Abstract
Group 1 innate lymphoid cells include natural killer (NK) cells and ILC1s, which mediate the response to intracellular pathogens. Thymic NK (tNK) cells were described with hybrid features of immature NK cells and ILC1 but whether these cells are related to NK cells or ILC1 has not been fully investigated. We report that murine tNK cells expressed the NK-cell associated transcription factor EOMES and developed independent of the essential ILC1 factor TBET confirming their placement within the NK lineage. Moreover, tNK cells resemble NK cells rather than ILC1 in their requirements for the E protein transcription factor inhibitor ID2. We provide further insight into the mechanisms governing tNK-cell development by showing that the transcription factor ETS1 prevented tNK cell acquisition of the conventional NK cell maturation markers CD11b and KLRG1. Our data reveal few ILC1 in the thymus and clarify the identity and developmental requirements of tNK cells.
Keywords: natural killer cells, innate lymphoid cells, ILC1, EOMES, Id2, Ets1
Introduction
The Group 1 innate lymphoid cells (ILC) include natural killer (NK) cells and ILC1 [1]. These cells function in the innate immune response to intracellular pathogens and tumors, and they share multiple features including expression of NK1.1 and NKp46, the β chain of the IL-2/IL-15 receptor (CD122), and a dependence on interleukin (IL)-15 for their survival [1]. All Group 1 ILC produce IFNγ but ILC1 differ from conventional (c)NK cells in the range of cytokines they produce, their transcription factor requirements, and in their expression of integrins and multiple NK-cell associated receptors [1]. ILC1s express and require the T-box transcription factor TBET but not EOMES whereas cNK cells express both TBET and EOMES [1]. In many tissues, ILC1 are distinguished from cNK cells by expression of the receptor of IL-7 (CD127) and integrin α1 (CD49a) and their lack of integrin α2 (DX5/CD49b) [2–6]. However, CD49a can be induced on cNK cells by activation of the transforming growth factor (TGF) beta-signaling pathway indicating that this adhesion molecule is not sufficient to distinguish ILC1 from NK cells [7].
The adult thymus has a unique subset of putative NK cells (tNK cells) that are similar to humans CD56bright NK cells [8]. These tNK cells are implicated in regulating dendritic cell function in lymph nodes and in tumor surveillance in the thymus. Thymic NK cells have many features associated with ILC1 including a dependence on the transcription factor GATA3, expression of CD127, and the absence of integrin αM (CD11b) and the Ly49 receptors that characterize cNK cells; however, tNK cells express the cNK cell integrin β2 DX5/CD49b [8]. Thymic NK cells are poor cytotoxic effectors but they have an enhanced ability to produce TNFα and IFNγ when compared to cNK cells [8]. Despite this knowledge, little is known about the transcription factor requirements for tNK cell development, an understanding of which could help to distinguish tNK cells from ILC1.
We report here that the thymic Lin−CD122+NK1.1+ ILC population is composed primarily of NK1.1+ T lymphocytes and tNK cells and that tNK cells have transcription factor requirements that are similar to cNK cells. In Rag1−/− mice tNK cells can develop in the absence of ID2 or ETS1 but have a phenotype similar to that of cNK cells that lack these factors. Indeed, ID2 promoted the tNK cell phenotype whereas ETS1 prevented acquisition of a cNK cell phenotype as measured by the expression of CD11b and the TNF receptor family member CD27 [9]. Our data provide insights into the identity and developmental requirements for tNK cells.
