Abstract
Non-dioxin-like polychlorinated biphenyls (NDL PCBs) activate ryanodine receptors (RyR), microsomal Ca2+ channels of broad significance. Teleost fish may be important models for NDL PCB neurotoxicity, and we used sequencing databases to characterize teleost RyR and FK506 binding protein 12 or 12.6 kDa (genes FKBP1A; FKBP1B), which promote NDL PCB-triggered Ca2+ dysregulation. Particular focus was placed on describing genes in the Atlantic killifish (Fundulus heteroclitus) genome and searching available RNA-sequencing datasets for single nucleotide variants (SNV) between PCB tolerant killifish from New Bedford Harbor (NBH) versus sensitive killifish from Scorton Creek (SC), MA. Consistent with the teleost whole genome duplication (tWGD), killifish have six RyR genes, corresponding to a and b paralogs of mammalian RyR1, 2 and 3. The presence of six RyR genes was consistent in all teleosts investigated including zebrafish. Killifish have four FKBP1; one FKBP1b and three FKBP1a named FKBP1aa, FKBP1ab, likely from the tWGD and a single gene duplicate FKBP1a3 suggested to have arisen in Atherinomorphae. The RyR and FKBP1 genes displayed tissue and developmental stage-specific mRNA expression, and the previously uncharacterized RyR3, herein named RyR3b, and all FKBP1 genes were prominent in brain. We identified a SNV in RyR3b encoding missense mutation E1458D. In NBH killifish, 57% were heterozygous and 28% were homozygous for this SNV, whereas almost all SC killifish (94%) lacked the variant (n≥39 per population). The outlined sequence differences between mammalian and teleost RyR and FKBP1 together with outlined population differences in SNV frequency may contribute to our understanding of NDL PCB neurotoxicity.
Keywords: Non-dioxin-like PCBs Ryanodine Receptor, FK Binding Protein 1, Fundulus heteroclitus
Introduction
Ortho-substituted polychlorinated biphenyl (PCBs) congeners, broadly termed non-coplanar or non-dioxin-like (NDL PCBs), have arisen as potential neurotoxic compounds due to observed changes in the behavior and cognition of children exposed during key stages of development (Lake et al., 1995; Lonky et al., 1996). Additionally, pure NDL PCB exposures in model organisms can cause performance deficits (Yang et al., 2009), hypoactivity and altered behavior (Schantz et al., 1997; Elnar et al., 2012). Several molecular mechanisms proposed to contribute to NDL PCB-induced neurotoxicity include congener potentiation of GABAA receptors (Fernandes et al., 2010), inhibition of dopamine transport via dopamine or vesicular monoamine transporters (Wigestrand et al., 2013), and induced activity of ryanodine receptors (RyR) (Wong and Pessah, 1996; Pessah et al., 2010). RyR Ca2+ channels are targets of particular interest because they are potentiated at pM to nM NDL PCB concentrations (Holland et al., 2016) and this induced receptor activation leads to altered neuronal growth and plasticity (Kenet et al., 2007; Wayman et al., 2012a; Wayman et al., 2012b; Lesiak et al., 2014) and marked deficits in cognitive function and behavior in mammals (Schantz et al., 1997; Roegge et al., 2006; Yang et al., 2009)
NDL PCB activation of the RyR also poses other significant health threats, as the receptor is a widely distributed Ca2+ release channel, participating in physiological processes in striated muscle, neuronal systems and endocrine health. In mammals, there are three RyR genes resulting in three main protein isoforms (RyR1, RyR2 and RyR3) of about 5,000 amino acids. RyR1 and RyR2 arefound primarily in skeletal and cardiac muscle, respectively, but are also located throughout the central and peripheral nervous system. The third form, RyR3 is often termed the brain RyR-isoform but has a wide tissue distribution (Giannini et al., 1995). It is thought that NDL PCBs interfere with RyR’s interaction with the accessory protein FK506 binding protein 12kDa and 12.6 kDa (protein name FKBP12 or FKBP12.6; hereafter referred to by their gene name FKBP1A and FKBP1B), which helps maintain the conformational state of the RyR (Zalk et al., 2007). Disruption of the FKBP1-RyR complex causes an increase in channel open probability (Bellinger et al., 2008; Zalk et al., 2007), which is similar to that caused by NDL PCBs (Holland et al., 2016). Additionally, pretreatment with the immunophilin drugs FK506 and rapamycin can eliminate NDL PCB toxicity at the RyR (Wong and Pessah, 1997). The importance of the Ca2+ channel to diverse physiological systems and demonstrated impacts of RyR over-activation support the idea that NDL PCBs pose specific threats to the health of human and wildlife populations. However, how these risks, together with other factors such as interactions with the FKBP1 accessory protein or genetic variability in RyR and FKBP1, lead to disease is not understood.
Small teleost species act as important models in toxicology due to their rapid development, high reproductive output, and both genomic similarities and dissimilarities with mammals. NDL PCBs and structurally related compounds are known to activate RyR1 found in the skeletal muscle of teleost fish, indicating a common mode of toxicity in vertebrates (Holland-Fritsch and Pessah, 2013; Holland-Fritsch et al., 2015). However, NDL PCBs may display different efficacy and potency in teleosts as compared to mammals, which raises mechanistic questions. In particular, teleosts have at least five RyR genes that display tissue and developmental stage-specific expression (Darbandi and Franck, 2009; Wu et al., 2011). Much less is known about FKBP1 in fish, but zebrafish (Danio rerio) have at least three FKBP1 genes (www.zfin.org; Somarelli and Herrera, 2007)), which may contribute to variable NDL PCB toxicity. There is a need to characterize potential genetic differences between mammalian and teleost species to develop teleost species as models for NDL PCB mediated toxicity at the RyR, and to understand toxic risks posed to fish.
The Atlantic killifish (Fundulus heteroclitus) is an estuarine fish species found along coastal regions of the Eastern Atlantic Ocean. The killifish has become an important model for environmental studies because they display tolerance to both natural and anthropogenic stressors, including tolerance to extreme chemical exposures (Burnett et al., 2007). Killifish from New Bedford Harbor (NBH), MA, a Superfund site highly contaminated by PCBs, contain tissue levels of NDL PCBs 100-times greater than levels known to cause RyR-activation (up to 300,000 ppm total ortho PCBs; Holland-Fritsch et al. 2015). We have recently shown that compared to a reference killifish population at Scorton Creek (SC), MA, adult NBH killifish have increased protein levels of RyR1 and FKBP1A, and embryos of F1 and F2 generations of NBH fish raised in the laboratory have increased mRNA levels of RyR2 (Holland-Fritsch et al., 2015). These findings suggest that NBH killifish may display compensatory responses and potentially heritable tolerance to NDL PCB toxicity.
The current work characterizes RyR and FKBP1 genes in teleost species with a particular focus on Atlantic killifish. We utilized the recently released Fundulus genome (fundulus.org) (Reid et al., 2016; Reid et al., 2017) together with multiple large sequencing data sets to 1) characterize RyR and FKBP1 sequences present in the Fundulus genome, 2) determine the age and tissue specific mRNA expression of RyR and FKBP1 genes in killifish 3) evaluate the molecular phylogenetic relationships and key residue conservation, as it relates to PCB toxicity, between the RyR and FKBP1 in killifish and that of other model teleost and mammalian species and 4) determine whether killifish from NBH display genetic variation, including single nucleotide variants (SNV), in RyR and FKBP1 genes as compared to the SC reference population.
Methods
Animal Collection
Adult killifish were collected from Scorton Creek (SC; Latitude and longitude; 41.7649 by 70.4800) using baited traps under permits from the Massachusetts Division of Marine Fisheries, and protocols approved by Animal Care and Use Committees at WHOI and the US EPA National Health and Environmental Effects Laboratory, Atlantic Ecology Division, Narragansett RI.. Upon capture, fish from SC were returned from the field and maintained in flow-through aquaria receiving 5 µM filtered sea water until dissection, at least 2h after collection. Here, fish were euthanized by immediately severing the spinal cord at the base of the head region. At this time fish were weighed and sex noted. Dissected tissue was flash frozen in liquid nitrogen and stored at −80°C.
Embryos and larvae used for qPCR and RNA-seq were obtained from the breeding stocks maintained in uncontaminated conditions at Atlantic Ecology Division, as described elsewhere (Nacci et al., 2002; Nacci et al., 2010; Holland-Fritsch et al., 2015). Specifically, the current study used embryo and larvae collected at 3, 9, or 10 days post fertilization (dpf) and 1 day post hatch (dph, approximately 14 dpf).
