Abstract
Background
Flos Chrysanthemi, as a part of Chinese culture for a long history, is valuable for not only environmental decoration but also the medicine and food additive. Due to their voluminously various breeds and extensive distributions worldwide, it is burdensome to make recognition and classification among numerous cultivars with conventional methods which still rest on the level of morphologic observation and description. As a fingerprint spectrum for parsing molecular information, surface-enhanced Raman scattering (SERS) could be a suitable candidate technique to characterize and distinguish the inter-cultivar differences at molecular level.
Results
SERS spectra were used to analyze the inter-cultivar differences among 26 cultivars of Chinese ornamental Flos Chrysanthemum. The characteristic peaks distribution patterns were abstracted from SERS spectra and varied from cultivars to cultivars. For the bands distributed in the pattern map, the similarities in general showed their commonality while in the finer scales, the deviations and especially the particular bands owned by few cultivars revealed their individualities. Since the Raman peaks could characterize specific chemical components, those diversity of patterns could indicate the inter-cultivar differences at the chemical level in fact.
Conclusion
In this paper, SERS technique is feasible for distinguishing the inter-cultivar differences among Flos Chrysanthemum. The Raman spectral library was built with SERS characteristic peak distribution patterns. A new method was proposed for Flos Chrysanthemum recognition and taxonomy.
Keywords: Flos Chrysanthemi, Raman spectra, SERS, Inter-cultivar differences, Plant taxonomy
Background
Flos Chrysanthemi, the flower of Chrysanthemum morifolium Ramat, was regarded as one of the most much-loved traditional flowers in China with a long history for its surpassingly beautiful look, richness of color, abundant of breeds and wide adaptability. The Flos Chrysanthemi recorded in ancient classic of China can be dated back to Spring and Autumn period of Zhou Dynasty in about 3000 years ago. In traditional Chinese and Korean medicines, Flos Chrysanthemi has been widely used as herbal medicine in clinical practice for the treatment of colitis, cancer, hypertension, pneumonia, pertussis, and stomatitis [1–7]. Additionally, Flos Chrysanthemi was also taken as conventional health food from beverages to spices such as Flos Chrysanthemi tea, a popular Chinese drink prepared by dry Flos Chrysanthemi in combination with boiling or hot water, and food additive for masking flavors in alcoholic beverages in Korea [8].
As a precious resource gifted by nature for human, Flos Chrysanthemi has a very abundant reserve, various type and extensive distribution through wide spread and cultivation around the world. According to investigation statistics, the total amount of Flos Chrysanthemum cultivars was estimated as 20,000–30,000 and at least 3000 in China alone [9]. Quite a few studies for Flos Chrysanthemi taxonomy in early time depended on relatively conventional methods at the level of morphologic observation and description [10–12]. Conventional taxonomy focused on macroscopic traits in quantitative (plant height, tubular flower diameter, receptacle size, etc.) and qualitative (stem form, petal distribution, receptacle shape, etc.) aspects. Japanese horticulturists usually classify Flos Chrysanthemum depend on the practical purpose into edibles and ornamentals two categories, and the ornamentals were divided into small, medium and big three types where the big type were further divided into about 20 subtypes. British gardeners tended to systematize the Flos Chrysanthemum into early, medium and late three categories based on flowering time, and in each category they were further divided into 10 subtypes. Flos Chrysanthemum classification in the United States was similar to that in Britain, which contained 15 categories.
Several studies that classifying Flos Chrysanthemum at chemical or molecular level were reported in recent years. Zhao Y et al. tried to make a classification for 20 varieties of Chrysanthemum by analyzing the peroxidase isozymes (PODs) using polyacrylamide gel electrophoresis (PAGE) [13]. Similarly, another study was reported to have analyzed the PODs in 20 cultivars of Dendranthema × grandiflorum hybrid groups and found the enzyme spectrums of distinct groups had significant difference, which reflected the genetic diversity of the Dendranthema × grandiflorum varieties [14]. Besides, a recent research used both PODs and esterase isozyme (EST) analysis to investigate the genetic diversity for 93 cultivars in Dendranthema × grandiflorum [15]. But the studies were performed with PAGE whose disadvantages of low efficiency and cumbersome operation are obviously: one new technique of high accuracy and efficiency was expected.