Results and Discussion
Lin−CD122+NK1.1+ thymocytes include tNK cells and other innate-like lymphoid cells
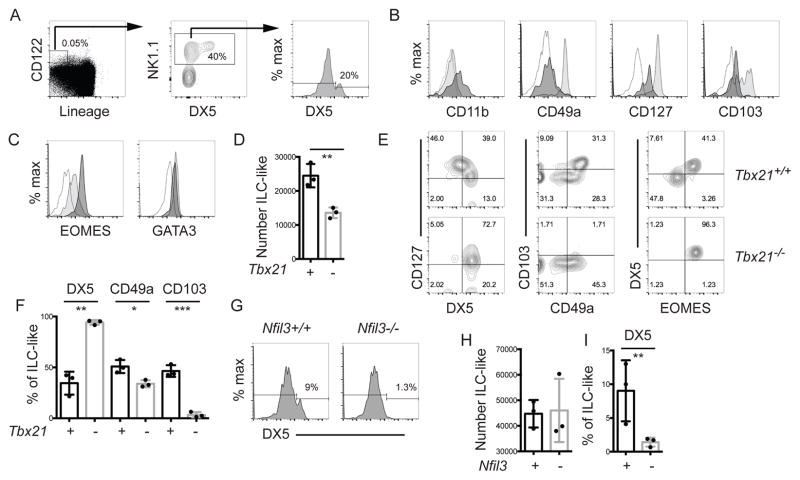
We characterized the surface markers and transcription factor requirements of Lineage negative (TCRβ, TCRγ, CD3ε, CD4, CD8/Lin−) CD122+NK1.1+ innate-like (ILC-like) cells in the thymus to gain insight into the identity of these cells. As reported previously [8], a minority of this population expressed the cNK cell marker DX5 (Fig. 1A). The DX5+ cells expressed CD127 and had low expression of CD11b (Fig. 1B), consistent with a previous study [8]. Most ILC-like cells were DX5− and expressed high levels of CD127 and CD49a (Fig. 1B), a marker associated with ILC1 [4, 10]. CD103, the αE integrin that is associated with tissue resident T cells, was expressed on approximately 50% of DX5− cells (Fig 1B) [11, 12]. In contrast, tNK cells in wild-type (WT) mice lacked these markers (Fig 1B). These data indicate that the thymic ILC-like population is heterogeneous with a majority of cells having an ILC1-like phenotype (CD122+NK1.1+CD127+CD49a+CD103+) and a minor population having the tNK cell phenotype (CD122+NK1.1+CD127+DX5+CD11blo) [9].
Figure 1. Characterization of the phenotype and transcription factor requirements of murine thymic ILC-like cells.
Wild-type C57BL/6 thymocytes were analyzed by FACS for (A) ILC-like cells (Lin−CD122+ NK1.1+). Lineage = TCRβ, TCRγ, CD3ε, CD4, and CD8. DX5 expression on ILC-like cells is also shown. (B) CD11b, CD49a, CD127 and CD103, and (C) EOMES and GATA3, expression on DX5+ (dark) and DX5− (light) ILC-like cells. The open profile is the FMO. (D) Mean number ± SEM of thymic ILC-like cells in Tbx21+/+ and Tbx21−/− mice ± SEM. (E) FACS analysis for CD127 versus DX5, CD103 versus CD49a, and DX5 versus EOMES on thymic ILC-like cells in Tbx21+/+ and Tbx21−/− mice. (F) Mean percent ± SEM of Tbx21+/+ (+, black) and Tbx21−/− (-, grey) thymic ILC-like cells expressing DX5, CD49a and CD103. (G) DX5 expression on thymic ILC-like cells from Nfil3+/+ and Nfil3−/− mice. (H) Thymic ILC-like numbers and (I) the percent DX5+ in Nfil3+/+ and Nfil3−/− mice. (A–C) Representative profile from > 7 experiments, (E, G) from 3 experiments with one mouse of each genotype/experiment. (D, F, H) Each dot represents one mouse. Unpaired t-test * p<0.05, **p<0.01, ***p<0.001.
