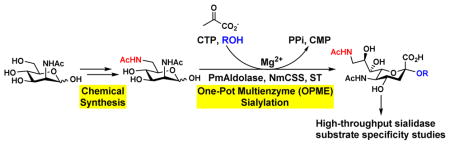
Abstract
O -Acetylation of sialic acid (Sia) modulates its recognition by sialic acid-binding proteins and plays an important role in biological and pathological processes. 9-O-Acetylation is the most common modification of sialic acid in human. However, study of O-acetylated sialoglycans is hampered due to the instability of O-acetyl group towards pH changes and sensitivity to esterases. Our previous studies demonstrated a chemical biology method to this problem by replacing the oxygen atom in the C9 ester group of sialic acid by a nitrogen to form an amide. Here, we synthesized a library of sixteen new 9-acetamido-9-deoxy-N-acetylneuraminic acid (Neu5Ac9NAc)-containing α2–3- and α2–6-linked sialosides with various underlying glycans using efficient one-pot three-enzyme (OP3E) sialylation systems. Neu5Ac9NAc-containing compounds with a para-nitrophenol aglycon have been used together with their 9-O-acetyl analogs in microtiter plate-based high-throughput substrate specificity studies of nine different sialidases including those from humans and bacteria. In general, similar to 9-O-acetylation, 9-N-acetyl modification of sialic acid in the substrates lowers sialic acid-cleavage activity of most sialidases. In most cases, Neu5Ac9NAc is a good analog of 9-O-acetyl sialic acid. However, exceptions do exist. For example, 9-N- and 9-O-acetyl modifications have different effects on the sialosides cleave efficiencies of a commercially available C. perfringens sialidase as well as recombinant Streptococcus pneumoniae sialidase SpNanC and Bifidobacterium infantis sialidase BiNanH2. The mechanism for the difference awaits further investigation.
Keywords: 9-N-acetyl sialic acid, 9-O-acetyl sialic acid, chemoenzymatic synthesis, one-pot multienzyme, sialic acid
Graphical Abstract
Sialic acids (Sia) are 9-carbon monosaccharides commonly presented at the glycan termini of glycoconjugates. They are directly involved in many molecular recognition events, including immune regulation, cell-cell interaction, inflammation, cancer metastasis, as well as bacterial and viral infection [1]. More than 50 structurally distinct sialic acid forms have been found in nature including three basic forms N-acetylneuraminic acid (Neu5Ac), N-glycolylneuraminic acid (Neu5Gc), 2-keto-3-deoxy-D-glycero-D-galacto-nonulosonic acid (Kdn), and their derivatives with modifications at various locations [2–4]. Among numerous modifications, O-acetylation is the most common which can occur at C-4, 7, 8, and 9 positions [5]. In nature, O-acetyl group spontaneously migrates from C7 and C8 to C9, which makes O-acetylation at C-4 and C-9 the most common naturally occurring sialic acid modifications [6, 7]. It has been shown that 9-O-acetylation is an effective biomarker in monitoring Acute Lymophoblastic Leukemia and Visceral Leishmaiasis [5]. It also regulates tissue morphogenesis during development, protects sialosides against sialidase cleavage, and inhibits binding by some proteins [8]. The 9-O-acetyl Neu5Ac (Neu5,9Ac2) is recognized by influenza C virus hemagglutinin [9] and by Factor H [10]. Nevertheless, study of the detailed biological significances of 9-O-acetyl sialic acids is hampered by the instability of O-acetyl ester group towards pH change and its sensitivity to esterases. As shown previously [11], a chemical biology strategy to partially solve this problem is to replace the oxygen atom in the ester group in Neu5,9Ac2 by an “NH” functionality in 9-acetamido-9-deoxy-N-acetylneuraminic acid (Neu5Ac9NAc) (Figure 1). The conformational resemblance of Neu5Ac9NAc and Neu5,9Ac2 in a sialyl trisaccharide has been confirmed by computational molecular dynamics, similar mammalian cell incorporation and surface expression, and similar interaction with Neu5,9Ac2-specific proteins by microarray studies. On the other hand, much improved stability of Neu5Ac9NAc compared to Neu5,9Ac2 has been demonstrated in binding and cell feeding studies [11]. A similar strategy of replacing sialic acid O-acetyl modification by an N-acetyl has also been successfully applied by others to investigate the roles of sialic acid O-acetylation [12, 13].
Figure 1.
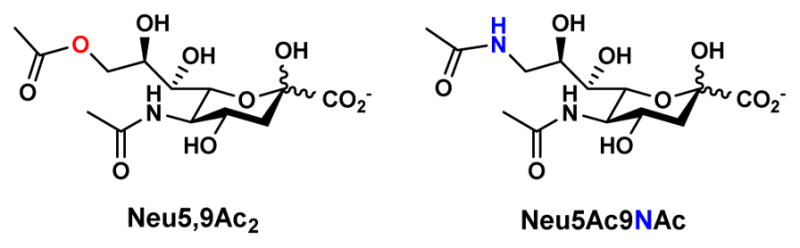
Structures of 9-O-acetyl-N-acetylneuraminic acid (Neu5,9Ac2) and its 9-N-acetyl analog, 9-acetamido-9-deoxy-N-acetylneuraminic acid (Neu5Ac9NAc).
Herein, we show that one-pot three-enzyme (OP3E) sialylation systems [14] are efficient for high-yield production of a library of Neu5Ac9NAc-containing α2–3- and α2–6-linked sialosides with diverse underlying glycans from chemically synthesized 6-acetamido-6-deoxy-N-acetylmannosamine (ManNAc6NAc). Among these, para-nitrophenylated α2–3- and α2–6-linked sialyl galactosides have been used in microtiter plate-based high-throughput substrate specificity studies of nine different sialidases including those from humans and bacteria [15–18].
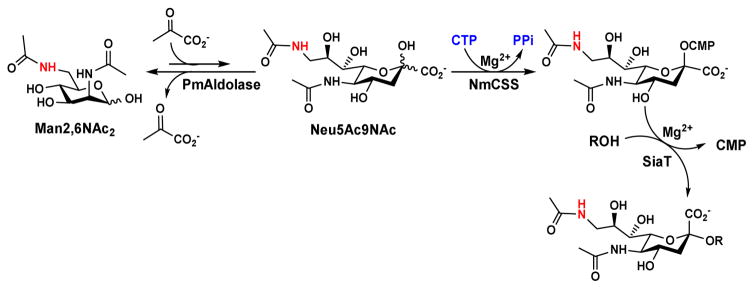
To synthesize a library of Neu5Ac9NAc-containing α2–3- and α2–6-linked sialosides, chemically synthesized ManNAc6NAc [11] was used directly in an efficient one-pot multienzyme (OPME) sialylation system [14] containing Pasteurella multocida sialic acid aldolase (PmAldolase) [19], Neisseria meningitidis CMP-sialic acid synthetase (NmCSS) [20, 21], and a sialyltransferase (Scheme 1). In this system, PmAldolase was used to catalyze the aldol addition reaction of ManNAc6NAc and sodium pyruvate (used in an excess to drive the reaction towards sialic acid formation) to obtain Neu5Ac9NAc. NmCSS was used for catalyzing the formation of the activated sugar nucleotide donor, cytidine 5′-monophosphate-Neu5Ac9NAc (CMP-Neu5Ac9NAc), for the subsequent sialyltransferase-catalyzed glycosylation reaction. Pasteurella multocida sialyltransferase 1 [22] M144D mutant (PmST1 M144D) [23] with decreased sialidase and donor hydrolysis activities was used for synthesizing α2–3-linked sialosides. Photobacterium damselae α2–6-sialyltransferase (Pd2,6ST) [24] or Photobacterium sp. α2–6-sialyltransferase [25] A366G mutant (Psp2,6ST A366G) with an improved expression level [26] was used for synthesizing α2–6-linked sialosides. The reactions were carried out in Tris-HCl buffer (100 mM) with a pH of 8.5 to optimize the activity of NmCSS while retain high activity of PmAldolase and sialyltransferases used. The products were obtained in good to excellent (61–98%) yields after purification using a gel filtration column and a C18 reverse phase column. The structures and the purities of the products were confirmed by high resolution mass spectrometry (HRMS) and nuclear magnetic resonance (NMR) spectroscopy.
