Receptor tyrosine kinases (RTKs) can signal via regulated intramembrane proteolysis, leading to release of an intracellular receptor fragment with functional activity. A system-wide screen covering 45 of the 55 human RTKs identified 12 novel gamma-secretase substrates and indicated that at least half of all RTKs are susceptible to cleavage.
Abstract
Receptor tyrosine kinases (RTKs) have been demonstrated to signal via regulated intramembrane proteolysis, in which ectodomain shedding and subsequent intramembrane cleavage by gamma-secretase leads to release of a soluble intracellular receptor fragment with functional activity. For most RTKs, however, it is unknown whether they can exploit this new signaling mechanism. Here we used a system-wide screen to address the frequency of susceptibility to gamma-secretase cleavage among human RTKs. The screen covering 45 of the 55 human RTKs identified 12 new as well as all nine previously published gamma-secretase substrates. We biochemically validated the screen by demonstrating that the release of a soluble intracellular fragment from endogenous AXL was dependent on the sheddase disintegrin and metalloprotease 10 (ADAM10) and the gamma-secretase component presenilin-1. Functional analysis of the cleavable RTKs indicated that proliferation promoted by overexpression of the TAM family members AXL or TYRO3 depends on gamma-secretase cleavage. Taken together, these data indicate that gamma-secretase–mediated cleavage provides an additional signaling mechanism for numerous human RTKs.
INTRODUCTION
Receptor tyrosine kinases (RTKs) are cell-surface receptors that regulate cellular processes such as cell cycle, survival, differentiation, and migration. Aberrant RTK signaling is frequently observed in cancer, and several cancer drugs targeting RTKs have been introduced into clinical practice. The human genome includes 55 RTKs, which are divided into 19 subfamilies (Wheeler and Yarden, 2015). The prototype RTKs consist of ligand-binding sequences in the extracellular domain, a single transmembrane helix, and a cytoplasmic domain that harbors the catalytically active tyrosine kinase domain along with additional C-terminal and juxtamembrane regulatory regions. Ligand binding at the RTK ectodomain results in receptor dimerization, transautophosphorylation, and activation of the tyrosine kinase domain. Activated RTKs initiate various downstream signaling cascades that indirectly translate the signal to the nucleus, where transcription of target genes is activated (Lemmon and Schlessinger, 2010).
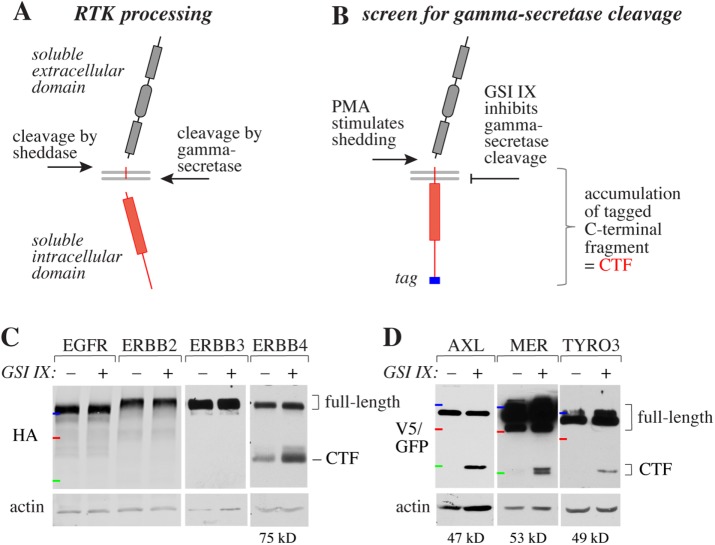
In addition to exhibiting the classical signaling mode involving the activation of pathways, RTKs have recently been demonstrated to modulate cell behavior by a process called regulated intramembrane proteolysis (RIP). RIP is a two-step process in which an integral cell-surface protein, such as an RTK, is first cleaved at the extracellular juxtamembrane region, resulting in the shedding of the ectodomain (Beel and Sanders, 2008; Ancot et al., 2009). This first cleavage step, which produces a short 12– to 35–amino acid stub of the ectodomain (Funamoto et al., 2013), exposes a secondary cleavage site in the receptor transmembrane domain. A second cleavage event then releases a soluble intracellular domain (ICD) fragment into the cytosol (Figure 1A). The first cleavage step is typically carried out by a member of the disintegrin and metalloprotease (ADAM) family of proteases, whereas the secondary site is cleaved by activity of the gamma-secretase complex (Beel and Sanders, 2008; Tomita, 2014).
FIGURE 1:
Screen for gamma-secretase–cleaved RTKs. (A) Two-step proteolytic processing of an RTK with a sheddase and gamma-secretase activity. (B) Outline of the screen. RTKs sensitive to gamma-secretase cleavage were identified on the basis of the accumulation of a C-terminal fragment in response chemical inhibition of gamma-secretase activity using GSI IX. (C) Western analysis of the gamma-secretase cleavage within the EGFR/ERBB RTK family in MCF-7 cells transfected with plasmids encoding EGFR, ERBB2, ERBB3, and ERBB4 with C-terminal HA-tags. Gamma-secretase was inhibited with 0 or 5 μM GSI IX for 4 h followed by stimulation of shedding for 20 min with 100 ng/ml PMA. Loading was controlled with anti-actin. (D) Western analysis of the gamma-secretase cleavage within the TAM RTK family in MCF-7 (AXL, TYRO3) or HEK293 (MER) cells transfected with plasmids encoding C-terminally tagged AXL (V5), MER (V5), or TYRO3 (GFP). Cells were treated as in C. Loading was controlled with anti-actin. The CTF is indicated in red and the C-terminal epitope tag in blue (A, B). The molecular weight markers are indicated by colored horizontal lines: blue, 150 kDa; red, 100 kDa; and green, 50 kDa. The estimated sizes of the cleaved CTFs excluding the tags are indicated below the respective Western blots (C, D).
When addressed for functionality, the generation of a soluble ICD by RIP has been shown to regulate RTK signaling. A well-studied example is ERBB4, which naturally exists in isoforms that are either susceptible to RIP or not (Määttä et al., 2006). The soluble ICD of ERBB4 traffics to the nucleus or mitochondria, where it can directly interact with multiple effector proteins such as transcription factors or regulators of apoptosis (Jones, 2008; Veikkolainen et al., 2011; Carpenter and Pozzi, 2012). In addition to its gain-of-function roles, it has been proposed that RTK RIP serves as a mechanism for down-regulation of classical RTK signaling. For example, the RIP of MET has been shown attenuate MET signaling by promoting efficient proteasomal degradation of the ICD (Foveau et al., 2009; Ancot et al., 2012; Montagne et al., 2015).
