Abstract
Carbohydrates constitute a structurally and functionally diverse group of biological molecules and macromolecules. In cells they are involved in, e.g., energy storage, signaling, and cell–cell recognition. All of these phenomena take place in atomistic scales, thus atomistic simulation would be the method of choice to explore how carbohydrates function. However, the progress in the field is limited by the lack of appropriate tools for preparing carbohydrate structures and related topology files for the simulation models. Here we present tools that fill this gap. Applications where the tools discussed in this paper are particularly useful include, among others, the preparation of structures for glycolipids, nanocellulose, and glycans linked to glycoproteins. The molecular structures and simulation files generated by the tools are compatible with GROMACS.
Introduction
Carbohydrates constitute a large and diverse class of chemical compounds involved in all aspects of life including energy storage and conversion, cellular signaling, and recognition. The function of carbohydrates is often based on highly specific interactions and seemingly tiny but extremely important structural details. Given this, atomistic simulations would be an exceptionally useful technique to explore such specific features. However, the progress in the field of computer simulations to address questions related to carbohydrates has been slow, and the reason is quite simple: the lack of accurate force fields. The force fields commonly used in biomolecular simulations, such as CHARMM36 or OPLS,1,2 have been optimized for proteins, and this situation has lasted for decades. Recently, lipids and nucleic acids have also been given the attention that they deserve, resulting in high-quality force fields for these molecule types.3−9 Meanwhile, the development of force fields for carbohydrates has not been given the same weight. For instance, carbohydrate parametrization in the OPLS force field is based on a single glucose unit only,10 which demonstrates how substandard the situation has been. Another example is the GROMOS force field, which has an extension for hexopyranoses only.11,12 Considerable improvement has taken place only through the recent development of GLYCAM,13 a force field based on AMBER, which includes parameters for describing a quite large set of carbohydrate types.
Thanks to the emerging progress in improving the quality of carbohydrate force fields, the next task is to develop practical tools for the design and preparation of carbohydrate structures. The first thing the tool should do is to define the molecule in terms of its chemical structure, thus constructing the molecular topology. This is not an issue with proteins and nucleic acids, for instance, since practically all packages for molecular dynamics (MD) simulations have tools for automatic preparation of topologies for these molecules. However, for carbohydrates found in, e.g., glycoproteins, the present state-of-the-art is less advanced. The only tools currently available for generating the molecular topology are GLYCAMWeb(14) and the Glycan Reader part of CHARMM-GUI.15,16 GLYCAM Web generates a topology file for glycoprotein simulations, but the files can only be used for MD simulations in the AMBER program,17 and converting the topology file for other MD simulation packages is not a simple feat. Meanwhile, the Glycan Reader tool (used with CHARMM) requires the input protein coordinate file to include carbohydrates for generating the molecular topology file. This is a severe limitation, since one of the main problems in glycolipid and glycoprotein modeling is the lack of three-dimensional (3D) structures of carbohydrate branches, which due to considerable thermal fluctuations are not resolved from crystal structures or they are removed from the protein prior to crystallization. The same issue concerns highly flexible protein loops. However, while for protein loops there are numerous tools allowing one to put them back to the protein structure, there is no analogical software for carbohydrate branches of glycoproteins, with one exception. The GlyProt tool enables addition of N-glycans to proteins; however, it lacks the feature of adding O-glycans, and it does not generate the molecular topology file for MD simulations.18
There are also other challenges. In particular, while there are several tools available for constructing and predicting 3D structures of glycans, such as SWEET-II,19Glydict,20POLYS,21 and Shape,22 none of these tools work in conjunction with proteins or generate the topology files required for MD simulations.
In the context of glycolipids, the only tool available is the Glycolipid Modeler part of CHARMM-GUI.15 However, this tool can use only a set of predefined glycolipids and glycan sequences. Possibilities to build glycolipids of ones own interest are therefore limited.