Scheme 1.
One-pot three-enzyme chemoenzymatic synthesis of Neu5Ac9NAc-containing α2–3- and α2–6-linked sialosides.
As shown in Table 1, using PmST1 M144D as the sialyltransferase, α2–3-linked sialosides (1–7) were obtained in 61–86% yields. These are comparable to previously reported synthesis of α2–3-sialyllactoside, Neu5Ac9NAcα2–3Galβ1–4GlcβProN3, which was obtained in 84% yield [11]. Two α2–6-sialyltransferases were used for synthesizing α2–6-linked sialosides (8–16). Psp2,6ST A366G mutant with an increased expression level and improved activities in sialylating Tn antigens [25, 26] was used for synthesizing Neu5Ac9NAcα2–6GalNAcαProN3 (16) with 71% yield. For other α2–6-linked Neu5Ac9NAc-containing sialosides (8–15) synthesized, both Pd2,6ST and Psp2,6ST A366G could provide similar yields in small scale reactions. Pd2,6ST was used for the synthesis of Neu5Ac9NAcα2–6GalβProN3 (9) and Neu5Ac9NAcα2–6Galβ1–3GalNAcαProN3 (12) with excellent 92% and 98% yields, respectively. Due to the higher expression level of Psp2,6ST A366G, it was used for the synthesis of the rest of the α2–6-linked sialoside targets (8, 10–11, 13–15). Good to excellent yields (64–96%) were obtained.
Table 1.
Neu5Ac9NAc-containing sialosides synthesized via the OP3E sialylation system.
| α 2–3/6-Sialosides | Yield (Comp#) | α 2–6-Sialosides | Yield (Comp#) |
|---|---|---|---|
|
Neu5Ac9NAcα2–3GalβpNP |
80% (1) |
|
64% (8) |
|
Neu5Ac9NAcα2–3GalβProN3 |
72% (2) |
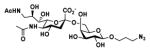 Neu5Ac9NAcα2–6GalβProN3 Neu5Ac9NAcα2–6GalβProN3
|
92% (9) |
|
Neu5Ac9NAcα2–3Galβ1–3GlcNAcβProN3 |
61% (3) |
|
68% (10) |
|
Neu5Ac9NAcα2–3Galβ1–4GlcNAcβProN3 |
86% (4) |
|
91% (11) |
|
Neu5Ac9NAcα2–3Galβ1–3GalNAcαProN3 |
64% (5) |
|
98% (12) |
|
Neu5Ac9NAcα2–3Galβ1–3GalNAcβProN3 |
84% (6) |
|
96% (13) |
|
Neu5Ac9NAcα2–3Galβ1–3GlcNAcαProN3 |
63% (7) |
|
76% (14) |
|
|
94% (15) |
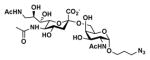 Neu5Ac9NAcα2–6GalNAcαProN3 Neu5Ac9NAcα2–6GalNAcαProN3
|
71% (16) |
The α2–3/6-sialosides obtained include Neu5Ac9NAcα2–3/6GalβpNP (1 and 8) and propyl azide (ProN3)-containing ones (2–7 and 9–16) such as disaccharides Neu5Ac9NAcα2–3/6GalβProN3 (2 and 9), sialyl type I glycans Neu5Ac9NAcα2–3/6Galβ1–3GlcNAcβProN3 (3 and 10), sialyl type II glycans Neu5Ac9NAcα2–3/6Galβ1–4GlcNAcβProN3 (4 and 11), sialyl type III glycans Neu5Ac9NAcα2–3/6Galβ1–3GalNAcαProN3 (5 and 12), sialyl type IV glycans Neu5Ac9NAcα2–3/6Galβ1–3GalNAcβProN3 (6 and 13), Neu5Ac9NAcα2–3/6Galβ1–3GlcNAcαProN3 (7 and 14), α2–6-sialyl type VI glycan [27] Neu5Ac9NAcα2–6Galβ1–4GlcβProN3 (15), and sialyl Tn antigen Neu5Ac9NAcα2–6GalNAcαProN3 (16). These represent common terminal sialyl glycan structures in vertebrates.
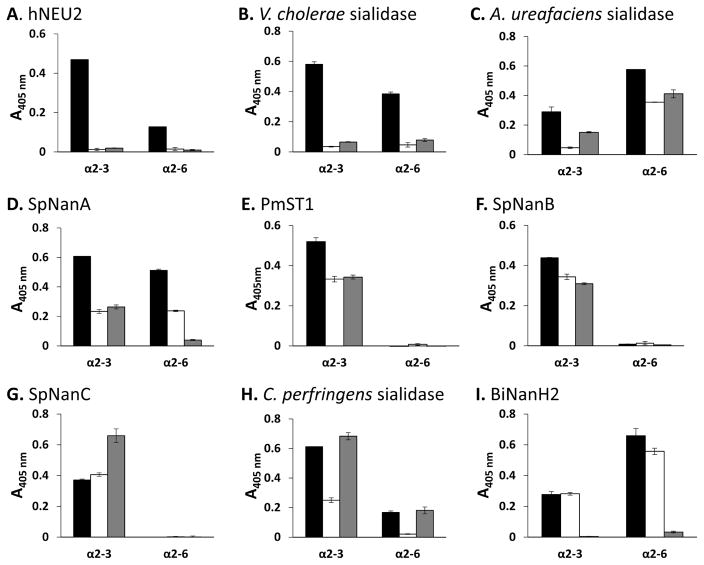
Among the compounds synthesized, two pNP-tagged Neu5Ac9NAc-containing sialosides Neu5Ac9NAcα2–3GalβpNP (1) and Neu5Ac9NAcα2–6GalβpNP (8) were used conveniently in a 384-well plate-based high-throughput colorimetric assay for substrate specificity studies [15] of nine sialidases. Four additional pNP-tagged sialosides including Neu5Ac-containing ones such as Neu5Acα2–3GalβpNP (17) and Neu5Acα2–6GalβpNP (18) as well as Neu5,9Ac2-cotaining ones such as Neu5,9Ac2α2–3GalβpNP (19) and Neu5,9Ac2α2–6GalβpNP (20) [15] (Figure 2) were also used as sialidase substrates for comparison purpose. Sialidases used include six recombinant sialidases and three commercially available sialidases. The recombinant sialidases used were human cytosolic sialidase hNEU2 [28], bacterial sialidases PmST1 (a multifunctional sialyltransferase which also has sialidase activity) [22], Bifidobacterium infantis sialidase NanH2 [29], and three Streptococcus pneumoniae sialidases SpNanA [30], SpNanB [30], and SpNanC [31, 32]. Three commercial bacterial sialidases used were those from Arthrobacter ureafaciens, Vibrio cholerae, and Clostridium perfringens. In this assay method, individual sialosides were incubated in duplicates at 37 °C for 30 min with an appropriate amount of a sialidase as well as an excess amount of β-galactosidase. The reactions were stopped by adding N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) buffer (0.5 M, pH 10.5) to adjust the pH value of the solution to higher than 9.5 to convert the para-nitrophenol formed in the enzymatic reactions to para-nitrophenolate which was quantified by a microplate reader at A405nm [15].
