Abstract
Elizabethkingia anophelis has become an emerging infection in humans. Recent research has shown that previous reports of E. meningoseptica infections might in fact be caused by E. anophelis. We aimed to investigate the genomic features, phylogenetic relationships, and comparative genomics of this emerging pathogen. Elizabethkingia anophelis strain EM361-97 was isolated from the blood of a cancer patient in Taiwan. The total length of the draft genome was 4,084,052 bp. The whole-genome analysis identified the presence of a number of antibiotic resistance genes, which corresponded with the antibiotic susceptibility phenotype of this strain. Based on the average nucleotide identity, the phylogenetic analysis revealed that E. anophelis EM361-97 was a sister group to E. anophelis FMS-007, which was isolated from a patient with T-cell non-Hodgkin’s lymphoma in China. Knowledge of the genomic characteristics and comparative genomics of E. anophelis will provide researchers and clinicians with important information to understand this emerging microorganism.
Introduction
Elizabethkingia is a genus of aerobic, nonfermenting, nonmotile, catalase-positive, oxidase-positive, indole-positive, and gram-negative bacilli that are usually distributed in soil and water environments1–3. Genus Elizabethkingia has been reported to cause human infection since Elizabeth O. King’s original work in 19594. However, this genus had rarely been responsible for infections in humans before. These microorganisms have been recently reported to cause life-threatening infections in immunocompromised patients, such as pneumonia, bacteraemia, meningitis, and neutropenic fever1–7.
Among genus Elizabethkingia, E. meningoseptica, previously known as Chryseobacterium meningosepticum, is the most well-known species that causes opportunistic infection in humans2,3. In contrast, little is known about E. anophelis. Elizabethkingia anophelis was first isolated from the midgut of a mosquito, Anopheles gambiae, in 20118 and has caused several outbreaks of infections in Africa7,9, Singapore10, Hong Kong11, and the USA5,6,12. The Centers for Disease Control and Prevention of the USA reported two outbreaks of infections caused by E. anophelis in the Midwest. A total of 63 patients in Wisconsin were confirmed to have E. anophelis infection between November 1, 2015 and April 12, 2017, and this outbreak caused 19 deaths13. Another cluster of 10 patients with E. anophelis infection was reported in Illinois, and six of the patients died of this infection5. Pulsed-field gel electrophoresis and whole-genome sequencing revealed that the strains of E. anophelis in these two outbreaks were genetically different6. However, recent research has shown that E. anophelis was frequently misidentified as E. meningoseptica, and previous reports of E. meningoseptica infections might in fact be caused by E. anophelis 9–12.
We previously published the draft whole-genome sequence of E. anophelis strain EM361-97 isolated in Taiwan (GenBank accession number, LWDS00000000.1)14. The whole-genome sequence could provide insights into the characteristics of the putative virulence factors, pathogenesis, and drug resistance of microorganisms. Comparison of genomes among different strains can be used in the analyses of phylogenetic relationships and epidemiological features. However, there has been little research investigating the genomic characteristics, global epidemiology, and genomic diversity of E. anophelis. In this study, we analysed the genomic features of E. anophelis strain EM361-97. We also compared the genomics and investigated the phylogenetic relationships with other strains of E. anophelis from other world regions.
Materials and Methods
Ethics and experimental biosafety statements
This study was approved by the Institutional Review Board of E-Da Hospital (EMRP-105-134). The need for patient’s informed consent was waived by the Institutional Review Board of E-Da Hospital as the retrospective analysis of anonymously clinical data posed no more than minimal risk of harm to subjects and involved no procedures for which written consent was normally required outside of the research context. The experiments in this study were approved by the Institutional Biosafety Committee of E-Da Hospital. All experiments were performed in accordance with relevant guidelines and regulations.
Isolate of E. anophelis
Elizabethkingia anophelis strain EM361-97 was isolated from the blood of a 46-year-old male patient with advanced nasopharyngeal carcinoma and lung cancer. During admission, the patient suffered from pneumonia, respiratory failure, and profound shock. He initially received empirical antibiotics with levofloxacin. Unfortunately, the patient died several days after this infection. One blood culture from the patient yielded a gram-negative bacillus that was initially identified as E. meningoseptica using API/ID32 GN (bioMérieux S.A., Marcy l’Etoile, France) by the clinical microbiology laboratory. This isolate was named strain EM361-97 and was stored at −80 °C as a glycerol stock for further experiments. We re-identified this isolate as E. anophelis using 16S ribosomal RNA (rRNA) gene sequencing as previously published15. The minimum inhibitory concentration (MIC) of this isolate was examined using the broth microdilution method. The susceptibilities were determined according to the interpretive standards for “other non-Enterobacteriaceae” as suggested by the Clinical and Laboratory Standards Institute (CLSI) guidelines16.