RNA Extractions and cDNA synthesis
RNA from adult killifish skeletal and cardiac muscle and whole brain tissue (n ≥ 8 per tissue type) was extracted using Stat-60™ (amsbio) and the aqueous phase clarified using the Aurum Total RNA Fatty and Fibrous Tissue Kit (Bio-Rad). RNA from embryos or larvae (n ≥ 6 per developmental stage) was extracted using Trizol following manufacturer’s protocols (Invitrogen). RNA concentrations were determined on a Nanodrop 1000 (Thermo Scientific) and quality of each sample assessed using 260/280 ratios and through gel electrophoresis. Complementary DNA was synthesized with 1 µg of total RNA using Random Primers and SuperScript Reverse Transcriptase III (Invitrogen).
Quantitative PCR
Expression of genes of interest (GOI) along with suitable reference genes (Table S1) were measured utilizing quantitative Polymerase Chain Reaction (qPCR) as described elsewhere (Connon et al., 2012). Primer probe combinations used for qPCR (Table S2) were designed using the Roche Universal Probe Library Assay Design Center and targeted to variable regions of the RyR and FKBP1 sequences from alignments conducted with Clustal Omega. Genes of interest were normalized to factors calculated from four reference genes, rpl8, rps20, β-actin and ef1α, using the geNorm algorithm (Vandesompele et al., 2002). These reference genes were previously determined to be stable across the tissue and developmental stages assessed in the current study (Holland-Fritsch et al., 2015). For visualization, expression-normalized mRNA levels were placed on a linear scale and potential tissue and age specific differences displayed using 95% confidence intervals.
Phylogenetic and Sequence Comparisons
Killifish genes of interest were identified within the annotated Fundulus genome (fundulus.org; KillishV2b) using BLAST run with known sequences from zebrafish (Danio rerio) or mammalian species. RyR and FKBP1 gene sequences for medaka (Oryzias latipes), stickleback (Gasterosteus aculeata) and fugu (Takifugu rubripes) were collected from Ensembl (Release 85). Due to the identification of variable gene numbers in teleost species (see Results), killifish RyR and FKBP1 sequences were also used to search model teleost genomes in Ensembl, including the zebrafish genome. All NCBI accession numbers and/or Ensemble Identification numbers utilized in the current work can be found in Table S2. Predicted RyR and FKBP1 were initially named based on the order of identification in the F. heteroclitus genome, or Ensembl database for other teleost species. Killifish sequences were then named based on sequence alignments and phylogenetic comparisons to zebrafish or other model fish species.
For phylogenetic comparisons, amino acid sequences were aligned using MUSCLE ([109]; Seaview4) or Clustal Omega. Maximum likelihood constructed phylogenetic trees, with 1000 rapid bootstrap iterations, were determined using RAxML v8.2, (Stamatakis, 2014) on the CIPRES portal (http://www.phylo.org/index.php) using sequences from Drosophilia melanogaster as the outgroup. RyR phylogenetic trees were constructed using amino acid sequences (~5000 aa) using maximum likelihood combined with an auto-selected substitution model with gamma rate heterogeneity. FKBP1 trees built from amino acid sequences often had branch low bootstrap support (~108 aa; data not shown) and therefore trees were determined with the open reading frame nucleotide sequences using maximum likelihood combined with a general time reversible nucleotide substitution model with gamma rate heterogeneity.
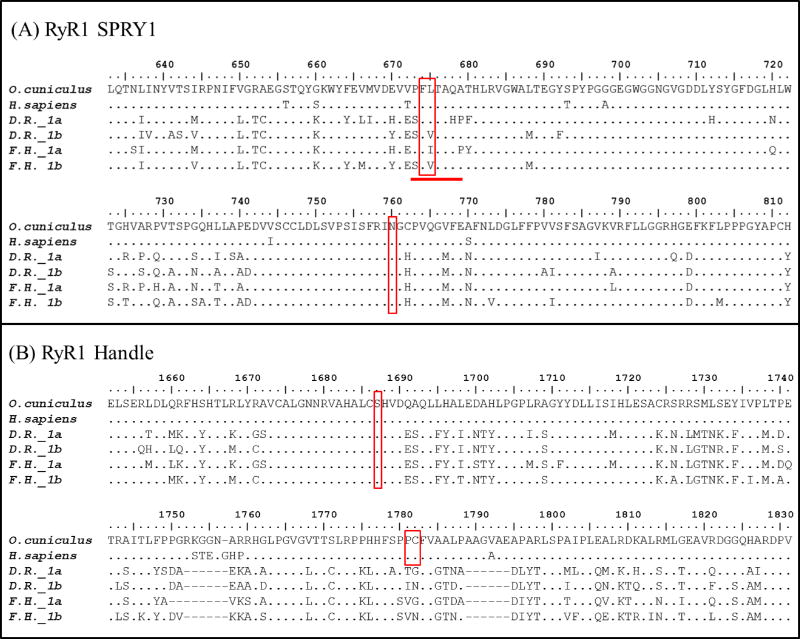
Amino acid alignments performed using Clustal Omega were used to define sequence similarities between entire RyR and FKBP sequences or select domains of interest hypothesized to be relevant to NDL PCB toxicity. Alignments of domains of interest were then visualized using BioEdit (BioEdit v7.2.5; Ibis Biosciences) and residues projected to be important for FKBP1:RyR interactions were compared between select species (human, rabbit and zebrafish) and F. heteroclitus. Regions of interest included areas suggested to be important for RyR:FKBP1 protein interactions based on high-resolution cryo-electron microscopy structures for the rabbit (Oryctolagus cuniculus) RyR1 homoprotein (Efremov et al., 2015; Yan et al., 2015; Zalk et al., 2015) and crystal solutions of the SPRY1 and tandem-repeat domains (Yuchi et al., 2015). For RyR1, studies by Yuchi et al. (2015) and Yan et al. (2015) specifically outlined RyR amino acids that likely interact with FKBP1. Yuchi et al. (2015) suggested that the SP1A kinase RyR domain 1 (SPRY1; O. cuniculus aa 632–826, 1466–1491 and 1615–1634; H. sapiens RyR1 aa 631–825, 1465–1490 and 1614–1633) was key to FKBP1’s regulation of RyR1, especially at F674 and L675 (H. sapiens; F673 L674) and the surrounding amino acids They also demonstrated that N760 (H. sapiens; N759) played a role in the SPRY1 domain folding and thereby significantly decreased FKBP1 maximum binding to RyR1 (Yuchi et al., 2015). In addition to the SPRY1 interaction, Yan et al. (2015) suggested that the so-called handle domain of RyR1 (O. cuniculus aa 1651–2145; H. sapiens RyR1 aa 1650–2144) was pertinent to FKBP1:RyR interactions, specifically at amino acids S1687, P1780 and C1781 (RyR1, O. cuniculus). Similarly, for important amino acids in FKBP1, the RyR based structural papers suggested that FKBP1 residues R40 and potentially R71 interact with the RyR1-SPRY1 domain, and Y26, F36, F99 and potentially I56 interact with the RyR-handle domain (Yan et al., 2015; Yuchi et al., 2015). Therefore, the SPRY1 and Handle domains of RyR proteins were aligned and the full-length FKBP1 sequences were aligned for amino acid comparisons with special emphasis on key residues.
Identification of Single Nucleotide Variation
Single nucleotide variants were screened using previously completed RNA-sequencing data (Illumina HiSeq 2500; Tufts University Genomics Core Facility; unpublished data). RNA was collected from NBH and SC embryos, F2 generation reared under non-contaminated conditions, that had been exposed to 28 µM PCB 153 or a DMSO control for 6 hours starting at 10 days post fertilization (N.Aluru, S.Karchner, D.Nacci, M.Hahn, manuscript in preparation). RNA was collected from 5 pools of 10 embryos per treatment per population. For the current study, chemical treatment was ignored and SNV identification was compared across population only (n = 10 per population). RNA-seq data were mapped to the recently released killifish genome (killifish version 2b, fundulus.org) and SAM files converted to BAM files, sorted and indexed using Samtools (Li et al., 2009). Bed files were created for the specific scaffold of interest (Table 1) and variant calls completed for full scaffold regions for both RyR and FKBP1 genes. Variants within NBH and SC were visualized against the reference genome using the Integrative Genomic Viewer (IGV; broadinstitute.org).
Table 1.