Figure 2.
Structures of Neu5Ac-sialosdies Neu5Acα2–3GalβpNP (17) and Neu5Acα2–6GalβpNP (18) as well as Neu5,9Ac2-sialosides Neu5,9Ac2α2–3GalβpNP (19) and Neu5,9Ac2α2–6GalβpNP (20) used as substrates for microtiter plate-based high-throughput sialidase assays.
As shown in Figure 3, substituting the 9-hydroxy group of the Neu5Ac in sialosides by an acetamido group led to significant reduction of the activities of hNEU2 (Fig. 3A) and V. cholerae sialidase (Fig. 3B), in which the 9-N-acetyl modification protected sialosides against sialidase cleavage. The effect was similar to that observed for Neu5Ac 9-O-acetyl modification (Fig. 3A and 3B). In comparison, both 9-N- and 9-O-acetyl modifications decreased the sialoside cleavage efficiencies of A. ureafaciens sialidase (Fig. 3C), SpNanA (Fig. 3D), PmST1 (Fig. 3E), and SpNanB (Fig. 3F) only moderately or slightly. Overall, these examples (Fig. 3A–3G) showed that Neu5Ac9NAc-sialosides are good mimics of Neu5,9Ac2-sialosides in probing the activities of these sialidases. Nevertheless, there were exceptions. Different effects were observed for 9-N- and 9-O-acetylation of Neu5Ac in affecting sialoside cleavage by SpNanC (Fig. 3G), C. perfringens sialidase (Fig. 3H), and BiNanH2 (Fig. 3I). While 9-N-acetylation of Neu5Ac did not alter the sialidase activity of SpNanC significantly, 9-O-acetylation of Neu5Ac improved the efficiency of sialoside cleavage by SpNanC (Fig. 3G). In comparison, 9-N-acetylation of Neu5Ac decreased the efficiency of sialoside cleavage by C. perfringens sialidase while 9-O-acetylation of Neu5Ac did not alter its activity significantly (Fig. 3H). On the other hand, Neu5Ac 9-N-acetylation did not have significant effect on the sialic acid cleavage efficiency of BiNanH2, but Neu5Ac 9-O-acetylation completely blocked its activity (Fig. 3I). A possible factor to consider for the differences is that “NH” in the amide in Neu5Ac9NAc can serve as both a hydrogen bond donor and a hydrogen bond acceptor while the oxygen atom in the ester in Neu5,9Ac2 can only serve as a hydrogen bond acceptor. The mechanism for these differences, however, needs further investigation.
Figure 3.
Sialidase substrate specificity studies using Neu5Ac-sialosides (black bars) Neu5Acα2–3GalβpNP (17) and Neu5Acα2–6GalβpNP (18), Neu5Ac9NAc-sialosdies (white bars) Neu5Ac9NAcα2–3GalβpNP (1) and Neu5Ac9NAcα2–6GalβpNP (8), as well as Neu5,9Ac2-sialosides (grey bars) Neu5,9Ac2α2–3GalβpNP (19) and Neu5,9Ac2α2–6GalβpNP (20) as substrates.
The results obtained by the microtiter plate-based assays for PmST1 (Fig. 3E), SpNanC (Fig. 3G), C. perfringens sialidase (Fig. 3H), and BiNanH2 (Fig. 3I) were further confirmed by high-performance liquid chromatography (HPLC)-based assays (Figures S1–S4, ESI). In addition, time course studies for BiNanH2 (Figure S5, ESI) and more detailed kinetics studies for SpNanC and BiNanH2 (Table 2) were carried out. As shown in Table 2, SpNanC catalyzes the cleavage of Neu5Ac9NAcα2–3GalβpNP (1) and Neu5Acα2–3GalβpNP (17) with similar efficiencies (kcat/KM = 137 s−1 mM− 1 and 130 s− 1 mM− 1, respectively). In contract, The catalytic efficiency of SpNanC for Neu5,9Ac2α2–3GalβpNP (19) (kcat/KM = 302 s− 1 mM− 1) is much (2.2–2.3 fold) higher. For BiNanH2, it has similar catalytic efficiencies towards Neu5Ac9NAcα2–3GalβpNP (1) and Neu5Acα2–3GalβpNP (17) (kcat/KM = 59.3 s− 1 mM− 1 and 66.7 s− 1 mM− 1, respectively). Its catalytic efficient towards Neu5Ac9NAcα2–6GalβpNP (8) (kcat/KM = 179 s− 1 mM− 1) is slightly lower than that of Neu5Acα2–6GalβpNP (18) (kcat/KM = 242 s− 1 mM− 1). In comparison, its activity towards Neu5,9Ac2α2–3GalβpNP (19) and Neu5,9Ac2α2–6GalβpNP (20) are not high enough for obtaining apparent kinetics parameters. These results validated those shown in Figure 3.
Table 2.
Apparent kinetics parameters for SpNanC and BiNanH2.
| Sialidases | Substrate | kcat (s− 1) | KM (mM) | kcat/KM (s− 1 mM− 1 ) |
|---|---|---|---|---|
| SpNanC | Neu5Acα2–3GalβpNP (17) | (3.65 ± 0.10)×102 | 2.66 ± 0.3 | 137 |
| Neu5Ac9NAcα2–3GalβpNP (1) | (3.38 ± 0.15)×102 | 2.60 ± 0.4 | 130 | |
| Neu5,9Ac2α β2–3Gal pNP (19) | (3.78 ± 0.12)×102 | 1.25 ± 0.2 | 302 | |
| BiNanH2 | Neu5Acα2–3GalβpNP (17) | (1.66 ± 0.02)×102 | 2.8 ± 0.1 | 59.3 |
| Neu5Ac9NAcα2–3GalβpNP (1) | (1.08 ± 0.02)×102 | 1.61 ± 0.2 | 66.7 | |
| Neu5Acα2–6GalβpNP (18) | (3.31 ± 0.06)×102 | 1.37 ± 0.1 | 242 | |
| Neu5Ac9NAcα2–6GalβpNP (8) | (8.07 ± 0.08)×102 | 4.51 ± 0.2 | 179 |
In summary, a library of sixteen new α2–3/6-linked Neu5Ac9NAc-containing sialosides have been successfully synthesized in good to excellent (61–98%) yields using highly efficient one-pot three-enzyme sialylation systems. Among the sialosides synthesized, para-nitrophenylated α2–3- and α2–6-linked Neu5Ac9NAc-containing sialyl galactosides have been used together with their Neu5Ac-, and Neu5,9Ac2-sialoside analogs in microtiter plate-based high-throughput substrate specificity studies of various sialidases. In general, Neu5Ac9NAc-sialosides are good mimics of Neu5,9Ac2-sialosides in probing the activities of most sialidases. Nevertheless, exceptions do exist and different effects were observed for Neu5Ac 9-N- and 9-O-acetylation in affecting sialoside cleavage by SpNanC, C. perfringens sialidase, and BiNanH2. Further investigation will be needed to elucidate the mechanism for these differences.