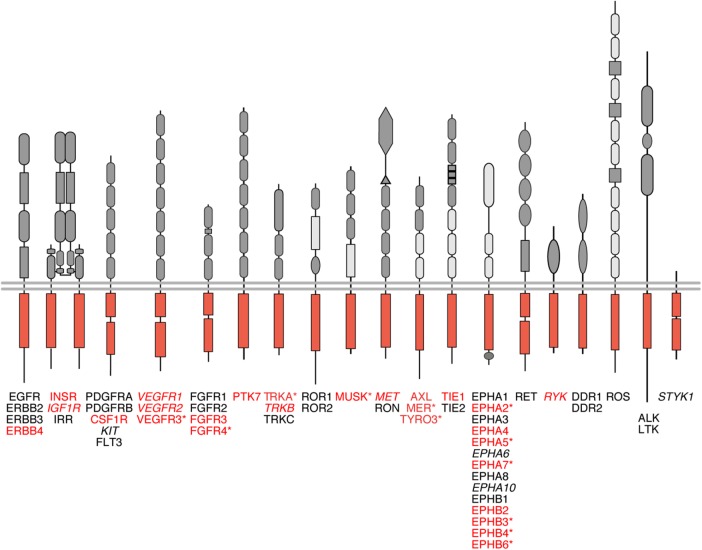
Despite the reported functional significance, for most human RTKs, there is no published information on whether they are susceptible to RIP-mediated cleavage. Here we set up a receptor kinome-wide screen to comprehensively characterize those human RTKs that can exploit RIP under standardized conditions. The screen identified all the nine previously identified gamma-secretase–sensitive RTKs that were covered by the screen (Ni et al., 2001; Wilhelmsen and van der Geer, 2004; Kasuga et al., 2007; Litterst et al., 2007; Marron et al., 2007; Inoue et al., 2009; Degnin et al., 2011; Na et al., 2012; Bae et al., 2015), which validated the method. In addition, 12 new gamma-secretase substrates (vascular endothelial growth factor receptor 3 [VEGFR3], fibroblast growth factor receptor 4 [FGFR4], TRKA, muscle-specific kinase [MUSK], MER, TYRO3, EPHA2, EPHA5, EPHA7, EPHB3, EPHB4, and EPHB6) were identified. Taken together, these findings indicate that at least half (27 out of 55) of all human RTKs can function as substrates for gamma-secretase cleavage and RIP.
RESULTS
Screen for gamma-secretase–cleaved RTKs
RTKs, such as ERBB4, have been reported to undergo RIP, which involves a two-step cleavage event. The cleavage of the ectodomain within the extracellular juxtamembrane domain by members of the ADAM family of proteases (sheddases) generates a substrate for the activity of the gamma-secretase complex that subsequently cleaves the RTK within the cytosolic half of the transmembrane domain (Figure 1A; Ancot et al., 2009). Activity of the sheddases is typically promoted by phorbol 12-myristate 13-acetate (PMA; Huovila et al., 2005), whereas cleavage by gamma-secretase is regulated by the accessibility of the enzyme complex to the sheddase-generated substrate sequence (Bolduc et al., 2016; Sannerud et al., 2016). The RIP process generates a soluble ICD, including the kinase domain with signaling activity (Figure 1A).
To identify human RTKs that are substrates for gamma-secretase, a receptor kinome-wide screen was set up. The screen was based on the effect of a chemical gamma-secretase inhibitor IX (GSI IX) on accumulating the membrane-anchored carboxy-terminal fragment (CTF) of the RTK after PMA-stimulated shedding of the RTK ectodomain. Accumulation of the CTF was detected by Western analysis of cells transfected with cDNAs encoding full-length human RTKs fused with carboxy-terminal epitope tags (Figure 1B). Only CTFs migrating in the Western gels at sizes consistent with the expected size of full-length CTF (whole ICD + transmembrane domain; see Figure 1B) were considered positive findings.
We initially validated the screen by analyzing the four members of the epidermal growth factor receptor (EGFR)/ERBB subfamily, which has been documented to include only one gamma-secretase substrate, ERBB4 (Ni et al., 2001; Blobel et al., 2009). As expected, GSI IX treatment of MCF-7 transfectants led to accumulation of a 75-kDa CTF of ERBB4, whereas the immunoblots representing other ERBB family members did not indicate CTF accumulation (Figure 1C).
Identification of novel gamma-secretase substrates
To identify new gamma-secretase targets, we expanded the screen to include 45 out of the 55 human RTKs. The screen excluded nine RTKs that were not adequately expressed using the experimental setup, as well as STYK1 because the approach was inefficient in detecting cleavage of a very short ectodomain (RTKs not analyzed are indicated by italics in Figure 2). The screen was carried out in both MCF-7 and HEK293 cells using RTKs tagged with either V5, hemagglutinin (HA), or green fluorescent protein (GFP) epitopes.
FIGURE 2:
Several but not all RTKs can be cleaved by gamma-secretase. Schematic representation of the 55 human RTKs divided into 19 subfamilies. Names of the RTKs that have been identified as substrates for gamma-secretase cleavage are written in red font, others in black. RTKs that were not analyzed in our screen are in italics. Asterisks (*) indicate RTKs identified in this paper as novel gamma-secretase substrates. RTK structures modified after Lemmon and Schlessinger (2010).
Altogether, 21 of the 45 analyzed RTKs were shown to react to GSI IX treatment with CTF accumulation (Figures 1, C and D, and 2; Supplemental Figure S1). In addition, six out of the 10 human RTKs not included in the screen had previously been shown to undergo gamma-secretase–regulated cleavage (Cai et al., 2006; McElroy et al., 2007; Lyu et al., 2008; Ablonczy et al., 2009; Foveau et al., 2009; Tejeda et al., 2016), suggesting that at least 27 of the total 55 of human RTKs can function as gamma-secretase substrates (indicated by red type in Figure 2). The analysis revealed 12 previously unreported substrates (marked with asterisks in Figure 2) and indicated that, for example, the whole TAM subfamily, encompassing TYRO3, AXL, and MER, is cleavable by gamma-secretase (Figure 1D). Strongly supporting the validity of these findings, the screen also identified all nine out of the nine RTKs that had been reported to be cleaved by gamma-secretase in previous publications (Ni et al., 2001; Wilhelmsen and van der Geer, 2004; Kasuga et al., 2007; Litterst et al., 2007; Marron et al., 2007; Inoue et al., 2009; Degnin et al., 2011; Na et al., 2012; Bae et al., 2015). The sizes of the GSI IX–induced CTF species varied between 35 and 75 kDa (Figure 1, C and D; Supplemental Figure S1).