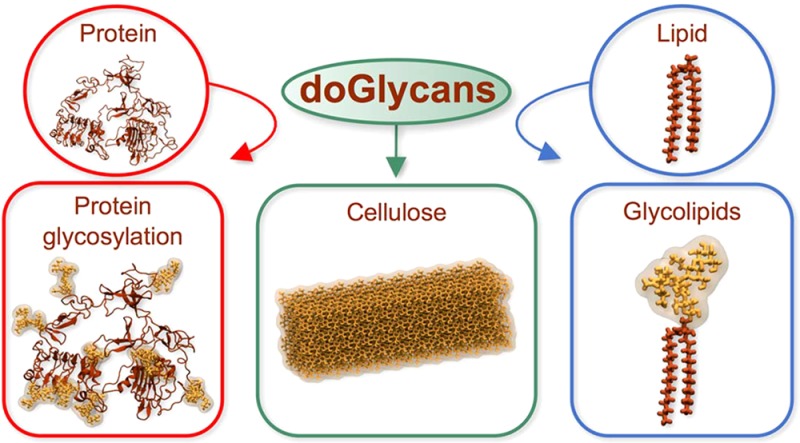
In this article, we present new Open Babel23 and python-based tools for preparation of carbohydrate structures that can be simulated with the GROMACS software package.24 The tools generate topologies of carbohydrates in the GROMACS format and then prepare 3D structures of carbohydrates covalently linked to the given lipids and proteins. The tools allow one to construct both linear and branched carbohydrates based on a user-defined glycan sequence. They have already been tested and validated in a number of case studies (see Figure 1).
Figure 1.
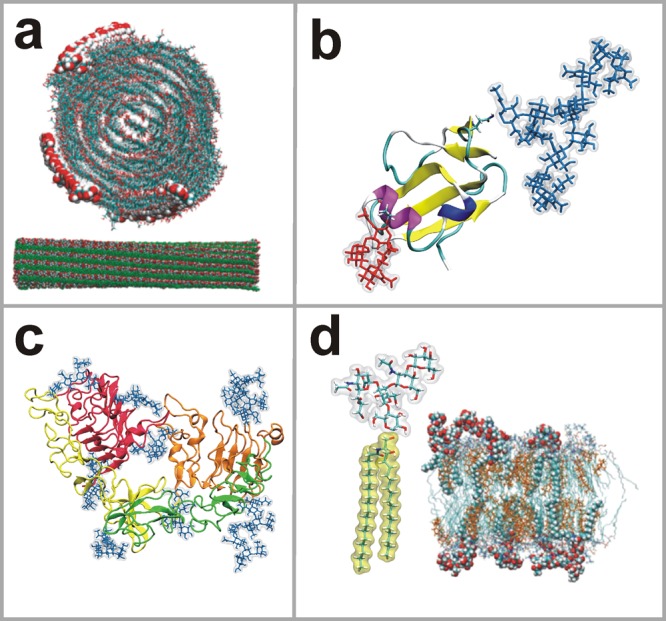
Snapshots of simulations of different carbohydrate systems that have been built with the present tool set, used as examples in this article. (a) Crystalline cellulose fragment (Reprinted with permission from ref (25). Copyright 2011 American Chemical Society.); (b) CD59 protein showing N- and O-glycan branches rendered as blue and red licorice, respectively;26 (c) EGFR monomer showing N-linked glycans rendered as blue licorice;27 and (d) structure of the GM1 glycolipid together with a snapshot of a lipid bilayer with 5 mol % of GM1 (Reprinted with permission from ref (28). Copyright 2011 Nature Publishing Group).
Software Functionalities
Here we present three illustrative examples of the main functionalities of the novel software package that we introduce in this work. We first discuss how the tools can be used to build elongated carbohydrate polymers. In this context, the molecule of interest is cellulose that is highly abundant in plant cells with a lot of potential applications. In the second example, we discuss how to construct carbohydrate branches of glycoproteins. Given that glycosylation is very common among the proteins found, e.g., in plasma membranes, and given that this topic has not been explored much until now, there is reason to assume that it will be paid considerable attention in future simulation studies. In the third and final example, we show how the tools can be employed to build carbohydrate units found in glycolipids. This topic is also one of the exceptionally important ones given that glycolipids modulate quite a few cellular functions. In all the three examples, we show every step needed to prepare a model for a given system.
