative subtomogram studies. These contradictions could be explained by nonmutually exclusive possibilities. The assembly level of partial spokeheads in ndk5 may be affected by culture conditions, as in the spokehead mutant pf26 (Huang et al., 1981; Wei et al., 2010). Early harvest of sufficient ndk5 flagella for Western blots may account for the seemingly normal abundance. Furthermore, moderate reduced abundance may not be revealed unequivocally by Western blots. Or, the mal-assembled components, at least RSP3’s C-terminal extension in the spokehead (Oda et al., 2014), may not confer substantial density above the background. Regardless, NDK5 is necessary for the assembly of intact spokes, especially the neck and head regions. Our tomographic data analysis clearly explains why the ndk5 mutants are paralyzed, similar to other headless RS mutants and cilia of PCD patients with the defective RSP1 gene (Lin et al., 2014) and unlike the jerky flagella that lack only HSP40 (Yang et al., 2008).
Therefore NDK5 has two structural effects. Diminished RSP1 and HSP40 in ndk5 flagella (Figure 3) support the predictions that flagellar NDKs are structural proteins (Munier et al., 2003) and RSP1 is assembled differently in comparison with the other four head proteins (Luck et al., 1977) and are consistent with the chemical cross-linking of NDK5 and RSP1 (Kohno et al., 2011). Because NDK5 is present in RSs in mutant flagella missing HSP40 or RSP1 (Patel-King et al., 2004; Yang et al., 2005 , 2008), it is NDK5 that tethers RSP1 and HSP40 to the RS scaffold, rather than vice versa. In addition, NDK5 is important for structural integrity of the RS head–neck regions as well as structural stability of the RS stalk. Without NDK5, the rest of the proteins in this region do not form stable bifurcated neck or spokehead, as shown in subtomogram average of ndk5 axonemes (Figure 4). NDK5, probably cooperating with HSP40 and RSP1, plays an essential role for stable formation of the bifurcated neck and spokehead.
NDK5 promotes phosphorylation-related assembly
Absence of entire spokes in some areas in short ndk5 flagella (Figures 5E and 3D) and hypophosphorylated RSs of reduced abundance in short DN flagella (Figure 8) revealed additional roles of NDK5. First, NDK5 promotes RS assembly, which correlates with protein phosphorylation. Phosphorylation of RSP3 has been implicated in the docking of RSs to axonemes (Gupta et al., 2012). While a major fraction of NDK5 and RSP3 in axonemes is phosphorylated (Figure 3B; Piperno et al., 1981; Gupta et al, 2012), both are largely hypophosphorylated in the cell body (Figure 9; Qin et al., 2004; Gupta et al., 2012). Importantly, hypophosphorylation of both molecules in DN flagella (Figure 8D) shows that NDK5 perturbation results in RSP3 hypophosphorylation and evidently reduces RS assembly. Therefore NDK5 promotes RSP3 phosphorylation and RS assembly.
Second, NDK5 also promotes full-length flagella. Chlamydomonas ndk5 (Figure 3D) and ndk5 mouse sperm (Vogel et al., 2012) have short flagella, which can be rescued by the NDK5 gene (Figure 6). Flagellar length is a multigenic trait ultimately determined by the balance between the rates of assembly and disassembly (Marshall and Rosenbaum, 2001). Flagella defective in one axonemal complex, including absence of the spokehead, are slightly shorter, presumably due to decreased microtubule stability (Kubo et al., 2015). However, ndk5 flagella are deficient in only three subunits at a similar region and are far shorter than the flagella lacking the entire spokehead. Because NDK5 and RSP3 had identical distributions (Figure 6E; Munier et al., 2003), the simplest explanation is that NDK5 in the RS participates in a distinct length-promotion mechanism. Alternatively, ndk5’s length phenotype is still due to a sensitive background in conjunction with the propensity of reduced RS abundance (Figures 5E and 7). Alternatively, NDK5 deficiency reduces the affinity of axonemal proteins and intraflagellar transport (IFT) trains and thus decreases the assembly rate.
We are partial to the possibility that NDK5 promotes axoneme stability or cargo–IFT train affinity via protein phosphorylation. Genetic studies have revealed a number of length-regulating protein kinases and phosphatases (Tam et al., 2013; reviewed by Wilson et al., 2008; Cao et al., 2009). Many other proteins near the RS base are phosphorylated (Lin et al., 2011). NDK5 may promote phosphorylation of multiple axonemal proteins during the flagellar assembly process, which in turn increases intermolecular affinity, leading to a higher IFT loading rate or greater axoneme stability, and thereby a faster assembly rate (Figure 3, D–F) and full-length flagella. The exacerbation of length phenotypes in the stationary phase (Figures 3, 7, and 8) may reflect declining assembly rates as nutrients are limited, while NDK5 is used to maintain full-length flagella regardless of nutrient availability for algae or sperm.