Raman scattering, a vibrational spectral technology, was widely applied in the field of material science for substance detection and analysis since it was found by Raman C.V. in 1928 [16]. Theorized as a kind of inelastic light scattering process, Raman scattering has the capacity of characterizing the vibration, rotation, phase transition and crystals structure of molecules specifically so that it could detect overall composition and analyze relative quantification as a fingerprint spectrum. With the development of laser technology and the discovery of enhancement effect by Fleischmann M. in 1974 [17], a breakthrough measurement science termed as surface enhanced Raman scattering (SERS) was born and got a quickly and widely application in material, chemistry and life science field owing to its getting rid of conventional Raman scattering’s shortcoming of rather low intensity. SERS enlarged Raman scattering’s usable scope and was regarded to be an accurate and efficient solution in substance detection and analysis.
In the past few decades, Raman spectroscopic technique was widely used for biological and medical researches, such as bacteria discrimination [18, 19], blood assay [20, 21], and cancer diagnosis [22–24]. While in phytological field, Raman spectroscopic technique was also used to plant identifying and analysis. According to a relevant study, Fourier-transform Raman spectroscopy with 1064 nm excitation was used to discriminate Green Arabica and Robusta coffee beans by monitoring the characteristic Raman bands of kahweol [25]. In another similar research, those two kinds of coffee beans were discriminated using the Fourier-transform Raman spectroscopy by analyzing the extracted lipid fractions [26]. Raman spectroscopy was also reported in discrimination for pollens of different plants. As reported, a set of allergy-relevant pollens, including common ragweed, white birch, English oak, and European linden, were studied using Raman spectroscopy with 633 nm excitation and were successfully distinguished by analyzing and comparing their specific chemical Raman peaks [27]. Raman spectroscopic technique was likely effective in discrimination for different Flos Chrysanthemum cultivars. Therefore, in this study, SERS with sliver nanoparticles colloid as enhancement substance was applied to observe the spectral differences among 26 cultivars of Chinese ornamental Flos Chrysanthemum and proposes a new method for classifying Flos Chrysanthemi by SERS technology.
Methods
Flos Chrysanthemi samples preparation
All the fresh Flos Chrysanthemi corollas, 26 in total, were gifted by the gardening center affiliated with logistics department of Shanghai University. After adequately grinded, the homogenate of each corolla was collected and transferred into a clean centrifuge tube. Then the tubes were centrifuged at 2000 rpm for 10 min and the upper aqueous phases were transferred to fresh tubes as samples for SERS measurement.
Silver nanoparticles preparation
Sliver colloid was synthesized referring to Lee’s sodium citrate reduction method [28]. Briefly, 1 mL of a 0.1 mol/L sliver nitrate (AgNO3, AP, Aladdin) solution was added into 99 mL deionized water (18.3 MΩ*cm, 25 °C) and heated to boiling. 1.9 mL of a 1% sodium citrate tribasic (Na3C6H5O7, AP, Aladdin) solution was then added with rapid stirring. The mixture was then kept boiling gently till the color turned celadon. All glassware used were cleaned carefully and thoroughly rinsed with deionized water. The synthesized sliver colloid was checked by Raman scattering before use so as to ensure its availability.
SERS measurement
Each aqueous phase sample extracted from Flos Chrysanthemi corolla was homogeneously mixed with the sliver colloid at the proportion of 1:2 (v/v) and then the mixture was standing for 5 min. A Raman microscope (LabRAM HR Evolution, Horiba, JP) equipped with a 785 nm He–Ne laser device was employed to excite and collect the Raman spectra, in which the initial parameters were configured as follows: exciting power of 3.3 mW (10% of the maximum power of the laser device), resolution of 1 cm−1, acquisition time of 10 s and scan range from 400 to 1800 cm−1. Every time before measuring, the system was calibrated by 520 cm−1 band from silicon reference sample. The laser was focused on the bottom surface of the liquid sample via a L50× microscope objective (N.A. 0.5). Each sample was measured for 6 parallel spectra from different spots to reduce the random error.
Data processing and analysis
All raw Raman spectra acquisitions were performed with LabSpec software (Horiba, Japan). The baselines of raw data were corrected by Subase V2.10 software (self-developed, HyStudio, Shanghai University, China) with integration of a Vancouver Raman Algorithm based on fifth-order polynomial fitting method [23] for fluorescence background removal.
