Abstract
A randomized controlled longitudinal field trial was undertaken to assess the effects of injectable ceftiofur crystalline-free acid (CCFA) versus in-feed chlortetracycline on the temporal dynamics of Salmonella enterica spp. enterica in feedlot cattle. Two replicates of 8 pens (total 176 steers) received one of 4 different regimens. All, or one, out of 11 steers were treated with CCFA on day 0 in 8 pens, with half of the pens later receiving three 5-day regimens of chlortetracycline from day 4 to day 20. Salmonella was isolated from faecal samples and antimicrobial susceptibility was analysed via microbroth dilution. Serotype was determined by whole-genome sequencing. On day 0, mean Salmonella prevalence was 75.0% and the vast majority of isolates were pansusceptible. Both antimicrobials reduced overall prevalence of Salmonella; however, these treatments increased the proportion of multi-drug resistant (MDR) Salmonella from day 4 through day 26, which was the last day of faecal collection. Only six Salmonella serotypes were detected. Salmonella serotype Reading isolates were extensively MDR, suggesting a strong association between serotype and resistance. Our study demonstrates that the selection pressures of a 3rd generation cephalosporin and chlortetracycline during the feeding period contribute to dynamic population shifts between antimicrobial susceptible and resistant Salmonella.
Introduction
Foodborne salmonellosis is estimated to cause more than 1.2 million illnesses annually in the United States, requiring 23,000 hospital admissions and resulting in 450 deaths1. Salmonellosis is usually self-limiting, and even severely affected patients generally recover in 5 to 7 days if given rehydration fluids. Antimicrobial treatment options for adults include ceftriaxone, a medically important third-generation cephalosporin, and fluoroquinolones. There are potential side effects of fluoroquinolones for paediatric patients, and hence the first choice of treatment is ceftriaxone2–4. Because fewer treatment options are available for antimicrobial-resistant Salmonella, infections with these strains are potentially more life threatening5. Use of antimicrobials can cause unintentional selection pressure for antimicrobial resistance in the gut microbiota of animals, and therefore can potentially lead to more severe cases of salmonellosis6–8.
The National Antimicrobial Resistance Monitoring System (NARMS) has previously reported an increase in ceftriaxone resistant Salmonella carrying bla CMY-2, a class C plasmid and chromosome encoded ampC gene, in human cases of salmonellosis9–11. The bla CMY-2 gene also confers resistance to ceftiofur, a third-generation cephalosporin approved for veterinary use, as well as ampicillin, amoxicillin-clavulanic acid, cephalothin, and cefoxitin9,10. Observed increases in ceftriaxone resistant Salmonella in humans may be due, at least in part, to the increased use of third-generation cephalosporins in food animals12,13. This is considered a high public health risk since ceftriaxone and ceftiofur belong to the same class of 3rd generation cephalosporins, which the World Health Organization (WHO) has classified as critically important for human medicine. A variety of risk management strategies have been employed to help maintain antimicrobial efficacy for human medicine and to reduce the spread of antimicrobial-resistant bacteria derived from food animals14. In 2008, the U.S. Food and Drug Administration (FDA) announced a plan to prohibit the extra-label use of all cephalosporins in food animals (with no exceptions); later, this was revoked due to concerns about overly broad restrictions and the potential for unintended negative consequences15. Following re-examination by the FDA, extra-label use of cephapirin, some extra-label uses for indications involving the same route of administration, dose, and duration, and the use in minor food-producing species were excluded from the 2012 prohibition on extra-label use of cephalosporins in food-producing animals15. The FDA also promoted judicious use of antimicrobials of importance to human medicine, by working to remove growth promotion labels as of January 1, 201716,17. The effects of such strategies on reducing human infections with resistant bacteria have yet to be determined18–20. In the current study, we investigated treatment strategies involving a 3rd generation cephalosporin and chlortetracycline in fed beef cattle and their effects on selecting for Salmonella enterica.
In the recent past, several studies have been conducted to investigate the selection of resistant E. coli and Salmonella with the use of ceftiofur in cattle, both in experimental and observational settings21–27. In one study, ceftiofur administration in beef cattle transiently increased ceftiofur-resistant E. coli; however, the bacterial population returned to the before-treatment level after 2 weeks22. Daniels and others showed that ceftiofur use in a dairy herd was not associated with the occurrence of ceftiofur-resistant Salmonella and E. coli 24. Singer and others reported that the therapeutic use of ceftiofur in dairy cattle opened the “window” to detect resistant E. coli, but it was not concluded that such use resulted in the emergence or amplification of resistant E. coli 23. Another dairy farm study reported that the ceftiofur use and E. coli with reduced susceptibility to ceftriaxone are associated at the herd level, but not at the individual cow level21. A 10-month long study by Schmidt et al. showed that the ceftiofur use in feedlot cattle did not increase the extended-spectrum cephalosporin resistant E. coli 26. These studies suggest that the therapeutic use of ceftiofur can transiently increase the detection of cephalosporin resistant E. coli; however, the bacterial population returns to susceptibility after a suitable washout period. Since E. coli and Salmonella belong to the same Enterobacteriaceae family, Salmonella seem likely to exhibit a similar response to ceftiofur as E. coli; however, it is not yet known since studies involving Salmonella require consistent presence. An observational study investigating varying levels of ceftiofur use and their association with resistant Salmonella isolated on swine farms showed that the barns with rare and common ceftiofur use had 4.1% and 6.0% recovery of Salmonella carrying the bla CMY gene, respectively, while only 0.15% recovery occurred in the barns with moderate uses of ceftiofur28. The results suggested that other factors, such as farm management or environmental factors may be more important than ceftiofur use.
Chlortetracycline (CTC) is a common feed additive used to treat and control bacterial pneumonia (bovine respiratory disease complex) in feedlot cattle; this is in addition to vaccination used to prevent respiratory disease and other injectable antimicrobials used for disease control purposes29. In previous work reported by Platt et al. from our group, CTC treatment paradoxically reduced the prevalence of ceftiofur resistant E. coli 30. However, contradictory results were found in a subsequent study by Kanwar et al. in which CTC treatment increased ceftiofur resistance, most likely due to co-selection27. In a longer term study, the effects of prophylactic use of CTC on antimicrobial-resistant E. coli in beef cattle were studied for 117 days by Agga et al.; their findings showed an increased level of tetracycline resistant E. coli on Day 5 post-treatment, but not on Days 27, 75, and 11731. Additionally, prevalence of cephalosporin-resistant E. coli remained same among CTC and control groups throughout the study period31. We have further investigated the antimicrobial resistance profiles of the Salmonella population dynamics in response to both ceftiofur and chlortetracycline administration in the very same cattle studied by Kanwar et al.27. Such a randomized controlled study has not previously been reported concerning antimicrobial resistant Salmonella enterica in feedlot cattle.
The current study is focused on the effects of ceftiofur crystalline-free acid (CCFA) and CTC on the prevalence of Salmonella in feedlot cattle, and on changes in the profile of antimicrobial susceptibilities of Salmonella resulting from the selective pressures of CTC and CCFA. Furthermore, we investigated and report on the temporal dynamics of the Salmonella population in response to the use of antimicrobials.