Whole-genome sequencing and genome annotation of E. anophelis EM361-97
The deoxyribonucleic acid (DNA) of this isolate was prepared using a Wizard Genomic DNA Purification Kit according to the manufacturer’s instructions (Promega, WI, USA). The genome was sequenced using an Illumina HiSeq. 2000 Sequencing Platform (Illumina, CA, USA). The short reads were assembled and optimized according to paired-end and overlap relationship via mapping reads to contig using SOAP de novo v. 2.0417. The assembled genome was then submitted to the NCBI Prokaryotic Genome Annotation Pipeline18 and the Rapid Annotations based on Subsystem Technology (RAST) Prokaryotic Genome Annotation Server (http://rast.nmpdr.org/) for gene function annotations19,20. The graphical map of the circular genome was generated using the CGView Server (http://stothard.afns.ualberta.ca/cgview_server/)21. The virulence factors of strain EM361-97 were analysed using the Virulence Factor Database (VFDB, http://www.mgc.ac.cn/VFs/)22,23. Antibiotic resistance genes were searched using the Antibiotic Resistance Genes Database BLAST Server (https://ardb.cbcb.umd.edu/)24, RAST Server19,20, and UniProtKB/Swiss-Prot database via OrthoVenn (http://probes.pw.usda.gov/OrthoVenn/)25.
Comparative genomic analysis
For comparison, the genome sequences of 34 available, nonduplicated, different genome sequences of E. anophelis in GenBank were downloaded from the National Center for Biotechnology Information (NCBI) genome sequence repository (https://www.ncbi.nlm.nih.gov/genome/). The genome-wide comparison and annotation of clusters of orthologous groups (COGs) were generated using the web server OrthoVenn25. The average nucleotide identity (ANI) values between two genome sequences were calculated using the original ANI function of OrthoANI26. The heat maps were generated using CIMminer (https://discover.nci.nih.gov/cimminer/). The in silico DNA-DNA hybridization (DDH)-analogous values between different strains were calculated using the Genome-to-Genome Distance Calculator (GGDC) 2 (http://ggdc.dsmz.de/distcalc2.php)27. A 70% similarity of in silico DDH value represents the cut-off value for species boundaries. The phylogenetic tree was constructed using CIMminer (https://discover.nci.nih.gov/cimminer/) based on ANI values.
Data Availability
The names of organisms, strains, biosample numbers, bioproject numbers, assembly numbers, isolated origins, and release dates of bacteria used in this study are shown in Supplementary Table S1.
Results and Discussion
General genome description of E. anophelis EM361-97
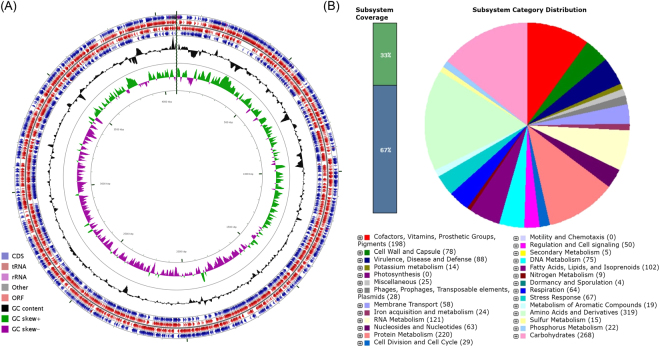
The statistics of assembly and annotation are shown in Table 1. The total length of the draft genome was 4,084,052 bp, with a mean GC content of 35.7%. This genome contained 3,774 genes that made up 87.9% of genome. The genomic features of E. anophelis EM361-97 are shown in Fig. 1. The number of tandem repeat sequence was 108. The assembly contained 18 scaffolds, 27 contigs, 3,743 coding sequences (CDSs), 53 minisatellite DNAs, 26 microsatellite DNAs, 51 transfer RNAs (tRNAs), and 15 rRNAs (Fig. 1A).
Table 1.
Assembly and annotation statistics.
| Total sequence length (bp) | 4,084,052 |
| Total assembly gap length (bp) | 6,353 |
| Gaps between scaffolds | 0 |
| Number of scaffolds | 18 |
| Scaffold N50 (bp) | 4,056,868 |
| Scaffold L50 | 1 |
| Number of contigs | 27 |
| Contig N50 (bp) | 1,882,703 |
| Contig L50 | 2 |
| Read length (bp) | 100 |
| Coverage depth | 95.95 |
| GC content (%) | 35.7 |
Figure 1.