NK cells are distinguished from ILC1 by their expression of EOMES and their ability to develop in the absence of TBET, which is required for ILC1 development (4, 5). Both GATA3 and EOMES were expressed in Lin−CD122+NK1.1+DX5+ cells, consistent with their designation as tNK, whereas DX5− cells expressed GATA3 but had low EOMES (Fig. 1C). In Tbx21−/− mice (TBET-deficient) there was an approximate 50% decrease in ILC-like thymocytes but >90% of the remaining cells were DX5+ (Fig. 1D, E and F). Indeed, in the absence of TBET there was a specific loss of CD49a+, CD127hi, CD103+, and DX5− cells (Fig. 1E, F). Therefore, tNK cells developed in TBET-deficient mice but DX5− ILC-like cells were TBET-dependent.
To confirm that the DX5+ tNK cells were related to NK cells, we tested whether they developed in the absence of NFIL3, a transcription factor that is essential for cNK cells and some ILC1 but not for innate-like T cells [13, 14]. In Nfil3−/− mice total ILC-like cell numbers were not altered but there was a near complete loss of the minor DX5+ tNK cell population (Fig. 1G, H, I). These data indicate that tNK cells are CD127+GATA3+EOMES+ cells that require NFIL3 but not TBET for their development, consistent with their designation as NK cells rather than ILC1, and consistent with the loss of tNK cells previously reported in Nfil3−/− mice [14]. Moreover, these data indicate that DX5− ILC-like thymocytes are TBET-dependent and NFIL3-independent.
Thymic NK cells in Rag1−/− mice acquire markers of tissue residency
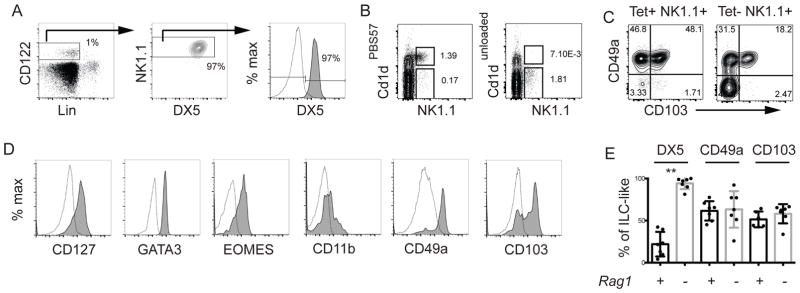
Given that a substantial portion of the ILC-like cells in WT mice are DX5−, we questioned whether these cells were ILC1s. To further test the identity of these cells we examined ILC-like cells in Rag1−/− mice, which lack adaptive lymphoid cells. Surprisingly, all Lin−CD122+NK1.1+ thymocytes in Rag1−/− mice expressed DX5 (Fig. 2A), suggesting that the major population of DX5− cells in WT mice were T lymphocytes. Consistent with this conclusion, a majority of NK1.1+ cells in the thymus of WT mice were NKT cells that stained positively with PBS57-loaded CD1d tetramers and some of these cells have low TCRβ staining (Fig. 2B, S1). Moreover, CD1d tetramer+ NK1.1+ cells expressed CD49a and approximately half were CD103+ (Fig. 2C), consistent with the phenotype of thymic Lin−CD122+NK1.1+DX5− cells. There may be additional NK1.1+ cells in the thymus that express these markers because we observed such cells in the CD1d tetramer negative NK1.1+ population (Fig. 2C). Some γδ T cells express NK1.1 and could fall into this gate [15]. In Rag1−/− mice the DX5+ tNK cells expressed EOMES, GATA3, and CD127 and had low expression of CD11b, similar to WT tNK cells (Fig. 2D) [8]. These cells also expressed EOMES confirming that they were tNK cells and not ILC1 that acquired DX5 expression (Fig. 2D). Interestingly however, Rag1−/− tNK cells expressed CD49a and CD103 indicating that, in the absence of T cells, they are impacted by factors that can drive the expression of these proteins (Fig. 2D and E) [7].
Figure 2. Thymic DX5+ ILC-like cells are RAG1-independent.