Methods
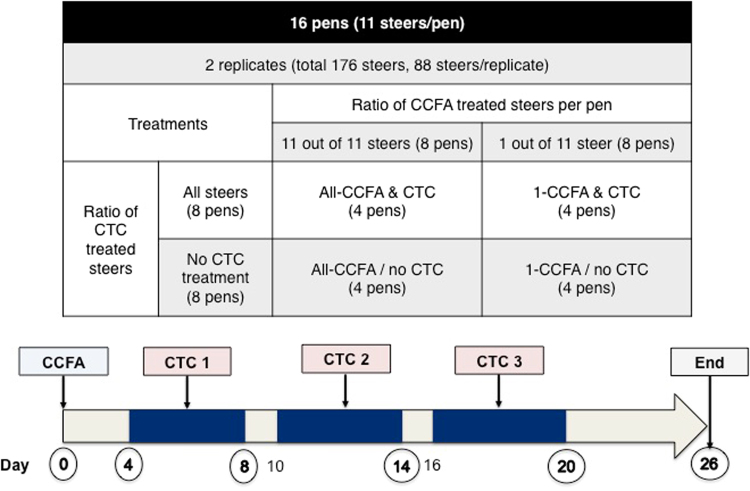
Experimental design
A randomized controlled longitudinal field trial was conducted in two sequential 26-day replicates in an experimental feedlot at West Texas A&M University in Canyon, Texas, USA. The first replicate began in early August and a second replicate began in the middle of September. The cattle were owned by a third party, and been purchased from a single operation in the far western United States. They were shipped directly to the experimental feed yard one month before the trial started. It is unknown if the cattle had previously been assembled at the source. They were yearling steers that were predominantly of the Angus breed and were fed diets typical of regional feedlots; that is, a flaked-corned based diet with added roughage, protein, vitamins and minerals. If any cattle became sick and required antimicrobial treatments, they were excluded from the study. In each replicate, 88 steers were assigned into 8 pens (n = 11 cattle) to distribute the body weights among the pens evenly in a two-by-two factorial design with four treatment regimens (Fig. 1), as described in a previous paper by our group27. Across both replicates, in 8 pens all 11 steers received 6.6 mg/kg of CCFA (Excede®, Zoetis Animal Health, Florham Park, NJ) treatment subcutaneously at the base of the ear (“All-CCFA & CTC” and “All-CCFA/no CTC” in Fig. 1; group metaphylaxis model), and in the remaining 8 pens, a single steer treated with CCFA on day 0 was co-housed (mixed) with 10 non-treated steers. Repeated within each of the two replicates, four of the pens receiving CCFA treatment group later received three 5-day pulses of 22 mg/kg CTC (Aureomycin®, chlortetracycline complex equivalent to 220.5 g/kg of chlortetracycline, Alpharma, Bridgewater, NJ). The CTC was top-dressed in feed with a one-day break between each 5-day pulse. The CTC feeding occurred during the period from day 4 until day 20 (“All-CCFA & CTC”, “1-CCFA & CTC”). The remaining 8 pens in each replicate did not receive CTC (“All-CCFA/no CTC” and “1-CCFA/no CTC”). Faecal samples were collected every other day per rectum as described previously27. These samples were mixed with glycerol at a 1:1 ratio and preserved at −80 °C. The animal experiments were approved by the Amarillo-Area Cooperative Research, Education, and Extension Triangle Animal Care and Use Committee (Protocol No. 2008-07), and by the Clinical Research Review Committee at Texas A&M University (CRRC # 09–35). All experiments were performed in accordance with institutional and United States Department of Agriculture (USDA) guidelines and regulations governing the oversight and conduct of experiments involving food producing animals in the U.S. Institutional biosafety committee approval # IBC 2014-043 permitted the microbiological laboratory experiments involving Salmonella enterica serotypes.
Figure 1.
Study design. Four pens were allocated to each treatment over two replicates. Samples were tested from the circled days on the arrow (0, 4, 8, 14, 20, and 26). Un-circled numbers represent the days when ‘pulsed’ CTC was added back into the feed bunks.
Salmonella isolation from faecal samples
A total of 1,040 faecal samples obtained on days 0, 4, 8, 14, 20, and 26 were tested for the recovery of Salmonella. The maximum effect of CCFA treatment for E. coli isolates being multidrug resistant was seen on day 4 in the previous study27, and we expected to observe a similar trend for Salmonella. Days 8, 14, and 20 were chosen because these were the last days of each of the 5-day CTC treatment pulses, and we predicted they would reflect the maximum effect on reducing Salmonella prevalence. The study was completed on day 26.
We isolated Salmonella by following a modified enrichment process as described previously32. Samples were thawed on ice and mixed thoroughly with a transfer pipette. In total, 500 mg of faeces were pre-enriched in 5 ml of tryptic soy broth (TSB) (Difco, Becton Dickinson, Franklin Lakes, NJ) for 2 hours at room temperature and then incubated 6 hours at 37 °C and then kept at 4 °C for 14 hours. A 1 ml aliquot of the enriched faeces in TSB was transferred into 9 ml of tetrathionate broth (Difco, Becton Dickinson) and incubated at 37 °C for 18 hours. After incubation, 100 μl of the tetrathionate broth culture was transferred into 10 ml of Rappaport-Vassiliadis R10 (RV) broth (Difco, Becton Dickinson) and incubated at 42 °C overnight. The next day, 50 μl of RV broth was spiral-plated onto Brilliant Green agar (BGA) (Difco, Becton Dickinson) using an Eddy Jet 2 spiral plater (Neutec Group Inc., Farmingdale, NY). Presumptive Salmonella isolates were plated on tryptic soy agar (TSA) with 5% sheep blood agar (RemelTM, Lenexa, KS) for isolation. Isolates were verified with Salmonella O-antiserum Poly A-I & Vi Factors 1–16, 19, 22–25, 34, Vi (Becton, Dickinson and Company, Franklin Lakes, NJ). Confirmed Salmonella isolates (n = 566) were preserved in cryobeads at −80 °C for further characterization.
Antimicrobial susceptibility testing
Minimum inhibitory concentrations (MIC) to 14 antimicrobials (9 antimicrobial classes) of Salmonella isolates were determined by the broth microdilution method using the Sensititre® system (TREK, Thermo Scientific Microbiology, Oakwood Village, OH). Tested antimicrobials were ampicillin (AMP), amoxicillin/clavulanic acid (AUG2), azithromycin (AZI), cefoxitin (FOX), ceftiofur (XNL), ceftriaxone (AXO), chloramphenicol (CHL), ciprofloxacin (CIP), gentamicin (GEN), nalidixic acid (NAL), streptomycin (STR), sulfisoxazole (FIS), tetracycline (TET), and trimethoprim/sulfamethoxazole (SXT). Briefly, isolates were plated onto TSA with 5% sheep blood agar and incubated at 37 °C for 18 hours. Then, 1 or 2 colonies were suspended in 4 ml of sterilized water adjusted to 0.5 McFarland standard and 50 µl of the culture suspension was transferred into 11 ml of Mueller-Hinton broth; thereafter, 50 ul of suspension was inoculated onto Gram Negative National Antimicrobial Resistance Monitoring System (NARMS) CMV3AGNF plates using the Sensititre® automated inoculation delivery system (TREK). Plates were incubated at 37 °C for 18 hours. Escherichia coli ATCC 25922, Escherichia coli ATCC 35218, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, and Enterococcus faecalis ATCC 29212 (American Type Culture Collection, Manassas, VA) were used as quality control strains for susceptibility testing. Plates were read on a Sensititre OptiRead™ (TREK, Thermo Scientific Microbiology). The results were interpreted as susceptible, intermediate, or resistant according to Clinical and Laboratory Standards Institute (CLSI) guidelines using SWIN software (TREK, Thermo Scientific Microbiology)33. When breakpoints were undetermined, we followed the breakpoints established by NARMS for Salmonella. Intermediate isolates were reclassified as being susceptible for further statistical analysis. Isolates resistant to three or more classes of antimicrobials were considered multidrug resistant (MDR) as defined by NARMS.
Salmonella DNA extraction for whole-genome sequencing
Salmonella DNA was isolated in a QIAcube HT robot using the QIAamp 96 DNA QIAcube HT Kit (Qiagen, Valencia, CA). A single Salmonella colony was suspended into 5 ml of TSB and incubated overnight at 37 °C. From the suspension culture, 1 ml was transferred into a 1.2 ml micro-collection tube and centrifuged at 4,000 rpm for 15 minutes at room temperature. After the supernatant was removed, the pellet was re-suspended in ATL buffer (Qiagen) mixed with reagent DX (Qiagen). One tube of small pathogen lysis beads (Qiagen) was mixed with the suspension, and disrupted with the Qiagen TissueLyser system (Qiagen) at 25 Hz, for 5 minutes. The tubes were briefly centrifuged and 40 μl of Proteinase K was added to each tube. The tubes were incubated at 56 °C for 1 hour at 900 rpm in a ThermoMixer (Eppendorf, Hauppauge, NY) followed by a heat shock for 10 minutes at 95 °C. The suspension was cooled to room temperature and 4 μl of RNAse A was added. The prepared samples were set in the QIAcube HT for DNA extraction using a modified protocol provided by Qiagen. The quality of the DNA was determined by the 260/280 ratios on the FLUOstar Omega Microplate Reader (BMG LABTECH, Cary, NC). The DNA quantity was measured with a Quant-iT™ Pico Green® ds DNA Assay kit (Thermo Fisher Scientific) and the DNA was stored at −20 °C until future use.
Whole-genome sequencing for serotyping and data analysis
To determine the serotypes of the Salmonella isolates, we conducted whole-genome sequencing (WGS) using the Illumina MiSeq platform (Illumina, San Diego, CA, USA) on 566 isolates (as described above). Libraries for 32 Salmonella DNA samples were multiplexed and pooled per sequence run using the Illumina Nextera XT kit and were run with the MiSeq Reagent Kit v3 with paired-end 2 × 300 bp reads (Illumina). Two Salmonella isolates were randomly chosen from each serotype determined by WGS and were sent to the National Veterinary Services Laboratories (NVSL), United States Department of Agriculture (USDA), for traditional serotyping to confirm the sequencing results.