1. Experimental
1.1. Materials
All chemicals were obtained from commercial suppliers and used without further purification. 1H NMR (400 or 800 MHz) and 13C NMR (400 or 800 MHz) spectra were recorded on a Bruker Avance-400 Spectrometer (400 MHz for 1H, 100 MHz for 13C) or a Avance-800 Spectrometer (800 MHz for 1H, 200 MHz for 13C). High resolution electrospray ionization (ESI) mass spectra were obtained using a Thermo Electron LTQ-Orbitrap Hybrid MS at the Mass Spectrometry Facility in the University of California, Davis. Column chromatography was performed using RediSep Rf silica columns or an ODS-SM column (51 g, 50 μm, 120 Å, Yamazen) on the CombiFlash® Rf 200i system. Analytical thin-layer chromatography was performed on silica gel plates 60 GF254 (Sorbent technologies) using anisaldehyde stain for detection. Gel filtration chromatography was performed using a column (100 cm × 2.5 cm) packed with BioGel P-2 Fine resins (Bio-Rad, Hercules, CA, USA). Arthrobacter ureafaciens sialidase, Vibrio cholerae sialidase, and Clostridium perfringens sialidase (CpNanH) were purchased from Prozyme (Hayward, CA, USA). Recombinant PmAldolase [19], NmCSS [20, 21], PmST1 M144D mutant [23], Pd2,6ST [24], Psp2,6ST A366G mutant [26], hNEU2 [28], PmST1 [22], SpNanA [30], SpNanB [30], SpNanC [32], and BiNanH2 [29], were expressed as described previously. Neu5Acα2–3GalβpNP (17), Neu5Acα2–6GalβpNP (18), Neu5,9Ac2α2–3GalβpNP (19), Neu5,9Ac2α2–6GalβpNP (20) [15], and ManNAc6NAc [11] were synthesized as reported previously.
1.2. One-pot three-enzyme synthesis of α2–3/6-linked sialosides
An acceptor (30–50 mg, 10 mM) and ManNAc6NAc (1.2–1.5 equiv.) were incubated at 37 °C in Tris-HCl buffer (100 mM) (pH 8.5 for synthesizing α2–3-sialosides or α2–6-sialosides using Psp2,6STA366G, pH 7.5 for synthesizing α2–6-sialosides using Pd2,6ST) containing sodium pyruvate (6.0–7.5 equiv.), CTP (1.5 equiv.), MgCl2 (20 mM), an appropriate amount of PmAldolase (1.5 mg), NmCSS (2.5 mg), and Pd2,6ST (2.5 mg), Psp2,6ST A366G mutant (2.5 mg), or PmST1 M144D mutant (2.5 mg). The reaction was monitored by thin-layer chromatography (TLC) using a developing solvent consisting of EtOAc:MeOH:H2O = 5:2:1 (by volume) and the TLC plates were stained with a p-anisaldehyde sugar stain. After 1–24 h, the reaction was quenched by adding the same volume of pre-chilled ethanol and the reaction mixture was centrifuged to remove precipitates. The supernatant was concentrated and passed through a BioGel P-2 gel filtration column eluting with water followed by a C18 column (H2O:CH3CN= 1:0 – 4:1) to obtain the target products.
1.2.1. 4-Nitrophenyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranoside (Neu5Ac9NAcα2–3GalβpNP, 1)
Yield 80%; 87 mg, White foam. 1H NMR (400 MHz, D2O) δ 8.32–8.15 (m, 2H), 7.32–7.17 (m, 2H), 5.29 (d, J = 7.8 Hz, 1H), 4.25 (dd, J = 3.2 and 9.8 Hz, 1H), 4.06 (d, J = 3.2 Hz, 1H), 3.98–3.84 (m, 4H), 3.81–3.64 (m, 4H), 3.61–3.46 (m, 2H), 3.29 (dd, J = 7.6 and 14.2 Hz, 1H), 2.80 (dd, J = 4.6 and 12.4 Hz, 1H), 2.04 (s, 3H), 1.95 (s, 3H), 1.84 (t, J = 12.1 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.95, 174.32, 173.75, 161.70, 142.47, 126.07(2C), 116.41(2C), 99.94, 99.70, 75.50, 75.45, 72.76, 70.00, 69.60, 68.77, 68.29, 67.29, 60.67, 51.67, 42.10, 39.67, 22.03, 21.75; HRMS (ESI) Anal. Calcd for C25H34N3O16 [M-H] −: 632.1945, Found: 632.1957.
1.2.2. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyrano-sylonic acid)-(2→3)-O-β-D-galactopyranoside (Neu5Ac9NAcα2–3Galβ ProN3, 2)
Yield 72%; 73 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.47 (d, J = 7.8 Hz, 1H), 4.08 (dd, J = 3.2 and 9.8 Hz, 1H), 4.05–3.63 (m, 10H), 3.59–3.45 (m, 5H), 3.32 (dd, J = 7.8 and 14.2 Hz, 1H), 2.76 (dd, J = 4.6 and 12.4 Hz, 1H), 2.04–2.03 (m, 6H), 1.96–1.89 (m, 2H), 1.81 (t, J = 12.2 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.93, 174.40, 173.80, 102.54, 99.93, 75.85, 74.88, 72.70, 69.95, 69.54, 69.13, 68.31, 67.52, 67.14, 60.93, 51.66, 47.90, 42.09, 39.61, 28.25, 22.02, 21.82; HRMS (ESI) m/z calcd for C22H36N5O14 [M-H] −: 594.2264, found 594.2282.