Suggested names and corresponding scaffold locations of ryanodine receptor and FK506 binding protein 1 genes in the Atlantic killifish genome.
| Suggested Name |
Protein Length (aa) |
Scaffold (Fundulus.org) | Transcript ID (NCBI) |
|---|---|---|---|
| RyR1a | 5,084 | Scaffold469:2114077-2182033 | XM_021312178 |
| RyR1b | 4,980 | Scaffold9971:315988,456989 | XM_021321451 |
| RyR2a | 4,923 | Scaffold10172:242755-455454 | XM_012882606 |
| RyR2b* | 2,871 | Scaffold9878:254646-441294 | XM_021317243 |
| 989 | Scaffold1755:82-22,0000 | XM_021314623 | |
| RyR3a | 4,848 | Scaffold10024:1227459-1358309 | XM_021323219 |
| RyR3b | 4,896 | Scaffold10114:385206-503355 | XM_021307330 |
| FKBP1aa | 109 | Scaffold10019:3016451-3022261 | XM_012875568 |
| FKBP1ab | 108 | Scaffold10096:92745-94803 | XM_012879414 |
| FKBP1a3 | 107 | Scaffold519:151369-153562 | XM_021312454 |
| FKBP1b | 108 | Scaffold9904: 479209-495284 | XM_012866692 |
Notes:
RyR2b likely represented by two partial sequences in the Fundulus genome based on sequence identity to zebrafish RyR2b and mammalian RyR2.
To confirm potential genetic differences identified in the RNA-sequencing data and quantify allele frequency in NBH versus SC killifish populations, FKBP1 and RyR sequences were also evaluated using Sanger sequencing. Forward and reverse primers used for PCR amplification (Table S3) were designed in the 5’ and 3’untranslated regions of each FKBP1 gene and regions flanking the RyR handle domain (aa1661–2145 Rabbit; approximately aa1556- 2100 killifish) projected to bind with FKBP1 (O. cuniculus; RyR1 S1685; P1779 and C1780; Yan et al. 2015). Sequences for killifish RyR3.1, herein named RyR3a, were gathered upstream of the handle due to the presence of SNVs in the RNA-sequencing data (see Results). The PCR products were amplified from the brain or skeletal muscle cDNA of at least eight individuals per killifish population and sequenced by Eurofins MWG Operon. If the initial sequences confirmed or identified a population specific SNV then corresponding PCR fragment from 48 individuals per population was subsequently evaluated.
Results
Sequence Identification and Phylogenetic Comparisons
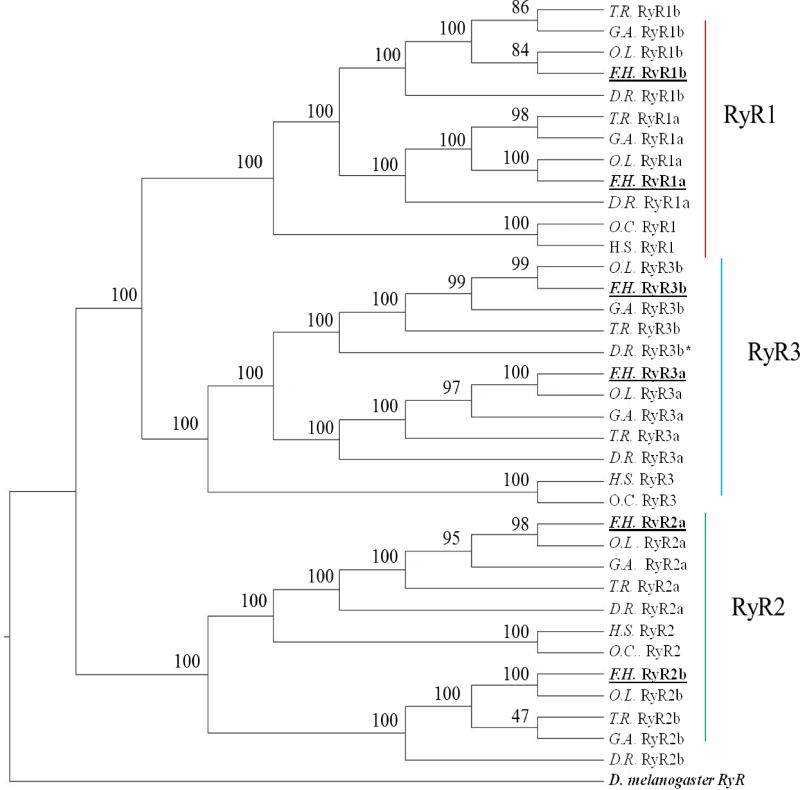
We identified six RyR genes in the killifish genome (Table 1). The six predicted RyR genes appeared to represent a and b paralogs for the mammalian RyR1, RyR2, and RyR3, as supported by sequence identity (Figure S1A) and phylogenetic comparisons (Figure 1). Five of the killifish RyRs, namely RyR1a, RyR1b, RyR2a, and the two RyR3 identified, suggested here as RyR3a (Scaffold10024) and RyR3b (Scaffold10114), represent full-length RyR proteins varying in length from 4,848aa (RyR3a) to 5,084aa (RyR1a), which is consistent with the approximate 5,000aa protein in human and rabbit. The two RyR3 were named according to their similarity and clustering to the single RyR3 (XM_009294773) currently recognized in zebrafish, herein suggested as RyR3a, and consequently the second RyR3 gene identified in the genome of killifish, and the corresponding second RyR3 in other species, be named RyR3b. The killifish RyR2a represented a full sequence with 4,923aa on Scaffold 10172 and shared high amino acid identity with the zebrafish RyR2a protein (82%). The killifish RyR2b appears to be represented on two genomic scaffolds (Table 1, 9878 and 1755). The RyR2b partial sequence on Scaffold9878 represented 2971aa residues that aligned with rabbit RyR2 from amino acid 1–91, 390–1537 and 3093–4968 with 64% protein identity. The RyR2b partial sequence on scaffold 9878 also had the highest amino acid identity with the zebrafish RyR2b protein (63%) compared to other zebrafish RyR sequences. Scaffold 1755 represented a 959aa partial protein that aligned with residues 2010–2965 of the rabbit RyR2 with 65% protein identity. The killifish RyR2b partial on Scaffold 1755 only shared 50% amino acid identity with the suggested killifish RyR2a and therefore, the predicted RyR2-like partial protein sequence located on Scaffold 1755 is suggested to represent part of the middle section of the RyR2b killifish protein. When combined, Scaffold 9878 and 1755 sequences lack several important regions of the RyR2 amino acid sequence. Namely, the two partial sequences are missing the section corresponding to rabbit cardiac amino acids 1538–2010, which represents the majority of the handle domain (O. cuniculus RyR2; aa1648–2109).
Figure 1.
Phylogenetic clustering of ryanodine receptor orthologs and paralogs in teleost and mammalian species. Phylogenetic analysis conducted using available amino acid sequences; branch labels are bootstrap values (%) for 1000 iterations. See Table S2 for NCBI sequence IDs used for tree development. Abbreviations and Notes: D.R. Danio rerio, F.H Fundulus heteroclitus, G.A. Gasterosteus aculeatus, H.S. Homo sapiens, O.L. Oryzias latipes, O.C. Oryctolagus cuniculus, and T.R. Takifugu rubripes. *Previously un-described RyR3 gene in Danio rerio.
The presence of 6 RyR genes was consistent in all teleosts investigated in the current study (Figure 1). This included an additional, previously uncharacterized, RyR3 predicted on chromosome 17 (location 519,900) in the zebrafish genome (Ensembl Zv10). The partial zebrafish sequence coded for a 1338aa protein sharing 79% identity with the C-terminal of rabbit RyR3 and 80 % and 86% identity with the C-terminal of killifish RyR3a and RyR3b sequences, respectively. Zebrafish RyR2b (XM_017351708.1) has also been placed on chromosome 17 (location 19,994,151) but the predicted additional zebrafish RyR3 only shares 66% amino acid identity with the zebrafish RyR2b C-terminal, supporting identification of a unique gene. As discussed for the killifish above, the previously recognized zebrafish RyR3 (XM_009294773) gene is suggested as RyR3a and the partial RyR3-like protein on chromosome 17 named RyR3b.