Raw fastq files obtained from the forward and reverse reads were uploaded to the web-based database SeqSero to determine the serotypes from WGS data (http://www.denglab.info/SeqSero)34. Multi-Locus Sequence Type (MLST) of each of the Salmonella isolates was determined by the combination of 7 gene alleles (aroC, dnaN, hemD, hisD, purE, sucA and thrA) using the PubMLST database by the SRST2 pipeline in the Illumina® BaseSpace® Sequence Hub35,36.
Statistical Analysis
Descriptive Statistics
Proportion of Salmonella positive (binary outcome) faecal samples and the proportion of isolates resistant to greater than or equal to 3 classes of antimicrobials, or else pansusceptible by sampling day and by treatment group were cross-tabulated in Stata® version 12.1 (StataCorp LLC, College Station, TX). Crude associations between the serotypes, sampling days, and treatment groups were initially tested via the Likelihood-ratio based Chi-Square test or the Fisher’s exact test for rare combinations. Graphics for descriptive statistics were created on Tableau Desktop Professional Edition 10.3.2.
Multivariable analyses
Prevalence of Salmonella (binary) was modelled using a multilevel mixed-effects logistic regression model (-melogit- in Stata® ver. 15.1) considering replicate (2 replicates), pen (16 pens) and animal identifier (176 animals) as potential clustering variables. Following initial assessment of these three potential sources of overdispersion, all three variables were included as significant random effects in the final statistical model. A 3-way full factorial statistical model incorporating fixed effects for 1-CCFA (mixing) and CTC antimicrobial regimens and sampling day was built.
Final model for the prevalence of MDR Salmonella (binary) was similarly modelled using multilevel mixed-effects logistic regression (-melogit- in Stata® ver. 15.1). Replicate, pen, and animal identifier were considered as potential clustering variables. Following the initial assessment, replicate was instead included as a fixed effect in the final model, while repeated observations within animal identifier fell out of significance in the presence of pen effects. In addition to replicate, a full factorial statistical model incorporating the fixed effects for each of the CCFA and CTC antimicrobial regimens, sampling day, and 2-way interactions between day and antimicrobial regimens was built.
Marginal mean estimates from the final models were produced and graphical representations of the temporal dynamics were plotted.
Data Availability
The datasets generated during and/or analysed during the current study are not publicly available due to on-going sequencing analyses at the time of publication; however, they will become available from the corresponding author upon request.
Results
Descriptive statistics of Salmonella among faecal samples of feedlot cattle
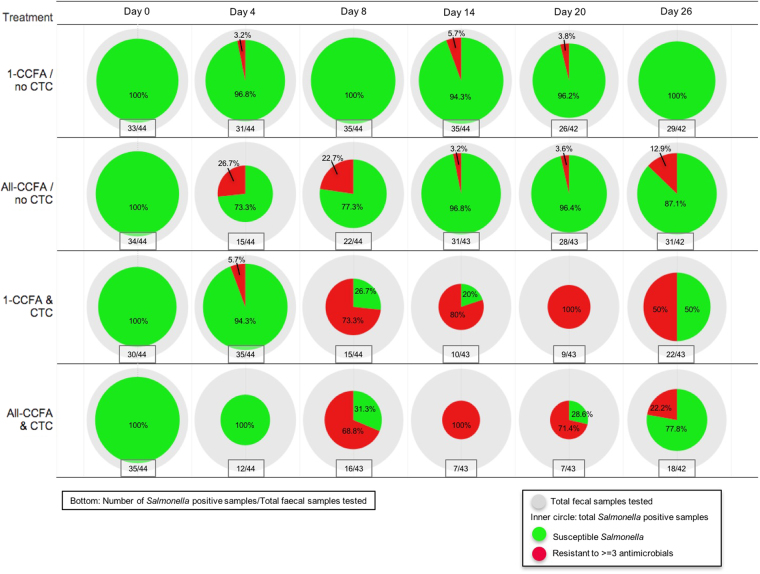
In total, Salmonella was recovered from 566 out of 1,040 faecal samples. The mean Salmonella prevalence before the antimicrobial treatments began (day 0) was 75.0% (95% CI: 67.9–81.2%), with 132 out of 176 samples testing positive. Among the two groups of all cattle treated with CCFA (All-CCFA/no CTC, All-CCFA & CTC), the prevalence of Salmonella was 34.1% (15 positive out of 44 samples) and 27.3% (12 positive out of 44 samples) on day 4, respectively. By day 14, the Salmonella prevalence in cattle receiving no subsequent CTC treatment (All-CCFA/no CTC; 31 positives out of 43 samples, 72.1%) was similar to that of those steers with least antimicrobial exposures (1-CCFA/no CTC; 35 positives out of 44 samples, 79.5%) as shown in Fig. 2 (second row). When CTC treatment followed CCFA treatment, the prevalence dropped even further to 16.3% by day 14 (7 out of 43 positive). By day 26, the prevalence in both CTC treatment groups (1-CCFA & CTC, All-CCFA & CTC) returned to 47.1% (40 out of 85 positive). By comparison, the overall mean prevalence in steers in those pens that received the least antimicrobial treatment (1-CCFA & no CTC) was estimated at 72.7% (95% CI: 66.8–78.0%) throughout the study period.
Figure 2.
Total number of isolates and proportion of MDR isolates per day and treatment. The outer circle (grey) reflects the number of faecal samples tested per pen each sampling day (n = 42 to 44). The size of the inner circle corresponds to the number of Salmonella isolates obtained each sampling day. The green piece of the inner circle represents pansusceptible Salmonella and the red piece represents resistance to 3 or more classes of antimicrobials (MDR). Total number of the Salmonella positive samples and total faecal samples tested are shown below each pie in the text box. Percentages of pansusceptible and MDR isolates are shown inside the circle.
Descriptive statistics on Salmonella isolates resistant to antimicrobials
The total number of Salmonella isolates and the percentage of MDR Salmonella (defined as resistant to ≥3 antimicrobial classes) for each sampling day by antimicrobial treatment groups are shown in an inner circle of the Fig. 2. Among the steers with the least exposure of antimicrobial treatment, the vast majority of isolates remained pansusceptible throughout the study period, although 3.2%, 5.7%, and 3.8% multidrug resistant Salmonella isolates were detected on days 4, 14 and 20, respectively (Fig. 2, first row). Among the group that all cattle treated with CCFA on day 0, 26.7% of Salmonella isolates were MDR on day 4 and 22.7% were MDR on day 8; however, MDR prevalence thereafter declined by day 14 (Fig. 2, second row). In the treatment group treated with CTC (1-CCFA & CTC) starting from day 4, pansusceptible isolates decreased and MDR isolates increased (73.3%) on day 8 and further increased to 80% and 100% on days 14 and 20, respectively (Fig. 2, third row). By the end of the study (day 26), 50% of the isolates in this latter group remained as MDR. Among animals sequentially receiving both CCFA and CTC treatment, the pattern of MDR dynamics resembled that of the treatment groups receiving CTC (Fig. 2, fourth row).
Mixed effects logistic regression model of Salmonella prevalence and MDR Salmonella prevalence
CCFA and CTC treatments were coded as binary variables, with 0 (no treatment) used as the referent category. Days were 0, 4, 8, 14, 20, and 26, with day 0 used as the referent. CCFA treatment effects were highly significant on days 4 (p < 0.004) and 8 (p < 0.003). CTC treatment significantly decreased the prevalence of Salmonella on days 8 (p < 0.004), 14 (p < 0.001), and 20 (p < 0.024) when compared to day 0 (Fig. 3).
Figure 3.
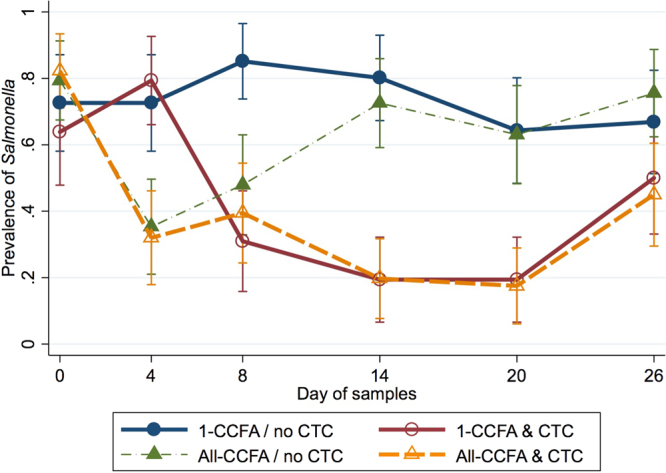
Modelled marginal mean prevalence of Salmonella by day and treatment. Modelled marginal mean prevalence of Salmonella and 95% confidence intervals in cattle faecal samples by sample day and treatment group. Solid lines represent treatment groups in which a single CCFA treated steer was mixed within an otherwise untreated group while dashed lines represent groups in which all steers received CCFA treatment.