1.2.3. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-β-D-glucopyranoside (Neu5Ac9NAcα2–3Galβ1–3GlcNAcβProN3, 3)
Yield 61%; 38 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.58 (d, J = 7.8 Hz, 1H), 4.50 (d, J = 7.8 Hz, 1H), 4.07 (dd, J = 3.2 and 9.8 Hz, 1H), 4.03–3.47 (m, 19H), 3.39 (t, J = 6.4 Hz, 2H), 3.27 (dd, J = 7.6 and 14.2 Hz, 1H), 2.76 (dd, J = 4.8 and 12.8 Hz, 1H), 2.13–2.03 (m, 9H), 1.89–1.77 (m, 3H); 13C NMR (100 MHz, D2O) δ 174.79, 174.35, 174.25, 173,79, 103.25, 100.76, 99.70, 82.41, 75.52, 75.25, 74.95, 72.54, 69.82, 69.40, 69.03, 68.64, 68.24, 67.17, 67.02, 60.89, 60.62, 54.33, 51.55, 47.67, 41.93, 39.56, 27.98, 22.14, 21.92, 21.69; HRMS (ESI) m/z calcd for C30H49N6O19 [M-H] −: 797.3058, found 797.3064.
1.2.4. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-2-acetamido-2-deoxy-β-D-glucopyranoside (Neu5Ac9NAcα2–3Galβ1–4GlcNAcβProN3, 4)
Yield 86%; 48 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.56–4.52 (m, 2H), 4.11 (dd, J = 3.2 and 10.0 Hz, 1H), 4.04–3.82 (m, 6H), 3.80–3.54 (m, 12H), 3.50 (dd, J = 1.8 and 9.0 Hz, 1H), 3.39 (td, J = 1.8 and 6.6 Hz, 2H), 3.28 (dd, J = 7.8 and 14.0 Hz, 1H), 2.76 (dd, J = 4.6 and 12.4 Hz, 1H), 2.12–1.98 (m, 9H), 1.92–1.74 (m, 3H); 13C NMR (100 MHz, D2O) δ 174.93, 174.46, 174.39, 173.79, 102.54, 101.13, 99.87, 78.23, 75.53, 75.14, 74.76, 72.74, 72.33, 69.95, 69.58, 69.37, 68.30, 67.46, 67.11, 61.01, 59.99, 55.06, 51.65, 47.76, 42.12, 39.61, 28.08, 22.14, 22.02, 21.81; HRMS (ESI) m/z calcd for C30H49N6O19 [M-H] −: 797.3058, found 797.3079.
1.2.5. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-α-D-galactopyranoside (Neu5Ac9NAcα2–3Galβ1–3GalNAcαProN3, 5)
Yield 64%; 57 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.92 (d, J = 3.8 Hz, 1H), 4.55 (d, J = 7.8 Hz, 1H), 4.32 (dd, J = 3.8 and 11.2 Hz, 1H), 4.28–4.23 (m, 1H), 4.16–3.39 (m, 20H), 3.28(dd, J = 7.6 and 14.2 Hz, 1H), 2.75 (dd, J = 4.6 and 12.4 Hz, 1H), 2.13–2.03 (m, 9H), 1.95–1.88 (m, 2H), 1.80 (t, J = 12.2 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.92, 174.51, 174.37, 173.90, 104.35, 99.88, 97.15, 77.44, 75.64, 74.72, 72.65, 70.60, 69.93, 69.49, 69.14, 68.51, 68.34, 67.43, 64.92, 61.20, 60.97, 51.66, 48.84, 48.66, 48.17, 42.03, 39.62, 27.94, 22.01, 21.82; HRMS (ESI) m/z calcd for C30H49N6O19 [M-H] −: 797.3058, found 797.3072.
1.2.6. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-β-D-galactopyranoside (Neu5Ac9NAcα2–3Galβ1–3GalNAcβProN3, 6)
Yield 84%; 14.8 mg, White foam. 1H NMR (600 MHz, D2O) δ 4.51–4.46 (m, 2H), 4.16 (d, J = 3.2 Hz, 1H), 4.03–3.93 (m, 3H), 3.97–3.50 (m, 15H), 3.47–3.45 (m, 1H), 3.36 (td, J = 1.5 and 6.8 Hz, 2H), 3.23 (dd, J = 7.6 and 14.1 Hz, 1H), 2.71 (dd, J = 4.6 and 12.3 Hz, 1H) 2.03–1.96 (m, 9H), 1.87–1.80 (m, 2H), 1.75 (t, J = 12.1 Hz, 1H); 13C NMR (151 MHz, D2O) δ 174.80, 174.56, 174.27, 173.85, 104.39, 101.28, 99.72, 79.95, 75.45, 74.63(2C), 72.53, 69.82, 69.42, 68.95, 68.26, 67.71, 67.29, 66.88, 60.88, 60.83, 51.55, 51.04, 48.74, 41.94, 39.53, 28.00, 22.16, 21.92, 21.71. HRMS (ESI) m/z calcd for C30H49N6O19 [M-H] −: 797.3058, found 797.3064
1.2.7. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-α-D-glucopyranoside (Neu5Ac9NAcα2–3Galβ1–3GlcNAcαProN3, 7)
Yield 63%; 85 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.88 (d, J = 3.6 Hz, 1H), 4.53 (d, J = 7.8 Hz, 1H), 4.09 (ddd, J = 3.4, 7.4 and 10.0 Hz, 2H), 4.01–3.42 (m, 20H), 3.28 (dd, J = 7.6 and 14.2 Hz, 1H), 2.76 (dd, J = 4.6 and 12.4 Hz, 1H), 2.04–2.03 (m, 9H), 1.95–1.89 (m, 2H), 1.80 (t, J = 12.2 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.91, 174.35(2C), 173.88, 103.26, 99.87, 97.04, 80.47, 75.67, 75.01, 72.66, 71.57, 69.94, 69.47, 69.22, 68.71, 68.34, 67.32, 64.96, 60.97, 60.51, 52.43, 51.66, 48.17, 42.01, 39.64, 27.93, 22.02, 21.99, 21.81; HRMS (ESI) m/z calcd for C30H49N6O19 [M-H] −: 797.3058, found 797.3071.
1.2.8. 4-Nitrophenyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-D-galactopyranoside (Neu5Ac9NAcα2–6GalβpNP, 8)
Yield 64%; 69 mg, White foam. 1H NMR (400 MHz, D2O) δ 8.26–8.15 (m, 2H), 7.26–7.12 (m, 2H), 5.09 (d, J = 7.6 Hz, 1H), 3.95 (dd, J = 4.0 and 7.6 Hz, 2H), 3.91–3.56 (m, 8H), 3.47 (dd, J = 2.8 and 14.2 Hz, 1H), 3.36 (dd, J = 1.6 and 8.8 Hz, 1H), 3.14 (dd, J = 8.0 and 14.2 Hz, 1H), 2.67 (dd, J = 4.4 and 12.4 Hz, 1H), 1.94 (s, 3H), 1.86 (s, 3H), 1.67–1.49 (m, 1H); 13C NMR (100 MHz, D2O) δ 174.94, 174.30, 173.48, 161.84, 142.51, 126.12(2C), 116.39(2C), 100.42, 99.88, 74.04, 72.47, 72.33, 70.25, 69.96, 69.69, 68.36, 68.15, 63.02, 51.80, 42.15, 40.14, 21.98, 21.72; HRMS (ESI) Anal. Calcd for C25H34N3O16 [M-H] −: 632.1945, Found: 632.1955.