(A) FACS analysis for ILC-like cells in Rag1−/− mice. (B) Thymocytes were enriched for CD8− cells and analyzed for expression of NK1.1 and the NKT-cell receptor using PBS57 (left) or unloaded (right) CD1d tetramers. (C) CD49a and CD103 on NK1.1+ and CD1dPBS57+ (NKT1) or CD1dPBS57− (Tet−NK1.1+) cells. (D) Expression of CD127, CD11b, CD49a, CD103, EOMES and GATA3 is shown for DX5+ tNK cells. Open profile is the FMO. (E) Mean percent ± SEM of thymic ILC-like cells expressing the indicated proteins in Rag1+/+ (+, black) or Rag1−/− (-, grey) mice. Each dot represents one mouse. (A, D) Data are representative of 4–7 experiments, (B, C) representative of 3 experiments with one mouse of each genotype. Unpaired t-test * p<0.05, **p<0.01.
We note that despite the loss of DX5− ILC-like cells in Tbx21−/− mice, tNK cells did not acquire expression of CD103. Therefore, the loss of TBET-dependent T lymphocytes (such as NKT cells) may not be sufficient to expose tNK cells to the factors that induce CD103. Alternatively, TBET may be required for CD103 expression on these cells.
Thymic NK cells develop in Rag1−/−Id2−/− mice
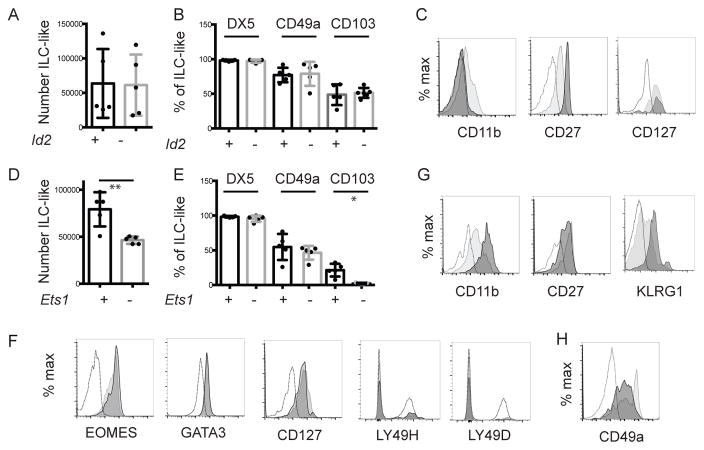
To gain further insight into the identity of tNK cells we examined their dependence on the transcription factor ID2. We examined tNK cell numbers in Rag1−/−GzmbCre+Id2f/f mice on a C57Bl/6 background (Rag1−/−Id2−/−), in which Id2 is deleted in all hematopoietic cells ([16], Fig. S2). In Rag1−/−Id2−/− mice, tNK cell numbers were similar to littermate control mice (LMC) indicating that tNK cells can develop independent of ID2 when T cells are absent (Fig. 3A and B). However, Rag1−/−Id2−/− mice had an altered phenotype; they failed to express even low levels of CD11b and had heightened CD27 expression (Fig. 3C). These cells also expressed DX5, CD127, CD103 and CD49a (Fig 3B, C).
Figure 3. ID2 and ETS1 regulate the phenotype of tNK-cells.
Thymocytes from the indicated mouse strains were analyzed by flow cytometry. (A) Mean number ± SEM of thymic ILC-like cells in Rag1−/−Id2+/+ and Rag1−/−Id2−/− mice. (B) Mean percent ± SEM of Rag1−/−Id2+/+ (+, black) or Rag1−/−Id2−/− (-, grey) thymic ILC expressing DX5, CD49a or CD103. (C) FACS analysis for CD11b, CD27, and CD127 on Rag1−/−Id2+/+ (light grey) and Rag1−/−Id2−/− (dark grey) tNK cells. Open histogram is FMO. (D) Mean number ± SEM of thymic ILC-like cells in Rag1−/−Ets1+/+ and Rag1−/−Ets1−/− mice. (E) Mean percent ± SEM of Rag1−/−Ets1+/+ (+, black) or Rag1−/−Ets1−/− (-, grey) thymic ILC expressing DX5, CD49a or CD103. Flow cytometry analysis for (F) CD11b, CD27, and CD127, (G) EOMES and GATA3, (H) Ly49H and Ly49D, or (I) CD49a on Rag1−/−Ets1+/+ (light) and Rag1−/−Ets1−/− (dark) tNK cells. Open histogram in (F, G, I) is FMO, in (H) is splenic cNK. Data are representative of 3–7 experiments one mouse of each genotype. Each dot represents one mouse. Unpaired t-test * p<0.05, **p<0.01.