The prevalence of MDR Salmonella was modelled similar to that of overall Salmonella prevalence, as above, except that day 4 was used as a referent category. This is because MDR isolates first appeared on day 4, yielding unstable model parameters for day 0. Interaction between CTC treatment and day was responsible for an increase of MDR Salmonella from day 8 (p < 0.008) to day 26 (p < 0.041) (Fig. 4). CCFA treatment on day 0 further increased MDR Salmonella probability on day 8 in the CCFA/CTC treated groups, compared with the group treated only with CTC. A slight increase of MDR Salmonella was seen on day 4 in the CCFA-only treated group, compared to the referent group (1-CCFA & no CTC); however, this was not significant (P > 0.05).
Figure 4.
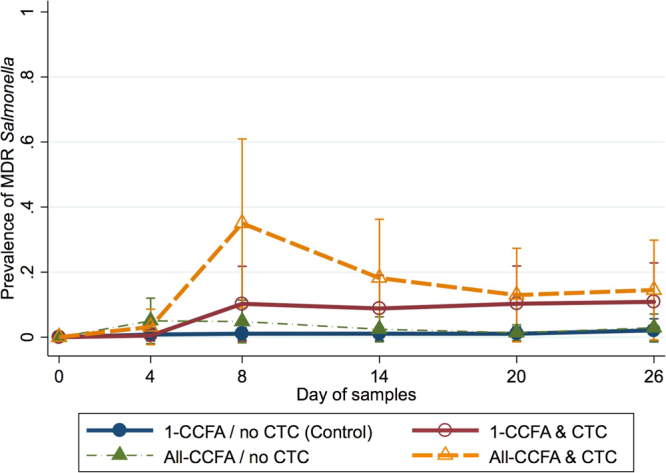
Modelled marginal mean prevalence of MDR Salmonella by day and treatment. Modelled marginal mean prevalence of Salmonella and 95% confidence intervals in cattle faecal samples by sample day and treatment group. Solid lines represent treatment groups in which a single CCFA treated steer was mixed within an otherwise untreated group while dashed lines represent groups in which all steers received CCFA treatment.
Identification of serotypes and MLST
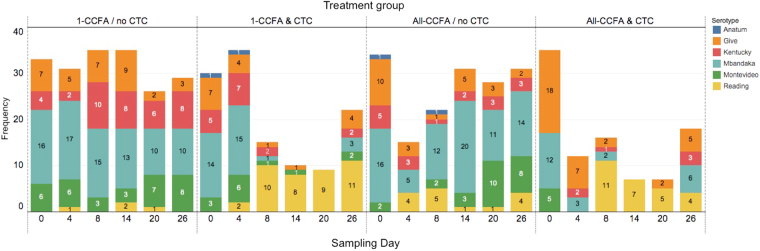
Salmonella serotypes were identified by whole-genome sequencing using the SeqSero pipeline34, which determines the serotype based on the sequence of the O-antigen gene cluster and H1 and H2 antigens. Six serotypes were detected; the most common serotype was Salmonella Mbandaka (38.0%), followed by S. Give (19.1%), S. Kentucky (13.6%), S. Reading (15.2%), S. Montevideo (13.4%), and S. Anatum (0.7%) as shown in Table 1. Serotypes and MLST were matching completely and no diverse serotypes were detected from any single MLST (Table 1). Salmonella Reading was detected starting on day 4, and increased greatly by day 8 in the All-CCFA/no-CTC groups, and especially in the 1-CCFA & CTC and All-CCFA & CTC treatment groups (Fig. 5) where it extended well past day 8. Five serotypes, except S. Reading, were identified on day 0. In the 1-CCFA/no CTC group, the composition of serotypes remained similar throughout the study. The S. Reading isolate detected on day 4 in the 1-CCFA/no CTC group was derived from the single steer that received CCFA and was mixed in the group comprised of 10 other non-CCFA-treated cattle; however, the isolates from day 14 and 20 were from the steers that were not treated with antimicrobials. In the All-CCFA/no CTC group, susceptible serotypes decreased on day 4 while S. Reading was increasingly present. In the 1-CCFA & CTC group, all isolates were S. Reading on day 20; however, susceptible serotypes had recovered by day 26. The All-CCFA & CTC group exhibited less serotype diversity. S. Montevideo was detected only on day 0 in this group. More S. Reading were isolated in the treatment groups that received CTC. Serotype distribution was similar between replicates 1 and 2, except that S. Anatum was identified only in replicate 1 (Fig. S1). S. Mbandaka were in the majority and S. Reading was the least detected serotype in the 1-CCFA & no CTC group (Table 2). S. Give were in the majority followed by S. Reading in All-CCFA & CTC group. In 1-CCFA & CTC group, 33.1% were S. Reading.
Table 1.
Multi-Locus Sequence Type (MLST) and serotypes of the Salmonella isolates.
| Serotype | Number of isolates | Antigenic profile from Seqsero | MLST | Gene Allele | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O | H1 (fliC) | H2 (fljB) | Predicted profile | aroC | dnaN | hemD | hisD | purE | sucA | thrA | |||
| Anatum | 4 (0.7%) | O-3,10 | e,h | 1,6 | 3,10:e,h:1,6 | 64 | 10 | 14 | 15 | 31 | 25 | 20 | 33 |
| Kentucky | 77 (13.6%) | O-8 | i | z6 | 8:i:z6 | 198 | 76 | 14 | 3 | 77 | 64 | 64 | 67 |
| Mbandaka | 215 (38.0%) | O-7 | z10 | e,n,z15 | 7:z10:e,n,z15 | 413 | 15 | 70 | 93 | 78 | 113 | 6 | 68 |
| Montevideo | 76 (13.4%) | O-7 | g,m,s | — | 7:g,m,s:- | 138 | 11 | 41 | 55 | 42 | 34 | 58 | 4 |
| Give | 108 (19.1%) | O-3,10 | l,v | 1,7 | 3,10:l,v:1,7 | 654 | 111 | 47 | 49 | 42 | 12 | 58 | 3 |
| Reading | 86 (15.2%) | O-4 | e,h | 1,5 | 4:e,h:1,5 | 1628 | 46 | 60 | 10 | 9 | 6 | 12 | 17 |
Figure 5.
Number of Salmonella enterica serotypes by treatment group and sample day. Six serotypes (Anatum, Give, Kentucky, Mbandaka, Montevideo, and Reading) were found among tested faecal samples. Numbers shown in the bars are the number of isolates for each serotype by treatment group across both trial replicates.
Table 2.
Proportion of Salmonella serotype and AMR phenotype by treatment group.
| Serotype | Serotype per Treatment Group | AMR Phenotype | |||
|---|---|---|---|---|---|
| 1-CCFA/no CTC | 1-CCFA & CTC | All-CCFA/no CTC | All-CCFA & CTC | ||
| Anatum (n = 4) | 0.0% (0) | 1.7% (2) | 1.2% (2) | 0.0% (0) | Pansusceptible: 100.0% (4) |
| Give (n = 108) | 17.5% (33) | 14.0% (17) | 14.9% (24) | 35.8% (34) | Pansusceptible: 100.0% (108) |
| Kentucky (n = 77) | 20.1% (38) | 13.2% (16) | 10.6% (17) | 6.3% (6) | Pansusceptible: 98.7% (76), STR-SUL-TET: 1.3% (1) |
| Mbandaka (n = 215) | 42.9% (81) | 27.3% (33) | 48.4% (78) | 24.2% (23) | Pansusceptible: 100.0% (215) |
| Montevideo (n = 76) | 17.5% (33) | 10.7% (13) | 15.5% (25) | 5.3% (5) | Pansusceptible: 100.0% (76) |
| Reading (n = 86) | 2.1% (4) | 33.1% (40) | 9.3% (15) | 28.4% (27) | AMP-AUG2-AXO-FOX-TIO-SOX-STR-CHL-TET: 96.5% (83), AMP-AUG2-AXO-FOX-TIO-STR-CHL-TET: 3.5% (3) |
| Total (n = 566) | 100.0% (189) | 100.0% (121) | 100.0% (161) | 100.0% (95) | |
For quality control purposes, 2 isolates randomly chosen from each of the 5 serotypes identified by whole-genome sequencing (WGS) were sent to the National Veterinary Services Laboratories for traditional serotyping. All traditional serotyping results matched exactly with sequence based serotyping.