Results
The preliminary pre-experiments were performed in order to optimize the experimental conditions and measurement parameters. The frequently-used excitation lasers in conventional Raman measurement for bio-samples were visible light (532 and 633 nm) and near-infrared light (785 nm). In most cases, the shorter the excitation wavelength is, the stronger the fluorescence background will be induced. However, trends are reverse for the thermal effects: longer-wavelength laser might heat the samples more strongly, especially the bio-samples. Therefore, although the shorter-wavelength lasers, like 532 and 633 nm light, were relatively poor in heating bio-samples, the fluorescence background they induced might be a non-negligible interference for the Raman spectral measurement. While, the longer-wavelength lasers, such as 785 nm light, were efficient with lower fluorescence background, but the robust thermal action on bio-samples might potentially limited their usage (irradiation power and irradiating time). For these reasons, an appropriate excitation wavelength and power were rather vital for achieving better measurements. We tentatively conducted the Raman spectral measurement for Marshal Flag cultivar using the 633 and 785 nm lasers to test and compare the fluorescence interferences. The result showed that the fluorescence background induced by 633 nm excitation light was stronger than that of 785 nm laser: the baseline was raised in the range from 1100 to 1700 cm−1 (Fig. 1). Since higher fluorescence background will bring about more difficulties for baseline correction and affect the reliability of spectral data, 785 nm light was chosen as the excitation laser for the Raman spectral measurement.
Fig. 1.
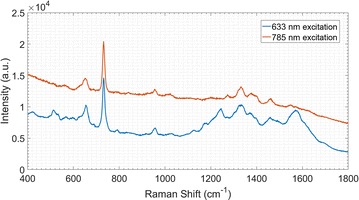
SERS spectra for Marshal Flag cultivar with excitation wavelength of 633 nm and 785 nm (10% of maximum laser device power)
To minimize the thermal damage to the samples, the 785 nm laser should be used in a proper power: higher power would destroy the active components in the samples while much lower power can’t acquire adequate spectral intensities. 0.033, 0.33, 1.65, 3.3, 6.6 and 16.5 mW excitation lasers of 785 nm (corresponding to the 0.1%, 1%, 5%, 10%, 20% and 50% of the maximum laser device output power, respectively) were used in Raman spectral measurement for Blue Eye cultivar as an example. The result showed that the overall Raman spectral intensities seemed to grow with the increase of laser power (Fig. 2a). In further analysis for individual peaks, this growth trends were not stable in all power level. In lower power levels (0.033 and 0.33 mW), the intensities were rather weak so that the random noise signals made them difficult to represent real spectra. And in higher power levels (6.6 and 16.5 mW), the intensity in peak 1461 cm−1 was slightly decrease at 6.6 mW power level, compared with the 3.3 mW level; the intensity in peak 959 cm−1 was slightly stronger at 6.6 mW power level than that at 16.5 mW power level (Fig. 2b). Moreover, the intensity growth trends of 959 and 1461 cm−1 at 6.6 and 16.5 mW power levels were not consistent with that at the middle power levels (1.65 and 3.3 mW). Those typical abnormal relationships between Raman spectra and the excitation power in higher levels might be attributed to that the thermal effect of higher power laser caused some certain impacts on some certain components in the bio-samples. In addition, the intensities at 3.3 mW power level were higher than that at 1.65 mW and Raman spectra were adequate for measurement. Therefore, in comprehensive consideration, the 3.3 mW 785 nm laser was used as the excitation light for the Raman spectral measurement.
Fig. 2.