Our observation that tNK cells developed independent of ID2 was surprising given that all ILCs and mature cNK cells are ID2-dependent [17–19]. However, CD27+CD11b− cNK cells are a minor portion of peripheral cNK cells and this subset was present in Rag1−/−Id2−/− mice and had a similar CD27hiCD11b− phenotype [20] (Fig. S2). Therefore, tNK cells resemble cNK cells in their requirement for ID2.
ETS1 maintains the tNK cell phenotype
ETS1 is required for the proper development of cNK cells and limits their activation [21, 22]. Therefore, we investigated the requirements for ETS1 in tNK cells using Rag1−/−Il7raCreEts1f/f mice (Rag1−/−Ets1−/−) [23]. In contrast to Rag1−/−Id2−/− mice, tNK cell numbers were decreased in the absence of ETS1 to approximately 50% of LMC (Fig. 3D). All of the ILC-like cells were tNK cells as assessed by expression of DX5, EOMES, GATA3, and CD127 and they had low expression of the cNK receptors Ly49H and Ly49D (Fig. 3E, F). Surprisingly, we observed that Rag1−/−Ets1−/− tNK cells had increased CD11b and decreased CD27 expression, a phenotype associated with cNK cells, and they expressed higher levels of another cNK cell receptor, KLRG1 (Fig. G). Interestingly, these cells failed to express CD103+ and had a lower intensity of CD49a+ (Fig. 3E, H). Our data indicate that some tNK cells developed in Rag1−/−Ets1−/− mice but that ETS1 prevented their acquisition of CD11b, KLRG1 and down modulation of CD27.
ETS1 is a signal-regulated factor whose DNA binding is controlled by Ca2+-dependent kinases and transactivation is dependent on mitogen-activated protein kinases. Therefore, the differences in tNK cell and cNK cell phenotype (CD27+CD11b− versus CD27−CD11b+) could be a consequence of the presence or absence of signals that control ETS1 function in the thymus. CD27+CD11b− tNK cells may be less differentiated cells, similar to what is proposed for CD27+CD11b− cNK cells, that have not been activated by IL-15 or other cytokines that are induced in dendritic cells by microbial products [24]. Therefore, the thymus may lack the cells that can stimulate tNK cell expression of cNK cell maturation markers; alternatively, tNK cells may be resistant to these signals. Thymic NK cells express CD127, which shares the common γ chain with the IL-15/IL2 receptor alpha chain, and expression of two receptors that share a common component may diminish responsiveness to either cytokine if the common component is limiting [25]. Thus, the expression of CD127 may reduce responsiveness to IL-15, an intriguing possibility since ETS1 limits the sensitivity of cNK cells to IL-15 [21]. Therefore, increased IL-15 sensitivity in Ets1−/− tNK cells could drive their maturation.
Concluding Remarks
We conclude that the thymus contained a heterogeneous population of Lin−CD122+NK1.1+ cells that included DX5−CD49a+ innate-like T lymphocytes and DX5+CD27+CD11blo NK cells. The innate-like T cells required the transcription factor TBET but not NFIL3 whereas tNK cell development required NFIL3 but was independent of TBET, consistent with the classification of the latter as NK cells rather than ILC1. Thymic NK cell development in Rag1−/− mice was independent of ID2 although ID2-deficiency resulted in a CD27hiCD11b− phenotype. Thymic NK cells also developed, albeit inefficiently, in the absence of ETS1 but these cells were primarily CD27lo and CD11b+ suggesting that ETS1 prevented tNK cells from acquiring a mature cNK cell phenotype. Our data demonstrate that tNK cells have developmental requirements consistent with the NK cell lineage and are not ILC1; however, they have a requirement for the transcription factor ETS1 that only partially overlaps with cNK cells.