Circular representation and subsystem category distribution of the genome of E. anophelis EM361-97. (A) Circles are numbered from 1 (the outermost circle) to 7 (the innermost circle). The outer four circles show the coding sequence (CDS), transfer ribonucleic acid (tRNA), ribosomal ribonucleic acid (rRNA), and open reading frame (ORF). The fifth circle represents the GC content (black). The sixth circle demonstrates the GC skew curve (positive GC skew, green; negative GC skew, violet). The genome position scaled in kb from base 1 is shown on the inner circle. (B) The genome of E. anophelis EM361-97 annotated using the Rapid Annotation System Technology (RAST) Server was classified into 356 subsystems and 27 categories. The green part in the bar chart at the leftmost position corresponds to the percentage of proteins included. The pie chart and the count of subsystem features in the right panel demonstrate the percentage distribution and category of the subsystems in E. anophelis EM361-97.
The genomic features of microorganism could be investigated according to the subsystem, a cluster of genes that function with a specific biological process or structural complex19,20. The genome of E. anophelis strain EM361-97 analysed by the RAST Server revealed 356 subsystems that could be classified into 27 categories (Fig. 1B). Among these, the “amino acid and derivatives” subsystem accounted for the largest number of 319 CDSs, followed by carbohydrate metabolism (268 CDSs), protein metabolism (220 CDSs), and RNA metabolism (121 CDSs). Regarding the 88 CDSs in the “virulence, disease, and defense” subsystem, 12 were related to invasion and intracellular resistance, and 76 were associated with resistance to antibiotics and toxic compounds. The high number of antibiotic resistance-associated CDSs suggests that E. anophelis EM361-97 might be resistant to multiple antibiotics.
Orthologous genes
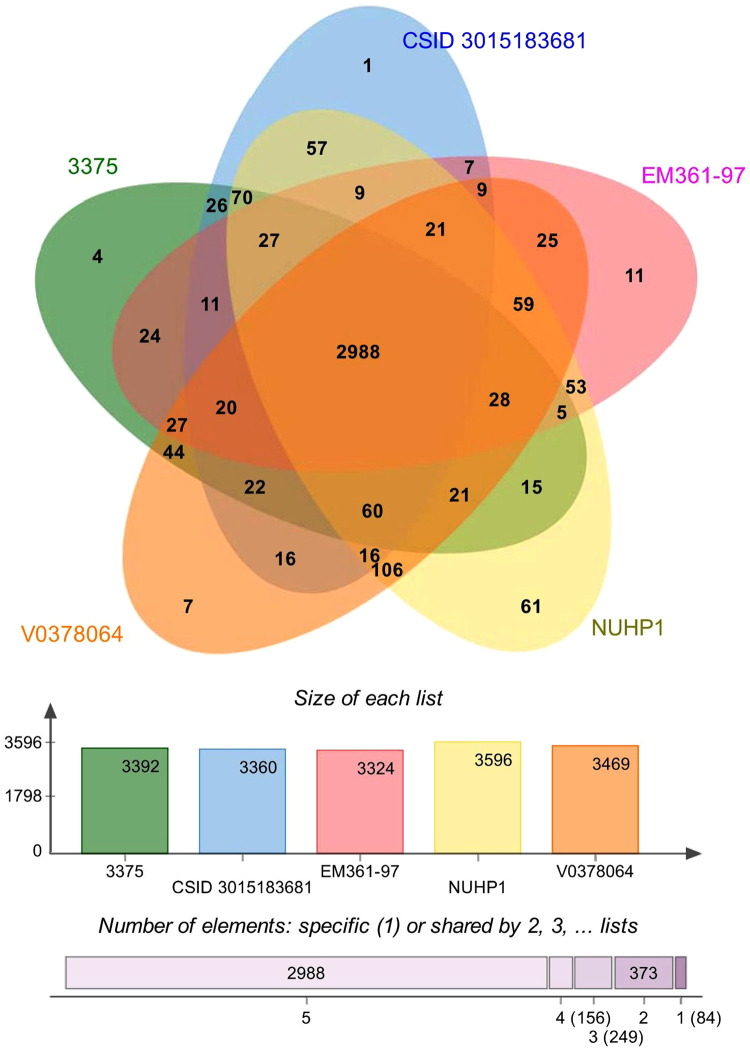
Orthologous genes are clusters of genes in different species that have evolved by vertical descent from a single ancestral gene. A genome-wide comparison of orthologous clusters in different isolates provides insight into the gene structure, gene function, and molecular evolution of genomes25. The COGs analysis of strain EM361-97 was compared with the other four genomes isolated from the USA (strains CSID_3015183681 and 3375), Africa (strain V0378064 [E18064]), and Singapore (strain NUHP1) (Fig. 2). The analysis shows that E. anophelis EM361-97 contained 3,611 proteins, 3,324 COGs, and 234 singletons. Among the 3,324 COGs in strain EM361-97, 2,988 COGs were shared by all five strains, and 11 COGs were only present in the strain EM361-97 genome. The unique COGs existing in EM361-97 involved genes functioning with transferase activity, cofactor binding, oxidoreductase activity, nucleotide binding, fatty acid elongation, and 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) activity. The representative meanings of these singular genes in E. anophelis EM361-97 are not clear. Further investigations to understand the features of these unique genes in E. anophelis EM361-97 are warranted.