It is not clear how NDK5 might promote protein phosphorylation. This ability does not require the canonical phosphotransfer pathway of NDK, because ndk5 is fully rescued by NDK5H121A incapable of the His-dependent phosphotransfer (Figure 6) or even by NDK5H121A lacking one of the additional four residues commonly involved in NTP catalysis. One possible explanation is that NDK5, or perhaps group II NDKs, evolved a distinctive phosphotransfer mechanism while maintaining the canonical one. This is supported by autophosphorylation of the mutated NDK whose conserved His is replaced (MacDonald et al., 1993); distinct residues critical for phosphotransfer in diverse NDKs (Tiwari et al., 2004); and sequence divergence of group I and group II NDKs (Desvignes et al., 2009). Alternatively, NDK5 recruits or activates a protein kinase. In any case, in ndk5 that lacks most NDK5, a surrogate kinase may phosphorylate RSP3 molecules, albeit less efficiently than NDK5 (Figures 3A, 6C, and 8D). This deficiency may become exacerbated as cultures are reaching stationary phase, leading to the absence of some RSs (Figure 5E). In the DN flagella, the presence of heterodimeric NDK5 may hinder the surrogacy, leading to largely hypophosphorylated RSP3 and, clearly, fewer RSs assembled (Figure 8D). This possibility is consistent with the association of a MAP kinase with the N-terminal extension unique to mammalian RSP3 (Jivan et al., 2009).
The most intriguing observation here is that, although NDK5H121A is fully functional in ndk5 cells, it elicits a DN effect in WT cells, perturbing phosphorylation of NDK5 and RSP3 and assembly of RSs and flagella. Likewise, NDKkpn that has DN effects in Drosophila is functional. The similar abundance of hypophosphorylated NDK5 polypeptides in the cell body of DN strains and pf14 (Figure 9) rules out protein degradation that is attributed to the DN effect of NDKkpn (Biggs et al., 1988; Lascu et al., 1992; Karlsson et al., 1996) or other common DN mechanisms (reviewed by Wilkie, 1994), such as overexpression of inactive molecules. Rather, we propose that NDK5—with or without His—operates with distinct molecular movements, but regardless, the two protomers must act in concert, and thus heterodimers with two functional but incompatible variants exhibit impaired activity. Uncoordinated heterodimers—not limited to phosphorylation—are consistent with NDK mutations in cancer cells (e.g., Chang et al., 1996) and NDK degradation in Drosophila embryos. It is likely that NDK5 heterodimers are unable or less efficient in promoting phosphorylation-related trafficking and assembly. Consistent with this, only a small fraction of NDK5H121A is present in P/S flagella (Figure 8, D and E). Varied rates of P strains among repeated transformations suggest that homo- and heterodimerization of NDK5 is not random.
In fact, intersubunit cross-talk between NDK monomers has been demonstrated (Tepper et al., 1994; Dar and Chakraborti, 2010) and is consistent with classic enzymology analysis that estimated only half of protomers in one NDK oligomer bind to phosphate at one time (reviewed by Parks and Agarwal, 1973). However, this notion has not yet gained the attention it deserves, even though it does not contradict the well-established ping-pong mechanism that depicts phosphotransfer occurring within individual monomers. It is time to reconsider intersubunit cross-talk as a common mechanism of oligomeric NDKs in His-dependent and His-independent actions that remain elusive.
In conclusion, NDK5 is crucial for the assembly of functional motile cilia and flagella, but not because of its canonical NDK activity, which could be redundant. It promotes proper assembly of the RS and full-length flagella as a structural protein and via phosphorylation-related mechanisms. These findings expand the versatility of this ancient molecular platform and provide new clues as to how NDK isoforms are involved in a wide array of cellular processes and how they become impaired.
MATERIALS AND METHODS
Chlamydomonas strains, culture conditions, and biochemistry
WT (cc124 and cc125) and pf14 allelic mutants—cc613, cc1032, and cc2496—were acquired from the Chlamydomonas Resource Center. The ndk5 insertional mutant (LMJ.SG0182.001362) and the parental strain (CMJ030) (www.chlamylibrary.org/) were provided by the Jonikas laboratory at the Carnegie Institute of Science (Li et al., 2016). Cells were grown in minimal M media or TAP media (Harris, 2009) as indicated. The light/dark cycle, preparation of flagella, and Western blot analysis were as described previously (Sivadas et al., 2012), except for the addition of protease inhibitor cocktail tablets (cOmplete Mini, EDTA-free; Roche, Basel, Switzerland).
Molecular biology
Characterization of the insertional site in ndk5.