Materials and methods
Mice
C57Bl/6, Rag1−/−, Rag1−/−Il7RaCreEts1f/f (Ets1−/−)[23], Rag1−/−GzmbCreId2f/f mice, and their Rag1−/−Cre+ littermate controls (LMC) were maintained in a specific pathogen free facility at the University of Chicago. Tbx21−/− mice were purchased from Jackson. Thymocytes from Nfil3−/− mice and LMC [26, 27] were provided by Dr. Joe Sun (Memorial Sloan Kettering Cancer Center, New York). All mice were on a C57BL/6 background. Rag1−/−GzmbCreId2f/f mice delete Id2 in all hematopoietic cells ([16] and manuscript in preparation). Mice were euthanized using CO2 asphyxiation followed by cervical dislocation.
Cell preparation and flow cytometry
Thymocytes were stained for flow cytometry using standard procedures. The antibodies used are available upon request. PBS57 loaded and unloaded CD1d tetramers were from the NIH Tetramer Facility (Atlanta, GA). The Foxp3 Transcription Factor Staining Buffer Set (eBioscience) was used for the intracellular staining with the EOMES and TBET antibodies. Flow cytometry was performed on a BD LSRIII Fortessa, and the data were analyzed using FlowJo software (Tree Star, Ashland, OR). All gates were set using negative controls. Cell numbers were calculated using Precision Count Beads (Biolegend)
CD8 lineage depletion was performed using CD8-biotin followed by streptavidin-magnetic beads (Miltenyi Biotech) prior to passing over an LS magnetic columns (Miltenyi Biotech).
Statistical analysis
Unpaired Student t-test’s were calculated using Prism 6 (GraphPad Software).
Supplementary Material
Acknowledgments
We thank Dr. Joe Sun for thymocytes from Nfil3−/− and their LMC mice and for critical reading of this manuscript, Dr. Hans-Reimer Rodewald for the Il7RaCre mice. We also thank the NIH Tetramer Facility for PBS57 loaded and unloaded CD1d tetramers and the Cytometry Antibody Technology Facility at the University of Chicago. This work was supported by NIH grants R01 AI106352, and R21 AI115338, to B.L.K.
Abbreviations
- NK
natural killer
- tNK
thymic natural killer
- NKT
Natural killer T cell
- ILC
innate lymphoid cell
- WT
wild-type
- IFN
interferon
- TNF
tumor necrosis factor
- TCR
T cell receptor
- FMO
fluorescence minus one
Footnotes
Conflict of interest: The authors declare no financial or commercial conflicts of interest
Author Contributions: S.G., M.S., and A.B performed experiments; E.C.Z. and R.F.deP. maintained mice and provided valuable advice, S.G. and B.L.K. analyzed data, S.G. and B.L.K. conceptualized the project and S.G., L.Z., and B.L.K. wrote the manuscript.