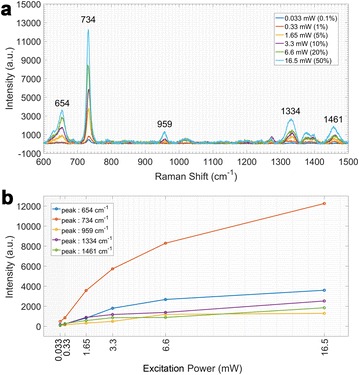
The tentative relationship between the laser excitation power and SERS spectra. a The SERS spectral intensity goes with the laser excitation power (for instance of the Blue Eye cultivar, 785 nm excitation). b The growing trends of intensities for individual peaks might be related to the laser excitation power (for instance of the Blue Eye cultivar, 785 nm excitation)
With the enhancement of sliver colloid, all SERS spectra of 26 cultivars were measured and their fluorescence backgrounds were removed by baseline subtraction. For each sample, the post-correction spectra signal data were averaged to be the mean spectrum and the characteristic peaks were extracted by picking out the distinct and tangible protuberances in spectral curves (height more than 400 a.u. and width less than 100 cm−1). All characteristic peaks of every sample were normalized by the max value among them so that the distribution patterns, including the relative heights (vertical axis) and positions (horizontal axis) in the measured range could be abstracted and recorded into a data library as the references for distinguishing different cultivars (Table 1). In order to make the characteristic peaks distribution patterns observed visually, the normalized peaks data of all the samples were mapped into a color block image, where the value range from 0 to 1 corresponded to the hue from violet to red (Fig. 4).
Table 1.
Raman spectral characteristic peaks and normalized intensities of all 26 Flos Chrysanthemum samples
| Common cultivar name | Characteristic peaks (Raman shift, cm−1) and normalized intensities | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Golden Queen | Peaks | 569 | 624 | 652 | 731 | 958 | 1324 | 1462 | ||||
| Normalized intensities | 0.049 | 0.046 | 0.155 | 1 | 0.136 | 0.171 | 0.078 | |||||
| Madame Guo | Peaks | 651 | 731 | 958 | 1018 | 1333 | 1460 | |||||
| Normalized intensities | 0.175 | 1 | 0.101 | 0.084 | 0.221 | 0.099 | ||||||
| Flying Ziyan | Peaks | 657 | 733 | 1026 | 1275 | 1335 | 1459 | |||||
| Normalized intensities | 0.208 | 1 | 0.084 | 0.095 | 0.220 | 0.135 | ||||||
| Fenghuang | Peaks | 647 | 731 | 1334 | ||||||||
| Normalized intensities | 0.150 | 1 | 0.198 | |||||||||
| Red Frost | Peaks | 654 | 733 | 954 | 1023 | 1272 | 1334 | 1459 | ||||
| Normalized intensities | 0.266 | 1 | 0.093 | 0.093 | 0.095 | 0.199 | 0.135 | |||||
| Red Jinbei | Peaks | 654 | 731 | 957 | 1017 | 1334 | 1461 | |||||
| Normalized intensities | 0.158 | 1 | 0.131 | 0.072 | 0.192 | 0.084 | ||||||
| Marshal Flag | Peaks | 567 | 654 | 732 | 959 | 1273 | 1333 | 1377 | 1461 | 1550 | ||
| Normalized intensities | 0.057 | 0.263 | 1 | 0.098 | 0.069 | 0.209 | 0.104 | 0.120 | 0.056 | |||
| Jade Feng | Peaks | 516 | 568 | 656 | 732 | 957 | 1174 | 1201 | 1331 | 1377 | 1455 | 1547 |
| Normalized intensities | 0.069 | 0.084 | 0.333 | 1 | 0.102 | 0.067 | 0.082 | 0.280 | 0.071 | 0.186 | 0.083 | |
| White Jade | Peaks | 512 | 564 | 654 | 732 | 958 | 1022 | 1329 | 1377 | 1457 | ||
| Normalized intensities | 0.065 | 0.065 | 0.266 | 1 | 0.114 | 0.0422 | 0.203 | 0.061 | 0.112 | |||
| White Lion | Peaks | 564 | 657 | 732 | 957 | 1020 | 1271 | 1333 | 1373 | 1459 | ||