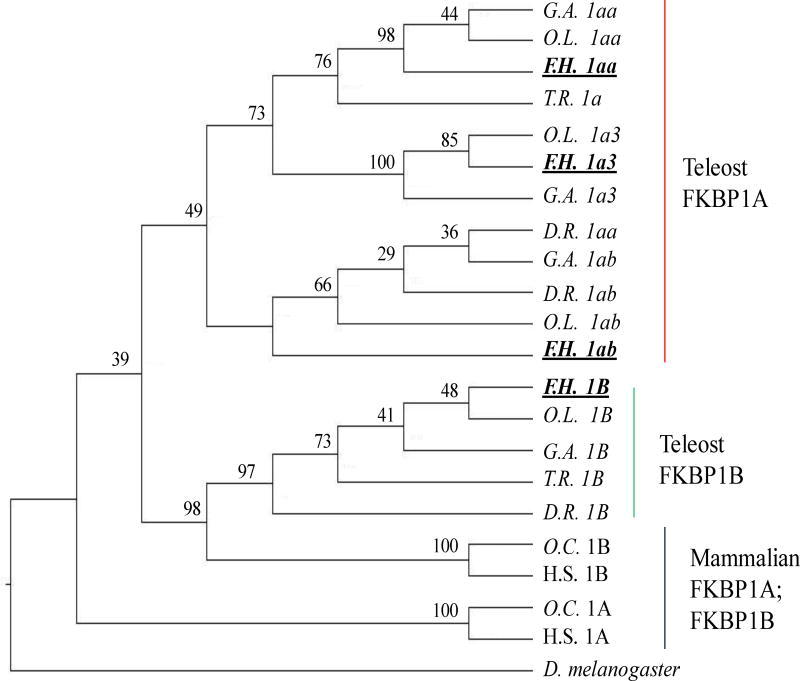
We also identified four FKBP1 genes (Table 1) in the killifish genome. The coded FKBP1 proteins ranged in length from 107 to 109 amino acids, which is similar to the 108 amino acid protein found in mammals. The number of FKBP1 genes present in teleost species varied between two and four. Takifugu rubripes appeared to have only two genes that clustered with the corresponding mammalian FKBP1A and FKBP1B (Figure 2); zebrafish were found to have three FKBP1 genes, corresponding to two FKBP1A (FKBP1aa and FKBP1ab) and one FKBP1B gene, but other teleosts investigated had four FKBP1.
Figure 2.
Phylogenetic clustering of FK506 binding protein 1 orthologs and paralogs in teleost and mammalian species. Phylogenetic analysis conducted using coding nucleotide sequences; branch labels are bootstrap values (%) for 1000 iterations. See Table S2 for NCBI sequence IDs used for tree development. Abbreviations D.R. Danio rerio, F.H. Fundulus heteroclitus, G.A. Gasterosteus aculeatus, H.S. Homo sapiens, O.L. Orysias latipes, O.C. Oryctolagus cuniculus, and T.R. Takifugu rubripes.
Of the four FKBP1 genes identified in the killifish, and other species, one likely represents an ortholog of mammalian FKBP1B (protein FKBP12.6) and three represent co-orthologs of mammalian FKBP1A (protein FKBP12). These suggestions are based on amino acid identity (Figure S1) and phylogenetic clustering (Figure 2). The suggested killifish FKBP1b shared 96% and 89% amino acid identity with zebrafish or rabbit FKBP1b, respectively and the presence of only one FKBP1b gene was consistent in all other model teleosts investigated. For the three co-orthologs of FKBP1A found in the killifish genome, we suggest the following naming FKBP1aa (Scaffold10119), FKBP1ab (Scaffold10096) and FKBP1a3 (Scaffold519) because killifish FKBP1aa and FKBP1ab shared greater than 82% amino acid sequence identity with zebrafish FKBP1aa and FKBP1ab, respectively. The killifish FKBP1aa or FKBP1ab also clustered with the corresponding genes in medaka and stickleback and the FKBP1aa or FKBP1ab of these two model organisms show shared synteny with the FKBP1aa or FKBP1ab in zebrafish (Ensembl). It should be noted that the spotted gar Lepisosteus oculatus, an ancestral ray finned fish, has only one FKBP1A gene that in Ensembl (v85; ENSLOCG0000006019) is currently named FKBP1ab and is similar to the FKBP1ab labeled in teleost species used in this study. The gar FKBP1ab likely represents the ortholog of tetrapod FKBP1A and therefore the FKBP1aa and FKBP1ab naming in teleost might need to be switched in teleost genome databases. However, for clarity the current study stayed consistent with the current naming in zebrafish.
The third killifish FKBP1a3 displayed 74% and 72% amino acid identity with zebrafish FKBP1aa and FKBP1ab and only 71% with zebrafish FKBP1B. The killifish FKBP1a3 amino acid sequence appeared to also have greater similarity to mammalian FKBP1A (71%), than to mammalian FKBP1B (67%). Similar to the killifish, the medaka and stickleback genomes also displayed a third FKBP1a3 but this third gene was not found in zebrafish.
Tissue and Age Specific Expression
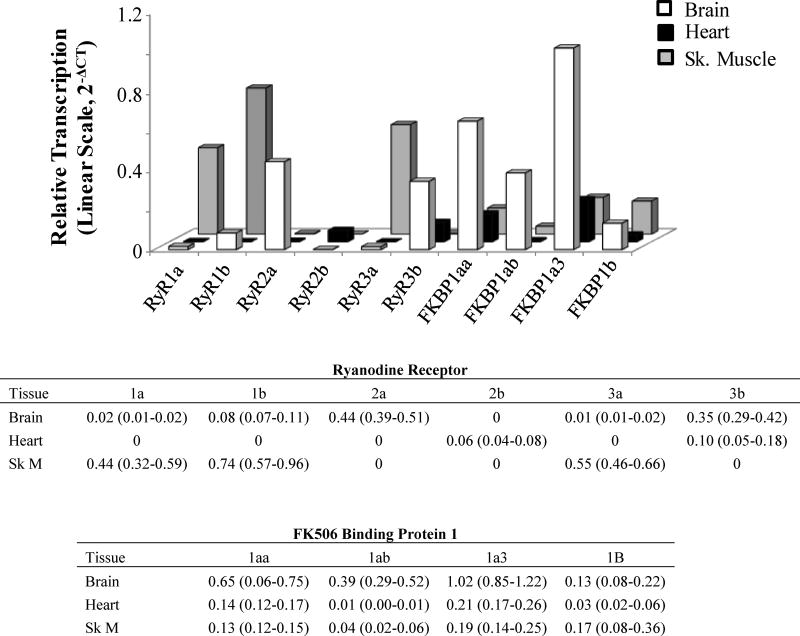
In adult killifish, RyR genes displayed tissue specific expression where RyR1a, RyR1b and RyR3a were primarily found in skeletal muscle, and RyR2a and RyR2b were expressed primarily in brain and heart, respectively. The mRNA expression of RyR3b was located primarily in the killifish brain. The killifish FKBP1 paralogs displayed wide tissue distribution (Figure 3), and all four were prominent in the killifish brain. In adults, FKBP1ab was the only FKBP1 that appeared to have tissue limiting mRNA expression, being located primarily in killifish brain with minimal or no expression in skeletal muscle or the heart.
Figure 3.
Relative transcript levels of ryanodine receptor and FK506 binding protein 1 genes in brain, heart and skeletal muscle (Sk. Muscle; Sk M) from adult killifish from Scorton Creek. Shown relative to the normalization factor developed from the reference genes rpl8, rps20, β-actin and ef1α and placed on the linear scale for visualization. Table insert shows Mean (Upper and Lower limits of the 95% Confidence Interval); n= 8.
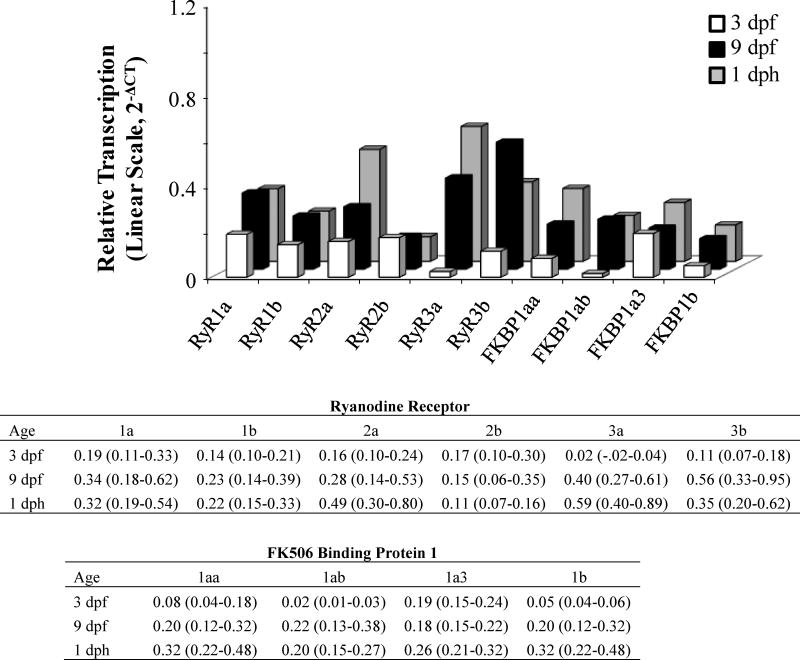
The RyR and FKBP genes were expressed at 3 dpf (stage 22), which in the killifish is just prior to the onset of circulation. Levels of RyR and FKBP1 mRNA subsequently increased at 9 dpf (stage 33), which is prior to coordinated movement, and at 1 dph (Figure 4) (Armstrong and Child, 1965). Of the genes investigated, RyR3a and FKBP1ab displayed the lowest mRNA levels at 3 dpf. The broad gene expression of FKBP1aa together with the more restricted expression of FKBP1ab in embryonic and adult killifish tissue, as discussed above, is consistent with the tissue distribution of FKBP1aa and FKBP1ab in the zebrafish (zfin.org).