Figure 2.
Proteome comparison among E. anophelis strains EM361-97 (origin, Taiwan), CSID_3015183681 (origin, USA), 3375 (origin, USA), V0378064 (E18064) (origin, Africa), and NUHP1 (origin, Singapore). The Venn diagram and bar chart represent the numbers of unique and shared orthologous genes of each strain.
Genomic comparison among Elizabethkingia species
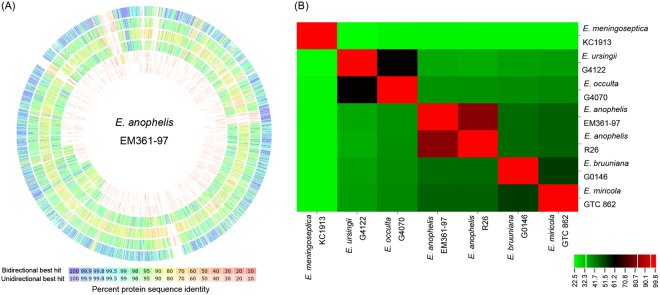
The genomic comparison among E. anophelis EM361-97, E. anophelis R26T, E. meningoseptica KC1913T, E. miricola GTC 862 T, E. bruuniana G0146T, E. ursingii G4122T, and E. occulta G4070T implemented using the RAST/SEED Server is shown in Fig. 3A. The genome of E. anophelis EM361-97 was apparently closer to that of E. anophelis R26T than the other Elizabethkingia species. The evolutionary relatedness among these strains was measured by in silico DDH-analogous values (Fig. 3B). The DDH value between E. anophelis EM361-97 and E. anophelis R26T was 82%. In contrast, the DDH value between E. anophelis EM361-97 and E. meningoseptica KC1913T was only 24.2%.
Figure 3.
Genomic comparison among Elizabethkingia species. (A) The genome of E. anophelis EM361-97 (center) compared to E. anophelis R26T (the outermost circle; ring 1), E. bruuniana G0146T (ring 2), E. meningoseptica KC1913T (ring 3), E. miricola GTC 862 T (ring 4), E. occulta G4070T (ring 5), and E. ursingii G4122T (the innermost circle; ring 6). The genome of E. anophelis EM361-97 was highly similar to the type strain of E. anophelis R26T. (B) The in silico DNA-DNA-hybridization (DDH) values between different strains calculated using the Genome-to-Genome Distance Calculator. The DDH value between E. anophelis EM361-97 and E. anophelis R26T was 82%. Elizabethkingia meningoseptica KC1913T demonstrated a relatively large phylogenetic distance from other strains of Elizabethkingia.
Genus Elizabethkingia previously comprised four species, namely E. meningoseptica, E. miricola, E. anophelis, and E. endophytica 28. However, the strain of E. endophytica was re-identified as an additional strain of E. anophelis based on in silico DDH of whole-genome sequencing (77% DDH value with regard to E. anophelis strain R26T)29. Recently, Nicholson et al.30 proposed three novel Elizabethkingia species, Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Our study showed that strain EM361-97 belonged to E. anophelis, with a DDH value of 82% between E. anophelis EM361-97 and the type strain of E. anophelis R26T. In addition, Elizabethkingia meningoseptica KC1913T demonstrated a relatively large phylogenetic distance from other strains of Elizabethkingia. These findings are consistent with the previous report of the taxonomic classification in genus Elizabethkingia 30.