Associations of phenotypic antimicrobial resistance profile and serotypes of Salmonella
All isolates (n = 566) were tested against a standard NARMS panel including 14 antimicrobials arising from 9 classes. Serotype and resistant phenotype were significantly associated (p < 0.05). All of the S. Anatum, S. Give, S. Mbandaka and S. Montevideo were phenotypically pansusceptible. Nearly all (96.5%) of the S. Reading isolates had at least the penta-resistant profile, ACSSuT (resistant to ampicillin, chloramphenicol, streptomycin, sulphonamides, and tetracycline), with additional resistance to 3rd generation cephalosporins ceftiofur and ceftriaxone (Table 2). The other 3.5% isolates detected from 1-CCFA & CTC group were not resistant to sulphonamides. The resistance profiles were AMP-AUG2-AXO-FOX-TIO-STR-CHL-TET for 8 antimicrobials and AMP-AUG2-AXO-FOX-TIO-FIS-STR-CHL-TET for 9 antimicrobials. One S. Kentucky isolate from 1-CCFA & CTC group was resistant to 3 antimicrobials: STR, FIS, and TET (Table 2). The proportion of S. Reading was significantly higher (p < 0.05) in the CTC treatment group than in the groups without CTC treatment. No resistance to azithromycin, ciprofloxacin, gentamicin, or nalidixic acid was detected.
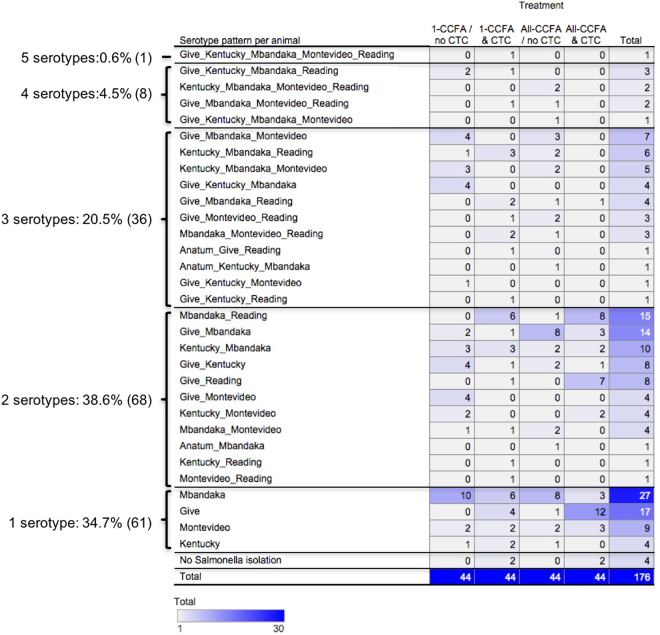
Serotype pattern per individual animal
We aggregated the Salmonella serotypes that were detected in the same animal from different sampling days to identify the serotype patterns (Fig. 6). In one steer, 5 serotypes (S. Give, Kentucky, Mbandaka, Montevideo, and Reading) were detected across the different days. In 4.5% (8 steers), 4 serotypes with different serotype patterns were detected. Three serotypes were detected from 20.5% (36 steers) and 2 serotypes were detected from 38.6% (68 steers). A single serotype was detected from 34.7% (61 steers). No steers were detected with S. Reading alone; that is, it was always detected with other serotypes across different days. Four steers were not detected as harbouring Salmonella throughout the study period.
Figure 6.
Total number of animals harboring each serotype pattern across all sampling days. Serotype patterns were clustered per treatment group across from different sampling days. Darker blue indicates more animals with specific serotype patterns; conversely, lighter blue shaded serotype combinations had fewer animals.
Discussion
This randomized controlled longitudinal field trial has clearly demonstrated that the use of antimicrobials shifts the antimicrobial resistance status of the Salmonella population by selecting for MDR Salmonella and against the pansusceptible Salmonella serotypes that are highly prevalent in beef cattle in this region of the USA. Since resistance phenotype and Salmonella serotype are so strongly associated, this effectively means that antimicrobial use selects for serotype. Chlortetracycline treatment alone, or CTC with prior treatment with CCFA, decreased the overall prevalence of Salmonella; further, each of these treatments increased the proportion of MDR Salmonella that remained. Ceftiofur treatment by itself did reduce the prevalence of Salmonella; however, such a reduction was relatively transient. The sole MDR phenotypic pattern was virtually identical among the S. Reading (Table 2). Six Salmonella serotypes, which have previously been reported in feedlot cattle and the feedlot environment, were detected in the faecal flora of these cattle37–40. As a point of reference, Salmonella Anatum, Montevideo, and Kentucky accounted for 50.4% of the serotypes detected in the National Animal Health Monitoring Systems (NAHMS) Feedlot 2011 study39.
We found a high prevalence of Salmonella in the faeces of feedlot cattle prior to antimicrobial treatment. Previous attempts to determine the dynamics of Salmonella in experimental studies were likely not successful due to a low prevalence of overall and resistant Salmonella in the study population; thus requiring a vast number of animals on trial to have enough power for the analyses22,25. Even in large-scale observational studies the prevalence can vary a great deal, illustrating that in many U.S. locations the prevalence is very low. The NAHMS Feedlot 2011 study had earlier shown that the overall pen-level prevalence of Salmonella in feedlot cattle across the United States was 35.6%, with 9.1% sample (cattle)-level prevalence39. Our study successfully showed that Salmonella population changes in response to antimicrobial treatments, in part due to the high prevalence at the beginning of the study (i.e., 70 + %). Some of the factors explaining the high initial prevalence in our study include geography (southern United States) and season (August-October). Several studies have shown a seasonal variation in Salmonella prevalence, such that sampling during the summer months yields the highest percentage of Salmonella positive samples41–44. One study from North Dakota found 62.2% prevalence of Salmonella in the tested herd during spring months, but only one serotype S. Typhimurium var. Copenhagen was isolated from the entire herd45. It is possible that if our study was conducted during colder weather, such an obvious change in the Salmonella population might not be observed. Regardless, the population dynamics of Salmonella associated with the antimicrobial treatments may be similar. The Salmonella prevalence and serotype populations differ by region, country, and ambient environment, which imply that the resistance phenotype will vary at the same time. Therefore, our results may not be necessarily be generalized to feedlots on other continents, as one example.
One-time CCFA treatment (as is typically used to control respiratory disease in beef feeder cattle) on day 0 reduced the overall prevalence of Salmonella in faeces. CCFA is not labelled for the control of Salmonella in cattle; however, its broad-spectrum nature, presence in multiple tissues, and effectiveness against Gram-negative bacteria appears to result in a temporal decrease of Salmonella prevalence immediately following treatment. The dose of CCFA used in the current study was for the treatment and control of bovine respiratory diseases such that the serum concentration is maintained over the minimum inhibitory concentration (0.2μg/ml) for up 10 days following a single-dose administration. A previous study illustrated that an extra-label regimen of ceftiofur decreased the detection and quantity of Salmonella and effectively treated salmonellosis in neonatal calves46. In our study, CCFA treatment also increased the proportion of MDR Salmonella on days 4 through 8. Despite only receiving a single dose of CCFA, by the end of the study (day 26) 12.5% of isolated Salmonella remained multidrug resistant (Fig. 2). Few observational or experimental studies have explored the association between antimicrobial use (including ceftiofur) and the temporal dynamics of resistant E. coli and Salmonella in cattle and pigs22,24,26,28,30,47–50. With the follow-up of CTC treatment to earlier CCFA treatment, MDR isolates increased further to 75% of total Salmonella by day 14. We chose to explore day 14 in order to examine the status of antimicrobial resistant Salmonella in cattle faeces on the first day post-treatment that the animals were eligible to be sent to a slaughterhouse based on residue avoidance; that is, because the labelled slaughter withholding time of CCFA is 13 days. Our findings indicate that at the point at which this compliance requirement is met, MDR Salmonella prevalence persists far above the baseline starting values, a finding that has not been previously reported. However, we are well aware that such a scenario is highly unlikely in real feedlot settings; that is, CCFA treatment at the whole pen level is extremely unlikely to occur anytime close to when cattle are sent to slaughterhouse. Typically, fed cattle would instead be sent to slaughter at least 6 months post-arrival. To understand longer-term dynamics, additional studies would be needed to further investigate the effects of CCFA and CTC treatments over periods of many months.