Figure 4.
Relative transcript levels of ryanodine receptor and FK506 binding protein 1 genes in embryo and larvae killifish from Scorton Creek. Shown relative to the normalization factor developed from the reference genes rpl8, rps20, β-actin and ef1α and placed on the linear scale for visualization. Table insert shows Mean (Upper and Lower limits of the 95% Confidence Interval); n= 6. Abbreviations; Days post fertilization (dpf) and Days Post Hatch (dph).
Key Residue Comparisons Between Mammals and Teleost Species
There were similarities and differences between mammalian and teleost RyR regions suggested to be important for FKBP1 interactions (see Methods for detailed list). Specifically, comparing the amino acids in the RyR1 SPRY1 domain showed that sites F674 and N760 are highly conserved between mammals and teleost species (F. heteroclitus; RyR1a F683 and RyR1b F610; RyR1a N769 and RyR1b N696; D. rerio; RyR1a F681 and RyR1b F6674; RyR1a N767 and RyR1b N760). However, amino acids surrounding F674, including L675, varied (Figure 5A) and may contribute to differences in NDL PCB toxicity among mammals and teleosts. In the handle domain of RyR1, sites S1687 and surrounding residues (Figure 5B) in mammals and the corresponding residues in teleost species were highly conserved (F. heteroclitus; RyR1a S1683 and Ry1b S1580; D. rerio RyR1a S1681 and RyR1b S1671). Conversely, handle site P1780 and C1781 varied widely (Figure 4B; F. heteroclitus; RyR1a VG1769–1770 and Ry1b VN1668–1669; D. rerio RyR1a TG1669–1670 and RyR1b IN1759–1760) suggesting these sites may also contribute to species-specific sensitivity to NDL PCBs.
Figure 5.
Partial amino acid sequence alignments of the SPRY1 and Handle domain of RyR1 orthologs and paralogs in teleost and mammalian species. Areas highlighted in red are suggested to be important for FKBP1 binding (Yan et al.2015 and Yuchi et al. 2015). Abbreviations D.R. Danio rerio, F.H Fundulus heteroclitus.
The amino acids in FKBP1 that interact with RyR1 SPRY1 and Handle domains were all highly conserved among mammalian and teleost FKBP1A and FKBP1B orthologs (Figure 6). Additional residues important for FKBP1’s action as a cis-trans peptidyl prolyl isomerase and/or for its binding to immunophilin drugs (Y26, D37, F48, V55, W59, Y82, H87 and F99) (see Ikura et al. and DeCenzo et al. 1996) were also highly conserved across taxa (Figure 6), supporting conserved function between teleost and mammal FKBP1 orthologs.
Figure 6.
Amino acid sequence alignments of FK506 binding protein 1 orthologs and paralogs in teleost and mammalian species. Areas highlighted in black and red are suggested to be important for FKBP1’s interaction with the RyR SPRY1 or Handle domain, respectively (Yan et al. 2015 and Yuchi et al. 2015). Abbreviations D.R. Danio rerio, F.H Fundulus heteroclitus.
Single Nucleotide Variation in New Bedford Harbor Killifish
RNA-sequencing data from NBH and SC killifish aligned with the killifish reference genome (Maine, USA) revealed a limited number of non-synonymous SNVs (Table S4) in the full-length sequences of RyR and FKBP1 genes. There were no non-synonymous SNVs identified in any of the killifish FKBP1 genes or the RyR2 genes; however, in the reference genome, killifish RyR2b was incomplete and SNVs in the missing sections could not be assessed. The majority of the non-synonymous SNVs found in RyR1a and RyR1b had a low-frequency, were not biased to one killifish population and/or were located far outside of the regions suggested to be important for FKBP1:RyR protein interactions. The dearth of non-synonymous SNVs found in RNA sequencing data for FKBP1, RyR1 and RyR2 genes was confirmed in targeted de novo sequencing (data not shown, n=12). Most variants identified in the RyR3a and RyR3b also appeared to have low frequency differences between populations and were outside regions of interest, with several exceptions. In RyR3a (Scaffold10024), one missense mutation identified at location 1286421 coded for a glycine (G) at killifish amino acid S1279 (O. cuniculus A1397) and was found to display a 55% allele frequency in NBH killifish but was not detected in the SC population. However, when this region was Sanger sequenced in the skeletal muscle tissue from additional adults from NBH and SC, the SNV was not detected (n=8 per population; data not shown).
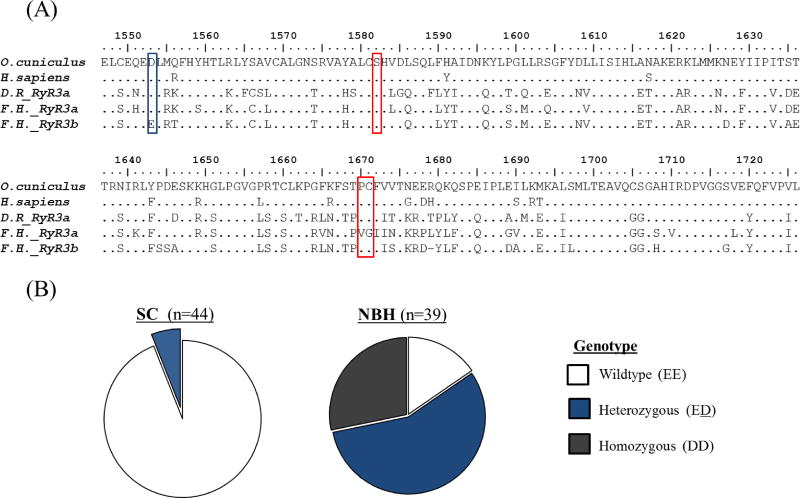
RNA-sequencing data also revealed several SNVs in the RyR3b (Scaffold 10114) sequence from NBH as compared to SC killifish. Of particular interest, NBH killifish displayed a missense mutation at scaffold location 464122 that coded for an aspartic acid (D) at killifish amino acid E1458 (O. cuniculus D1553, RyR3). Amino acid E1458 is upstream of the conserved serine residue, killifish S1487 (O. cuniculus S1582, RyR3), suggested to interact with FKBP1 (Figure 7A). In the RNA-sequencing data, the E1458D SNV displayed 60% and 15% frequencies in NBH versus SC killifish, respectively. When the handle domain for RyR3b was Sanger sequenced in additional killifish, E1458D was found to display 60% allele frequency in NBH but only 6% allele frequency in SC killifish (n ≥ 39 per population). Interestingly, in NBH killifish, 57% of the individuals were heterozygous for the SNV (DE), 28% were homozygous for the SNV (DD) and only 15% were EE (Figure 7B), where EE is the genotype found in the Maine reference genome. Conversely, almost all individuals sequenced from SC were similar to Maine (EE; 94%), only 6% heterozygous (ED) and no homozygous (DD) individuals were identified. The genotype frequencies in NBH and SC are both suggested to be Hardy-Weinberg equilibrium (NBH; χ2 =0.81; df=1; α=0.05; SC; χ2 =0.16; df=1; α=0.05).
Figure 7.
Population frequency of a missense mutation identified in the Handle domain of Ryanodine Receptor 3b in New Bedford Harbor versus Scorton Creek killifish. (A) Amino acid sequence alignment of a portion of the RyR3 handle domain in mammals and teleosts. The killifish RyR3b amino acid sequence shown in the alignment is from the reference killifish genome (Maine, USA fundulus.org) and is considered ‘wildtype’ (EE) in the current work; area highlighted in blue represents the location of the identified missense mutation coding for E1458D in the New Bedford Harbor killifish. (B) Frequency of individuals in the Scorton Creek or New Bedford Harbor killifish populations containing the missense mutation E1458D. Abbreviations D.R. Danio rerio, F.H Fundulus heteroclitus. Note: The sequence for Danio rerio RyR3b Handle was not available.