Whole-genome phylogenetic analysis of E. anophelis
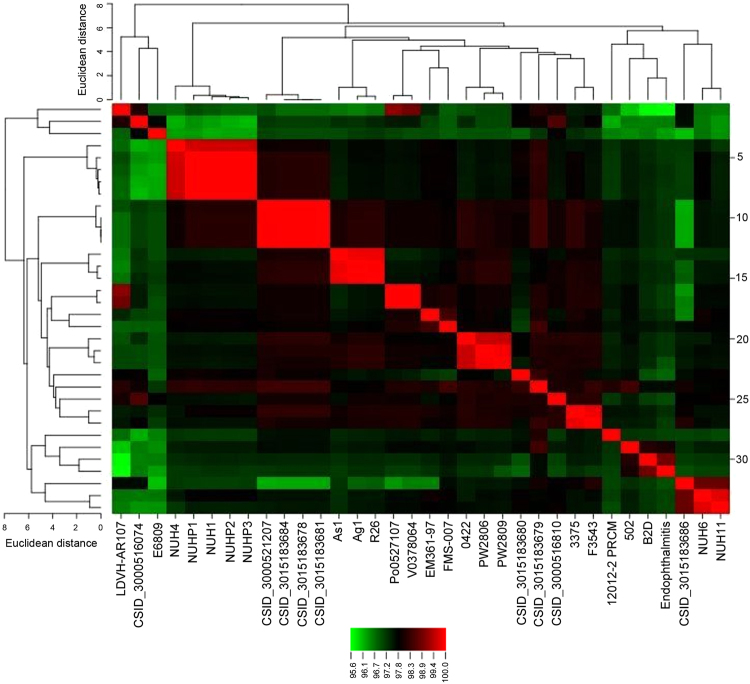
The phylogeny of the 34 available strains of E. anophelis based on ANI is shown in Fig. 4. The phylogenetic analysis revealed that E. anophelis EM361-97 was a sister group to E. anophelis FMS-007, which was isolated from a patient with T-cell non-Hodgkin’s lymphoma in China. The sister group of E. anophelis strains EM361-97 and FMS-007 was a clade sister of strains Po0527107 (E27017) and V0378064 (E18064) isolated from two neonates with meningitis in the Central African Republic7. The seven strains isolated from Singapore were divided into two clusters (NUHP1, NUHP2, NUHP3, NUH1, NUH4; and NUH6, NUH11). The 13 strains isolated from the USA clustered in four groups, and the four strains that caused the outbreak of E. anophelis infection in Wisconsin (strains CSID_3015183678, CSID_3015183681, CSID_3015183684, CSID_3000521207) were in the same clade.
Figure 4.
The phylogenetic tree of the 34 available strains of E. anophelis in GenBank based on average nucleotide identity (ANI) values. The phylogenetic analysis revealed that E. anophelis EM361-97 was a sister group to E. anophelis FMS-007, which was a clade sister of strains Po0527107 (E27017) and V0378064 (E18064) isolated in the Central African Republic.
Virulence factors
Elizabethkingia anophelis infections in humans have shown a mortality rate of 24% to 60%5,6,11, and this high mortality rate may be in part correlated with the virulence of this pathogen and also the preexisting conditions of the patients (e.g., old age, neonates, and immunosuppression). In this study, homologs of 25 virulence factors were identified in E. anophelis EM361-97 using VFDB22,23 (Supplementary Table S2). These virulence factors included products of the capsule, lipopolysaccharide, endopeptidase, lipid biosynthesis and metabolism, magnesium transport protein, macrophage infectivity, heat shock protein, catalase, peroxidase, superoxide dismutase, two-component regulatory system, and others.
According to the VFDB classification scheme, virulence factors are divided into offensive, defensive, nonspecific, and virulence-associated regulatory genes22. In our study, 13 of 25 pathogen-associated virulence factors homologs were identified to play offensive functional roles, eight were associated with defensive functions, three were nonspecific virulence factors, and one was related to regulation of virulence-associated genes. In strains Po0527107 (E27017) and V0378064 (E18064), Breurec et al.7 identified several offensive virulence factors that were found in strain EM361-97, including clpC, kdtB, pilR, sodB, galE, bplC, katA, clpP, fleQ, and htpB. These virulence factors were also detected in the Wisconsin strains12.
Pathogenic genomes were identified to have more offensive virulence factors, such as toxin and type III/IV secretion systems, than non-pathogenic genomes. In contrast, defensive, nonspecific, and regulatory virulence factors, such as iron uptake, motility, and antiphagocytosis, were found more frequently in non-pathogenic genomes than in pathogenic genomes31. Ho Sui et al.32 carried out a large-scale study to analyse the virulence factors of multiple bacteria and found over-presentation of offensive virulence factors, such as type III/IV secretion systems or toxins, within genomic islands of invasive pathogens. The manifestation of many offensive virulence factors in E. anophelis suggests this microorganism may severely damage the host. However, this hypothesis lacks validity. More experiments are warranted to test the hypothesis of offensive virulence factors in E. anophelis.