PCR was conducted to amplify the PMM-resistance cassette–genome junctions, the upstream SPL4 gene, and the NDK5 gene. Templates were genomic DNA freshly prepared from ndk5 and its parental strain CMJ030. Briefly, cells (∼1 μl) from TAP plates were resuspended in 20 μl 10 mM EDTA and boiled for 5 min. Following a 30-s vortex and 3-min centrifugation, the supernatant was used in PCR. The primer pairs were as follows:
P1S (GCTCGTGGAGCTCTGAATCT) and P1As (GATACACGAACTCCTGCGC) (Li et al., 2016);
P2S (GCTGCATGTAGGAGGCGCCACCTACC) and P2As (GGTGGCATTCTGGGTGCCGTCCGTTCC); and
P3S (GGCAAATGGAGCTGTTTGGA) and P3As (CCTTAAAGCGTGCTCCTGAC).
Engineering of cDNA constructs.
Two consecutive rounds of PCR were performed using the pMAL-c2 vector (Patel-King et al., 2004) that harbors a partial cDNA encoding aa 8–586 of NDK5 as a template to create a full-length cDNA construct. Two sense primers and one antisense primer are as follows:
NDKS (GCTAGAAAAGACTTTCGCGTTAATCAAGCCAGATGC),
NdeS (TCCATATGGCAGAGCTAGAAAAGACTTTCG), and
KpnAS (GCGCATCACCGCCACCTCCTTCCG).
The final 1101–base pair NdeS-KpnAS PCR product and the 600–base pair KpnI-HindIII 3′ fragment released from the pMAL-c2 plasmid were cloned into the pET28 vector between the NdeI and HindIII sites. This pET28-NDK5 plasmid was converted into pCRS23.2 to express soluble MBP-tagged recombinant NDK51-201 in bacteria (Donnelly et al. 2006). For expression in Chlamydomonas, the coding sequence for NDK5 and the His tag was PCR amplified, and the DNA fragment was used to replace the coding sequence of the LC8 genomic construct that also harbored a PMM-resistance cassette (Yang et al., 2009). The H121 codon in the NDK5 minigene was mutated to an alanine codon using site-directed mutagenesis and the following primer pair:
NDKH2AS (GGCACCCAGAATGCCACCGCGGGCAGCGACTCGCC) and
NDKH2AAS (GTAGGCGAGTCGCTGCCCGCGGTGGCATTCTGGGTGCC).
Engineering of genomic constructs.
For generation of genomic DNA constructs, an NdeI-NdeI fragment containing the NDK5 gene was released from the 21I22 BAC clone (Clemson University Genome Institute) and ligated into the pGEM-T easy vector. The plasmid was further digested with EcoRV and SmaI, and blunt end–ligated to eliminate most of the neighboring SLP4 gene. The resulting pNDK5 plasmid was converted into pNDK5H121A using site-directed mutagenesis and a primer pair:
NDK5MutS (CCCAGAATGCCACCGCGGGCAGCGACTCGCCTATCAGC) and
NDK5MutAs (GCTGCCCGCGGTGGCATTCTGGGTGCCGTCCGTTCC).
An HYG-resistance cassette was PCR-amplified using pHyg3 as a template (Berthold et al., 2002) and inserted into the NdeI site in both genomic constructs. The final constructs were 11.3 kb. The primers were as follows:
NdeHygS (GCATATGGATTACGAATTCGATATCAAGCTTCTTTCTTGC) and
NdeHygAs (GCATATGCGCTTCAAATACGCCCAGC).
To express fluorescent RSPs in Chlamydomonas, mNeonGreen (Shaner et al., 2013) codons modified in accordance with the Chlamydomonas codon bias (Harris et al., 2016) were PCR amplified and cloned into the XhoI site inserted before the stop codons of the NDK5 gene and RSP3 gene (Sivadas et al., 2012).
Purification of bacterial recombinant proteins
For His-luciferase production, the firefly luciferase gene was amplified from pTRE2-Luc (Clontech, Mountainview, CA) and cloned into the pET28 vector. The plasmids for Hi1-luciferase and His-HSP40 (Yang et al., 2005) were transformed into BL21 (DE3) for protein expression. MBP-His-NDK51-201 was expressed as previously described (Donnelly et al., 2006). All His-tagged recombinant proteins were purified with Ni-NTA matrix following the manufacturer’s instruction (Qiagen, Hilden, Germany).
Chlamydomonas experiments
Backcross.
ndk5 mt(−) was crossed with cc620 or cc125 mt(+) as indicated, using the synchronous gametogenesis method (Snell, 1976).
Transformation.
All transformations were conducted using a previously described glass-bead method (Yang et al., 2006) with minor modifications. NDK5 minigenes at a concentration of 2–8 μg were transformed into WT cc124 cells, and the transformants were selected on TAP plates containing 10 μg/ml PMM. NDK5 genomic constructs at a concentration of 1–2 μg were transformed into ndk5 or its parental strain CMJ030, and the transformants were selected on TAP plates containing 10 μg/ml HYG. RSP3 genomic constructs were transformed into pf14 cells similarly but the transformants were selected for PMM resistance. Fractions of single colonies were resuspended in 200 μl double-distilled water in 96-well plates for motility analysis under a stereomicroscope.