Cleavage of endogenous AXL
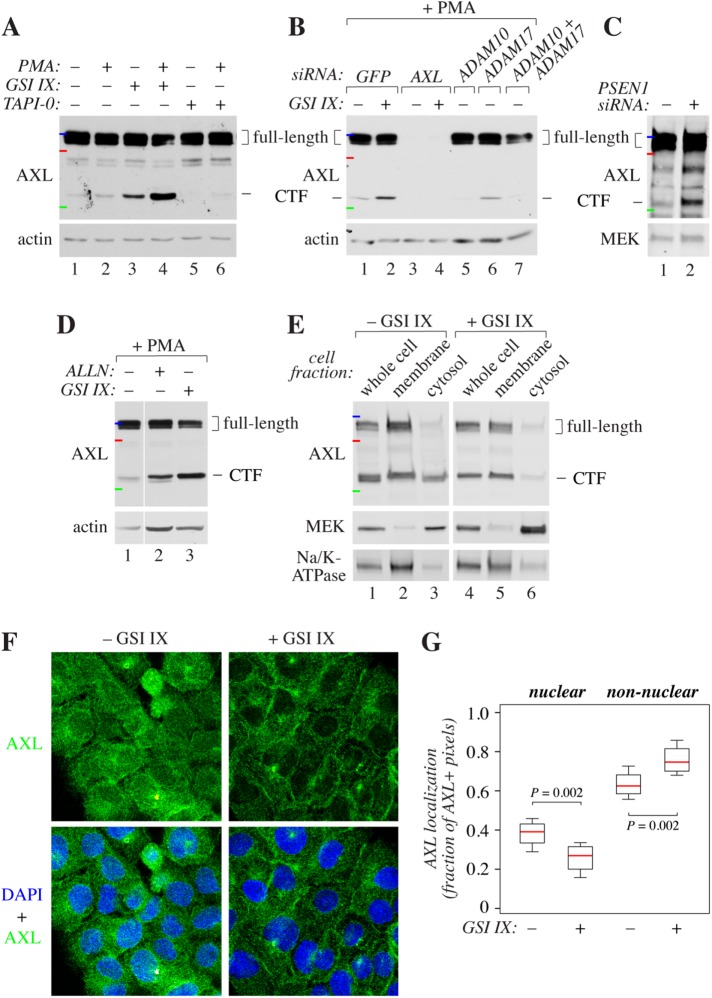
To further address the relevance of the findings for endogenously expressed RTKs, we chose AXL from the TAM subfamily for additional experimentation. PC-3 prostate cancer (Figure 3, A–C), MDA-MB-231 breast cancer (Figure 3, D and E), and A431 epidermoid cancer (Figure 3, F and G) cells expressed AXL endogenously. Silencing of AXL expression by RNA interference confirmed that both the full-length and CTF species recognized by the AXL antibody were indeed products of AXL transcripts (Figure 3B, lanes 3 and 4 vs. lanes 1 and 2). In both PC-3 and MDA-MB-231 cells, treatment with either PMA or GSI IX promoted accumulation of the CTF (Figure 3, A and D), indicating induced shedding and inhibition of gamma-secretase–mediated cleavage of the membrane-anchored CTF, respectively. PMA-promoted cleavage of endogenous AXL was suppressed by a chemical ADAM inhibitor, TAPI-0 (Figure 3A), as well as by RNA interference targeting ADAM10 but not RNA interference targeting ADAM17 (Figure 3B). As expected from findings observed by using the chemical inhibitor of gamma-secretase, also RNA interference–mediated targeting of presenilin-1, a key component in the gamma-secretase complex (De Strooper, 2003), resulted in CTF accumulation (Figure 3C).
FIGURE 3:
Biochemical validation of the cleavage of endogenous AXL. (A) Western analysis of PC3 cells treated for 4 h with or without 5 μM GSI IX or 10 μM TAPI-0 followed by 20-min treatment with 100 ng/ml PMA, when indicated. Loading was controlled with anti-actin. (B) Western analysis of PC3 cells transfected with 25 nM of esiRNAs targeting ADAM10 and/or ADAM17. In parallel, GFP was silenced as a negative control and AXL to control the specificity of the AXL antibody. Cells were treated or not for 4 h with 5 μM GSI IX followed by 20-min treatment with 100 ng/ml PMA. Loading was controlled with anti-actin. ADAM10- and ADAM17-specific silencing resulted in 50 and 85% reduction in protein expression, respectively, when compared with the GFP control with densitometric analysis. (C) Western analysis of PC3 cells transfected with 25 nM of esiRNAs targeting PSEN1 or GFP. Loading was controlled with anti-MEK1/2. PSEN1-specific silencing resulted in a 70% reduction in protein expression when compared with the GFP control with densitometric analysis. (D) Western analysis of MDA-MB231 treated for 4 h with or without 10 μM of the proteasome inhibitor ALLN or 5 μM GSI IX followed by 20-min treatment with 100 ng/ml PMA. Loading was controlled with anti-actin. (E) Western analysis of MDA-MB231 cells treated or not for 4 h with 5 μM GSI IX and subjected to subcellular fractionation. Expression of MEK1/2 and Na/K-ATPase were analyzed as markers of cytosolic and membrane fractions, respectively. (F) Confocal microscopy analysis of the localization of AXL in A431 cells treated for 4 h with or without 5 μM GSI IX. Cells were stained with an antibody against an intracellular epitope of AXL (green) and DAPI to visualize nuclei (blue). (G) Quantification of the confocal immunofluorescence analysis of AXL localization shown in F. Nuclear localization is presented as the percentage of AXL-specific signals colocalizing with DAPI of all AXL-specific signals within the cells. At least 100 cells were analyzed per treatment. The molecular weight markers in A–E are indicated by colored horizontal lines: blue, 150 kDa; red, 100 kDa; and green, 50 kDa.
Gamma-secretase generates a soluble AXL fragment
Gamma-secretase–released RTK ICDs have been reported to become degraded in the proteasomes or form soluble intracellular signaling units (Sardi et al., 2006; Marron et al., 2007; Foveau et al., 2009; Xu et al., 2009). To characterize the subcellular distribution of endogenous AXL CTF, MDA-MB-231 cells were treated with the proteasome inhibitor ALLN or subjected to subcellular fractionation separating membrane and cytosolic fractions. Proteasome inhibition indeed led to accumulation of an AXL CTF fragment similar in size to fragments induced by PMA or GSI IX (Figure 3D). Moreover, AXL CTF was present in a cytosolic fraction including soluble proteins, and its presence in the cytosol was suppressed by GSI IX treatment (Figure 3E). Interestingly, the cytosolic GSI IX–sensitive soluble CTF species migrated at a slightly smaller relative size in the gradient SDS–PAGE gel used for the fractionation analyses as compared with the membrane-associated CTF species (Figure 3E, lane 3 vs. lane 2). This finding is consistent with the expected smaller molecular weight of an ICD fragment cleaved by gamma-secretase at the transmembrane domain as opposed to a fragment cleaved by sheddase at the extracellular juxtamembrane domain (Figure 1A). GSI IX treatment further suppressed nuclear localization of an intracellular AXL epitope in a confocal immunofluorescence analysis of A431 cells that naturally demonstrate relatively high nuclear AXL immunoreactivity (Figure 3, F and G). Taken together, these validation experiments indicate that endogenous AXL can produce a soluble ICD that can be targeted for degradation or translocate into the cytosol or nucleus.