| Normalized intensities | 0.064 | 0.182 | 1 | 0.078 | 0.060 | 0.089 | 0.227 | 0.082 | 0.138 | |||
| Golden Lotus | Peaks | 656 | 733 | 957 | 1333 | 1374 | 1461 | |||||
| Normalized intensities | 0.275 | 1 | 0.097 | 0.220 | 0.083 | 0.142 | ||||||
| Green Zhaoyun | Peaks | 570 | 654 | 732 | 959 | 1022 | 1270 | 1333 | 1376 | 1399 | 1457 | |
| Normalized intensities | 0.051 | 0.273 | 1 | 0.115 | 0.055 | 0.049 | 0.203 | 0.104 | 0.087 | 0.138 | ||
| Gushui Liuxia | Peaks | 563 | 657 | 733 | 957 | 1021 | 1272 | 1333 | 1379 | 1401 | 1457 | |
| Normalized intensities | 0.065 | 0.287 | 1 | 0.102 | 0.084 | 0.081 | 0.209 | 0.106 | 0.082 | 0.155 | ||
| Green Window | Peaks | 515 | 567 | 657 | 732 | 958 | 1173 | 1245 | 1334 | 1457 | 1578 | |
| Normalized intensities | 0.133 | 0.062 | 0.351 | 1 | 0.134 | 0.064 | 0.180 | 0.148 | 0.142 | 0.342 | ||
| Fireworks | Peaks | 649 | 731 | 958 | 1331 | 1403 | ||||||
| Normalized intensities | 0.112 | 1 | 0.136 | 0.218 | 0.069 | |||||||
| Golden Needle | Peaks | 733 | 1331 | 1375 | 1459 | 1576 | ||||||
| Normalized intensities | 1 | 0.216 | 0.104 | 0.107 | 0.145 | |||||||
| Blue Eye | Peaks | 654 | 734 | 959 | 1273 | 1334 | 1461 | 1549 | ||||
| Normalized intensities | 0.288 | 1 | 0.080 | 0.126 | 0.198 | 0.157 | 0.076 | |||||
| Clean Water Hehua | Peaks | 567 | 653 | 732 | 957 | 1274 | 1333 | 1372 | 1462 | |||
| Normalized intensities | 0.065 | 0.249 | 1 | 0.118 | 0.064 | 0.202 | 0.059 | 0.115 | ||||
| Mini Ju | Peaks | 655 | 733 | 957 | 1018 | 1278 | 1331 | 1463 | ||||
| Normalized intensities | 0.260 | 1 | 0.113 | 0.098 | 0.086 | 0.211 | 0.156 | |||||
| Gold Ball | Peaks | 513 | 595 | 656 | 733 | 1223 | 1333 | 1377 | 1459 | 1576 | ||
| Normalized intensities | 0.115 | 0.077 | 0.183 | 1 | 0.088 | 0.227 | 0.076 | 0.119 | 0.211 | |||
| Beautiful Purple | Peaks | 732 | ||||||||||
| Normalized intensities | 1 | |||||||||||
| Beijing Red | Peaks | 733 | ||||||||||
| Normalized intensities | 1 | |||||||||||
| Mini Red Peach | Peaks | 731 | 1335 | |||||||||
| Normalized intensities | 1 | 0.188 | ||||||||||
| Afric Ju | Peaks | 649 | 731 | 957 | 1335 | |||||||
| Normalized intensities | 0.230 | 1 | 0.130 | 0.212 | ||||||||
| White Powder | Peaks | 568 | 651 | 732 | 896 | 957 | 1272 | 1334 | 1372 | 1457 | ||
| Normalized intensities | 0.073 | 0.188 | 1 | 0.078 | 0.092 | 0.065 | 0.213 | 0.077 | 0.117 | |||
| Golden Mudan | Peaks | 651 | 732 | 959 | 1326 | 1383 | 1399 | 1459 | ||||
| Normalized intensities | 0.187 | 1 | 0.132 | 0.223 | 0.081 | 0.082 | 0.115 | |||||
Fig. 4.
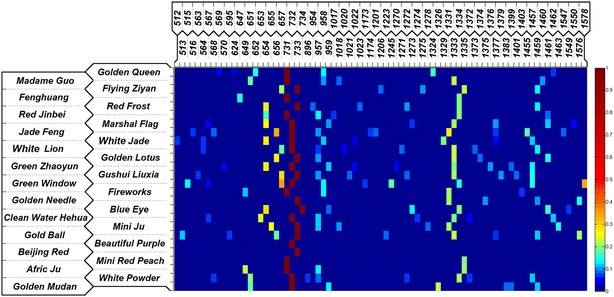
Characteristic peaks distribution patterns map for all 26 Flos Chrysanthemum samples
Golden Queen and Mini Red Peach breed are taken as examples to illustrate how to parse the inter-cultivar differences by SERS spectra. In spite of the similarity lying in the trend and shape between these two spectra (Fig. 3a, b), their detailed traits including peak heights make obvious differences (Fig. 3c). In addition, their characteristic peak distribution patterns could contribute to distinguishing and matching the different cultivars via less dimensionality (Figs. 3d, 4).