Flagellar-length quantification.
Cells fixed with 2.5% glutaraldehyde were placed on poly-l-lysine–coated slides for imaging. The lengths of 50 randomly selected flagella were measured using ImageJ software. For flagellar regeneration experiments, pH shock with acetate was used to excise flagella of log-phase cells from TAP medium cultures. After confirmation of deflagellation microscopically, an aliquot of the cells was fixed with 2.5% glutaraldehyde every 15 min for subsequent imaging. The lengths of 20 randomly chosen flagella were measured at each time point.
Light microscopy
Bright-field and fluorescent images were captured with a Nikon Eclipse E600W compound microscope at 400× magnification using a CoolSNAP-ES charge-coupled device camera (Photometrics, Tucson, AZ) and a MetaMorph imaging system (Molecular Devices, Sunnyvale, CA).
Quick-plunge freezing and electron microscopy
Isolated axonemes were quickly frozen in liquid ethane at liquid nitrogen temperatures with the help of a vitrification device (Cryo-plunge 3, Gatan, Pleasanton, CA). Holey carbon grids (200 mesh, R3.5/1; Quantifoil Micro Tools GmbH, Großlöbichau, Germany) were used. Gold colloid particles (10 nm) were applied to the sample before freezing as fiducial markers for tomographic reconstruction. Data collection was performed using a JEM2200FS transmission electron microscope (JEOL, Tokyo, Japan) equipped with an in-column energy filter and a field-emission gun. Micrographs were recorded with a 4K × 4K CMOS camera (F416 from TVIPS, Gauting, Germany). Tomographic image series were collected using a previously described procedure (Pigino et al., 2011), except for the use of SerialEM software (Mastronarde, 2005). Image analysis was performed as described previously (Pigino et al., 2011; Bui and Ishikawa, 2013). Initially, 239 particles (subvolumes) were picked from reconstructed tomograms, aligned, and averaged based on 24-nm and 96-nm periodic units. Final average was obtained from 184 best-selected particles. Individual particles included in the final average were used for calculating the 3D variance map using SPIDER software (Frank et al., 1996). Each of the represented individual MTD subaverages contains 12 aligned particles following 96-nm periodic units.
NDK activity assay
The assay measured luminescence emitted from luciferin and was catalyzed by luciferase using ATP generated by NDKs that transferred the γ-phosphate from GTP to ADP (Karamohamed et al., 1999). An aliquot of 60 ng purified recombinant proteins or 1 μg axonemes was added to the 0.2 ml assay buffer (0.1 M Tris-acetate, pH 7.8, 10 mM MgSO4, 2 mM EDTA, 1 mM dithiothreitol, 1 mg/ml bovine serum albumin, 0.1 mg/ml d-luciferin, 10 ng/ml Ni-NTA–purified His-firefly luciferase, 0.5 mM ADP, and 0.5 mM GTP in a cuvette placed in a Monolight 2010 luminometer [Analytical Luminescence Laboratory, Ann Arbor, MI]). The change in light emission was measured for 10 s and expressed as RLU (light emission/s), as recommended by the manufacturer. Each sample was analyzed in triplicate. The RLU of axonemes for each experimental group was then normalized to that of the WT control and denoted as relative kinase activity (%).
Antibodies
NDK5 Western blots were probed with the rabbit polyclonal antibody raised against Ni-NTA–purified His-tagged NDK51-201 unless indicated otherwise. The affinity-purified rabbit polyclonal antibodies for NDK58-586 and the antibodies for the other proteins were described in previous studies (LeDizet and Piperno, 1995; Patel-King et al., 2004; Yang et al., 2006). The 3G3 was a mouse mAb raised against 120-kDa spoke proteins spot-purified from two-dimensional gels of axonemes (Williams et al., 1989). This study showed that 3G3 recognized NDK5. Anti-IC1 was a mouse mAb (King et al., 1991).
Acknowledgments
We thank the research team of Martin Jonikas (Carnegie Institute for Science) for providing the insertional mutants; the research team of Mark Donnelly and Andrzej Joachimiak (Argonne National Laboratory) for generating recombinant NDK5 expression constructs; the Electron Microscopy Facility (EMF) at the Paul Scherrer Institut (Switzerland) for the technical assistance during electron microscopy data collection; David Mitchell (SUNY Upstate Medical University) for providing cpc1 and pf14cpc1 double mutants and anti-CPC1 antibody; Elizabeth Richey and Hongmin Qin (Texas A&M University), Ursula Goodenough (Washington University), and Xingshan Jiang and William Snell (University of Maryland) for advice on Chlamydomonas mating and tetrad dissection; and Steve Munroe and Kathleen Karrer (Marquette University) for providing a luciferase plasmid and fruitful discussion. This work is supported by Marquette University Startup for P.Y. X.Z. is supported by Marquette University fellowships. S.M.K. is supported by grant GM051293 from the National Institutes of Health.