Gamma-secretase–sensitive growth promoted by the cleavable RTKs
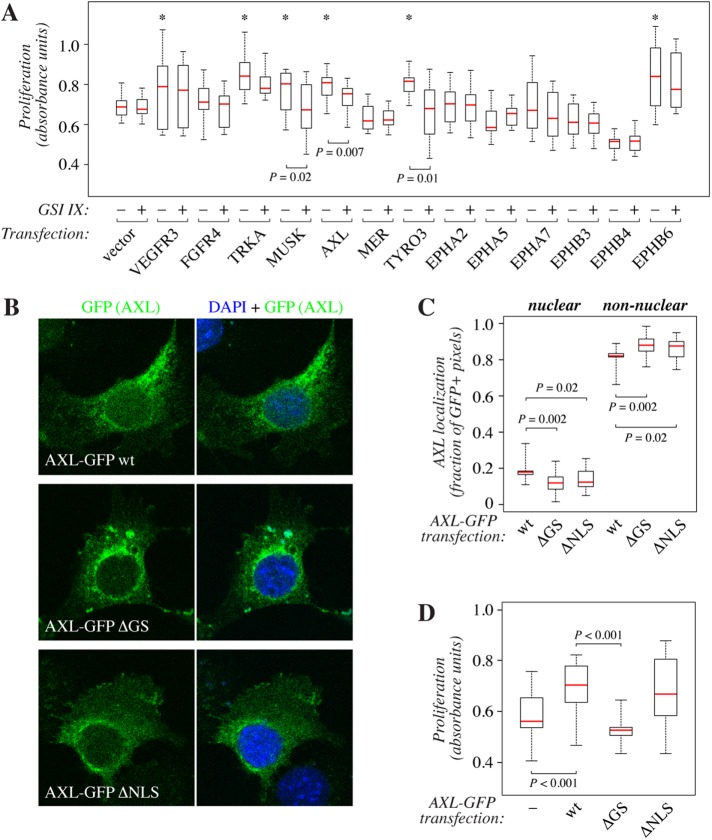
Ectopic expression of AXL has been shown to promote growth of NIH-3T3 fibroblasts (O’Bryan et al., 1991; Burchert et al., 1998). To characterize the functional significance of the gamma-secretase–mediated cleavage, NIH-3T3 cells were transfected with vectors encoding AXL or each of the 12 newly characterized cleavable RTKs and cultured in the presence or absence of 5 µM GSI IX for 72 h. Cell growth was analyzed by measuring the number of viable cells using the WST-8 reagent (Figure 4A). Overexpression of VEGFR3, TRKA, MUSK, AXL, TYRO3, or EPHB6 significantly promoted growth. However, the enhanced growth was significantly suppressed by inhibition of gamma-secretase activity only in the case of the two TAM family members AXL and TYRO3, as well as MUSK (Figure 4A). Blocking the gamma-secretase activity had no significant effect on control samples without ectopic RTK expression or on the growth of cells overexpressing other cleavable RTKs, indicating specificity of the effect.
FIGURE 4:
Functional analysis of gamma-secretase–mediated RTK cleavage. (A) WST-8 cell proliferation analysis of NIH-3T3 transfectants expressing novel cleavable RTKs VEGFR3, FGFR4, TRKA, MUSK, AXL, MER, TYRO3, EPHA2, EPHA5, EPHA7, EPHB3, EPHB4, or EPHB6. The cells were treated for 72 h in the presence or absence of 5 μM GSI IX. Data from at least six parallel samples from three independent experiments are shown. An asterisk (*) on top of a box plot indicates significantly (p < 0.05) increased proliferation as compared with the vector control transfectant in the absence of GSI IX. (B) Confocal microscopy analysis of NIH-3T3 cells overexpressing C-terminally GFP-tagged wild-type AXL (wt), AXL with a mutated gamma-secretase cleavage site (∆GS), or AXL with a mutated nuclear localization signal (∆NLS). GFP signal is shown in green, DAPI-stained nuclei in blue. (C) Quantification of the confocal immunofluorescence analysis of AXL-GFP localization shown in B. Nuclear localization is presented as the percentage of GFP-specific signals colocalizing with DAPI of all GFP-specific signals within the cells. At least 20 cells were analyzed per construct. (D) A 72-h WST-8 cell proliferation analysis of NIH-3T3 transfectants expressing wild-type AXL (wt) or AXL ∆GS or AXL ∆NLS mutant constructs.
Growth promoted by AXL or TYRO3 is suppressed by mutating the gamma-secretase cleavage site
To validate the functional data derived from analyses with a chemical GSI IX, GFP-fusion constructs encoding AXL or TYRO3 with mutated gamma-secretase cleavage sites or nuclear localization signals (NLSs) were generated. Mutations at the AXL gamma-secretase cleavage site (amino acids 453YVLLGAVV459 replaced by 453IIIGPLIF459) and NLS (AXL R474A/R475A) have been previously described and functionally verified (Lu et al., 2017). An I449A mutation near the cytoplasmic end of the transmembrane domain of TYRO3 generated a receptor insensitive to gamma-secretase cleavage, as no CTF accumulated from the mutant receptor in response to treatment with GSI IX (Supplemental Figure S2A). The TYRO3 ICD contains a putative NLS closely resembling that of AXL (Migdall-Wilson et al., 2012) and was mutated at the respective position to generate the TYRO3 NLS mutant R452A/K453A.
Both mutations at the gamma-secretase cleavage site as well as at the NLS resulted in a decrease in the amount of nuclearly localized AXL-GFP signal in confocal fluorescence analysis of the NIH-3T3 cells (Figure 4, B and C). However, only mutation at the gamma-secretase site reduced the growth of the transfectants as compared with control cells expressing the wild-type construct (Figure 4D). Similarly to AXL, the TYRO3 I449A mutant with impaired gamma-secretase cleavage was significantly less efficient than the wild-type receptor in promoting growth, whereas TYRO3 R452A/K453A with mutations at the putative NLS demonstrated no difference (Supplemental Figure S2D). However, there was no significant difference between the different TYRO3 constructs in their localization in the nucleus, and neither the wild-type nor the mutant TYRO3 proteins were abundant in the nuclei (Supplemental Figure S2, B and C). These observations are consistent with the findings obtained using chemical gamma-secretase inhibition and suggest that generation of the soluble ICD by gamma-secretase, but not the translocation of the ICD into the nucleus, is necessary for the efficient AXL- or TYRO3-promoted growth in the NIH-3T3 model.
DISCUSSION
Our findings represent the first comprehensive analysis of gamma-secretase–regulated RTK signaling. We examined 45 out of the 55 human RTKs for GSI IX–sensitive accumulation of the CTF and identified 12 new RTKs as gamma-secretase substrates complementing, for example, the whole TAM subfamily (TYRO3, AXL, and MER) as cleavable RTKs. Furthermore, all previously published gamma-secretase–sensitive RTKs that were tested were also identified as cleavable in our analysis, validating the screen. Taken together with previously published data, our results indicate that a total of at least 27 out of 55 human RTKs are cleavable by gamma-secretase activity. These data demonstrate that the phenomenon is common but not universally exploited by all human RTKs.
Despite the selective presence of gamma-secretase–mediated RTK cleavage in some but not all RTKs, we were not able to identify a conserved consensus sequence exclusively present in the cleavable RTKs. It is plausible that factors other than the primary sequence of the membrane-proximal extracellular or the transmembrane domain of gamma-secretase substrate are the major determinants of recognition by the gamma-secretase complex. Such factors may include the length of the remaining ectodomain after shedding of the substrate (Funamoto et al., 2013), the strength of the interaction between the transmembrane domain of the substrate and the gamma-secretase complex (Bolduc et al., 2016), and subcellular localization of the substrate in relation to the gamma-secretase activity (Sannerud et al., 2016).