Discussion
The RyR is a common target of NDL PCBs in mammalian and teleost species (Holland-Fritsch and Pessah, 2013) and small fish species have been suggested as useful models for understanding the physiological ramifications of NDL PCB disruption of the RyR. The killifish has been shown to thrive in severally polluted locations, including a population in PCB laden NBH; therefore providing a unique model particularly suited for NDL PCBs toxic assessments. Here, we identified six RyR and four FKBP1 genes in the Atlantic killifish and phylogenetic comparisons and amino acid sequence alignments highlight several differences that may contribute to species-specific responses to NDL PCBs. Together this work can help support the use of teleost to understand the impact of NDL PCBs, or related compounds in vertebrate species.
Here, we report the presence of six RyRs in all common model teleost species investigated that corresponded to an a and b paralog for each of mammalian RyR1, RyR2 and RyR3 monophyletic clades. This includes a previously undescribed RyR3 gene, RyR3b, found in the zebrafish genome. These findings are in contrast to that outlined in previous studies (Darbandi and Franck, 2009; Wu et al., 2011). Darbandi and Franck (2009) suggested that only RyR1 in zebrafish displayed an a and b paralog. They suggested that the RyR1 duplication took place prior to the teleost whole genome duplication (tWGD) because they showed that bichir (Polypterus ornatipinnis), a basal ray finned fish, also possess an RyR1a and an RyR1b with only one RyR2 and one RyR3. When the bichir RyR1a and RyR1b (NCBI, FJ976726 and FJ976727) are aligned (Needle, pairwise alignment) they display 96.5% nucleotide identity and may represent transcript variants but the two sequences were shown to display variable mRNA expression in red versus white skeletal muscle. Searching the genome of the spotted gar (Lepisosteus oculatus; Ensembl 85), another basal ray finned species, we found only three RyR genes (data not shown) supporting the presence of teleost RyR1a/b, RyR2 a/b and RyR3a/b as a result of the tWGD (Glasauer and Neuhauss, 2014).
We identified 2–4 FKBP1 genes in common model teleost species. This included three FKBP1a genes in killifish, medaka and stickleback. The FKBP1aa and FKBP1ab were also present in zebrafish and are suggested to have arisen as a result of the tWGD. The third FKBP1A, FKBP1a3, is lacking in the zebrafish and is suggested to have arisen as a single gene duplication event at the base of Atherinomorphae (~100 million years ago), which includes Fundulus, Oryzias and Gasterosteus species. Supporting this suggestion, FKBP1a3 was found in Atherinomorpha species tilapia (Oreochromis niloticus) and amazon molly (Poecilia Formosa; Genomicus v89.01). Currently, it is unclear if FKBP1a3 is a duplicate of FKBP1aa or FKBP1ab.
The RyR genes in the killifish have distinct tissue-specific expression patterns suggesting tissue-specific roles in Ca2+ signaling. Expression is consistent with other teleost, where RyR1a and RyR1b are associated with red and white skeletal muscle (O'Brien et al., 1995), respectively, RyR2a and RyR2b are associated with the brain and cardiac tissue, respectively, and RyR3 (likely RyR3a) is associated with skeletal muscle (Wu et al., 2011). The additional RyR3 in teleost, RyR3b, appeared to have the strongest expression in the brain. The role of RyR3 in vertebrate neuronal signaling is not completely understood but RyR3 is present in the hippocampus, thalamus, and the olfactory system of mammals (Giannini et al., 1995). Studies outlining the roles of RyR3b in the brain of teleosts may further our understanding of RyR3’s role in neuronal Ca2+ signaling similar to recent studies (Perni et al., 2015) in zebrafish that demonstrated that RyR3a is the main contributor to Ca2+ spark generation in skeletal muscle fibers and the formation of para-junctional feet in the sarcoplasmic reticulum (SR). Findings suggested that RyR3a, or RyR3 in other vertebrates, may contribute to Ca2+ release from SR stores and muscle contraction, which is supported by other studies (Essin and Gollasch, 2009; Murayama and Kurebayashi, 2011). RyR3b may play a similar role in zebrafish neuronal tissue and may contribute to NDL PCB neurotoxicity.
To date research has only addressed NDL PCB toxicity in fish skeletal muscle (Holland-Fritsch and Pessah, 2013), and whether variable RyR paralogs identified in teleost have differential sensitivity to NDL PCBs is unknown. Research in mammals has shown that both RyR1 and RyR2 display similar NDL PCB structure activity relationships, such that PCBs with high ortho substitution have increased potency toward both receptors (Pessah et al., 2006; Pessah et al., 2009). Research indicates that NDL PCBs may be less potent and efficacious at RyR2, compared to RyR1 (Wong and Pessah, 1996; Pessah et al., 2009), but these studies were done in different species. To our knowledge there have been no studies addressing NDL PCB toxicity directly towards RyR3in vertebrates but RyR3 is likely sensitive to NDL PCBs because it is altered by known RyR modulators (Fessenden et al., 2000). Work addressing RyR3 based toxicity in general would further our understanding of NDL PCB risks.
Variable RyR to FKBP1 paralog interactions may contribute to differential RyR-based toxic outcomes because in mammals FKBP1A and FKBP1B display different affinities for RyR1 and RyR2, they may compete in regulation of RyR1 and RyR2 channel gating behavior (Galfré et al., 2012; Venturi et al., 2014), and this regulation may vary between mammalian species alone (Zissimopoulos et al., 2012). RyR3 also binds four FKBP1A or FKBP1B (Qi et al., 1998) but likely with decreased apparent affinity (Fessenden et al. 2000). We observed amino acid differences between the RyR SPRY1 and Handle domains in mammalian and killifish orthologs and paralogs (e.g. Figure 5 and Figure 7), specifically at regions thought to be important for FKBP1 interactions. These differences might contribute to potential toxicity differences between taxa. The RyRs in skeletal muscle of rainbow trout (Oncorhyncus mykiss) can bind up to four FKBP1As or FKBP1Bs, both 1A and 1B regulate skeletal muscle RyR function (Qi et al., 1998) but similar studies are limited for fish RyR2 and RyR3 proteins. Studies of the direct changes in RyR:FKBP1 paralog interactions in teleosts are necessary to better address paralog-specific NDL PCB toxicity.
Finally, we observed a limited occurrence of SNV in RyR and FKBP1 genes of killifish from PCB-laden NBH versus the reference population at SC, which is consistent with studies that suggest that RyR proteins are under strong purifying selection (McKay and Griswold, 2014). There were no non-synonymous SNVs observed in killifish RyR2 genes, and evolutionarily RyR2 is suggested to undergo the strongest purifying selection due to its essential role in cardiac and neuronal function. Consistent with this hypothesis, knockout of either RyR1 or RyR2 results in embryonic lethality in mammals (Takeshima et al., 1994; Takeshima et al., 1998). In contrast, RyR3 is thought to be under the least purifying selection (McKay and Griswold, 2014), and RyR3 knockout does not result in embryonic lethality in mammals (Matsuo et al., 2009). We found the largest number of non-synonymous SNVs in the killifish RyR3 sequences including a high frequency SNV in NBH killifish leading to missense mutation E1458D. Glutamic acid (E) and aspartic acid (D) are negatively charged, acidic amino acids that are considered to be conserved and the corresponding amino acid location in RyR3 of mammals, and other RyR teleost sequences, codes for D (e.g. Figure 7; O. cuniculus RyR3 D1553 or Figure 5 O. cuniculus RyR1 D1658). The SNV detected in NBH RyR3b was also observed in a recent scan of genome wide sequence variation in killifish populations from severely polluted locations compared to populations from specific reference sites (Reid et al., 2016). The E1458D SNV was present at approximately 56% in NBH but was also present at a reasonably high frequency in killifish from the Block Island (MA) reference site (40%) suggesting that this locus may not be under strong directional selection for pollution tolerance. Thus the physiological implication of this common variant on the function of NBH killifish RyR3b, specifically, is unknown.
Conclusion
Polychlorinated biphenyls remain one of the most common classes of environmental pollutants, and there are still questions regarding their impact of exposure on human and wildlife health. The Atlantic killifish is an important model for pollutants, including dioxin like PCBs, and the current work further establishes this species for use in understanding NDL PCB toxicity. Specifically, the phylogenetic and sequence based comparisons presented in the current work address genetic similarities and differences between the killifish, other teleosts, and mammals to provide a framework for comparable or divergent NDL PCB neurotoxic responses across taxa. Ryanodine receptors are involved in countless physiological pathways; genetic or pharmacological disruption of the receptor, and related Ca2+-signaling partners, contributes to diseases such as cardiac arrhythmias, malignant hypothermia or central core disease, and neurodegeneration (Berridge, 2012; Lanner, 2012). Deregulation of FKBP1 can contribute to RyR based-disease states (Bellinger et al., 2008), and the peptidyl-prolyl isomerase has a number of other cellular binding partners that may contribute to toxic outcomes in addition to RyR-based endpoints.