Abbreviations used:
- CIAP
calf intestine alkaline phosphatase
- DN
dominant negative
- HYG
hygromycin
- IFT
intraflagellar transport
- mAb
monoclonal antibody
- MBP
maltose-binding protein
- MTDs
microtubule doublets
- NDK
nucleoside diphosphate kinase
- NG
NeonGreen
- P
Paralyzed
- PCD
primary ciliary dyskinesia
- PMM
paromomycin
- RS
radial spoke
- S
Swimmers
- TAP
Tris-acetate-phosphate
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-03-0156) on September 6, 2017.
REFERENCES
- Attwood PV, Wieland T. Nucleoside diphosphate kinase as protein histidine kinase. Naunyn Schmiedebergs Arch Pharmacol. 2015;388:153–160. doi: 10.1007/s00210-014-1003-3. [DOI] [PubMed] [Google Scholar]
- Berg P, Joklik WK. Transphosphorylation between nucleoside polyphosphates. Nature. 1953;172:1008–1009. doi: 10.1038/1721008a0. [DOI] [PubMed] [Google Scholar]
- Berthold P, Schmitt R, Mages W. An engineered Streptomyces hygroscopicus aph 7" gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist. 2002;153:401–412. doi: 10.1078/14344610260450136. [DOI] [PubMed] [Google Scholar]
- Biggs J, Tripoulas N, Hersperger E, Dearolf C, Shearn A. Analysis of the lethal interaction between the prune and Killer of prune mutations of Drosophila. Genes Dev. 1988;2:1333–1343. doi: 10.1101/gad.2.10.1333. [DOI] [PubMed] [Google Scholar]
- Boissan M, Montagnac G, Shen Q, Griparic L, Guitton J, Romao M, Sauvonnet N, Lagache T, Lascu I, Raposo G, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 2014;344:1510–1515. doi: 10.1126/science.1253768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Ishikawa T. 3D structural analysis of flagella/cilia by cryo-electron tomography. Methods Enzymol. 2013;524:305–323. doi: 10.1016/B978-0-12-397945-2.00017-2. [DOI] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol. 2012;198:913–25. doi: 10.1083/jcb.201201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Srivastava S, Surindran S, Li Z, Skolnik EY. Regulation of the epithelial Ca2+ channel TRPV5 by reversible histidine phosphorylation mediated by NDPK-B and PHPT1. Mol Biol Cell. 2014;25:1244–1250. doi: 10.1091/mbc.E13-04-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Li G, Pan J. Regulation of cilia assembly, disassembly, and length by protein phosphorylation. Methods Cell Biol. 2009;94:333–346. doi: 10.1016/S0091-679X(08)94017-6. [DOI] [PubMed] [Google Scholar]
- Carotenuto M, Pedone E, Diana D, de Antonellis P, Dzeroski S, Marino N, Navas L, Di Dato V, Scoppettuolo MN, Cimmino F, et al. Neuroblastoma tumorigenesis is regulated through the Nm23-H1/h-Prune C-terminal interaction. Sci Rep. 2013;3:1351. doi: 10.1038/srep01351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Strahler JR, Thoraval DH, Qian MG, Hinderer R, Hanash SM. A nucleoside diphosphate kinase A (nm23-H1) serine 120→glycine substitution in advanced stage neuroblastoma affects enzyme stability and alters protein-protein interaction. Oncogene. 1996;12:659–667. [PubMed] [Google Scholar]
- D’Angelo A, Garzia L, Andre A, Carotenuto P, Aglio V, Guardiola O, Arrigoni G, Cossu A, Palmieri G, Aravind L, Zollo M. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell. 2004;5:137–149. doi: 10.1016/s1535-6108(04)00021-2. [DOI] [PubMed] [Google Scholar]
- Dar HH, Chakraborti PK. Intermolecular phosphotransfer is crucial for efficient catalytic activity of nucleoside diphosphate kinase. Biochem J. 2010;430:539–549. doi: 10.1042/BJ20100026. [DOI] [PubMed] [Google Scholar]
- Desvignes T, Pontarotti P, Fauvel C, Bobe J. Nme protein family evolutionary history, a vertebrate perspective. BMC Evol Biol. 2009;9:256. doi: 10.1186/1471-2148-9-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di L, Srivastava S, Zhdanova O, Sun Y, Li Z, Skolnik EY. Nucleoside diphosphate kinase B knock-out mice have impaired activation of the K+ channel KCa3.1, resulting in defective T cell activation. J Biol Chem. 2010;285:38765–38771. doi: 10.1074/jbc.M110.168070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Ang LH, Rosenbaum JL. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J Cell Biol. 1993;123:183–190. doi: 10.1083/jcb.123.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Yang P, Geimer S, Cole DG, Sale WS, Rosenbaum JL. Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken) 2011;68:389–400. doi: 10.1002/cm.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly MI, Zhou M, Millard CS, Clancy S, Stols L, Eschenfeldt WH, Collart FR, Joachimiak A. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr Purif. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duriez B, Duquesnoy P, Escudier E, Bridoux AM, Escalier D, Rayet I, Marcos E, Vojtek AM, Bercher JF, Amselem S. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. Proc Natl Acad Sci USA. 2007;104:3336–3341. doi: 10.1073/pnas.0611405104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez A, Kourembanas S, Wyatt TA, Mitsialis SA. Mutation of murine adenylate kinase 7 underlies a primary ciliary dyskinesia phenotype. Am J Respir Cell Mol Biol. 2009;40:305–313. doi: 10.1165/rcmb.2008-0102OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li YH, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gupta A, Diener DR, Sivadas P, Rosenbaum JL, Yang P. The versatile molecular complex component LC8 promotes several distinct steps of flagellar assembly. J Cell Biol. 2012;198:115–126. doi: 10.1083/jcb.201111041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook, Chapter 8. Vol. I. San Diego, CA: 2009. Introduction to Chlamydomonas and its laboratory use. In: [Google Scholar]
- Harris JA, Liu Y, Yang P, Kner P, Lechtreck KF. Single-particle imaging reveals intraflagellar transport-independent transport and accumulation of EB1 in Chlamydomonas flagella. Mol Biol Cell. 2016;27:295–307. doi: 10.1091/mbc.E15-08-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Piperno G, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J Cell Biol. 1981;88:80–88. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanson L, Copin B, Papon JF, Dastot-Le Moal F, Duquesnoy P, Montantin G, Cadranel J, Corvol H, Coste A, Desir J, et al. RSPH3 mutations cause primary ciliary dyskinesia with central-complex defects and a near absence of radial spokes. Am J Hum Genet. 2015;97:153–162. doi: 10.1016/j.ajhg.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jivan A, Earnest S, Juang YC, Cobb MH. Radial spoke protein 3 is a mammalian protein kinase A-anchoring protein that binds ERK1/2. J Biol Chem. 2009;284:29437–29445. doi: 10.1074/jbc.M109.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamohamed S, Nordstrom T, Nyren P. Real-time bioluminometric method for detection of nucleoside diphosphate kinase activity. Biotechniques. 1999;26:728–734. doi: 10.2144/99264rr02. [DOI] [PubMed] [Google Scholar]
- Karlsson A, Mesnildrey S, Xu Y, Morera S, Janin J, Veron M. Nucleoside diphosphate kinase. Investigation of the intersubunit contacts by site-directed mutagenesis and crystallography. J Biol Chem. 1996;271:19928–19934. doi: 10.1074/jbc.271.33.19928. [DOI] [PubMed] [Google Scholar]
- King SM, Wilkerson CG, Witman GB. The Mr 78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with alpha-tubulin in situ. J Biol Chem. 1991;266:8401–8407. [PubMed] [Google Scholar]
- Kohno T, Wakabayashi K, Diener DR, Rosenbaum JL, Kamiya R. Subunit interactions within the Chlamydomonas flagellar spokehead. Cytoskeleton (Hoboken) 2011;68:237–246. doi: 10.1002/cm.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Hirono M, Aikawa T, Kamiya R, Witman GB. Reduced tubulin polyglutamylation suppresses flagellar shortness in Chlamydomonas. Mol Biol Cell. 2015;26:2810–2822. doi: 10.1091/mbc.E15-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascu I, Chaffotte A, Limbourg-Bouchon B, Veron M. A Pro/Ser substitution in nucleoside diphosphate kinase of Drosophila melanogaster (mutation Killer of prune) affects stability but not catalytic efficiency of the enzyme. J Biol Chem. 1992;267:12775–12781. [PubMed] [Google Scholar]
- Lascu I, Gonin P. The catalytic mechanism of nucleoside diphosphate kinases. J Bioenerg Biomembr. 2000;32:237–246. doi: 10.1023/a:1005532912212. [DOI] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol Biol Cell. 1995;6:697–711. doi: 10.1091/mbc.6.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre P, Fitz-Gibbon ST, et al. An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell. 2016;28:367–387. doi: 10.1105/tpc.15.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Tritschler D, Song K, Barber CF, Cobb JS, Porter ME, Nicastro D. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J Biol Chem. 2011;286:29175–29191. doi: 10.1074/jbc.M111.241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Yin W, Smith MC, Song K, Leigh MW, Zariwala MA, Knowles MR, Ostrowski LE, Nicastro D. Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat Commun. 2014;5:5727. doi: 10.1038/ncomms6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Choi YK, Qi RZ. NME7 is a functional component of the gamma-tubulin ring complex. Mol Biol Cell. 2014;25:2017–2025. doi: 10.1091/mbc.E13-06-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck D, Piperno G, Ramanis Z, Huang B. Flagellar mutants of Chlamydomonas: studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc Natl Acad Sci USA. 1977;74:3456–3460. doi: 10.1073/pnas.74.8.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald NJ, De la Rosa A, Benedict MA, Freije JM, Krutsch H, Steeg PS. A serine phosphorylation of Nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J Biol Chem. 1993;268:25780–25789. [PubMed] [Google Scholar]