The gamma-secretase cleavage of RTKs and the solubilization of the CTF can add diversity to intracellular signaling by either creating additional signaling fragments or representing a mechanism of turnover and degradation of a transmembrane protein by the proteasome. Several cleavable RTKs, including AXL, have recently been reported to regulate cellular mechanosensing (Prager-Khoutorsky et al., 2011; Yang et al., 2016), a process that may also involve modification by proteolytic processing of the membrane-anchored RTKs. Soluble RTK ICDs have been reported to translocate to the nucleus (ERBB4, FGFR3, PTK7, and RYK; Ni et al., 2001; Lyu et al., 2008; Degnin et al., 2011; Na et al., 2012), interact with transcription factors (ERBB4; Komuro et al., 2003; Williams et al., 2004; Arasada and Carpenter, 2005; Linggi and Carpenter, 2006; Paatero et al., 2012), and actively regulate differentiation (ERBB4; Sardi et al., 2006) or angiogenesis (VEGFR1; Cai et al., 2006). In addition, the ICD of ERBB4 has been reported to localize to mitochondria and promote apoptosis (Naresh et al., 2006; Vidal et al., 2007) and the ICD of EPHB2 to phosphorylate other membrane receptors and modify their subcellular localization (Xu et al., 2009). On the other hand, the soluble ICD of MET has been shown to be quickly degraded after constitutive gamma-secretase cleavage (Foveau et al., 2009), and macrophage activation by lipopolysaccharide, a major component of Gram-negative bacterial cell walls, results in gamma-secretase–mediated cleavage and down-regulation of CSF1R by rapid degradation of CTF (Glenn and van der Geer, 2008). In any case, it is conceivable that as long as the ICD retains an active kinase domain, it can affect signal transduction by posttranslational modification of substrates and by phoshotyrosine-dependent interactions. Consistent with a functional role, our analysis with NIH-3T3 transfectants indicated that the released ICDs of AXL and TYRO3 may participate in regulation of cell proliferation with a mechanism that is dependent on gamma-secretase cleavage.
Several RTKs are key oncogenes, and a number of RTK-targeted cancer drugs are currently in clinical use. On the basis of our screen and earlier reports, it has been determined that several of these cancer drug targets are also substrates for gamma-secretase, raising the hypothesis that gamma-secretase–mediated RIP may modify the tumor-promoting activity of the RTKs. Indeed, chemical gamma-secretase inhibitors have demonstrated anti-tumor activity in tumor models, and multiple clinical cancer trials testing gamma-secretase inhibitors have been carried out (Golde et al., 2013; https://clinicaltrials.gov). Most of the activities of gamma-secretase inhibitors, however, have been attributed to their ability to block Notch signaling (Golde et al., 2013), and there is currently very little information about the role of RTKs in mediating either the anti-tumor activity or adverse effects of the gamma-secretase inhibitors (Rahimi et al., 2009; Golde et al., 2013). In addition to improving the understanding of the mechanism of action of the gamma-secretase inhibitors developed to target non-RTK targets, our findings may have implications for the development of therapeutic approaches to specifically modulate gamma-secretase–regulated RTK cleavage.
MATERIALS AND METHODS
Plasmids
cDNA inserts encoding 50 out of 55 human RTKs (not including EGFR, ERBB2, ERBB3, ERBB4, and MUSK) were obtained from the Human Kinase Open Reading Frame Collection (Addgene #1000000014; Johannessen et al., 2010; Yang et al., 2011) and from the Mammalian Gene Collection (GE Healthcare Dharmacon and Source Biosciences). The inserts were cloned from pDONR223, pDONR221, or pENTR223 plasmids into pLX302 (Addgene #25896; Yang et al., 2011), pMAX-DEST (Addgene #37631; Klezovitch et al., 2008) or pDEST-eGFP-N1 (Addgene #31796; Hong et al., 2010) expression plasmids using Gateway Cloning Technology with LR Clonase II Enzyme Mix (Life Technologies) to allow expression of C-terminally V5-tagged and eGFP-tagged proteins, respectively.
The full-length cDNAs encoding human EGFR, ERBB2, ERBB3, or MUSK were PCR amplified with oligonucleotides TTTTTTCTCGAGACCATGCGACCCTCCGGGACGGCCGGGGCA and TTTTTTCGCGGCCGCTGCTCCAATAAATTCACTGCTTTGTGGC (for EGFR), TTTTTTCTCGAGACCATGGAGCTGGCGGCCTTGTGCCGCTGGGGGCTC and TTTTTTCGCGGCCGCCACTGGCACGTCCAGACCCAGGTAC (for ERBB2), TTTTTTTCTAGAACCATGAGGGCGAACGACGCTCTGCAGGTGCTG and TTTTTTCGCGGCCGCCGTTCTCTGGGCATTAGCCTTGGG (for ERBB3), or GCCGTCTAGAACCATGAGAGAGCTCGTCAAC and TATAGCGGCCGCGACACTCACAGT TCC (for MUSK) using pEGFP-based EGFR construct, pcDNA3.1-based ERBB2 and ERBB3 constructs, and pCR-Blunt II-TOPO–based MUSK construct as templates, respectively. The PCR fragments were digested with XhoI and NotI and ligated into XhoI- and NotI-digested pcDNA3.1-Hygro(-)–based vector where a region encoding an HA tag (GCGGCCGCGTACCCATACGATGTTCCAGATTACGCGTAAAAGCTT) had been engineered between the NotI and HindIII sites. The resulting plasmids used to express a C-terminally HA-tagged EGFR, ERBB2, ERBB3, or MUSK were designated as pcDNA3.1-EGFR-HA, pcDNA3.1-ERBB2-HA, pcDNA3.1-ERBB3-HA, or pcDNA3.1-MUSK-HA, respectively. The expression plasmid encoding ERBB4-HA has been previously described (Määttä et al., 2006).
Constructs encoding TYRO3 and AXL with mutated gamma-secretase cleavage sites or NLSs were generated using pDEST-eGFP-N1–based TYRO3 and AXL expression plasmids. To generate a plasmid encoding TYRO3 with a mutated gamma-secretase cleavage site (TYRO3∆GS), an I449A mutation was introduced. To generate a plasmid encoding TYRO3 with a mutated nuclear localization signal (TYRO3∆NLS), a R452A/K453A double mutation was introduced. AXL gamma-secretase cleavage site mutant (AXL ∆GS) and NLS mutant (AXL ∆NLS) have been previously described (mut1 and R474A/R475A, respectively; Lu et al., 2017). For cloning, the pDEST-eGFP-N1–based plasmids were digested with HindIII and SmaI (TYRO3∆GS), EcoNI and SmaI (TYRO3∆NLS), or BsaBI (AXL∆GS and AXL∆NLS). DNA sequences containing the mutations were ordered as synthetic DNA fragments (Integrated DNA Technologies) and ligated to the digested plasmids using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs) according to the manufacturer’s instructions. Mutations were verified by sequencing and expression of constructs by Western blotting.