Fig. 3.
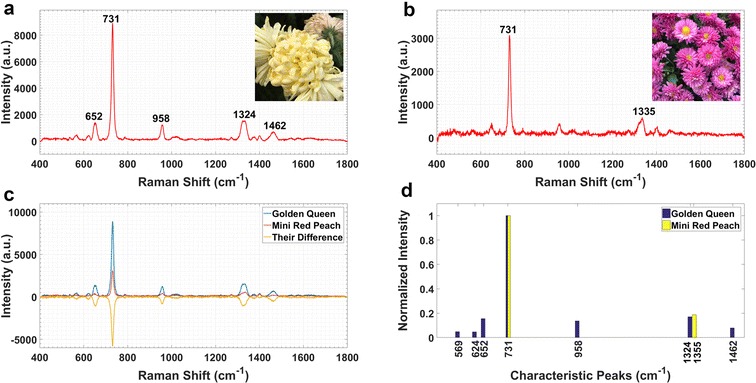
SERS spectra reveal the differences among cultivars of Flos Chrysanthemum. a Spectrum and image of Golden Queen cultivar. b Spectrum and image of Mini Red Peach cultivar. c The differences between these two spectra. d The contrast between their distribution patterns of normalized characteristic peaks
Except for these two cultivars, the rest in the spectral library are shown in Figs. 4, 5 and Table 1. It can be viewed in the characteristic peaks distribution patterns map (Fig. 4) and spectral library (Fig. 5) that the general outlines and trends of spectra from all samples were similar in a broad scale; after all, they belonged to the same species and contained semblable chemical constituents. Almost all spectra contained a remarkable peak with the strongest scattering intensity in the range of 731–734 cm−1, which might be assigned to the gallates [29], a kind of antioxidant phenolic esters contained in Chrysanthemum [30]. In addition to the 731–734 cm−1 bands, the characteristic peaks distribution patterns map (Fig. 4) showed another 4 obvious bands of peaks in the vertical angle of view (651–657, 954–959, 1324–1135 and 1455–1462 cm−1). The characteristic peaks in range of 651–657 cm−1 could be assigned to the 1,8-Cineol (δ-ring vibration), a kind of disaccharides in Chrysanthemum [31] and the weaker bands in 954–959 cm−1 might be assigned to the carotenoid [32]. Bands from 1324 to 1135 cm−1 might be attributed to the chlorophyll a and b, which were found in most parts of the plants [33, 34]. The peaks distributed in range of 1455–1463 might be assigned to the sucrose, which widely existed in plants [35, 36].
Fig. 5.
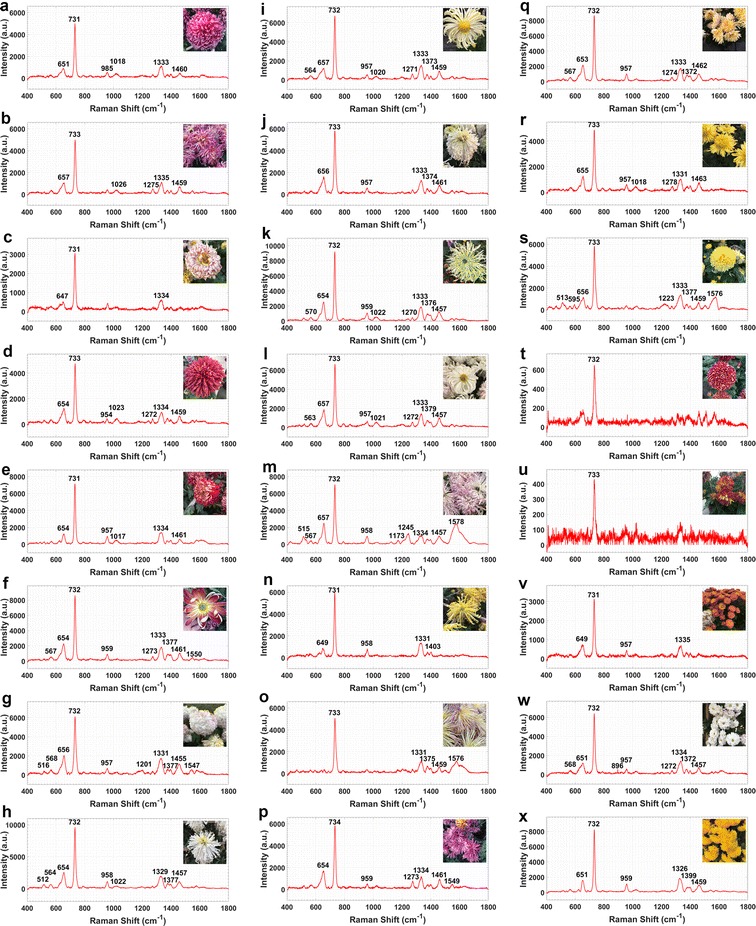
Part of SERS spectra and images from the spectral library of Chinese ornamental Flos Chrysanthemum. a Madame Guo. b Flying Ziyan. c Fenghuang. d Red Frost. e Red Jinbei. f Marshal Flag. g Jade Feng. h White Jade. i White Lion. j Golden Lotus. k Green Zhaoyun. l Gushui Liuxia. m Green Window. n Fireworks. o Golden Needle. p Blue Eye. q Clean Water Huhua. r Mini Ju. s Gold Ball. t Beautiful Purple. u Beijing Red. v Afric Ju. w White Powder. x Golden Mudan
Apart from the comparatively universal peaks in most cultivars described above, there also existed other peaks which were unique for specific cultivars and that might be just the reason why they differed. The cultivar White Lion had a characteristic peak (1020 cm−1) in it, which could be assigned to the crocetin, making itself distinct from others. If take measurement errors in consideration, Red Jinbei (1017 cm−1), Madam Guo (1018 cm−1), Mini Ju (1018 cm−1), Gushui Liuxia (1021 cm−1), Green Zhaoyun (1022 cm−1), White Jade (1022 cm−1) and Red Frost (1023 cm−1) could also be tentatively inferred to contain crocetin because the peaks were close to 1020 cm−1 in an acceptable range. The characteristic peaks distribution patterns abstracted from the SERS spectra varied from cultivars to cultivars. For the bands distributed in the pattern map (Fig. 4), the