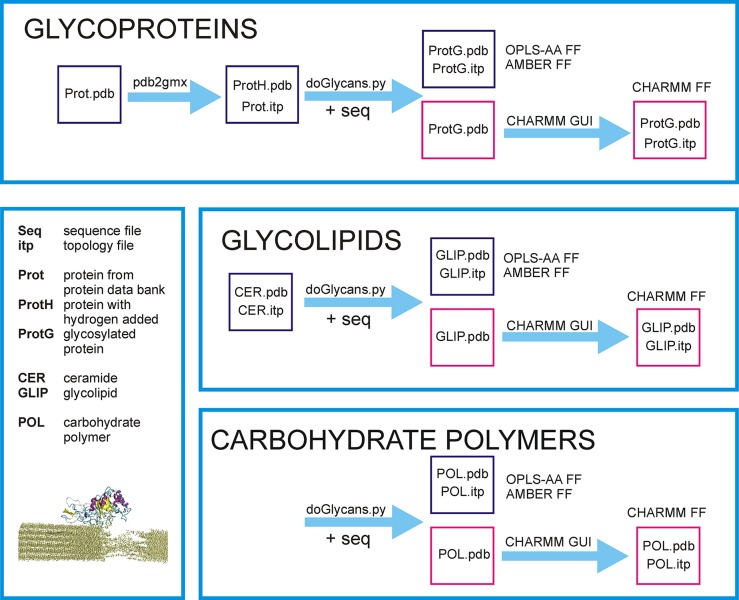
Below, for each of the topics discussed in this paper, we first briefly outline the biological relevance to study these molecules or their molecular complexes. Then, we show concretely how the application prepreader.py (using only lowercase font in the command name) can be used to prepare carbohydrate chains for polymer simulations, and the tool doglycans.py (again using only lowercase) to prepare models for glycoproteins and glycolipids. Together, these constitute the doGlycans tool set.
Carbohydrate Polymers
Long carbohydrate polymers play an important role in the functions of animals, plants, and microorganisms. Key examples in this context are cellulose and hemicellulose that are the main components of rigid plant cell walls. Cellulose is composed of long unbranched glucose chains having up to thousands of monomers, while hemicellulose chains that include small numbers of other hexose units may be moderately branched. Cellulose is the most common organic polymer on earth and an important means for storing glucose. It therefore has a lot of relevance in ruminant animals feeding, and it is also a potential source of fuel and renewable energy. Cellulose has numerous applications in paper and textile industries, pharmaceutical industry, and food processing,29 and it is also a promising material in nanotechnology.30,31
In our previous atomistic MD simulation studies of cellulose nanofibers, where we used the present tool to build the carbohydrate units, we identified the cause of cellulose twisting,25 elucidated the role of amorphous cellulose regions in the elastic properties of cellulose nanofibers,32 and showed how cellulose enzymes interact with differently ordered regions of cellulose nanofibers.33
To set up related cellulose simulations, all one has to do is to prepare a sequence file with the required number of d-glucose units and then run the prepreader.py script. Detailed instructions are given in section 3 in the doGlycans Manual. The procedure to do it is shown schematically in Figure 2.
Figure 2.
Functionality of the doGlycans tool set described in a schematic manner for glycoproteins, glycolipids, and carbohydrate polymers that are the main application targets. Part of the diagram shown in pink refers to the use of the CHARMM force field, which required the use of CHARMM GUI. (bottom-left inset) Cellulose Cel5A complex with cellulose nanofiber (Reprinted with permission from ref (33). Copyright 2015 Springer.).
Glycosylation of Proteins
Glycosylation is a complex cotranslational and posttranslational process that takes place in all species in which glycans (oligosaccharides) are covalently attached to proteins.34 A majority of proteins (>50%) secreted by eukaryotic cells undergo glycosylation, and in general, all cells in nature are coated with distinct glycans that are biologically important for cell function.35 There are five different classes of glycosylation, namely (a) N-linked glycosylation, (b) O-linked glycosylation, (c) phospho-serine glycosylation, (d) C-mannosylation, and (e) glypiation (formation of glycosylphosphatidylinositol (GPI) anchors).36 Each glycan added to a protein is involved in a wide range of functions discussed elsewhere.37−41
Here we focus on glycosylation in the context of signaling. Membrane receptors governing many signaling processes have been studied quite extensively through atomistic simulations, yet the role of glycosylation in receptor function has been paid exceptionally little attention through simulations. This is largely due to the lack of structural information about the carbohydrate chains. Most of the glycoprotein structures that have been determined are based on extensively manipulated proteins, where in particular the carbohydrate chains are often truncated from the studied receptor before its 3D structure is being determined (see, e.g., ref (39)), since the soft carbohydrates often hinder or even block crystallization. Given this issue, there is considerable demand for a tool that would prepare carbohydrate structures to be attached to the glycosylation sites of glycoproteins. The present tool renders this possible and thereby fosters means to bridge simulations closer to systems explored in experiments.