Antibodies
Anti-AXL (8661), anti-HA (2367), and anti-MEK1/2 (4694) antibodies were purchased from Cell Signaling Technology. Anti-GFP (sc-9996), anti-V5 (sc-81594), anti-actin (sc-1616), and anti-RNA polymerase II (sc-900) antibodies were purchased from Santa Cruz Biotechnology. Anti-Hsp90 (ab13495) and anti-Na/K-ATPase (ab76020) antibodies were purchased from Abcam. Anti-PSEN1 (MAB5232) antibody was purchased from Millipore.
Cell culture and transfections
MCF-7 human breast cancer cells were cultured in RPMI 1640 medium (Life Technologies) supplemented with 10% (wt/vol) fetal calf serum (FCS) (Promocell), 50 U/ml penicillin and 50 μg/ml streptomycin solution (Sigma-Aldrich), 1 nM 17-β-estradiol (Sigma-Aldrich), and 1 μg/ml insulin (Sigma-Aldrich). PC-3 human prostate cancer cells were cultured in RPMI 1640 supplemented with 10% (wt/vol) FCS, 50 U/ml penicillin, and 50 μg/ml streptomycin. A431 human epidermoid carcinoma cells, MDA-MB-231 human breast cancer cells, HEK293 human embryonic kidney cells, and NIH-3T3 mouse fibroblasts were cultured in DMEM (Life Technologies) supplemented with 10% (wt/vol) FCS, 50 U/ml penicillin, and 50 μg/ml streptomycin. Cells were transfected using FuGENE 6 (Promega), HilyMAX (Dojindo), or jetPRIME (Polyplus-transfection) transfection reagents according to the manufacturers’ instructions.
Screening of gamma-secretase substrates
MCF-7 or HEK293 cells growing on six-well plates were transfected with expression plasmids encoding RTKs. After 24 h, cells were treated with or without 5 μM GSI IX (Calbiochem) for 4 h, and gamma-secretase cleavage was induced with 100 ng/ml PMA (Sigma-Aldrich) for 20 min. The generation of C-terminal fragments was analyzed by Western blotting.
Analysis of endogenous cleavage of AXL
MDA-MB-231 or PC-3 cells were treated with the proteasome inhibitor ALLN (10 μM; Calbiochem), GSI IX (5 μM; Calbiochem) or the ADAM inhibitor TAPI-0 (5 μM; Calbiochem) for 4 h, followed by stimulation of cleavage by 0 or 100 ng/ml PMA for 20 min. Effects of the treatments on the levels of different RTK protein species were analyzed by Western blotting.
Western blotting
Cells were lysed in lysis buffer (0.1% Triton X-100, 10 mM Tris-Cl [pH 7.4], 1 mM EDTA, 5 mM NaF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM Na3VO4, 2 mM phenylmethane sulfonyl fluoride, and 10 mM Na4P2O7), the lysates were centrifuged, and the supernatants were used for analyses. Equal amounts of samples were separated by SDS–PAGE and transferred to nitrocellulose membranes. Antibodies against Actin, Hsp90, MEK1/2, or RNA polymerase II were used to control loading. Signals were visualized using the Odyssey CLx imaging system (LI-COR) or using enhanced chemiluminescence by SuperSignal West Pico chemiluminescent substrate (ThermoFischer Scientific).
Gene silencing
Endoribonuclease-prepared small interfering RNA (esiRNA) oligonucleotides targeting human ADAM10 (EHU129311), ADAM17 (EHU075381), AXL (EHU081461), or PSEN1 (EHU073361) were purchased from Sigma-Aldrich. PC-3 cells growing on 12-well plates were transfected with esiRNAs at final concentrations of 25 nM using Lipofectamine 2000 (Thermo Fischer Scientific) according to the manufacturer’s instructions. EsiRNA against GFP (EHUEGFP; Sigma-Aldrich) was used as a control. Knockdown efficacy was determined by Western blotting and ImageStudio Lite v5.2 (LI-COR) software.
Subcellular fractionation
MDA-MB-231 cells growing on 10-cm plates were treated with or without 5 μM GSI IX. Subcellular fractionation was carried out using a subcellular fractionation kit (Cell Signaling Technologies) according to the manufacturer’s instructions except that 1 mg/ml trypsin inhibitor (Sigma-Aldrich) was added to the samples after collecting the cells from the plates. Samples were analyzed by Western blotting.
Cell proliferation assay
NIH-3T3 transfectants were plated on 96-well plates in 12 replicates at a density of 3000 cells per well and cultured for 72 h in the presence or absence of 5 µM GSI IX in DMEM + 1% FCS. Fresh medium with or without GSI IX was replaced every 24 h. After 72 h, the number of viable cells was estimated by adding WST-8 reagent (Nippon Genetic) and measuring absorbance at 450 nm with a Thermo Scientific Multiskan FC Microplate Photometer.
Immunofluorescence and confocal microscopy
To detect endogenous AXL, A431 cells were cultured on coverslips and treated for 4 h in the presence or absence of 5 μM GSI IX. The cells were fixed with methanol and stained with anti-AXL (8661; Cell Signaling Technologies) and AlexaFluor 488 goat anti-rabbit (Molecular Probes). To detect ectopically expressed AXL or TYRO3, NIH-3T3 transfectants expressing GFP-tagged constructs were cultured on coverslips, fixed with 3% paraformaldehyde, and permeabilized with 0.1% Triton X-100. After labeling the nuclei with 4’,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich), all cells were mounted on glass slides with Mowiol 40-88 (Sigma-Aldrich). Images were acquired with a Zeiss LSM 780 confocal microscope and Zen software (Zeiss). Representative 1.1-μm-section images of A431 cells and 1.6-μm-section images of the transfected NIH-3T3 cells are shown in Figures 3F and 4B and Supplemental Figure S2B. Microscopy images were analyzed with ImageJ software version 1.51, and the Coloc2 plug-in was used to analyze colocalization. Nuclear localization of AXL or TYRO3 was estimated by measuring the percentage of AXL- or GFP-specific signals colocalizing with DAPI of all AXL- or GFP-specific signals within the cells.
Statistical analyses
WST-8 cell proliferation data analyses and confocal microscopy colocalization data analyses were performed using R software version 3.3.2 and RStudio version 1.0.136 as the user interface. Data from cell proliferation assays were processed by the ComBat algorithm to remove the batch effect between repeated samples (Johnson et al., 2007). p values were calculated using a Mann-Whitney nonparametric U-test or Kruskal-Wallis test and adjusted using false discovery rate correction. p values smaller than 0.05 were considered significant. Box plot presentations indicate the median (red horizontal line), the second and third quartiles (the box), and the range (whiskers) of the data.
Supplementary Material
Acknowledgments
We thank Minna Santanen and Maria Tuominen for skillful technical assistance and Katri Vaparanta for data analysis support. This work was supported by the Academy of Finland, Cancer Foundation Finland, the Sigrid Juselius Foundation, the Turku University Central Hospital, the Turku Doctoral Programme of Biomedical Sciences, the Cancer Society of Southwestern Finland, the Jenny and Antti Wihuri Foundation, and the Finnish Cultural Foundation.