Supplementary Material
Highlights.
Teleosts have a and b parologs for each mammalian ryanodine receptor (RyR1, 2, 3)
Teleost have 2–4 FKBP1; proteins involved in NDL PCB toxicity through RyR
Sequence differences in RyR and FKBP1 may describe species sensitivity to NDL PCBs
Killifish from PCB polluted New Bedford Harbor have a missense mutation in RyR3b
Supports teleost as models for NDL PCB toxicity induced by the RyR
Acknowledgments
This research was supported by the KC Donnelly Research Externship made possible by the National Institute of Environmental Health Sciences’ Superfund Research Program (EBH) and the Superfund Research Programs at UC Davis (INP and EBH; P42ES004699) and Boston University (JJS, JVG, MEH, SIK; P42ES007381). Additional support was provided by the National Institute of Health (INP; R01 ES014901; and P01 AR052354) and by National Science Foundation collaborative research grants (MEH and SIK; DEB-1265282 and DEB-1120263). This research was also supported in part by an appointment (to BC) with the Postdoctoral Research Program at the U.S. Environmental Protection (US EPA) Office of Research and Development administered by the Oak Ridge Institute for Science and Education (ORISE) through Interagency Agreement No. DW92429801 between the U.S. Department of Energy and the US EPA. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the US EPA. The funding agencies were not involved in study design or performance or in the decision to publish the manuscript. The U.S. Government is authorized to produce and distribute reprints for governmental purposes notwithstanding any copyright notation that may appear hereon.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong PB, Child JS. Stages in the Normal Development of Fundulus heteroclitus. Biological Bulletin. 1965;128:143–168. [Google Scholar]
- Bellinger AM, Mongillo M, Marks AR. Stressed out: the skeletal muscle ryanodine receptor as a target of stress. The Journal of Clinical Investigation. 2008;118:445–453. doi: 10.1172/JCI34006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ. Cell SIgnaling Pathways. Cell Signaling Biology Module. 2012:2. [Google Scholar]
- Burnett KG, Bain LJ, Baldwin WS, Callard GV, Cohen S, Di Giulio RT, Evans DH, Gómez-Chiarri M, Hahn ME, Hoover CA, Karchner SI, Katoh F, MacLatchy DL, Marshall WS, Meyer JN, Nacci DE, Oleksiak MF, Rees BB, Singer TD, Stegeman JJ, Towle DW, Van Veld PA, Vogelbein WK, Whitehead A, Winn RN, Crawford DL. Fundulus as the premier teleost model in environmental biology: Opportunities for new insights using genomics. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics. 2007;2:257–286. doi: 10.1016/j.cbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connon RE, D’Abronzo LS, Hostetter NJ, Javidmehr A, Roby DD, Evans AF, Loge FJ, Werner I. Transcription Profiling in Environmental Diagnostics: Health Assessments in Columbia River Basin Steelhead (Oncorhynchus mykiss) Environmental Science & Technology. 2012;46:6081–6087. doi: 10.1021/es3005128. [DOI] [PubMed] [Google Scholar]
- Darbandi S, Franck JPC. A comparative study of ryanodine receptor (RyR) gene expression levels in a basal ray-finned fish, bichir (Polypterus ornatipinnis) and the derived euteleost zebrafish (Danio rerio) Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2009;154:443–448. doi: 10.1016/j.cbpb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Efremov RG, Leitner A, Aebersold R, Raunser S. Architecture and conformational switch mechanism of the ryanodine receptor. Nature. 2015;517:39–43. doi: 10.1038/nature13916. [DOI] [PubMed] [Google Scholar]
- Elnar AA, Diesel B, Desor F, Feidt C, Bouayed J, Kiemer AK, Soulimani R. Neurodevelopmental and behavioral toxicity via lactational exposure to the sum of six indicator non-dioxin-like-polychlorinated biphenyls (Σ6 NDL-PCBs) in mice. Toxicology. 2012;299:44–54. doi: 10.1016/j.tox.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Essin K, Gollasch M. Role of Ryanodine Receptor Subtypes in Initiation and Formation of Calcium Sparks in Arterial Smooth Muscle: Comparison with Striated Muscle. Journal of Biomedicine and Biotechnology. 2009:2009. doi: 10.1155/2009/135249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes ECA, Hendriks HS, van Kleef RG, Reniers A, Andersson PL, van den Berg M, Westerink RH. Activation and Potentiation of Human GABAA Receptors by Non-Dioxin-Like PCBs Depends on Chlorination Pattern. Toxicological Sciences. 2010;118:183–190. doi: 10.1093/toxsci/kfq257. [DOI] [PubMed] [Google Scholar]
- Fessenden JD, Wang Y, Moore RA, Chen SRW, Allen PD, Pessah IN. Divergent Functional Properties of Ryanodine Receptor Types 1 and 3 Expressed in a Myogenic Cell Line. Biophysical Journal. 2000;79:2509–2525. doi: 10.1016/S0006-3495(00)76492-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galfré E, Pitt SJ, Venturi E, Sitsapesan M, Zaccai NR, Tsaneva-Atanasova K, O'Neill S, Sitsapesan R. FKBP12 Activates the Cardiac Ryanodine Receptor Ca2+-Release Channel and Is Antagonised by FKBP12.6. PLOS ONE. 2012;7:e31956. doi: 10.1371/journal.pone.0031956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini G, Conti A, Mammarella S, Scrobogna M, Sorrentino V. The ryanodine receptor/calcium channel genes are widely and differentially expressed in murine brain and peripheral tissues. The Journal of Cell Biology. 1995;128:893–904. doi: 10.1083/jcb.128.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasauer SM, Neuhauss SC. Whole-genome duplication in teleost fishes and its evolutionary consequences. Molecular Genetics and Genomics. 2014;289:1045–1060. doi: 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- Holland-Fritsch EB, Pessah IN. Structure-activity relationship of non-coplanar polychlorinated biphenyls toward skeletal muscle ryanodine receptors in rainbow trout (Oncorhynchus mykiss) Aquatic Toxicology. 2013;140–141:204–212. doi: 10.1016/j.aquatox.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland-Fritsch EB, Stegeman JJ, Goldstone JV, Nacci DE, Champlin D, Jayaraman S, Connon RE, Pessah IN. Expression and function of ryanodine receptor related pathways in PCB tolerant Atlantic killifish (Fundulus heteroclitus) from New Bedford Harbor, MA, USA. Aquatic Toxicology. 2015;159:156–166. doi: 10.1016/j.aquatox.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EB, Feng W, Zheng J, Dong Y, Li X, Lehmler H-J, Pessah IN. An extended structure-activity relationship of non-dioxin-like PCBs evaluates and supports modeling predictions and identifies picomolar potency of PCB 202 towards ryanodine receptors. Toxicological Sciences. 2016 doi: 10.1093/toxsci/kfw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenet T, Froemke RC, Schreiner CE, Pessah IN, Merzenich MM. Perinatal exposure to a noncoplanar polychlorinated biphenyl alters tonotopy, receptive fields, and plasticity in rat primary auditory cortex. Proceedings of the National Academy of Sciences. 2007;104:7646–7651. doi: 10.1073/pnas.0701944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JL, McKinney R, Lake CA, Osterman FA, Heltshe J. Comparisons of patterns of polychlorinated biphenyl congeners in water, sediment, and indigenous organisms from New Bedford Harbor, Massachusetts. Archives of Environmental Contamination and Toxicology. 1995;29:207–220. [Google Scholar]
- Lanner J. Ryanodine Receptor Physiology and Its Role in Disease. In: Islam MS, editor. Calcium Signaling. Springer; Netherlands: 2012. pp. 217–234. [DOI] [PubMed] [Google Scholar]
- Lesiak A, Zhu M, Chen H, Appleyard SM, Impey S, Lein PJ, Wayman GA. The Environmental Neurotoxicant PCB 95 Promotes Synaptogenesis via Ryanodine Receptor-Dependent miR132 Upregulation. The Journal of Neuroscience. 2014;34:717–725. doi: 10.1523/JNEUROSCI.2884-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing, S The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonky E, Reihman J, Darvill T, Mather J, Daly H. Neonatal behavioral assessment scale performance in humans influenced by maternal consumption of environmentally contaminated Lake Ontario fish. Journal of Great Lakes Research. 1996;22:198–212. [Google Scholar]