In our recent studies on epidermal growth factor receptor (EGFR), we used the tool to elucidate the role of N-linked glycans in the structural arrangement and interactions of EGFR with a lipid membrane.27 We glycosylated EGFR with Man3GlcNAc2, a minimal N-linked glycan core which is independent of cell-type and is essential for protein folding in the endoplasmic reticulum.42,43 We found that the presence of the Man3GlcNAc2 glycan on the EGFR ectodomain significantly altered the receptor subdomains alignment on the membrane surface by lifting the subdomains from the membrane surface. Glycosylation-induced elevation of EGFR from the membrane surface therefore exposes the extracellular subdomains for ligand binding, which is a necessary step in subsequent EGFR dimerization and signal transduction processes.
In section 4 (see the Manual), we show how doglycans.py can be used to glycosylate EGFR by adding N-linked glycans to the receptor. The description demonstrates the glycosylation in three steps (see Figure 2), where one first processes the protein data bank (PDB) file, then prepares the sequence file, and finally runs the script for glycosylation.
The script can also be used to covalently link O-linked glycans. As a test case we demonstrate (see the SI) the linking of both N- and O-glycans to the human CD59 glycoprotein, which is a small GPI-anchored glycoprotein, whose function is to protect host cells by interfering with membrane attack complex (MAC) structure formation.44
Glycolipids
Glycolipids constitute a large and important class of lipids. However, they are poorly characterized from the biophysical point of view. MD simulations are not an exception here as the number of simulation studies of glycolipids is very limited.45,46 The three main classes of glycolipids are glycerol-based galactolipids typical for plants, ceramide-based glycolipids typical for animals, and lipopolysaccharides (LPSs) typical for Gram-negative bacteria. Galactolipids are the main component of photosynthesizing organelles thylakoids, where their content is as large as 75 mol %. Effectively 50% of all lipids in plant cells are galactolipids. Ceramide-based glycolipids in animals are present in small concentrations (of a few mole percent) and are located predominately in the extracellular leaflet of cell membranes. Their functions are mostly related to cell signaling as they act as receptors on the cell surface. For instance, ganglioside GM1 known to be a receptor for bacterial toxins such as cholera toxin regulates the function of membrane proteins such as EGFR.47,48 Gangliosides are known to take part in cell–cell recognition and to play an important role particularly in the development of the central nervous system.49 Meanwhile, LPS is a complex molecule with three functional parts: lipid A, a carbohydrate core, and O antigen. Lipid A is based on a phosphorylated glucosamine disaccharide with 6–8 hydrocarbon chains attached, while the carbohydrate core is attached to lipid A and is conserved within the species. O antigen is the most variable part of the molecule and is characteristic for a given strain of bacteria, being therefore of high relevance in medical diagnostics. LPS and its fragments are recognized by the innate immunological system, thus inducing a strong immunological response.
To facilitate the preparation of related glycolipid structures (see Figure 2), in the software package we provide previously constructed topologies and structure files for the most relevant glycolipid (GM1) as well as examples of commands and sequence files needed to prepare them. Predefined lipid bases are also provided. Instructions to this end are given in section 5 in the doGlycans Manual.
Force Field and Software Limitations
Although the doGlycans tool set is highly flexible, it has certain limitations. Perhaps most importantly, only the sugars units defined in GLYCAM can be used. For instance, in the current version of GLYCAM, common bacterial sugars present in LPS such as KDO and amino sugars are still missing.
Moving on, the topologies generated by doGlycans for glycosylated proteins are currently compatible only with the OPLS and AMBER force fields, the topologies generated for glycolipids are compatible with the OPLS force field, and the topology for carbohydrate polymers is based on the GLYCAM force field. There is however an extension to AMBER: to generate glycolipids topology and structure files for simulations with the AMBER force field, the only input that the user has to provide to the doGlycans tool set is the ceramide topology.
The carbohydrate polymer topology for simulations with OPLS can be generated based on a structure file built by doGlycans, using for example the MKTOP script.50 Structure files generated by doGlycans for glycoproteins, glycolipids, and carbohydrate polymers can be used in combination with CHARMM GUI to generate topologies for simulations with the CHARMM force field. This is, however, limited to the components defined in CHARMM GUI.