Abbreviations used:
- ADAM
disintegrin and metalloprotease
- CTF
carboxy-terminal fragment
- DAPI
4’,6-diamidino-2-phenylindole
- EGFR
epidermal growth factor receptor
- FCS
fetal calf serum
- FGFR4
fibroblast growth factor receptor 4
- GFP
green fluorescent protein
- GSI IX
gamma-secretase inhibitor IX
- HA
hemagglutinin
- ICD
intracellular domain
- MUSK
muscle-specific kinase
- NLS
nuclear localization signal
- PMA
phorbol 12-myristate 13-acetate
- RIP
regulated intramembrane proteolysis
- RTK
receptor tyrosine kinase
- VEGFR1 and 3
vascular endothelial growth factor receptors 1 and 3.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-04-0261) on September 13, 2017.
REFERENCES
- Ablonczy Z, Prakasam A, Fant J, Fauq A, Crosson C, Sambamurti K. Pigment epithelium-derived factor maintains retinal pigment epithelium function by inhibiting vascular endothelial growth factor-R2 signaling through gamma-secretase. J Biol Chem. 2009;284:30177–30186. doi: 10.1074/jbc.M109.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancot F, Leroy C, Muharram G, Lefebvre J, Vicogne J, Lemiere A, Kherrouche Z, Foveau B, Pourtier A, Melnyk O. Shedding-generated met receptor fragments can be routed to either the proteasomal or the lysosomal degradation pathway. Traffic. 2012;13:1261–1272. doi: 10.1111/j.1600-0854.2012.01384.x. [DOI] [PubMed] [Google Scholar]
- Ancot F, Foveau B, Lefebvre J, Leroy C, Tulasne D. Proteolytic cleavages give receptor tyrosine kinases the gift of ubiquity. Oncogene. 2009;28:2185–2195. doi: 10.1038/onc.2009.88. [DOI] [PubMed] [Google Scholar]
- Arasada RR, Carpenter G. Secretase-dependent tyrosine phosphorylation of Mdm2 by the ErbB-4 intracellular domain fragment. J Biol Chem. 2005;280:30783–30787. doi: 10.1074/jbc.M506057200. [DOI] [PubMed] [Google Scholar]
- Bae SY, Hong JY, Lee HJ, Park HJ, Lee SK. Targeting the degradation of AXL receptor tyrosine kinase to overcome resistance in gefitinib-resistant non-small cell lung cancer. Oncotarget. 2015;6:10146–10160. doi: 10.18632/oncotarget.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel AJ, Sanders CR. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell Mol Life Sci. 2008;65:1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel CP, Carpenter G, Freeman M. The role of protease activity in ErbB biology. Exp Cell Res. 2009;315:671–682. doi: 10.1016/j.yexcr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc DM, Montagna DR, Gu Y, Selkoe DJ, Wolfe MS. Nicastrin functions to sterically hinder γ-secretase–substrate interactions driven by substrate transmembrane domain. Proc Natl Acad Sci USA. 2016;113:E509–E518. doi: 10.1073/pnas.1512952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchert A, Attar EC, McCloskey P, Fridell YW, Liu ET. Determinants for transformation induced by the Axl receptor tyrosine kinase. Oncogene. 1998;16:3177–3187. doi: 10.1038/sj.onc.1201865. [DOI] [PubMed] [Google Scholar]
- Cai J, Jiang WG, Grant MB, Boulton M. Pigment epithelium-derived factor inhibits angiogenesis via regulated intracellular proteolysis of vascular endothelial growth factor receptor 1. J Biol Chem. 2006;281:3604–3613. doi: 10.1074/jbc.M507401200. [DOI] [PubMed] [Google Scholar]
- Carpenter G, Pozzi A. Cell responses to growth factors: the role of receptor tyrosine kinase intracellular domain fragments. Sci Signal. 2012;5:pe42. doi: 10.1126/scisignal.2003526. [DOI] [PubMed] [Google Scholar]
- De Strooper B. Aph-1, Pen-2, and nicastrin with presenilin generate an active γ-secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- Degnin CR, Laederich MB, Horton WA. Ligand activation leads to regulated intramembrane proteolysis of fibroblast growth factor receptor 3. Mol Biol Cell. 2011;22:3861–3873. doi: 10.1091/mbc.E11-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foveau B, Ancot F, Leroy C, Petrelli A, Reiss K, Vingtdeux V, Giordano S, Fafeur V, Tulasne D. Down-regulation of the met receptor tyrosine kinase by presenilin-dependent regulated intramembrane proteolysis. Mol Biol Cell. 2009;20:2495–2507. doi: 10.1091/mbc.E08-09-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S, Sasaki T, Ishihara S, Nobuhara M, Nakano M, Watanabe-Takahashi M, Saito T, Kakuda N, Miyasaka T, Nishikawa K. Substrate ectodomain is critical for substrate preference and inhibition of γ-secretase. Nat Commun. 2013;4:2529. doi: 10.1038/ncomms3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn G, van der Geer P. Toll-like receptors stimulate regulated intramembrane proteolysis of the CSF-1 receptor through Erk activation. FEBS Lett. 2008;582:911–915. doi: 10.1016/j.febslet.2008.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde TE, Koo EH, Felsenstein KM, Osborne BA, Miele L. γ-Secretase inhibitors and modulators. Biochim Biophys Acta. 2013;1828:2898–2907. doi: 10.1016/j.bbamem.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong TT, Smyth JW, Gao D, Chu KY, Vogan JM, Fong TS, Jensen BC, Colecraft HM, Shaw RM. BIN1 localizes the L-type calcium channel to cardiac T-tubules. PLoS Biol. 2010;8:e1000312. doi: 10.1371/journal.pbio.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huovila A-PJ, Turner AJ, Pelto-Huikko M, Kärkkäinen I, Ortiz RM. Shedding light on ADAM metalloproteinases. Trends Biochem Sci. 2005;30:413–422. doi: 10.1016/j.tibs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Inoue E, Deguchi-Tawarada M, Togawa A, Matsui C, Arita K, Katahira-Tayama S, Sato T, Yamauchi E, Oda Y, Takai Y. Synaptic activity prompts gamma-secretase-mediated cleavage of EphA4 and dendritic spine formation. J Cell Biol. 2009;185:551–564. doi: 10.1083/jcb.200809151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jones FE. HER4 intracellular domain (4ICD) activity in the developing mammary gland and breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:247–258. doi: 10.1007/s10911-008-9076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga K, Kaneko H, Nishizawa M, Onodera O, Ikeuchi T. Generation of intracellular domain of insulin receptor tyrosine kinase by gamma-secretase. Biochem Biophys Res Commun. 2007;360:90–96. doi: 10.1016/j.bbrc.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, Nelson PS, Vasioukhin V. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci USA. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linggi B, Carpenter G. ErbB-4 s80 intracellular domain abrogates ETO2-dependent transcriptional repression. J Biol Chem. 2006;281:25373–25380. doi: 10.1074/jbc.M603998200. [DOI] [PubMed] [Google Scholar]