Antimicrobial resistance and associated genes of E. anophelis EM361-97
The MIC and susceptibility of E. anophelis EM361-97 are shown in Table 2. This isolate was only susceptible to piperacillin-tazobactam and minocycline. The MIC of tigecycline was 2 mg/L. However, there are no interpretive criteria of the susceptibility for E. anophelis to tigecycline in the CLSI16 and European Committee on Antimicrobial Susceptibility Testing33.
Table 2.
The minimum inhibitory concentration, susceptibility, and genes associated with antibiotic resistance in E. anophelis EM361-97.
| Antibiotic Group† | Antibiotics | MIC | Interpretation* | Resistant Gene/Protein/Mechanism† |
|---|---|---|---|---|
| Penicillins | Piperacillin | 32 | I | β-lactamase (BRO-1, 2)Class A β-lactamaseGOB-1 β-lactamaseSubclass B3 metallo-β-lactamaseSubclass B1 metallo-β-lactamase |
| β-lactam/β-lactamase inhibitor combinations | Piperacillin-tazobactam | 16/4 | S | |
| Ticarcillin-clavulanic acid | >64/2 | R | ||
| Cephems | Ceftazidime | >16 | R | |
| Cefepime | 32 | R | ||
| Ceftriaxone | >32 | R | ||
| Monobactams | Aztreonam | >16 | R | |
| Carbapenems | Imipenem | >8 | R | Carbapenem antibiotics biosynthesis protein CarD |
| Meropenem | >8 | R | ||
| Aminoglycosides | Gentamicin | >8 | R | Resistance-nodulation-cell division transporter system Multidrug resistance efflux pumpAminoglycoside N-acetyltransferaseAPH(3’) family aminoglycoside O-phosphotransferaseelongation factor Tu |
| Tobramycin | >8 | R | ||
| Amikacin | >32 | R | ||
| Tetracyclines | Tetracycline | >8 | R | Tetracycline resistance protein TetXMajor facilitator superfamily transporterTetracycline efflux pumpNADP-requiring oxidoreductase |
| Minocycline | <1 | S | ||
| Tigecycline | 2 | — | ||
| Fluoroquinolones | Ciprofloxacin | >2 | R | DNA gyrase subunit A and subunit BTopoisomerase IV |
| Levofloxacin | >8 | R | ||
| Folate pathway inhibitors | Trimethoprim-sulfamethoxazole | >4/76 | R | Group A drug-insensitive dihydrofolate reductaseSulfonamide-resistant dihydropteroate synthase Sul1 |
| Macrolides | — | — | — | Macrolide export protein MacA, MacBErythromycin resistance ATP-binding protein MsrA |
| Vancomycin | — | — | — | Vancomycin B-type resistance protein VanW |
| Clindamycin | — | — | — | Lincomycin resistance protein |
| Chloramphenicol | — | — | — | Resistance-nodulation-cell division transporter system Multidrug resistance efflux pumpGroup B chloramphenicol acetyltransferase (xenobiotic acetyltransferase) |
MIC, minimum inhibitory concentration.
†Associated with multidrug resistance: membrane component of tripartite multidrug resistance system, multidrug resistance efflux pumps (CmeB, TolC, MATE family efflux pump, OML, AcrA, AcrB), outer membrane efflux protein BepC, multidrug resistance proteins (MdtA, MdtB, MdtC, MdtD, MdtE, MdtK, MdtL), multidrug resistance protein EmrK, multidrug export protein EmrA, multidrug resistance outer membrane protein MdtQ, ABC transporter, MFS transporter, transcription-repair coupling factor, acriflavin resistance protein, and isoleucine-tRNA ligase.
*Susceptibility was determined according to the interpretive standards for other non-Enterobacteriaceae of CLSI.