Conclusions
In spite of the exceptional variety of carbohydrates and their functions, the progress in atomistic simulation studies of carbohydrates has been slowed down due to practical limitations. One of the key issues has been the limited availability of user-friendly tools to generate the structures and topology files needed in atomistic MD simulations of carbohydrate polymers, glycoproteins, and glycolipids. To overcome these problems, we developed tools for atomistic MD simulations of carbohydrates and carbohydrate conjugates (glycoproteins and glycolipids). These tools allow their users to easily build and simulate carbohydrates with varying complexity.
The structures and simulation files generated by the tools are compatible with the GROMACS package. The functionalities of the tools include the preparation of 3D structures of carbohydrates and the generation of topologies that are consistent with the GROMACS format. The most important function of the tools is the preparation of glycoproteins, where the glycans missing in protein crystal structures are added to the protein in question.
We extensively tested, validated, and applied the tools in our previous simulation studies.27 Given this, the doglycans.py application (together with prepreader.py) will help scientists to foster their work in exploring the rich variety of carbohydrate functions through atomistic MD simulations. Given that ∼50% of all eukaryotic proteins are glycosylated, the tool may have a quite considerable impact in strengthening the progress in the field.
Acknowledgments
We wish to thank CSC–IT Centre for Science (Espoo, Finland) for computing resources. The Academy of Finland (Center of Excellence in Biomembrane Research), the Sigrid Juselius Foundation, and the European Research Council (Advanced Grant project CROWDED-PRO-LIPIDS) are acknowledged for financial support.
Glossary
Abbreviations
- GROMACS
GROningen MAchine for Chemical Simulations
- MD
molecular dynamics
- OPLS
Optimized Parameters for Liquid Simulations
- CHARMM
Chemistry at HARvard Macromolecular Mechanics
- AMBER
Assisted Model Building with Energy Refinement
- GUI
graphical user interface
- GM1
Gal5-β1,3-GalNAc4-β1,4-(NeuAc3-α2,3)-Gal2-β1,4-Glc1-β1,1-Cer
- GM2
GalNAc4-β1,4-(NeuAc3-α2,3)-Gal2-β1,4-Glc1-β1,1-Cer
- GM3
NeuAc3-α2,3-Gal2-β1,4-Glc1-β1,1-Cer
- PDB
protein data bank
- GPCR
G protein-coupled receptor
- EGFR
epidermal growth factor receptor
- Man
mannose
- Glc
glucose
- Gal
galactose
- Fuc
Fucose
- GlcNAc
N-acetyl-d-glucosamine
- GalNAc
N-acetyl-d-galactosamine
- NeuNAc
N-acetylneuraminic acid
- LPS
lipopolysaccharide
- KDO
3-deoxy-d-manno-oct-2-ulosonic acid
- MAC
membrane attack complex
- SI
Supporting Information
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jcim.7b00237.
This includes doglycans.py and prepreader.py tools. We also provide input sequences, structure files, and molecular topology files for all the examples described in this article. These files can be directly used in MD simulations (ZIP)
Author Contributions
⊥ R.D. and C.P. contributed equally to this work.
The authors declare no competing financial interest.
Notes
An updated version of the doglycan.py tool with track changes and the manual will be stored in a public repository at https://bitbucket.org/biophys-uh/doglycans/.