1.2.9. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-D-galactopyranoside (Neu5Ac9NAcα2–6GalβProN3, 9)
Yield 92%; 107 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.39 (d, J = 7.8 Hz, 1H), 4.05–3.88 (m, 4H), 3.87–3.57 (m, 8H), 3.54–3.44 (m, 4H), 3.28 (dd, J = 7.8 and 14.0 Hz, 1H), 2.73 (dd, J = 4.8 and 12.4 Hz, 1H), 2.04–2.02 (m, 6H), 1.95–1.89 (m, 2H), 1.70 (t, J = 12.2 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.98, 174.37, 173.41, 102.90, 100.47, 73.39, 72.57, 72.48, 70.65, 69.94, 69.69, 68.54, 68.17, 67.45, 63.29, 51.83, 47.87, 42.13, 40.17, 28.30, 22.01, 21.82; HRMS (ESI) m/z calcd for C22H36N5O14 [M-H] −: 594.2264, found 594.2285.
1.2.10. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-D-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-β-D-glucopyranoside (Neu5Ac9NAcα2–6Galβ1–3GlcNAcβProN3, 10)
Yield 68%; 34 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.58 (d, J = 8.4 Hz, 1H), 4.39 (d, J = 7.8 Hz, 1H), 4.05–3.45 (m, 20H), 3.39 (td, J = 1.2 and 6.6 Hz, 2H), 3.30 (dd, J = 7.6 and 14.2 Hz, 1H), 2.70 (dd, J = 4.8 and 12.4 Hz, 1H), 2.10–1.89 (m, 9H), 1.89–1.83 (m, 2H), 1.70 (t, J = 12.2 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.84, 174.59, 174.38, 173.46, 103.89, 100.88, 100.11, 84.01, 75.46, 73.61, 72.37, 72.28, 70.48, 69.93, 68.91(2C), 68.48, 68.25, 67.14, 63.56, 60.87, 54.30, 51.78, 47.78, 42.15, 40.12, 28.08, 22.19, 22.03, 21.83; HRMS (ESI) m/z calcd for C30H49N6O19 [M-H] −: 797.3058, found 797.3065.
1.2.11. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-D-galactopyranosyl-(1→4)-2-acetamido-2-deoxy-β-D-glucopyranoside (Neu5Ac9NAcα2–6Galβ1–4GlcNAcβProN3, 11)
Yield 91%; 56 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.62–4.52 (m, 1H), 4.45 (d, J = 7.8 Hz, 1H), 4.04–3.89 (m, 5H), 3.88–3.51(m, 14H), 3.45 (dd, J = 1.8 and 9.0 Hz, 1H), 3.39 (td, J = 1.4 and 6.6 Hz, 2H), 3.30 (dd, J = 7.8 and 14.0 Hz, 1H), 2.76 (dd, J = 4.6 and 12.4 Hz, 1H), 2.16–1.98 (m, 9H), 1.89–1.83 (m, 2H), 1.71 (t, J = 12.2 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.83, 174.51, 174.39, 173.49, 103.46, 100.95, 100.18, 80.74, 74.45, 73.68, 72.39(2C), 70.70, 69.87(2C), 68.39, 68.17, 67.09, 63.44, 61.75, 60.34, 54.83, 51.85, 47.77, 42.14, 40.06, 28.09, 22.27, 22.01, 21.83; HRMS (ESI) m/z calcd for C30H49N6O19 [M-H] −: 797.3058, found 797.3063.
1.2.12. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-D-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-α-D-galactopyranoside (Neu5Ac9NAcα2–6Galβ1–3GalNAcαProN3, 12)
Yield 98%; 86 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.91 (d, J = 3.6 Hz, 1H), 4.46 (d, J = 7.8 Hz, 1H), 4.33 (dd, J = 3.8 and 11.2 Hz, 1H), 4.29–4.24 (m, 1H), 4.10–3.41 (m, 20H), 3.31 (dd, J = 7.6 and 14.0 Hz, 1H), 2.74 (dd, J = 4.8 and 12.4 Hz, 1H), 2.13–1.99 (m, 9H), 1.95–1.89 (m, 2H), 1.66 (t, J = 12.2 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.98, 174.51, 174.40, 173.40, 104.52, 100.39, 97.16, 77.27, 73.20, 72.43, 72.37, 70.73, 70.48, 69.81, 69.65, 68.67, 68.56, 68.20, 64.90, 63.48, 61.42, 51.86, 48.72, 48.19, 42.08, 40.19, 27.96, 22.00, 21.98, 21.82; HRMS (ESI) m/z calcd for C30H49N6O19 [M-H] −: 797.3058, found 797.3079.
1.2.13. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-D-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-β-D-galactopyranoside (Neu5Ac9NAcα2–6Galβ1–3GalNAcβProN3, 13)
Yield 96%; 16.8 mg, White foam. 1H NMR (600 MHz, D2O) δ 4.47–4.38 (m, 2H), 4.17 (dd, J = 3.2 Hz, 1H), 3.98–3.55 (m, 17H), 3.51–3.42 (m, 2H), 3.35 (td, J = 4.5 and 6.5 Hz, 2H), 3.26–3.21 (m, 1H), 2.68 (dd, J = 4.6 and 12.4 Hz, 1H), 2.11–1.89 (m, 9H), 1.84–1.77 (m, 2H), 1.64 (t, J = 12.2 Hz, 1H); 13C NMR (151 MHz, D2O) δ 174.85, 174.58, 174.28, 173.31, 104.79, 101.38, 100.35, 79.80, 74.85, 73.00, 72.40, 72.38, 70.43, 69.81, 69.60, 68.48, 68.08, 67.72, 67.11, 63.26, 60.85, 52.24, 51.70, 51.07, 42.07, 40.11, 28.06, 22.11, 21.90, 21.72, HRMS (ESI) m/z calcd for C30H49N6O19 [M-H] −: 797.3058, found 797.3067
1.2.14. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-D-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-α-D-glucopyranoside (Neu5Ac9NAcα2–6Galβ1–3GlcNAcαProN3, 14)
Yield 76%; 51mg, White foam. 1H NMR (400 MHz, D2O) δ 4.87 (d, J = 3.6 Hz, 1H), 4.39 (d, J = 7.8 Hz, 1H), 4.10 (dd, J = 3.6 and 10.6 Hz, 1H), 4.03–3.37 (m, 21H), 3.30 (dd, J = 7.7 and 14.1 Hz, 1H), 2.70 (dd, J = 4.7 and 12.4 Hz, 1H), 2.09–1.94 (m, 9H), 1.94–1.90 (m, 2H), 1.71 (t, J = 12.2 Hz, 1H); 13C NMR (101 MHz, D2O) δ 174.83, 174.42, 174.38, 173.48, 103.71, 100.12, 96.91, 81.81, 73.60, 72.42, 72.29, 71.58, 70.53, 69.93, 69.91, 68.79, 68.50, 68.26, 64.91, 63.51, 60.64, 52.27, 51.78, 48.14, 42.15, 40.14, 27.96, 22.03, 21.93, 21.82; HRMS (ESI) m/z calcd for C30H49N6O19 [M-H] −: 797.3058, found 797.3078.