similarities in general showed their commonality while in the finer scales, the deviations and especially the particular bands owned by few cultivars revealed their individualities. Since the Raman peaks could characterize specific chemical components, those diversity of patterns could indicate the inter-cultivar differences at the chemical level in fact. Furthermore, a SERS spectral library, containing the images, spectra and the corresponding characteristic peak distribution patterns of all 26 cultivars of Chinese ornamental Flos Chrysanthemum within this study, was built in order to provide convenience for Flos Chrysanthemum finding, referring, recognition and taxonomy researches.
Discussion
Raman scattering, a vibrational molecular spectroscopic technique, has emerged as one of the major tools in biological application, especially the components detection and identification. In Raman spectra analysis, the Raman shift is unique to individual molecules topically while globally the Raman spectrum, the plot of scattering intensity as a function of wavenumber, varies for different components. In finer spatial scales, the peaks in spectrum could always correspond to particular chemical bonds in the molecule, for which the Raman spectrum was regarded as a molecular specific fingerprint spectrum to provide direct information from chemical components. Compared with other conventional methods used for substance and material analysis, Raman scattering technique was relatively simple, reproducible, nondestructive and only a small part of samples were needed. In view of these advantages, we tried to use this technique to analyze the inter-cultivar differences in Flos Chrysanthemum.
The surface-enhanced Raman scattering (SERS), an enhancing effect for Raman scattering by rough metal nanoparticles [17], could overcome the shortcoming of low sensitivity in conventional Raman spectroscopy and became a rather mature technology for substance detection and analysis. In this study, we used sliver colloids as enhancement substrate for SERS measurement and achieved relative good enhancement effects.
Since a clear and narrow spectral peak corresponds to a specific vibrational bond in molecules, in many cases a combination of several peaks could represent a certain molecule. However, probably not all the bonds in a molecule would be sensitive to Raman scattering, even only one spectral peak being observed mostly. In general, bio-samples such as cells, tissues and secretions, contained numerous kinds of molecular components liked proteins, nucleic acids, lipids, amino acids and etc.. Wherein the macromolecules were synthesized by a combination of small molecules; for instance, the proteins were made of a variety of amino acids. And moreover, many cases existed that the same bond distributed in different molecules; for example the π-bond vibration of aromatic ring laid in both tryptophan and tyrosine. Therefore, it was essential to select a unique bond vibration as the characteristic peak to make the similar molecules distinct.