Supplementary Material
References
- Best R. B.; Zhu X.; Shim J.; Lopes P. E. M.; Mittal J.; Feig M.; MacKerell A. D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone phi, psi and Side-Chain chi(1) and chi(2) Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder E.; Damm W.; Maple J.; Wu C. J.; Reboul M.; Xiang J. Y.; Wang L. L.; Lupyan D.; Dahlgren M. K.; Knight J. L.; Kaus J. W.; Cerutti D. S.; Krilov G.; Jorgensen W. L.; Abel R.; Friesner R. A. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- Botan A.; Favela-Rosales F.; Fuchs P. F. J.; Javanainen M.; Kanduc M.; Kulig W.; Lamberg A.; Loison C.; Lyubartsev A.; Miettinen M. S.; Monticelli L.; Maatta J.; Ollila O. H. S.; Retegan M.; Rog T.; Santuz H.; Tynkkynen J. Toward Atomistic Resolution Structure of Phosphatidylcholine Headgroup and Glycerol Backbone at Different Ambient Conditions. J. Phys. Chem. B 2015, 119, 15075–15088. 10.1021/acs.jpcb.5b04878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubartsev A. P.; Rabinovich A. L. Force Field Development for Lipid Membrane Simulations. Biochim. Biophys. Acta, Biomembr. 2016, 1858, 2483–2497. 10.1016/j.bbamem.2015.12.033. [DOI] [PubMed] [Google Scholar]
- Hart K.; Foloppe N.; Baker C. M.; Denning E. J.; Nilsson L.; MacKerell A. D. Optimization of the CHARMM Additive Force Field for DNA: Improved Treatment of the BI/BII Conformational Equilibrium. J. Chem. Theory Comput. 2012, 8, 348–362. 10.1021/ct200723y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning E. J.; Priyakumar U. D.; Nilsson L.; Mackerell A. D. Impact of 2 ′-Hydroxyl Sampling on the Conformational Properties of RNA: Update of the CHARMM All-Atom Additive Force Field for RNA. J. Comput. Chem. 2011, 32, 1929–1943. 10.1002/jcc.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares T. A.; Hunenberger P. H.; Kastenholz M. A.; Krautler V.; Lenz T.; Lins R. D.; Oostenbrink C.; Van Gunsteren W. F. An Improved Nucleic Acid Parameter Set for the GROMOS Force Field. J. Comput. Chem. 2005, 26, 725–737. 10.1002/jcc.20193. [DOI] [PubMed] [Google Scholar]
- Zgarbova M.; Otyepka M.; Sponer J.; Mladek A.; Banas P.; Cheatham T. E.; Jurecka P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theory Comput. 2011, 7, 2886–2902. 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivani I.; Dans P. D.; Noy A.; Perez A.; Faustino I.; Hospital A.; Walther J.; Andrio P.; Goni R.; Balaceanu A.; Portella G.; Battistini F.; Gelpi J. L.; Gonzalez C.; Vendruscolo M.; Laughton C. A.; Harris S. A.; Case D. A.; Orozco M. Parmbsc1: a Refined Force Field for DNA Simulations. Nat. Methods 2015, 13, 55–58. 10.1038/nmeth.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kony D.; Damm W.; Stoll S.; van Gunsteren W. F. An Improved OPLS-AA Force Field for Carbohydrates. J. Comput. Chem. 2002, 23, 1416–1429. 10.1002/jcc.10139. [DOI] [PubMed] [Google Scholar]
- Pol-Fachin L.; Rusu V. H.; Verli H.; Lins R. D. GROMOS 53A6GLYC, an Improved GROMOS Force Field for Hexopyranose-Based Carbohydrates. J. Chem. Theory Comput. 2012, 8, 4681–4690. 10.1021/ct300479h. [DOI] [PubMed] [Google Scholar]
- Plazinski W.; Lonardi A.; Hünenberger P. H. Revision of the GROMOS 56A6CARBO Force Field: Improving the Description of Ring-Conformational Equilibria in Hexopyranose-Based Carbohydrates Chains. J. Comput. Chem. 2016, 37, 354–365. 10.1002/jcc.24229. [DOI] [PubMed] [Google Scholar]
- Kirschner K. N.; Yongye A. B.; Tschampel S. M.; Gonzalez-Outeirino J.; Daniels C. R.; Foley B. L.; Woods R. J. GLYCAM06: A Generalizable Biomolecular Force Field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods Group. GLYCAM Web. Complex Carbohydrate Research Center. http://glycam.org/ (accessed 01.07.16).