- MacDonald NJ, Freije JM, Stracke ML, Manrow RE, Steeg PS. Site-directed mutagenesis of nm23-H1. Mutation of proline 96 or serine 120 abrogates its motility inhibitory activity upon transfection into human breast carcinoma cells. J Biol Chem. 1996;271:25107–25116. doi: 10.1074/jbc.271.41.25107. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Rosenbaum JL. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BF, Pedersen LB, Feely M, Rosenbaum JL, Mitchell DR. ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol Biol Cell. 2005;16:4509–4518. doi: 10.1091/mbc.E05-04-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv Exp Med Biol. 2007;607:130–140. doi: 10.1007/978-0-387-74021-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munier A, Feral C, Milon L, Pinon VP, Gyapay G, Capeau J, Guellaen G, Lacombe ML. A new human nm23 homologue (nm23-H5) specifically expressed in testis germinal cells. FEBS Lett. 1998;434:289–294. doi: 10.1016/s0014-5793(98)00996-x. [DOI] [PubMed] [Google Scholar]
- Munier A, Serres C, Kann ML, Boissan M, Lesaffre C, Capeau J, Fouquet JP, Lacombe ML. Nm23/NDP kinases in human male germ cells: role in spermiogenesis and sperm motility. Exp Cell Res. 2003;289:295–306. doi: 10.1016/s0014-4827(03)00268-4. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Yagi T, Kikkawa M. Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J Cell Biol. 2014;204:807–819. doi: 10.1083/jcb.201312014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Takai H, Ogiwara A, Yokota E, Shimizu T, Inaba K, Mohri H. Is outer arm dynein intermediate chain 1 multifunctional. Mol Biol Cell. 1996;7:1895–1907. doi: 10.1091/mbc.7.12.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padma P, Hozumi A, Ogawa K, Inaba K. Molecular cloning and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene. 2001;275:177–183. doi: 10.1016/s0378-1119(01)00661-8. [DOI] [PubMed] [Google Scholar]
- Parks RE Jr, Agarwal RP. Nucleoside diphosphokinases. In: Boyer P.D., editor. The Enzymes, vol. VIII. Academic; 1973. pp. 307–333. [Google Scholar]
- Patel-King RS, Gorbatyuk O, Takebe S, King SM. Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol Biol Cell. 2004;15:3891–3902. doi: 10.1091/mbc.E04-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Bui KH, Maheshwari A, Lupetti P, Diener D, Ishikawa T. Cryoelectron tomography of radial spokes in cilia and flagella. J Cell Biol. 2011;195:673–687. doi: 10.1083/jcb.201106125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Huang B, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J Cell Biol. 1981;88:73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Child FM. Flagellar regeneration in protozoan flagellates. J Cell Biol. 1967;34:345–364. doi: 10.1083/jcb.34.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengard AM, Krutzsch HC, Shearn A, Biggs JR, Barker E, Margulies IM, King CR, Liotta LA, Steeg PS. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature. 1989;342:177–180. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- Sadek CM, Jimenez A, Damdimopoulos AE, Kieselbach T, Nord M, Gustafsson JA, Spyrou G, Davis EC, Oko R, van der Hoorn FA, Miranda-Vizuete A. Characterization of human thioredoxin-like 2. A novel microtubule-binding thioredoxin expressed predominantly in the cilia of lung airway epithelium and spermatid manchette and axoneme. J Biol Chem. 2003;278:13133–13142. doi: 10.1074/jbc.M300369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivadas P, Dienes JM, St Maurice M, Meek WD, Yang P. A flagellar A-kinase anchoring protein with two amphipathic helices forms a structural scaffold in the radial spoke complex. J Cell Biol. 2012;199:639–651. doi: 10.1083/jcb.201111042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell WJ. Mating in Chlamydomonas: a system for the study of specific cell adhesion. I. Ultrastructural and electrophoretic analyses of flagellar surface components involved in adhesion. J Cell Biol. 1976;68:48–69. doi: 10.1083/jcb.68.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Li Z, Ko K, Choudhury P, Albaqumi M, Johnson AK, Yan Y, Backer JM, Unutmaz D, Coetzee WA, Skolnik EY. Histidine phosphorylation of the potassium channel KCa3.1 by nucleoside diphosphate kinase B is required for activation of KCa3.1 and CD4 T cells. Mol Cell. 2006;24:665–675. doi: 10.1016/j.molcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, Sobel ME. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- Steeg PS, Zollo M, Wieland T. A critical evaluation of biochemical activities reported for the nucleoside diphosphate kinase/Nm23/Awd family proteins: opportunities and missteps in understanding their biological functions. Naunyn Schmiedebergs Arch Pharmacol. 2011;384:331–339. doi: 10.1007/s00210-011-0651-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. A highly specific complementary lethal system in Drosophila melanogaster. Genetics. 1956;41:118–123. doi: 10.1093/genetics/41.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs-Vellai K, Vellai T, Farkas Z, Mehta A. Nucleoside diphosphate kinases (NDPKs) in animal development. Cell Mol Life Sci. 2015;72:1447–1462. doi: 10.1007/s00018-014-1803-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei GL, Miyashiro D, Mukai C, Okuno M. Glycolysis plays an important role in energy transfer from the base to the distal end of the flagellum in mouse sperm. J Exp Biol. 2014;217:1876–1886. doi: 10.1242/jeb.090985. [DOI] [PubMed] [Google Scholar]
- Tam LW, Ranum PT, Lefebvre PA. CDKL5 regulates flagellar length and localizes to the base of the flagella in Chlamydomonas. Mol Biol Cell. 2013;24:588–600. doi: 10.1091/mbc.E12-10-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper AD, Dammann H, Bominaar AA, Veron M. Investigation of the active site and the conformational stability of nucleoside diphosphate kinase by site-directed mutagenesis. J Biol Chem. 1994;269:32175–32180. [PubMed] [Google Scholar]
- Thakur RK, Kumar P, Halder K, Verma A, Kar A, Parent JL, Basundra R, Kumar A, Chowdhury S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S, Kishan KV, Chakrabarti T, Chakraborti PK. Amino acid residues involved in autophosphorylation and phosphotransfer activities are distinct in nucleoside diphosphate kinase from Mycobacterium tuberculosis. J Biol Chem. 2004;279:43595–43603. doi: 10.1074/jbc.M401704200. [DOI] [PubMed] [Google Scholar]
- Vogel P, Read RW, Hansen GM, Payne BJ, Small D, Sands AT, Zambrowicz BP. Congenital hydrocephalus in genetically engineered mice. Vet Pathol. 2012;49:166–181. doi: 10.1177/0300985811415708. [DOI] [PubMed] [Google Scholar]
- Wagner PD, Steeg PS, Vu ND. Two-component kinase-like activity of nm23 correlates with its motility-suppressing activity. Proc Natl Acad Sci USA. 1997;94:9000–9005. doi: 10.1073/pnas.94.17.9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Sivadas P, Owen HA, Mitchell DR, Yang P. Chlamydomonas mutants display reversible deficiencies in flagellar beating and axonemal assembly. Cytoskeleton (Hoboken) 2010;67:71–80. doi: 10.1002/cm.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AO. The molecular basis of genetic dominance. J Med Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Velleca MA, Curry AM, Rosenbaum JL. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14 is an ochre allele. J Cell Biol. 1989;109:235–245. doi: 10.1083/jcb.109.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NF, Iyer JK, Buchheim JA, Meek W. Regulation of flagellar length in Chlamydomonas. Semin Cell Dev Biol. 2008;19:494–501. doi: 10.1016/j.semcdb.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Compton MM, Yang P. Dimeric novel HSP40 is incorporated into the radial spoke complex during the assembly process in flagella. Mol Biol Cell. 2005;16:637–648. doi: 10.1091/mbc.E04-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Owen HA, Yang P. Dimeric heat shock protein 40 binds radial spokes for generating coupled power strokes and recovery strokes of 9 + 2 flagella. J Cell Biol. 2008;180:403–415. doi: 10.1083/jcb.200705069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, et al. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Yang C, Sale WS. Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryot Cell. 2004;3:72–81. doi: 10.1128/EC.3.1.72-81.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Yang C, Wirschell M, Davis S. Novel LC8 mutations have disparate effects on the assembly and stability of flagellar complexes. J Biol Chem. 2009;284:31412–31421. doi: 10.1074/jbc.M109.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Qi Y, Zhou K, Zhang G, Linask K, Xu H. The cAMP phosphodiesterase Prune localizes to the mitochondrial matrix and promotes mtDNA replication by stabilizing TFAM. EMBO Rep. 2015;16:520–527. doi: 10.15252/embr.201439636. [DOI] [PMC free article] [PubMed] [Google Scholar]