- Litterst C, Georgakopoulos A, Shioi J, Ghersi E, Wisniewski T, Wang R, Ludwig A, Robakis NK. Ligand binding and calcium influx induce distinct ectodomain/gamma-secretase-processing pathways of EphB2 receptor. J Biol Chem. 2007;282:16155–16163. doi: 10.1074/jbc.M611449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wan J, Yang Z, Lei X, Niu Q, Jiang L, Passtoors WM, Zang A, Fraering PC, Wu F. Regulated intramembrane proteolysis of the AXL receptor kinase generates an intracellular domain that localizes in the nucleus of cancer cells. FASEB J. 2017;31:1382–1397. doi: 10.1096/fj.201600702R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu J, Yamamoto V, Lu W. Cleavage of the Wnt receptor Ryk regulates neuronal differentiation during cortical neurogenesis. Dev Cell. 2008;15:773–780. doi: 10.1016/j.devcel.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Määttä JAJ, Sundvall M, Junttila TT, Peri L, Laine VJO, Isola J, Egeblad M, Elenius K. Proteolytic cleavage and phosphorylation of a tumor-associated ErbB4 isoform promote ligand-independent survival and cancer cell growth. Mol Biol Cell. 2006;17:67–79. doi: 10.1091/mbc.E05-05-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marron MB, Singh H, Tahir TA, Kavumkal J, Kim HZ, Koh GY, Brindle NP J. Regulated proteolytic processing of Tie1 modulates ligand responsiveness of the receptor-tyrosine kinase Tie2. J Biol Chem. 2007;282:30509–30517. doi: 10.1074/jbc.M702535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy B, Powell JC, McCarthy J V. The insulin-like growth factor 1 (IGF-1) receptor is a substrate for γ-secretase-mediated intramembrane proteolysis. Biochem. Biophys Res Commun. 2007;358:1136–1141. doi: 10.1016/j.bbrc.2007.05.062. [DOI] [PubMed] [Google Scholar]
- Migdall-Wilson J, Bates C, Schlegel J, Brandão L, Linger RMA, DeRyckere D, Graham DK. Prolonged exposure to a Mer ligand in leukemia: Gas6 favors expression of a partial Mer glycoform and reveals a novel role for Mer in the nucleus. PLoS One. 2012;7:e31635. doi: 10.1371/journal.pone.0031635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne R, Berbon M, Doublet L, Debreuck N, Baranzelli A, Drobecq H, Leroy C, Delhem N, Porte H, Copin MC. Necrosis- and apoptosis-related Met cleavages have divergent functional consequences. Cell Death Dis. 2015;6:e1769. doi: 10.1038/cddis.2015.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na HW, Shin WS, Ludwig A, Lee S-T. The cytosolic domain of protein-tyrosine kinase 7 (PTK7), generated from sequential cleavage by a disintegrin and metalloprotease 17 (ADAM17) and γ-secretase, enhances cell proliferation and migration in colon cancer cells. J Biol Chem. 2012;287:25001–25009. doi: 10.1074/jbc.M112.348904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naresh A, Long W, Vidal GA, Wimley WC, Marrero L, Sartor CI, Tovey S, Cooke TG, Bartlett JMS, Jones FE. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006;66:6412–6420. doi: 10.1158/0008-5472.CAN-05-2368. [DOI] [PubMed] [Google Scholar]
- Ni CY, Murphy MP, Golde TE, Carpenter G. Gamma-secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- O’Bryan JP, Frye RA, Cogswell PC, Neubauer A, Kitch B, Prokop C, Espinosa R, Le Beau MM, Earp HS, Liu ET. Axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol. 1991;11:5016–5031. doi: 10.1128/mcb.11.10.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paatero I, Jokilammi A, Heikkinen PT, Iljin K, Kallioniemi OP, Jones FE, Jaakkola PM, Elenius K. Interaction with ErbB4 promotes hypoxia-inducible factor-1α signaling. J Biol Chem. 2012;287:9659–9671. doi: 10.1074/jbc.M111.299537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager-Khoutorsky M, Lichtenstein A, Krishnan R, Rajendran K, Mayo A, Kam Z, Geiger B, Bershadsky AD. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- Rahimi N, Golde TE, Meyer RD. Identification of ligand-induced proteolytic cleavage and ectodomain shedding of VEGFR-1/FLT1 in leukemic cancer cells. Cancer Res. 2009;69:2607–2614. doi: 10.1158/0008-5472.CAN-08-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R, Esselens C, Ejsmont P, Mattera R, Rochin L, Tharkeshwar AK, De Baets G, De Wever V, Habets R, Baert V. Restricted location of PSEN2/γ-secretase determines substrate specificity and generates an intracellular Aβ Pool. Cell. 2016;166:193–208. doi: 10.1016/j.cell.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell. 2006;127:185–197. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- Tejeda GS, Ayuso-Dolado S, Arbeteta R, Esteban-Ortega GM, Vidaurre OG, Díaz-Guerra M. Brain ischaemia induces shedding of a BDNF-scavenger ectodomain from TrkB receptors by excitotoxicity activation of metalloproteinases and γ-secretases. J Pathol. 2016;238:627–640. doi: 10.1002/path.4684. [DOI] [PubMed] [Google Scholar]
- Tomita T. Molecular mechanism of intramembrane proteolysis by γ-secretase. J Biochem. 2014;156:195–201. doi: 10.1093/jb/mvu049. [DOI] [PubMed] [Google Scholar]
- Veikkolainen V, Vaparanta K, Halkilahti K, Iljin K, Sundvall M, Elenius K. Function of ERBB4 is determined by alternative splicing. Cell Cycle. 2011;10:2647–2657. doi: 10.4161/cc.10.16.17194. [DOI] [PubMed] [Google Scholar]
- Vidal GA, Clark DE, Marrero L, Jones FE. A constitutively active ERBB4/HER4 allele with enhanced transcriptional coactivation and cell-killing activities. Oncogene. 2007;26:462–466. doi: 10.1038/sj.onc.1209794. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Yarden Y. Receptor Tyrosine Kinases: Family and Subfamilies. Cham, Switzerland: Springer International Publishing; 2015. [Google Scholar]
- Wilhelmsen K, van der Geer P. Phorbol 12-myristate 13-acetate-induced release of the colony-stimulating factor 1 receptor cytoplasmic domain into the cytosol involves two separate cleavage events. Mol Cell Biol. 2004;24:454–464. doi: 10.1128/MCB.24.1.454-464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CC, Allison JG, Vidal GA, Burow ME, Beckman BS, Marrero L, Jones FE. The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J Cell Biol. 2004;167:469–478. doi: 10.1083/jcb.200403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Litterst C, Georgakopoulos A, Zaganas I, Robakis NK. Peptide EphB2/CTF2 generated by the γ-secretase processing of EphB2 receptor promotes tyrosine phosphorylation and cell surface localization of N -Methyl-d-aspartate receptors. J Biol Chem. 2009;284:27220–27228. doi: 10.1074/jbc.M109.048728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Lieu ZZ, Wolfenson H, Hameed FM, Bershadsky AD, Sheetz MP. Mechanosensing controlled directly by tyrosine kinases. Nano Lett. 2016;16:5951–5961. doi: 10.1021/acs.nanolett.6b02995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.