1.2.15. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside (Neu5Ac9NAcα2–6Galβ1–4GlcβProN3, 15)
Yield 94%; 73 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.50 (d, J = 8.0 Hz, 1H), 4.44 (d, J = 7.8 Hz, 1H), 4.04–3.24 (m, 23H), 2.71(dd, J = 4.8 and 12.4 Hz, 1H), 2.14–2.03 (m, 6H), 1.96–1.89 (m, 2H), 1.74 (t, J = 12.2 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.82, 174.39, 173.43, 103.18, 101.98, 100.30, 79.62, 74.62, 74.59, 73.68, 72.71, 72.33, 70.75, 69.93, 69.89, 68.52, 68.33, 67.31, 63.62, 60.24, 51.75, 48.85, 47.86, 42.16, 40.06, 28.22, 22.04, 21.84; HRMS (ESI) m/z calcd for C28H46N5O19 [M-H] −: 756.2792, found 756.2812.
1.2.16. 3-Azidopropyl O-(5,9-diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→6)-2-acetamido-2-deoxy-α-D-galactopyranoside (Neu5Ac9NAcα2–6GalNAcαProN3, 16)
Yield 71%; 78 mg, White foam. 1H NMR (400 MHz, D2O) δ 4.89 (d, J = 3.6 Hz, 1H), 4.15 (dd, J = 3.8 and 11.2 Hz, 1H), 4.06–4.00 (m, 2H), 3.96–3.89 (m, 3H), 3.84–3.78 (m, 2H), 3.74–3.41 (m, 8H), 3.29 (dd, J = 7.8 and 14.0 Hz, 1H), 2.73 (dd, J = 4.8 and 12.4 Hz, 1H), 2.13–2.03 (m, 9H), 1.94–1.88 (m, 2H), 1.69 (t, J = 12.2 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.94, 174.51, 174.37, 173.37, 100.37, 97.04, 72.40, 69.96, 69.74, 69.49, 68.49, 68.21, 67.46, 65.15, 63.79, 51.82, 49.92, 48.14, 42.14, 40.23, 27.91, 22.01, 21.92, 21.82; HRMS (ESI) m/z calcd for C24H39N6O14 [M-H] −: 635.2530, found 635.2554.
1.3. Microtiter plate-based sialidase substrate specificity assays
Assays were carried out in duplicates. For each reaction in a final volume of 20 μL, a sialoside was incubated with an appropriate amount of a sialidase and an excess amount of β-galactosidase (12 μg) in a buffer solution in a 384-well plate at 37 °C for 30 min. The sialidase amounts and buffers used were: A. ureafaciens sialidase (0.5 mU), NaOAc buffer (100 mM, pH 5.5); C. perfringens sialidase (0.75 mU), MES buffer (100 mM, pH 5.0); V. cholerae sialidase (1.5 mU), NaCl (150 mM), CaCl2 (10 mM), NaOAc buffer (100 mM, pH 5.5); SpNanA (1.5 ng), NaOAc buffer (100 mM, pH 6.0); SpNanB (3 ng), NaOAc buffer (100 mM, pH 6.0); SpNanC (20 ng), MES buffer (100 mM, pH 6.5); PmST1 (0.4 μg), NaOAc buffer (100 mM, pH 5.5), CMP (0.4 mM); hNEU2 (1.3 μg), MES buffer (100 mM, pH 5.0); BiNanH2 (4 ng), NaOAc buffer (100 mM, pH 5.0). The reactions were stopped by adding 40 μL of N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) buffer (0.5 M, pH 10.5) to adjust pH to higher than 9.5 and A405 nm values of samples were read by a microplate reader [15].
1.4. HPLC-based assays for PmST1, SpNanC, C. perfringens sialidase, and BiNanH2
Assays were carried out as described above for microtiter plate-based assays. The reactions were stopped by adding 40 μL of pre-chilled ethanol. The mixtures were then centrifuged and the supernatants were analyzed by Agilent 1290 Infinity HPLC system at 315 nm. A C14 reverse phase Rapid Resolution High Definition column (BONUS RP RRHD 1.8 μm, 2.1 × 150 mm, Agilent) was used for analyzing samples with Neu5Acα2–3GalβpNP (17), Neu5,9Ac2α2–3GalβpNP (19), Neu5Ac9NAcα2–6GalβpNP (8), or Neu5Acα2–6GalβpNP (18). A C18 reverse phase Rapid Resolution High Definition column (EclipsePlusC18 RRHD 1.8 μm, 2.1 × 50 mm, Agilent) was used for analyzing samples with Neu5Ac9NAcα2–3GalβpNP (1) or Neu5,9Ac2α2–6GalβpNP (20). The mobile phases used were acetonitrile (ACN) in H2O mixed solvent with varied percentages of acetonitrile: 4.5% for Neu5Acα2–6GalβpNP (18); 6.5% for Neu5Ac9NAcα2–3GalβpNP (1) or Neu5,9Ac2α2–6GalβpNP (20); 9% for Neu5Ac9NAcα2–6GalβpNP (8); and12% for Neu5Acα2–3GalβpNP (17) or Neu5,9Ac2α2–3GalβpNP (19).
1.5. Time course studies for BiNanH2
Time course studies for BiNanH2 were carried out in duplicate at 37 °C in reaction mixtures (200 μL each) containing NaOAc buffer (100 mM, pH 5.0), BiNanH2 (220 ng/mL), and a sialidase substrate (0.3 mM) selected from Neu5Acα2–3GalβpNP (17), Neu5Acα2–6GalβpNP (18), Neu5Ac9NAcα2–3GalβpNP (1), and Neu5Ac9NAcα2–6GalβpNP (8). Aliquots (20 μL each) were taken at 5, 10, 15, 20, 30, 45, or 60 min intervals and added to microcentrifuge tubes (500 μL) containing 40 μL of pre-chilled ethanol. The mixtures were centrifuged on a bench-top centrifuge (13,000 rpm × 3 min). The supernatants (45 μL) were analyzed by Agilent 1290 Infinity HPLC system at 315 nm as described above.
1.6. Kinetic studies for BiNanH2
The kinetic studies for BiNanH2 were performed in duplicates at 37 °C for 10 min in a total volume of 20 μL each containing NaOAc buffer (100 mM, pH 5.0), a sialidase substrate [selected from Neu5Ac9NAcα2–3GalβpNP (1), Neu5Ac9NAcα2–6GalβpNP (8), Neu5Acα2–3GalβpNP (17), and Neu5Acα2–6GalβpNP (18)], and BiNanH2 (5.8 ng when compound 1 or 17 was used as the substrate and 1.5 ng when compound 8 or 18 was used as the substrate). The reactions were stopped by adding 40 μL of pre-chilled ethanol. The mixtures were then centrifuged and the supernatants were analyzed by the HPLC system described above for HPLC-based assays. Apparent kinetic parameters were obtained by varying substrate concentrations from 0.1–40 mM (0.1, 0.2, 0.4, 1, 2, 4, 10, 20, and 40 mM) and fitting the data (the average values of duplicate assay results) into the Michaelis–Menten equation using Grafit 5.0.