Little information is known about the antimicrobial susceptibility of E. anophelis. Han et al.34 reported the susceptibilities of 51 E. anophelis isolates from South Korea. The susceptibility rates to piperacillin-tazobactam, piperacillin, levofloxacin, ciprofloxacin, gentamicin, and trimethoprim-sulfamethoxazole were 92%, 82%, 29%, 22%, 22%, and 22%, respectively. All the isolates were resistant to ceftazidime and imipenem. However, the MICs of minocycline and tigecycline were not examined in that study34. Perrin et al.12 used the disk diffusion method to examine antimicrobial susceptibilities of 29 E. anophelis isolates in the Wisconsin outbreak. Most of these isolates were resistant to ceftazidime, imipenem, amikacin, tobramycin, gentamicin, but susceptible to cefepime, piperacillin, piperacillin-tazobactam, ciprofloxacin, and levofloxacin. Minocycline was also not tested in the study of Perrin et al.12. The antibiogram of isolates in the Wisconsin outbreak was different from that of isolates in Singapore by macrolides and isepamycin10,35.
Gene functions annotated using the RAST/SEED Server recognised 76 genes of E. anophelis EM361-97 that were related to antibiotic resistance, including 12 for β-lactamase resistance, one for vancomycin resistance (vanW), four for fluoroquinolone resistance (parC, parE, gyrA, gyrB), nine for the membrane component of the tripartite multidrug resistance system, and 16 for multidrug resistance efflux pumps (six CmeB, one TolC, two MATE family efflux pumps, five OML, and two AcrB) (Table 2). The protein function annotations based on UniProtKB/Swiss-Prot demonstrated a number of proteins that played the role of antibiotic resistance, including multidrug resistance proteins (MdtA, MdtB, MdtC, MdtD, MdtE, MdtK, MdtL), probable multidrug resistance protein EmrK, multidrug export protein EmrA, macrolide export protein MacA, macrolide export ATP-binding/permease protein MacB, multidrug resistance outer membrane protein MdtQ, outer membrane efflux protein BepC, carbapenem antibiotics biosynthesis protein CarD, β-lactamase (BRO-1, 2), multidrug efflux pump subunit AcA, lincomycin resistance protein, DNA gyrase subunit A and subunit B, erythromycin resistance ATP-binding protein MsrA, and vancomycin B-type resistance protein VanW (Table 2). A replacement of serine by isoleucine at position 83 of DNA gyrase subunit A (Ser83Ile; AGC → ATC) was identified in E. anophelis strain EM361-97. Perrin et al.12 also found the same mutation of DNA gyrase subunit A in the Wisconsin outbreak strain CSID_3000521792. These findings are in agreement with the resistance of these two strains to fluoroquinolones.
Conclusions
In this work, the genomic features of the E. anophelis strain EM361-97 were constructed and compared with the genomes of other Elizabethkingia strains. Functional studies of this pathogen are required to validate these findings.
Electronic supplementary material
Acknowledgements
This work was supported by grants EDPJ105082 from E-Da Hospital and MOST 105-2314-B-214-008 and 106-2314-B-214-009-MY2 from the Ministry of Science and Technology, Taiwan.
Author Contributions
All authors provided significant contributions, and all authors are in agreement regarding the content of the manuscript. Conception/design: Jiun-Nong Lin and Hsi-Hsun Lin; provision of study materials: Chung-Hsu Lai; collection and assembly of data: Jiun-Nong Lin, Chung-Hsu Lai, Chih-Hui Yang, and Yi-Han Huang; data analysis and interpretation: all authors; manuscript writing: Jiun-Nong Lin and Chih-Hui Yang; and final approval of the manuscript: all authors.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14841-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Henriques IS, et al. Prevalence and diversity of carbapenem-resistant bacteria in untreated drinking water in Portugal. Microb. Drug Resist. 2012;18:531–537. doi: 10.1089/mdr.2012.0029. [DOI] [PubMed] [Google Scholar]
- 2.da Silva PSL, Pereira GH. Elizabethkingia meningoseptica: Emergent bacteria causing pneumonia in a critically ill child. Pediatr. Int. 2013;55:231–234. doi: 10.1111/j.1442-200X.2012.03650.x. [DOI] [PubMed] [Google Scholar]
- 3.Jean SS, Lee WS, Chen FL, Ou TY, Hsueh PR. Elizabethkingia meningoseptica: an important emerging pathogen causing healthcare-associated infections. J. Hosp. Infect. 2014;86:244–249. doi: 10.1016/j.jhin.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 4.King EO. Studies on a group of previously unclassified bacteria associated with meningitis in infants. Am. J. Clin. Pathol. 1959;31:241–247. doi: 10.1093/ajcp/31.3.241. [DOI] [PubMed] [Google Scholar]
- 5.CDC. Recent Outbreaks, Elizabethkingia. https://www.cdc.gov/elizabethkingia/outbreaks/ (2016).