- Jo S.; Song K. C.; Desaire H.; MacKerell A. D.; Im W. Glycan Reader: Automated Sugar Identification and Simulation Preparation for Carbohydrates and Glycoproteins. J. Comput. Chem. 2011, 32, 3135–3141. 10.1002/jcc.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S.; Kim T.; Iyer V. G.; Im W. Software News and Updates - CHARMM-GUI: A Web-Based Grraphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Cornell W. D.; Cieplak P.; Bayly C. I.; Gould I. R.; Merz K. M.; Ferguson D. M.; Spellmeyer D. C.; Fox T.; Caldwell J. W.; Kollman P. A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1996, 118, 2309–2309. 10.1021/ja955032e. [DOI] [Google Scholar]
- Bohne-Lang A.; von der Lieth C. W. GlyProt: in Silico Glycosylation of Proteins. Nucleic Acids Res. 2005, 33, W214–W219. 10.1093/nar/gki385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohne A.; Lang E.; von der Lieth C. W. W3-SWEET: Carbohydrate Modeling by Internet. J. Mol. Model. 1998, 4, 33–43. 10.1007/s008940050068. [DOI] [Google Scholar]
- Frank M.; Bohne-Lang A.; Wetter T.; von der Lieth C. W. Rapid Generation of a Representative Ensemble of N-Glycan Conformations. In Silico Biol. 2002, 2, 427–439. [PubMed] [Google Scholar]
- Engelsen S. B.; Hansen P. I.; Perez S. POLYS 2.0: An Open Source Software Package for Building Three-Dimensional Structures of Polysaccharides. Biopolymers 2014, 101, 733–743. 10.1002/bip.22449. [DOI] [PubMed] [Google Scholar]
- Rosen J.; Miguet L.; Perez S. Shape: Automatic Conformation Prediction of Carbohydrates Using a Genetic Algorithm. J. Cheminf. 2009, 1, 16. 10.1186/1758-2946-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N. M.; Banck M.; James C. A.; Morley C.; Vandermeersch T.; Hutchison G. R. Open Babel: an Open Chemical Toolbox. J. Cheminf. 2011, 3, 33. 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B.; Kutzner C.; van der Spoel D.; Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Paavilainen S.; Rog T.; Vattulainen I. Analysis of Twisting of Cellulose Nanofibrils in Atomistic Molecular Dynamics Simulations. J. Phys. Chem. B 2011, 115, 3747–3755. 10.1021/jp111459b. [DOI] [PubMed] [Google Scholar]
- Rudd P. M.; Morgan B. P.; Wormald M. R.; Harvey D. J.; vandenBerg C. W.; Davis S. J.; Ferguson M. A. J.; Dwek R. A. The Glycosylation of the Complement Regulatory Protein, Human Erythrocyte CD59. J. Biol. Chem. 1997, 272, 7229–7244. 10.1074/jbc.272.11.7229. [DOI] [PubMed] [Google Scholar]
- Kaszuba K.; Grzybek M.; Orlowski A.; Danne R.; Rog T.; Simons K.; Coskun U.; Vattulainen I. N-Glycosylation as Determinant of Epidermal Growth Factor Receptor Conformation in Membranes. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 4334–4339. 10.1073/pnas.1503262112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D.; Binnington B.; Rog T.; Vattulainen I.; Grzybek M.; Coskun U.; Lingwood C. A.; Simons K. Cholesterol Modulates Glycolipid Conformation and Receptor Activity. Nat. Chem. Biol. 2011, 7, 260–262. 10.1038/nchembio.551. [DOI] [PubMed] [Google Scholar]
- Kamel S.; Ali N.; Jahangir K.; Shah S. M.; El-Gendy A. A. Pharmaceutical Significance of Cellulose: A Review. eXPRESS Polym. Lett. 2008, 2, 758–778. 10.3144/expresspolymlett.2008.90. [DOI] [Google Scholar]
- Abdul Khalil H.; Bhat A. H.; Yusra A. F. I. Green Composites from Sustainable Cellulose Nanofibrils: A Review. Carbohydr. Polym. 2012, 87, 963–979. 10.1016/j.carbpol.2011.08.078. [DOI] [Google Scholar]
- Eichhorn S. J.; Dufresne A.; Aranguren M.; Marcovich N. E.; Capadona J. R.; Rowan S. J.; Weder C.; Thielemans W.; Roman M.; Renneckar S.; Gindl W.; Veigel S.; Keckes J.; Yano H.; Abe K.; Nogi M.; Nakagaito A. N.; Mangalam A.; Simonsen J.; Benight A. S.; Bismarck A.; Berglund L. A.; Peijs T. Review: Current International Research into Cellulose Nanofibres and Nanocomposites. J. Mater. Sci. 2010, 45, 1–33. 10.1007/s10853-009-3874-0. [DOI] [Google Scholar]
- Paavilainen S.; McWhirter J. L.; Rog T.; Jarvinen J.; Vattulainen I.; Ketoja J. A. Mechanical Properties of Cellulose Nanofibrils Determined Through Atomistic Molecular Dynamics Simulations. Nord. Pulp Pap. Res. J. 2012, 27, 282–286. 10.3183/NPPRJ-2012-27-02-p282-286. [DOI] [Google Scholar]