- Matsuo N, Tanda K, Nakanishi K, Yamasaki N, Toyama K, Takao K, Takeshima H, Miyakawa T. Comprehensive behavioral phenotyping of ryanodine receptor type3 (RyR3) knockout mice: Decreased social contact duration in two social interaction tests. Frontiers in behavioral neuroscience. 2009;3:3. doi: 10.3389/neuro.08.003.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay PB, Griswold CK. A comparative study indicates both positive and purifying selection within ryanodine receptor (RyR) genes, as well as correlated evolution. Journal of Experimental Zoology Part A: Ecological Genetics and Physiology. 2014;321:151–163. doi: 10.1002/jez.1845. [DOI] [PubMed] [Google Scholar]
- Murayama T, Kurebayashi N. Two ryanodine receptor isoforms in nonmammalian vertebrate skeletal muscle: Possible roles in excitation-contraction coupling and other processes. Progress in Biophysics and Molecular biology. 2011;105:134–144. doi: 10.1016/j.pbiomolbio.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Nacci DE, Champlin D, Coiro L, McKinney R, Jayaraman S. Predicting the occurrence of genetic adaptation to dioxinlike compounds in populations of the estuarine fish Fundulus heteroclitus. Environmental Toxicology and Chemistry. 2002;21:1525–1532. [PubMed] [Google Scholar]
- Nacci DE, Champlin D, Jayaraman S. Adaptation of the estuarine fish Fundulus heteroclitus (Atlantic killifish) to polychlorinated biphenyls (PCBs) Estuaries and Coasts. 2010;33:853–864. [Google Scholar]
- O'Brien J, Valdivia HH, Block BA. Physiological differences between the alpha and beta ryanodine receptors of fish skeletal muscle. Biophysical Journal. 1995;68:471–482. doi: 10.1016/S0006-3495(95)80208-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perni S, Marsden KC, Escobar M, Hollingworth S, Baylor SM, Franzini-Armstrong C. Structural and functional properties of ryanodine receptor type 3 in zebrafish tail muscle. The Journal of general physiology. 2015;145:173–184. doi: 10.1085/jgp.201411303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessah Cherednichenko G, Lein PJ. Minding the Calcium Store: Ryanodine Receptor Activation as a Convergent Mechanism of PCB Toxicity. Pharmacology & Therapeutics. 2010;125:260–285. doi: 10.1016/j.pharmthera.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessah IN, Hansen LG, Albertson TE, Garner CE, Ta TA, Do Z, Kim KH, Wong PW. Structure-Activity Relationship for Noncoplanar Polychlorinated Biphenyl Congeners toward the Ryanodine Receptor-Ca2+ Channel Complex Type 1 (RyR1) Chemical Research in Toxicology. 2006;19:92–101. doi: 10.1021/tx050196m. [DOI] [PubMed] [Google Scholar]
- Pessah IN, Lehmler H-J, Robertson LW, Perez CF, Cabrales E, Bose DD, Feng W. Enantiomeric Specificity of (−)−2,2′,3,3′,6,6′-Hexachlorobiphenyl toward Ryanodine Receptor Types 1 and 2. Chemical Research in Toxicology. 2009;22:201–207. doi: 10.1021/tx800328u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Ogunbunmi EM, Freund EA, Timerman AP, Fleischer S. FK-binding Protein Is Associated with the Ryanodine Receptor of Skeletal Muscle in Vertebrate Animals. Journal of Biological Chemistry. 1998;273:34813–34819. doi: 10.1074/jbc.273.52.34813. [DOI] [PubMed] [Google Scholar]
- Reid NM, Jackson CE, Gilbert D, Minx P, Montague MJ, Hampton TH, Helfrich LW, King BL, Nacci DE, Aluru N. The landscape of extreme genomic variation in the highly adaptable Atlantic killifish. Genome Biology and Evolution. 2017;9:659–676. doi: 10.1093/gbe/evx023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid NM, Proestou DA, Clark BW, Warren WC, Colbourne JK, Shaw JR, Karchner SI, Hahn ME, Nacci D, Oleksiak MF. The genomic landscape of rapid repeated evolutionary adaptation to toxic pollution in wild fish. Science. 2016;354:1305–1308. doi: 10.1126/science.aah4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roegge CS, Morris JR, Villareal S, Wang VC, Powers BE, Klintsova AY, Greenough WT, Pessah IN, Schantz SL. Purkinje cell and cerebellar effects following developmental exposure to PCBs and/or MeHg. Neurotoxicology and Teratology. 2006;28:74–85. doi: 10.1016/j.ntt.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Schantz S, Seo B, Wong P, Pessah I. Long-term effects of developmental exposure to 2, 2', 3, 5', 6-pentachlorobiphenyl (PCB 95) on locomotor activity, spatial learning and memory and brain ryanodine binding. Neurotoxicology. 1997;18:457. [PubMed] [Google Scholar]
- Somarelli JA, Herrera RJ. Evolution of the 12 kDa FK506-binding protein gene. Biology of the Cell. 2007;99:311–321. doi: 10.1042/BC20060125. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Komazaki S, Hirose K, Nishi M, Noda T, Iino M. Embryonic lethality and abnormal cardiac myocytes in mice lacking ryanodine receptor type 2. The EMBO journal. 1998;17:3309–3316. doi: 10.1093/emboj/17.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Takekura H, Nishi M, Kuno J, Minowa O, Takano H, Noda T. Excitation-contraction uncoupling and muscular degeneration in mice lacking functional skeletal muscle ryanodine-receptor gene. 1994 doi: 10.1038/369556a0. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002:3. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi E, Galfré E, O’Brien F, Pitt Samantha J, Bellamy S, Sessions Richard B, Sitsapesan R. FKBP12.6 Activates RyR1: Investigating the Amino Acid Residues Critical for Channel Modulation. Biophysical Journal. 2014;106:824–833. doi: 10.1016/j.bpj.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Bose DD, Yang D, Lesiak A, Bruun D, Impey S, Ledoux V, Pessah IN, Lein PJ. PCB-95 Modulates the Calcium-Dependent Signaling Pathway Responsible for Activity-Dependent Dendritic Growth. Environmental Health Perspectives. 2012a:120. doi: 10.1289/ehp.1104833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Yang D, Bose DD, Lesiak A, Ledoux V, Bruun D, Pessah IN, Lein PJ. PCB-95 Promotes Dendritic Growth via Ryanodine Receptor–Dependent Mechanisms. Environmental Health Perspectives. 2012b:120. doi: 10.1289/ehp.1104832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigestrand MB, Stenberg M, Walaas SI, Fonnum F, Andersson PL. Non-dioxin-like PCBs inhibit [3H]WIN-35,428 binding to the dopamine transporter: A structure-activity relationship study. Neurotoxicology. 2013;39:18–24. doi: 10.1016/j.neuro.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Wong PW, Pessah IN. Ortho-substituted polychlorinated biphenyls alter calcium regulation by a ryanodine receptor-mediated mechanism: structural specificity toward skeletal- and cardiac-type microsomal calcium release channels. Molecular Pharmacology. 1996;49:740–751. [PubMed] [Google Scholar]
- Wu HH, Brennan C, Ashworth R. Ryanodine receptors, a family of intracellular calcium ion channels, are expressed throughout early vertebrate development. BMC research notes. 2011;4:541. doi: 10.1186/1756-0500-4-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Bai X-c, Yan C, Wu J, Li Z, Xie T, Peng W, Yin C-c, Li X, Scheres SH. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2015;517:50–55. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Kim KH, Phimister A, Bachstetter AD, Ward TR, Stackman RW, Mervis RF, Wisniewski AB, Klein SL, Kodavanti PRS. Developmental exposure to polychlorinated biphenyls interferes with experience-dependent dendritic plasticity and ryanodine receptor expression in weanling rats. Environmental Health Perspectives. 2009;117:426. doi: 10.1289/ehp.11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuchi Z, Yuen SMWK, Lau K, Underhill AQ, Cornea RL, Fessenden JD, Van Petegem F. Crystal structures of ryanodine receptor SPRY1 and tandem-repeat domains reveal a critical FKBP12 binding determinant. Nature communications. 2015:6. doi: 10.1038/ncomms8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalk R, Clarke OB, Des Georges A, Grassucci RA, Reiken S, Mancia F, Hendrickson WA, Frank J, Marks AR. Structure of a mammalian ryanodine receptor. Nature. 2015;517:44–49. doi: 10.1038/nature13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalk R, Lehnart SE, Marks AR. Modulation of the ryanodine receptor and intracellular calcium. Annual Review of Biochemistry. 2007;76:367–385. doi: 10.1146/annurev.biochem.76.053105.094237. [DOI] [PubMed] [Google Scholar]
- Zissimopoulos S, Seifan S, Maxwell C, Williams AJ, Lai FA. Disparities in the association of the ryanodine receptor and the FK506-binding proteins in mammalian heart. Journal of Cell Science. 2012;125:1759–1769. doi: 10.1242/jcs.098012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.