While CTC has been used for several decades in animal agriculture for prevention and growth promotion purposes, the effects of therapeutic doses of CTC on the prevalence and resistance of Salmonella in cattle have not been studied extensively. CTC was added to the feed (as a top-dress) for 3 pulses of 5 continuous days, with a one-day interval in between. This regimen was designed to observe the maximum effect of CTC, to follow the product label in the U.S., and to be consistent with a previous study published by Platt et al.30. CTC treatment alone reduced Salmonella prevalence to the same levels as cattle first injected with CCFA and then subsequently treated with CTC. This further suggests that initial CCFA treatment did not have a significant long-term effect on Salmonella prevalence. However, it remains possible that the initial CCFA may have selected for MDR Salmonella, which will be discussed later. The effects of CTC given at therapeutic or subtherapeutic doses on pathogens including Salmonella have been reported in pigs51,52. Although not statistically significant, pulsed CTC feeding during the finishing period lowered Salmonella prevalence in pigs51. Wells et al. have shown that CTC supplemented in the diet reduced the prevalence of both Campylobacter and Shiga-toxin producing E. coli 52. Prevalence of Salmonella was at a negligible level in their study. A study by Agga et al. found that one-set of 5-day CTC in-feed treatments transiently increased tetracycline resistant E. coli concentration in the faecal swabs of feedlot cattle. Importantly, generic E. coli concentration in the CTC treatment group remained the same, thus indicating that tetracycline resistant E. coli effectively replaced the susceptible E. coli population31.
Previous work by our group illustrated an inexplicable paradoxical reduction of ceftiofur resistant E. coli via CTC treatment (despite mechanistic potential for co-location via tet(A) and bla cmy-2 genes housed on a single IncA/C plasmid in many cattle E. coli). We explored the potential that this might serve as a potential intervention to reduce ceftiofur resistance by instead favouring singly resistant E. coli strains harbouring the tet(B) gene on the bacterial chromosome27,30. In our current study, three pulsed (intermittent) CTC treatments displayed a stronger selection pressure on MDR Salmonella than CCFA treatment alone, which was completely the opposite of the previous published work exploring the effects of these same regimens in E. coli populations27. In that study, faecal E. coli from CCFA administrated pens showed more resistance to a full range of antimicrobials than in cattle from the pens administered only CTC (the latter effect was negligible)27. As mentioned, while Agga et al. showed that a 5 day single-pulsed CTC treatment increased tetracycline resistant E. coli in faecal swabs of beef cattle at 5 days-post-treatment, no differences in cephalosporin resistant E. coli were found31. It is possible that the resistant E. coli population were different between these two studies. Comparatively phenotypic resistance patterns and sequence type (or PFGE patterns) of E. coli in earlier studies of food animals are often diverse, while Salmonella are often either pansusceptible or else MDR (assuming they are present at all)26,27,38,53. Importantly, in Salmonella, drug resistance phenotypes and serotypes are much more highly associated than is seen across the more prevalent and diverse populations of E. coli 38,54,55. These differences in resistance patterns likely indicate that Salmonella and E. coli are not readily sharing their resistance elements in the cattle gut microbiome53; conversely, they also might not be sharing the same ecological niches in the varying cross-sections of intestinal regions56. One previous experimental study suggested that the use of ceftiofur in dairy cattle did not promote the transfer of bla CMY-2 coding plasmids among Salmonella and commensal E. coli in calves inoculated with plasmid-bearing bacteria and in dairy herds using ceftiofur24. However, another study found that the presence of an inflammatory condition in the gut could boost horizontal gene transfer between Salmonella and E. coli 57, something that is often lacking in studies involving healthy animals. Although limited to phenotypic resistance, our study reveals that Salmonella and E. coli isolated from the same gut microbiome did not share similar resistant patterns, either at baseline or during specified treatment periods and across treatment regimens27.
We detected 6 serotypes in our study population; in decreasing prevalence, these were Salmonella Mbandaka, Give, Kentucky, Reading, Montevideo, and Anatum. The serotypes detected in this study were consistent with those published in previous studies and isolated from the lymph nodes, hides, and faeces of feedlot and dairy cattle from the same region of Texas38,43,58–61. Salmonella serotypes often appear to adapt to specific animal hosts. Among human clinical isolates reported to the CDC, S. Montevideo was 10th among frequently reported serotypes in 201362. In the current study, S. Mbandaka (serogroup C1) was the dominant serotype followed by Give (serogroup E1). The dominance of certain serotypes has been observed in other studies in feedlot cattle as well60,61. However, it is possible that Salmonella enrichment via RV media may bias the detection towards serogroups C1, C2, and E63. We detected S. Reading, which belongs to serogroup B, but none of these were pansusceptible. It remains a possibility that pansusceptible S. Reading isolates were not detected in our study due to enrichment bias or else the resistant S. Reading population was stable in the gut microbiota but were not able to outcompete the susceptibles prior to antimicrobial pressure. Our study has illustrated that serotype and MDR may be strongly associated. Most (96.5%) S. Reading isolates had the ACSSuT resistance profile, while the other serotypes did not; this agrees with previous work that suggests that S. Reading is more likely to be of the ACSSuT resistance phenotype than other serotypes43,59. It remains unclear why certain serotypes are more highly resistant to antimicrobials, while others are not. However, all of these cited studies suggest that the antimicrobial susceptibility of the various Salmonella serotypes might have been determined not only by their genetic predispositions, but also by coexisting serotypes as well as environmental factors, such as antimicrobial selection pressure.
The first and second replicates of these cattle trials were conducted at the same feedlot with a one-month interval between the studies. Cattle were housed in porous-fenced pens through which contact could be made with cattle in adjacent pens. Treatments were randomly assigned to balance the contact potential across pens; however, sterility could not be maintained between pens in such a field setting as is standard in the beef feeding industry. The serotype distribution of the first and second replicates was similar throughout the study and no S. Reading was found on day 0, even in the second replicate (Supplementary Fig. S1). This latter point strongly suggests that MDR Salmonella transiently detected solely due to the antimicrobial pressure applied during this trial. All the steers that were detected with S. Reading on at least one sampling day also had other serotypes isolated during at least one other sampling day (Fig. 6). Since these serotypes were not detected at the same time point (i.e., since we limited the number of colonies we assayed from each plate), it does not necessarily mean that the steers were infected with 2 or more serotypes at once; however, this point does support the idea that S. Reading likely co-resided with other serotypes under “normal” non-antimicrobial selection pressure conditions, and was more readily detected after the antimicrobial treatments since other pansusceptible Salmonella serotypes were selected against by antimicrobials.
The steer isolated with 5 different serotypes on 5 different days illustrates the complexity of Salmonella colonization in cattle; this is consistent with another study in dairy cattle that showed multiple serotype colonizations per animal38,64,65. In agreement with our study, confirmation of Salmonella infection or colonization sometimes requires multiple testing methods, enrichment, and sequential sampling time-points. Four steers in our study were negative for Salmonella throughout the whole study period (26-days); even after this, these can be interpreted still as being of rare or intermittent shedding of Salmonella or as being truly negative for Salmonella colonization.
One limitation of this study is that we terminated the study on day 26 and thus did not track the cattle to the age at which they would be sent to the slaughterhouse, and whose faecal, hide, and lymph node Salmonella populations would represent a greater potential threat to public health. The feedlot is the final production stage for beef cattle, and thus represents a more proximate source of MDR Salmonella contamination at the slaughterhouse than occurs earlier in the beef production cycle66. In the future, we plan to conduct a similarly designed randomized controlled study with follow-up extending to the slaughterhouse; that is, approximately 150 days post-treatment. We currently hypothesize that any temporal antimicrobial treatment effects will wane over such an extended period of time, yielding few, if any, differences among the treatment groups by the time animals are ready for slaughter as shown in previous studies in E. coli population26,31.
In conclusion, we found that CTC and CCFA treatments dramatically decreased overall Salmonella prevalence in feedlot cattle; however, multi-drug resistant Salmonella strains were selected (or, detected) instead of those that comprised the original antimicrobial-susceptible Salmonella population. Salmonella serotypes and antimicrobial resistance phenotypes displayed strong associations and suggest that specific serotypes may be more likely to carry MDR genes. Although it was transient, this study indicates that the use of CCFA and CTC exerts a strong selection pressure on the Salmonella populations in the gut of feedlot cattle; therefore, judicious use of antimicrobials is necessary for beef cattle feeding operations to aid in preventing increased levels of MDR Salmonella being present in cattle destined for slaughter.