References
- 1.Spits H, Bernink JH, Lanier L. NK cells and type 1 innate lymphoid cells: partners in host defense. Nat Immunol. 2016;17(7):758–764. doi: 10.1038/ni.3482. [DOI] [PubMed] [Google Scholar]
- 2.Robinette ML, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16(3):306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortez VS, Fuchs A, Cella M, Gilfillan S, Colonna M. Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. J Immunol. 2014;192(10):4487–4491. doi: 10.4049/jimmunol.1303469. [DOI] [PubMed] [Google Scholar]
- 4.Sojka DK, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doisne JM, et al. Composition, Development, and Function of Uterine Innate Lymphoid Cells. J Immunol. 2015;195(8):3937–3945. doi: 10.4049/jimmunol.1500689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erick TK, Anderson CK, Reilly EC, Wands JR, Brossay L. NFIL3 Expression Distinguishes Tissue-Resident NK Cells and Conventional NK-like Cells in the Mouse Submandibular Glands. J Immunol. 2016;197(6):2485–2491. doi: 10.4049/jimmunol.1601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortez VS, et al. Transforming Growth Factor-beta Signaling Guides the Differentiation of Innate Lymphoid Cells in Salivary Glands. Immunity. 2016;44(5):1127–1139. doi: 10.1016/j.immuni.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vosshenrich CA, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7(11):1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 9.Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113(22):5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 10.Daussy C, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211(3):563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crozat K, et al. Cutting edge: expression of XCR1 defines mouse lymphoid-tissue resident and migratory dendritic cells of the CD8alpha+ type. J Immunol. 2011;187(9):4411–4415. doi: 10.4049/jimmunol.1101717. [DOI] [PubMed] [Google Scholar]
- 12.Woodberry T, et al. Alpha E beta 7 (CD103) expression identifies a highly active, tonsil-resident effector-memory CTL population. J Immunol. 2005;175(7):4355–4362. doi: 10.4049/jimmunol.175.7.4355. [DOI] [PubMed] [Google Scholar]
- 13.Gascoyne DM, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10(10):1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 14.Seillet C, et al. Differential requirement for Nfil3 during NK cell development. J Immunol. 2014;192(6):2667–2676. doi: 10.4049/jimmunol.1302605. [DOI] [PubMed] [Google Scholar]
- 15.Stewart CA, Walzer T, Robbins SH, Malissen B, Vivier E, Prinz I. Germ-line and rearranged Tcrd transcription distinguish bona fide NK cells and NK-like gammadelta T cells. Eur J Immunol. 2007;37(6):1442–1452. doi: 10.1002/eji.200737354. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Evaristo C, Alegre ML, Gurbuxani S, Kee BL. Analysis of GzmbCre as a Model System for Gene Deletion in the Natural Killer Cell Lineage. PLoS One. 2015;10(4):e0125211. doi: 10.1371/journal.pone.0125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delconte RB, et al. The Helix-Loop-Helix Protein ID2 Governs NK Cell Fate by Tuning Their Sensitivity to Interleukin-15. Immunity. 2016;44(1):103–115. doi: 10.1016/j.immuni.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Satoh-Takayama N, Lesjean-Pottier S, Vieira P, Sawa S, Eberl G, Vosshenrich CA, Di Santo JP. IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+ cell subsets from Id2-dependent precursors. J Exp Med. 2010;207(2):273–280. doi: 10.1084/jem.20092029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klose CS, Hoyler T, Kiss EA, Tanriver Y, Diefenbach A. Transcriptional control of innate lymphocyte fate decisions. Curr Opin Immunol. 2012;24(3):290–296. doi: 10.1016/j.coi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204(5):1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, Kee BL. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36(6):921–932. doi: 10.1016/j.immuni.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9(4):555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 23.Zook EC, et al. The ETS1 transcription factor is required for the development and cytokine-induced expansion of ILC2. J Exp Med. 2016;213(5):687–696. doi: 10.1084/jem.20150851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamimura Y, Lanier LL. Homeostatic control of memory cell progenitors in the natural killer cell lineage. Cell Rep. 2015;10(2):280–291. doi: 10.1016/j.celrep.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cotari JW, Voisinne G, Dar OE, Karabacak V, Altan-Bonnet G. Cell-to-cell variability analysis dissects the plasticity of signaling of common gamma chain cytokines in T cells. Sci Signal. 2013;6(266):ra17. doi: 10.1126/scisignal.2003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiger TL, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med. 2014;211(9):1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kashiwada M, et al. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc Natl Acad Sci U S A. 2010;107(2):821–826. doi: 10.1073/pnas.0909235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.