1.7. Kinetic studies for SpNanC
The kinetic studies for SpNanC were performed in duplicates at 37 °C for 10 min in a total volume of 20 μL each containing MES buffer (100 mM, pH 6.5), a sialidase substrate [selected from Neu5Ac9NAcα2–3GalβpNP (1), Neu5Acα2–3GalβpNP (17), and Neu5,9Ac2α2–3GalβpNP (19)], and SpNanC (2.5 ng when compound 1 or 17 was used as the substrate and 1.5 ng when compound 19 was used as the substrate). After stopping the reactions by by adding 40 μL of pre-chilled ethanol, the mixtures were centrifuged and the supernatants were analyzed by the HPLC system as described above. Apparent kinetic parameters were obtained by varying substrate concentrations from 0.1–40 mM (0.1, 0.2, 0.4, 1, 2, 4, 10, 20, and 40 mM) and fitting the data (the average values of duplicate assay results) into the Michaelis–Menten equation using Grafit 5.0.
Supplementary Material
Highlights.
Sixteen new Neu5Ac9NAc-containing sialosides are successfully synthesized
One-pot multienzyme (OPME) sialylation systems are highly efficient for the synthesis
High-throughput microtiter plate assay results were confirmed by HPLC-based methods
9-N-Acetyl Neu5Ac is a good mimic of 9-O-acetyl Neu5Ac to probe most sialidases
Exceptions exist and mechanism needs further investigation
Acknowledgments
This work was partially supported by National Institutes of Health (NIH) Common Fund grant U01GM120419. Bruker Avance-800 NMR spectrometer was funded by National Science Foundation grant DBIO-722538. Y.L., H.Y., and X.C. are co-founders of Glycohub, Inc., a company focusing on the development of carbohydrate-based reagents, diagnostics, and therapeutics. Glycohub, Inc. played no role in the design, execution, interpretation, or publication of this study.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/...
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Varki A, Gagneux P. Ann N Y Acad Sci. 2012;1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, Varki A. ACS Chem Biol. 2010;5:163–176. doi: 10.1021/cb900266r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angata T, Varki A. Chem Rev. 2002;102:439–470. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 4.Schauer R. Glycoconj J. 2000;17:485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein A, Roussel P. Biochimie. 1998;80:49–57. doi: 10.1016/s0300-9084(98)80056-4. [DOI] [PubMed] [Google Scholar]
- 6.Varki A, Diaz S. Anal Biochem. 1984;137:236–247. doi: 10.1016/0003-2697(84)90377-4. [DOI] [PubMed] [Google Scholar]
- 7.Kamerling JP, Schauer R, Shukla AK, Stoll S, Halbeek H, Vliegenthart J. Eur J Biochem. 1987;162:601–607. doi: 10.1111/j.1432-1033.1987.tb10681.x. [DOI] [PubMed] [Google Scholar]
- 8.Klotz FW, Orlandi PA, Reuter G, Cohen SJ, Haynes JD, Schauer R, Howard RJ, Palese P, Miller LH. Mol Biochem Parasitol. 1992;51:49–54. doi: 10.1016/0166-6851(92)90199-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers GN, Herrler G, Paulson J, Klenk H. J Biol Chem. 1986;261:5947–5951. [PubMed] [Google Scholar]
- 10.Varki A, Kornfeld S. J Exp Med. 1980;152:532–544. doi: 10.1084/jem.152.3.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khedri Z, Xiao A, Yu H, Landig CS, Li W, Diaz S, Wasik BR, Parrish CR, Wang LP, Varki A, Chen X. ACS Chem Biol. 2017;12:214–224. doi: 10.1021/acschembio.6b00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrler G, Gross HJ, Imhof A, Brossmer R, Milks G, Paulson JC. J Biol Chem. 1992;267:12501–12505. [PubMed] [Google Scholar]
- 13.Bakkers MJ, Zeng Q, Feitsma LJ, Hulswit RJ, Li Z, Westerbeke A, van Kuppeveld FJ, Boons GJ, Langereis MA, Huizinga EG, de Groot RJ. Proc Natl Acad Sci U S A. 2016;113:E3111–3119. doi: 10.1073/pnas.1519881113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu H, Chokhawala HA, Huang S, Chen X. Nat Protoc. 2006;1:2485–2492. doi: 10.1038/nprot.2006.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chokhawala HA, Yu H, Chen X. Chembiochem. 2007;8:194–201. doi: 10.1002/cbic.200600410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khedri Z, Muthana MM, Li Y, Muthana SM, Yu H, Cao H, Chen X. Chem Commun. 2012;48:3357–3359. doi: 10.1039/c2cc17393j. [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Cao H, Tiwari VK, Li Y, Chen X. Bioorg Med Chem Lett. 2011;21:5037–5040. doi: 10.1016/j.bmcl.2011.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Cao H, Dao N, Luo Z, Yu H, Chen Y, Xing Z, Baumgarth N, Cardona C, Chen X. Virology. 2011;415:12–19. doi: 10.1016/j.virol.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Yu H, Cao H, Lau K, Muthana S, Tiwari VK, Son B, Chen X. Appl Microbiol Biotechnol. 2008;79:963–970. doi: 10.1007/s00253-008-1506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Yu H, Karpel R, Chen X. Bioorg Med Chem. 2004;12:6427–6435. doi: 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 21.Sun M, Li Y, Chokhawala HA, Henning R, Chen X. Biotechnol Lett. 2008;30:671–676. doi: 10.1007/s10529-007-9588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. J Am Chem Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- 23.Sugiarto G, Lau K, Qu J, Li Y, Lim S, Mu S, Ames JB, Fisher AJ, Chen X. ACS Chem Biol. 2012;7:1232–1240. doi: 10.1021/cb300125k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, Huang S, Chokhawala H, Sun M, Zheng H, Chen X. Angew Chem Int Ed Engl. 2006;45:3938–3944. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding L, Yu H, Lau K, Li Y, Muthana S, Wang J, Chen X. Chem Commun. 2011;47:8691–8693. doi: 10.1039/c1cc12732b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding L, Zhao C, Qu J, Li Y, Sugiarto G, Yu H, Wang J, Chen X. Carbohydr Res. 2015;408:127–133. doi: 10.1016/j.carres.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye J, Liu X, Peng P, Yi W, Chen X, Wang F, Cao H. ACS Catal. 2016;6:8140–8844. [Google Scholar]
- 28.Li Y, Cao H, Yu H, Chen Y, Lau K, Qu J, Thon V, Sugiarto G, Chen X. Mol BioSyst. 2011;7:1060–1072. doi: 10.1039/c0mb00244e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, Chen X, Lebrilla CB, Mills DA. J Biol Chem. 2011;286:11909–11918. doi: 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tasnima N, Yu H, Li Y, Santra A, Chen X. Org Biomol Chem. 2016;15:160–167. doi: 10.1039/c6ob02240e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owen CD, Lukacik P, Potter JA, Sleator O, Taylor GL, Walsh MA. J Biol Chem. 2015;290:27736–27748. doi: 10.1074/jbc.M115.673632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu G, Kiefel MJ, Wilson JC, Andrew PW, Oggioni MR, Taylor GL. J Am Chem Soc. 2011;133:1718–1721. doi: 10.1021/ja110733q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.