- 6.Navon L, et al. Notes from the Field: Investigation of Elizabethkingia anophelis cluster - Illinois, 2014–2016. MMWR. 2016;65:1380–1381. doi: 10.15585/mmwr.mm6548a6. [DOI] [PubMed] [Google Scholar]
- 7.Breurec S, et al. Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci. Rep. 2016;6:30379. doi: 10.1038/srep30379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kämpfer P, et al. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int. J. Syst. Evol. Microbiol. 2011;61:2670–2675. doi: 10.1099/ijs.0.026393-0. [DOI] [PubMed] [Google Scholar]
- 9.Frank T, et al. First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet. 2013;381:1876. doi: 10.1016/S0140-6736(13)60318-9. [DOI] [PubMed] [Google Scholar]
- 10.Teo J, et al. First case of E anophelis outbreak in an intensive-care unit. Lancet. 2013;382:855–856. doi: 10.1016/S0140-6736(13)61858-9. [DOI] [PubMed] [Google Scholar]
- 11.Lau SKP, et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci. Rep. 2016;6:26045. doi: 10.1038/srep26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrin A, et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat. Commun. 2017;8:15483. doi: 10.1038/ncomms15483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wisconsin Department of Health Services. Elizabethkingia. https://www.dhs.wisconsin.gov/disease/elizabethkingia.htm (2017).
- 14.Lin, J. N., Yang, C. H., Lai, C. H., Huang, Y. H. & Lin, H. H. Draft genome sequence of Elizabethkingia anophelis strain EM361-97 isolated from the blood of a cancer patient. Genome Announc. 4 (2016). [DOI] [PMC free article] [PubMed]
- 15.Hantsis-Zacharov E, Shakéd T, Senderovich Y, Halpern M. Chryseobacterium oranimense sp. nov., a psychrotolerant, proteolytic and lipolytic bacterium isolated from raw cow’s milk. Int. J. Syst. Evol. Microbiol. 2008;58:2635–2639. doi: 10.1099/ijs.0.65819-0. [DOI] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, M100-S26 (Clinical and Laboratory Standards Institute, 2016).
- 17.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinforma. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 18.Tatusova T, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:D325–328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 2016;44:D694–697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B, Pop M. ARDB–Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37:D443–447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Coleman-Derr D, Chen G, Gu YQ. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2015;43:W78–84. doi: 10.1093/nar/gkv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, I., Kim, Y. O., Park, S. C. & Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 10.1099/ijsem.0.000760 [Epub ahead of print] (2015). [DOI] [PubMed]
- 27.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kämpfer P, Busse HJ, McInroy JA, Glaeser SP. Elizabethkingia endophytica sp. nov., isolated from Zea mays and emended description of Elizabethkingia anophelis Kämpfer et al. 2011. Int. J. Syst. Evol. Microbiol. 2015;65:2187–2193. doi: 10.1099/ijs.0.000236. [DOI] [PubMed] [Google Scholar]
- 29.Doijad S, Ghosh H, Glaeser S, Kämpfer P, Chakraborty T. Taxonomic reassessment of the genus Elizabethkingia using whole-genome sequencing: Elizabethkingia endophytica Kämpfer et al. 2015 is a later subjective synonym of Elizabethkingia anophelis Kämpfer et al. 2011. Int. J. Syst. Evol. Microbiol. 2016;66:4555–4559. doi: 10.1099/ijsem.0.000821. [DOI] [PubMed] [Google Scholar]
- 30.Nicholson, A. C. et al. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie Van Leeuwenhoek [Epub ahead of print] (2017). [DOI] [PMC free article] [PubMed]
- 31.Che D, Hasan MS, Chen B. Identifying pathogenicity islands in bacterial pathogenomics using computational approaches. Pathogens. 2014;3:36–56. doi: 10.3390/pathogens3010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho Sui SJ, Fedynak A, Hsiao WWL, Langille MGI, Brinkman FSL. The association of virulence factors with genomic islands. PloS One. 2009;4:e8094. doi: 10.1371/journal.pone.0008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.EUCAST: Clinical breakpoints. http://www.eucast.org/clinical_breakpoints/ (2017).
- 34.Han MS, et al. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J. Clin. Microbiol. 2017;55:274–280. doi: 10.1128/JCM.01637-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teo J, et al. Comparative genomic analysis of malaria mosquito vector-associated novel pathogen Elizabethkingia anophelis. Genome Biol. Evol. 2014;6:1158–1165. doi: 10.1093/gbe/evu094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The names of organisms, strains, biosample numbers, bioproject numbers, assembly numbers, isolated origins, and release dates of bacteria used in this study are shown in Supplementary Table S1.