Electronic supplementary material
Acknowledgements
We acknowledge funding support by USDA-NIFA-AFRI (formerly CSREES; grant numbers 2008-35201-30235 and 2008-35201-04682) entitled: “Novel pre-harvest interventions to protect antimicrobials of critical importance in human and veterinary medicine” for the cattle field trial, while the subsequent phenotypic analysis was funded by USDA-NIFA-NIFSI (grant number 2010-51110-21083) entitled: “Practical interventions to effectively manage antibiotic resistance in beef and dairy cattle systems: a fully integrated approach.” Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the U.S. Department of Agriculture. Whole-genome sequencing of Salmonella isolates was funded by the National Cattlemen’s Beef Association, a contractor to the Beef Checkoff. Technical assistance in the laboratory was provided by Ms. Roberta Pugh and Ms. Anisa Wakil. Dr. Barbara Gastel and Dr. Colin Young provided important feedback on earlier versions of this manuscript.
Author Contributions
H.M.S., B.N., G.H.L. conceived and designed the study. N.O., H.M.S., K.N.N., B.N., J.V., and G.H.L. performed the experiments. N.O., H.M.S., K.N.N., S.D.L., H.B., and G.H.L., analysed and interpreted data. N.O., K.N.N., and H.M.S. drafted the manuscript. All authors revised manuscript for critically important intellectual content and approved the final version to be published.
Competing Interests
Dr. Guy H. Loneragan has provided scientific consulting services to Zoetis Inc. (manufacturer of the CCFA product). This does not alter the authors’ adherence to all the journal policies on sharing data and materials.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14751-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Scallan E, et al. Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin TY, et al. Short-term ceftriaxone therapy for treatment of severe non-typhoidal Salmonella enterocolitis. Acta paediatrica. 2003;92:537–540. doi: 10.1111/j.1651-2227.2003.tb02502.x. [DOI] [PubMed] [Google Scholar]
- 3.Grady RW. Systemic quinolone antibiotics in children: a review of the use and safety. Expert opinion on drug safety. 2005;4:623–630. doi: 10.1517/14740338.4.4.623. [DOI] [PubMed] [Google Scholar]
- 4.Soo-Han Choi EYK. and Yae-Jean Kim. Systemic use of fuoroquinolone in children. Korean J Pediatr. 2013;56:196–201. doi: 10.3345/kjp.2013.56.5.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krueger AL, et al. Clinical outcomes of nalidixic acid, ceftriaxone, and multidrug-resistant nontyphoidal salmonella infections compared with pansusceptible infections in FoodNet sites, 2006–2008. Foodborne Pathog Dis. 2014;11:335–341. doi: 10.1089/fpd.2013.1642. [DOI] [PubMed] [Google Scholar]
- 6.Holmberg SD, Osterholm MT, Senger KA, Cohen ML. Drug-resistant Salmonella from animals fed antimicrobials. The New England journal of medicine. 1984;311:617–622. doi: 10.1056/NEJM198409063111001. [DOI] [PubMed] [Google Scholar]
- 7.Cohen ML, Tauxe RV. Drug-resistant Salmonella in the United States: an epidemiologic perspective. Science. 1986;234:964–969. doi: 10.1126/science.3535069. [DOI] [PubMed] [Google Scholar]
- 8.Travers K, Barza M. Morbidity of infections caused by antimicrobial-resistant bacteria. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2002;34(Suppl 3):S131–134. doi: 10.1086/340251. [DOI] [PubMed] [Google Scholar]
- 9.Whichard JM, et al. Human Salmonella and concurrent decreased susceptibility to quinolones and extended-spectrum cephalosporins. Emerg Infect Dis. 2007;13:1681–1688. doi: 10.3201/eid1311.061438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacoby, G. A. AmpC beta-lactamases. Clin Microbiol Rev22, 161–182, Table of Contents, 10.1128/CMR.00036-08 (2009). [DOI] [PMC free article] [PubMed]
- 11.Hamilton RD, Hulsebus HJ, Akbar S, Gray JT. Increased resistance to multiple antimicrobials and altered resistance gene expression in CMY-2-positive Salmonella enterica following a simulated patient treatment with ceftriaxone. Appl Environ Microbiol. 2012;78:8062–8066. doi: 10.1128/AEM.02077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fey PD, et al. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. The New England journal of medicine. 2000;342:1242–1249. doi: 10.1056/NEJM200004273421703. [DOI] [PubMed] [Google Scholar]
- 13.Doyle MP, Loneragan GH, Scott HM, Singer RS. Antimicrobial Resistance: Challenges and Perspectives. Comprehensive Reviews in Food Science and Food Safety. 2013;12:234–248. doi: 10.1111/1541-4337.12008. [DOI] [Google Scholar]
- 14.Maron DF, Smith TJ, Nachman KE. Restrictions on antimicrobial use in food animal production: an international regulatory and economic survey. Global Health. 2013;9:48. doi: 10.1186/1744-8603-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.FDA. New Animal Drugs; Cephalosporin Drugs; Extralabel Animal Drug Use; Oreder of Prohibition. https://www.gpo.gov/fdsys/pkg/FR-2012-01-06/pdf/2012-35.pdf (2012).
- 16.FDA. Guidance for Industry #213 New Animal Drugs and New Animal Drug Combination Products Administered in or on Medicated Feed or Drinking Water of Food-Producting Animals: Recommendations for Drug Sponsors for Voluntarily Aligning Product Use Conditions with GFI #209. https://www.fda.gov/downloads/AnimalVeterinary/GuidanceComplianceEnforcement/GuidanceforIndustry/UCM299624.pdf (2013).
- 17.FDA. TheJudicious Use of Medically Important Antimicrobial Drugs in Food-Producing Animals, Guidance for Industry #209 (2012).
- 18.Phillips I. Withdrawal of growth-promoting antibiotics in Europe and its effects in relation to human health. International journal of antimicrobial agents. 2007;30:101–107. doi: 10.1016/j.ijantimicag.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 19.Carol Cogliani, H. G. and Christina Greko. Restricting Antimicrobial Use in Food Animals: Lessons fromEurope. Microbe5 (2011).
- 20.Aarestrup F. Sustainable farming: Get pigs off antibiotics. Nature. 2012;486:465–466. doi: 10.1038/486465a. [DOI] [PubMed] [Google Scholar]
- 21.Tragesser LA, Wittum TE, Funk JA, Winokur PL, Rajala-Schultz PJ. Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am J Vet Res. 2006;67:1696–1700. doi: 10.2460/ajvr.67.10.1696. [DOI] [PubMed] [Google Scholar]
- 22.Lowrance, T. C. et al. Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. American Journal of Veterinary Research68, 501–507, 10.2460/ajvr.68.5.501 [doi] (2007). [DOI] [PubMed]
- 23.Singer RS, Patterson SK, Wallace RL. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl Environ Microbiol. 2008;74:6956–6962. doi: 10.1128/AEM.01241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniels JB, et al. Role of ceftiofur in selection and dissemination of blaCMY-2-mediated cephalosporin resistance in Salmonella enterica and commensal Escherichia coli isolates from cattle. Appl Environ Microbiol. 2009;75:3648–3655. doi: 10.1128/AEM.02435-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heider LC, et al. Identification of Escherichia coli and Salmonella enterica organisms with reduced susceptibility to ceftriaxone from fecal samples of cows in dairy herds. Am J Vet Res. 2009;70:389–393. doi: 10.2460/ajvr.70.3.389. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt JW, Griffin D, Kuehn LA, Brichta-Harhay DM. Influence of therapeutic ceftiofur treatments of feedlot cattle on fecal and hide prevalences of commensal Escherichia coli resistant to expanded-spectrum cephalosporins, and molecular characterization of resistant isolates. Appl Environ Microbiol. 2013;79:2273–2283. doi: 10.1128/AEM.03592-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanwar, N. et al. Effects of ceftiofur and chlortetracycline treatment strategies on antimicrobial susceptibility and on tet(A), tet(B), and bla CMY-2 resistance genes among E. coli isolated from the feces of feedlot cattle. PloS one8, e80575, 10.1371/journal.pone.0080575 [doi] (2013). [DOI] [PMC free article] [PubMed]
- 28.Lutz EA, et al. Ceftiofur use in finishing swine barns and the recovery of fecal Escherichia coli or Salmonella spp. resistant to ceftriaxone. Foodborne Pathog Dis. 2011;8:1229–1234. doi: 10.1089/fpd.2011.0925. [DOI] [PubMed] [Google Scholar]
- 29.USDA, A. F 2011 Part I: Management Practices on U.S. Feedlots with a Capacity of 1,000 or More Head. (USDA 2013).