- Orlowski A.; Rog T.; Paavilainen S.; Manna M.; Heiskanen I.; Backfolk K.; Timonen J.; Vattulainen I. How Endoglucanase Enzymes Act on Cellulose Nanofibrils: Role of Amorphous Regions Revealed by Atomistic Simulations. Cellulose 2015, 22, 2911–2925. 10.1007/s10570-015-0705-0. [DOI] [Google Scholar]
- Spiro R. G.Advances in Protein Chemistry. In Glycoproteins; Anfinsen C. B., Edsall J. T., Richards F. M., Eds.; Academic Press Inc., Elsevier Inc, 1973; pp 349–467. [DOI] [PubMed] [Google Scholar]
- Meyer B.; Möller H.. Glycopeptides and Glycoproteins. In Conformation of Glycopeptide and Glycoproteins; Wittmann V., Ed.; Springer-Verlag: Berlin, Heidelberg, 2006; pp 187–251. [Google Scholar]
- Spiro R. G. Protein Glycosylation: Nature, Distribution, Enzymatic Formation, and Disease Implications of Glycopeptide Bonds. Glycobiology 2002, 12, 43R–56R. 10.1093/glycob/12.4.43R. [DOI] [PubMed] [Google Scholar]
- Wormald M. R.; Rudd P. M.; Harvey D. J.; Chang S. C.; Scragg I. G.; Dwek R. A. Variations in Oligosaccharide-Protein Interactions in Immunoglobulin G Determine the Site-Specific Glycosylation Profiles and Modulate the Dynamic Motion of the Fc Oligosaccharides. Biochemistry 1997, 36, 1370–1380. 10.1021/bi9621472. [DOI] [PubMed] [Google Scholar]
- Shental-Bechor D.; Levy Y. Folding of Glycoproteins: Toward Understanding the Biophysics of the Glycosylation Code. Curr. Opin. Struct. Biol. 2009, 19, 524–533. 10.1016/j.sbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Shental-Bechor D.; Levy Y. Effect of Glycosylation on Protein Folding: A Close Look at Thermodynamic Stabilization. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 8256–8261. 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintegaard T. V.; Thygesen P.; Rahbek-Nielsen H.; Levery S. B.; Kristensen C.; Clausen H.; Bolt G. N-Glycosylation Increases the Circulatory Half-Life of Human Growth Hormone. Endocrinology 2010, 151, 5326–5336. 10.1210/en.2010-0574. [DOI] [PubMed] [Google Scholar]
- van Kooyk Y.; Rabinovich G. A. Protein-Glycan Interactions in the Control of Innate and Adaptive Immune Responses. Nat. Immunol. 2008, 9, 593–601. 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- Kornfeld S. Structure and Function of the Mannose 6-Phosphate/Insulinlike Gowth Factor II Receptors. Annu. Rev. Biochem. 1992, 61, 307–330. 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- Helenius A.; Aebi M. Intracellular Functions of N-linked Glycans. Science 2001, 291, 2364–2369. 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Ruiz-Arguelles A.; Llorente L. The Role of Complement Regulatory Proteins (CD55 and CD59) in the Pathogenesis of Autoimmune Hemocytopenias. Autoimmun. Rev. 2007, 6, 155–161. 10.1016/j.autrev.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Rog T.; Vattulainen I. Cholesterol, Sphingolipids, and Glycolipids: What Do We Know about Their Role in Raft-Like Membranes?. Chem. Phys. Lipids 2014, 184, 82–104. 10.1016/j.chemphyslip.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Manna M.; Rog T.; Vattulainen I. The Challenges of Understanding Glycolipid Functions: an Open Outlook Based on Molecular Simulations. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2014, 1841, 1130–1145. 10.1016/j.bbalip.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Sandvig K.; Bergan J.; Kavaliauskiene S.; Skotland T. Lipid Requirements for Entry of Protein Toxins into Cells. Prog. Lipid Res. 2014, 54, 1–13. 10.1016/j.plipres.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Coskun U.; Grzybek M.; Drechsel D.; Simons K. Regulation of Human EGF Receptor by Lipids. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 9044–9048. 10.1073/pnas.1105666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood C. A. Glycosphingolipid Functions. Cold Spring Harbor Perspect. Biol. 2011, 3, a011874. 10.1101/cshperspect.a011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A. A. S. T.; Horta B. A. C.; de Alencastro R. B. MKTOP: a Program for Automatic Construction of Molecular Topologies. J. Braz. Chem. Soc. 2008, 19, 1433–1435. 10.1590/S0103-50532008000700031. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.