- 30.Platt, T. M. et al. Antimicrobial susceptibility of enteric bacteria recovered from feedlot cattle administered chlortetracycline in feed. American Journal of Veterinary Research69, 988–996, doi:10.2460/ajvr.69.8.988 [doi] (2008). [DOI] [PubMed]
- 31.Agga GE, Schmidt JW, Arthur TM. Effects of In-Feed Chlortetracycline Prophylaxis in Beef Cattle on Animal Health and Antimicrobial-Resistant Escherichia coli. Appl Environ Microbiol. 2016;82:7197–7204. doi: 10.1128/AEM.01928-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barkocy-Gallagher GA, et al. Development of methods for the recovery of Escherichia coil O157:H7 and Salmonella from beef carcass sponge samples and bovine fecal and hide samples. J Food Prot. 2002;65:1527–1534. doi: 10.4315/0362-028X-65.10.1527. [DOI] [PubMed] [Google Scholar]
- 33.CLSI. Performance standards for antimicrobial disk susceptibility tests; Approved standard-Twelfth Edition. Clinical and Laboratory Standards Institute, Wayne, PA (2015).
- 34.Zhang S, et al. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol. 2015;53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inouye M, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome medicine. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jolley KA. & Maiden, M. C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frye JG, Fedorka-Cray PJ. Prevalence, distribution and characterisation of ceftiofur resistance in Salmonella enterica isolated from animals in the USA from 1999 to 2003. International journal of antimicrobial agents. 2007;30:134–142. doi: 10.1016/j.ijantimicag.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Loneragan GH, et al. Salmonella diversity and burden in cows on and culled from dairy farms in the Texas High Plains. Foodborne Pathog Dis. 2012;9:549–555. doi: 10.1089/fpd.2011.1069. [DOI] [PubMed] [Google Scholar]
- 39.USDA-APHIS. Salmonella in U. S. Cattle Feedlots. https://www.aphis.usda.gov/animal_health/nahms/feedlot/downloads/feedlot2011/Feed11_is_Salm.pdf (2014).
- 40.Dargatz DA, Kopral CA, Erdman MM, Fedorka-Cray PJ. Prevalence and Antimicrobial Resistance of Salmonella Isolated from Cattle Feces in United States Feedlots in 2011. Foodborne Pathog Dis. 2016;13:483–489. doi: 10.1089/fpd.2016.2128. [DOI] [PubMed] [Google Scholar]
- 41.USDA-APHIS. Salmonella in United States Feedlots. https://www.aphis.usda.gov/animal_health/nahms/feedlot/downloads/feedlot99/Feedlot99_is_Salmonella.pdf (2001).
- 42.Fossler CP, et al. Cattle and environmental sample-level factors associated with the presence of Salmonella in a multi-state study of conventional and organic dairy farms. Prev Vet Med. 2005;67:39–53. doi: 10.1016/j.prevetmed.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Kunze DJ, et al. Salmonella enterica burden in harvest-ready cattle populations from the southern high plains of the United States. Appl Environ Microbiol. 2008;74:345–351. doi: 10.1128/AEM.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Likavec T, Pires AF, Funk JA. Association between thermal environment and Salmonella in fecal samples from dairy cattle in midwestern United States. Can J Vet Res. 2016;80:183–188. [PMC free article] [PubMed] [Google Scholar]
- 45.Khaitsa ML, et al. A longitudinal study of Salmonella shedding and antimicrobial resistance patterns in North Dakota feedlot cattle. J Food Prot. 2007;70:476–481. doi: 10.4315/0362-028X-70.2.476. [DOI] [PubMed] [Google Scholar]
- 46.Fecteau ME, et al. Efficacy of ceftiofur for treatment of experimental salmonellosis in neonatal calves. Am J Vet Res. 2003;64:918–925. doi: 10.2460/ajvr.2003.64.918. [DOI] [PubMed] [Google Scholar]
- 47.Beyer A, et al. Effects of ceftiofur treatment on the susceptibility of commensal porcine E. coli–comparison between treated and untreated animals housed in the same stable. BMC Vet Res. 2015;11:265. doi: 10.1186/s12917-015-0578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleury MA, et al. Impact of ceftiofur injection on gut microbiota and Escherichia coli resistance in pigs. Antimicrob Agents Chemother. 2015;59:5171–5180. doi: 10.1128/AAC.00177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agga GE, Schmidt JW, Arthur TM. Antimicrobial-Resistant Fecal Bacteria from Ceftiofur-Treated and Nonantimicrobial-Treated Comingled Beef Cows at a Cow-Calf Operation. Microbial drug resistance. 2016;22:598–608. doi: 10.1089/mdr.2015.0259. [DOI] [PubMed] [Google Scholar]
- 50.Helke KL, et al. Effects of antimicrobial use in agricultural animals on drug-resistant foodborne salmonellosis in humans: A systematic literature review. Crit Rev Food Sci Nutr. 2017;57:472–488. doi: 10.1080/10408398.2016.1230088. [DOI] [PubMed] [Google Scholar]
- 51.Wagner BA, Straw BE, Fedorka-Cray PJ, Dargatz DA. Effect of antimicrobial dosage regimen on Salmonella and Escherichia coli isolates from feeder swine. Appl Environ Microbiol. 2008;74:1731–1739. doi: 10.1128/AEM.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells JE, Kalchayanand N, Berry ED, Oliver WT. Effects of antimicrobials fed as dietary growth promoters on faecal shedding of Campylobacter, Salmonella and shiga-toxin producing Escherichia coli in swine. J Appl Microbiol. 2013;114:318–328. doi: 10.1111/jam.12065. [DOI] [PubMed] [Google Scholar]
- 53.Daniels JB, Call DR, Besser TE. Molecular epidemiology of blaCMY-2 plasmids carried by Salmonella enterica and Escherichia coli isolates from cattle in the Pacific Northwest. Appl Environ Microbiol. 2007;73:8005–8011. doi: 10.1128/AEM.01325-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dargatz DA, et al. Antimicrobial susceptibility patterns of Salmonella isolates from cattle in feedlots. J Am Vet Med Assoc. 2002;221:268–272. doi: 10.2460/javma.2002.221.268. [DOI] [PubMed] [Google Scholar]
- 55.Hong S, et al. Serotypes and Antimicrobial Resistance in Salmonella enterica Recovered from Clinical Samples from Cattle and Swine in Minnesota, 2006 to 2015. PLoS One. 2016;11:e0168016. doi: 10.1371/journal.pone.0168016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stecher B, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci USA. 2012;109:1269–1274. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beach JC, Murano EA, Acuff GR. Serotyping and antibiotic resistance profiling of Salmonella in feedlot and nonfeedlot beef cattle. J Food Prot. 2002;65:1694–1699. doi: 10.4315/0362-028X-65.11.1694. [DOI] [PubMed] [Google Scholar]
- 59.Edrington TS, Callaway TR, Anderson RC, Nisbet DJ. Prevalence of multidrug-resistant Salmonella on commercial dairies utilizing a single heifer raising facility. J Food Prot. 2008;71:27–34. doi: 10.4315/0362-028X-71.1.27. [DOI] [PubMed] [Google Scholar]
- 60.Brichta-Harhay DM, et al. Diversity of multidrug-resistant salmonella enterica strains associated with cattle at harvest in the United States. Appl Environ Microbiol. 2011;77:1783–1796. doi: 10.1128/AEM.01885-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dodd CC, et al. Prevalence and persistence of Salmonella in cohorts of feedlot cattle. Foodborne Pathog Dis. 2011;8:781–789. doi: 10.1089/fpd.2010.0777. [DOI] [PubMed] [Google Scholar]
- 62.CDC. National Enteric Disease Surveillance: Salmonella Annual Report, 2013 (2013).
- 63.Gorski L. Selective enrichment media bias the types of Salmonella enterica strains isolated from mixed strain cultures and complex enrichment broths. PLoS One. 2012;7:e34722. doi: 10.1371/journal.pone.0034722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edrington TS, et al. Investigation into the seasonal salmonellosis in lactating dairy cattle. Epidemiol Infect. 2008;136:381–390. doi: 10.1017/S0950268807008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edrington TS, et al. Evaluation of the potential antimicrobial resistance transfer from a multi-drug resistant Escherichia coli to Salmonella in dairy calves. Curr Microbiol. 2013;66:132–137. doi: 10.1007/s00284-012-0249-6. [DOI] [PubMed] [Google Scholar]
- 66.Gragg SE, et al. Substantial within-animal diversity of Salmonella isolates from lymph nodes, feces, and hides of cattle at slaughter. Appl Environ Microbiol. 2013;79:4744–4750. doi: 10.1128/AEM.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are not publicly available due to on-going sequencing analyses at the time of publication; however, they will become available from the corresponding author upon request.