Thanks to the set of tentative peaks assigned for part of bio-components in plants provided by the published reports (Table 2), it is convenient for us to infer the underlying specific molecules according to the peaks in Raman spectrum. It should be noted that the peak was in a narrow range rather than an exact value because the system errors and random errors could not be avoided. The system errors resulted from instrumental effects might lead to the difference between the value measured in this study and in the referenced literature. The difference of instruments and the calibration methods might impact on the Raman shifts of the same material [37]. And the random errors caused by sample effects might make the value fluctuant in a range. For instance, the condition temperature could influence the Raman shifts to some extent [38]. Besides, the Raman spectrum was a comprehensive and superposed plot for all scattering lights from a mixture of all bio-matters participating in the interaction. Scattering spectrum of each active vibrational bond or molecule superposed other spectra, making the resultant spectral peaks offset from the original position. Therefore the peak gap between the same bonds in the mixed bio-matters and the pure substances sounded reasonable.
Table 2.
Tentative Raman spectral peak assignments for metabolites in plants [50]
| Compounds | Representatives | Peaks (cm−1) | Assignment |
|---|---|---|---|
| Amino acids | Cysteine | 512 | ν(S–S) g – g – g |
| Methionine | 630–670 | ν(C–S) g | |
| 700–745 | ν(C–S) t | ||
| Proteins | Disordered structure (solvated) | 1245 | |
| Lipids/fatty acids | 800–1100 | ν(C–C) | |
| Disaccharides | Sucrose | 1462 | δ(CH2) |
| Bicyclic monoterpenes | 1,8-Cineol | 652 | δ(ring) |
| Sabinene | 652 | δ(ring) | |
| Tetraterpenes | Crocetin | 1020 | ρ(C–C) |
| Chlorophyll | Chlorophyll a, b | 1326 | |
| Chlorophyll a | 1549 | ||
| Carotenoid | 956–957 [32] |
Vibrations: ν—stretching; δ—deformation; ρ—in-plane rocking. Conformations: g—gauche; t—trans
The inter-cultivar differences among Flos Chrysanthemum naturally resulted from the differences of matters they contained, and these differences involved in both substances types and their proportions. Ingredients and their proportions in Flos Chrysanthemum varied from cultivars to cultivars, leading to offsets of peaks in spectra. This might be the part reason why the peak of same vibrational bond fluctuated within a narrow range instead of keeping the same value, which made the characteristic peaks distribution patterns in the map looked irregular (Fig. 4). Although the ingredients and chemical components of the 26 cultivars were similar as the same species, their relative proportions differed and the differences were embodied in the distinctions of characteristic peaks distribution patterns. Along with the specificity and uniqueness for several peaks in individual cultivars, the diversity of patterns jointly resulted in the inter-cultivar differences.
Currently, vibration spectral techniques, mainly the infrared spectroscopy and Raman spectroscopy, were being widely applied in plant-related identification, analysis and characterization. In a recent study, the infrared spectroscopy was used as an inexpensive and rapid method to characterization plants by their pollens. The spectroscopic-based methodology was able to detect the phylogenetic variations, including the separation of confamiliar and congeneric species [39]. Another similar research was reported to characterize the aeroallergens by measuring the spore and pollen samples using single reflectance attenuated total reflectance Fourier transform infrared spectroscopy (SR-ATR FTIR) [40]. Also as a kind of vibrational spectroscopic technique, Raman spectra could provide complementary information to that obtained by infrared spectra.
The fingerprint Raman spectrum has capacity of charactering the specific vibration, rotation, phase transition and crystals structure information for individual molecules by their unique vibrational spectra. Based on such features, Raman spectroscopic analyses in principle allow to discriminate different species, and even to make classifications among the same species. In the past few decades, numerous studies have been reported on the application of Raman spectroscopy in botanical researches, including the content measurement for fatty oils [41–43], proteins [44], carbohydrates [45], phenolic [46], alkaloids [47], essential oils [48], carotenoids [49], polyacetylenes [50] and etc.. According to a relevant research, the combinational method of Fourier-transform Raman and Fourier-transform infrared was used to study the plant cell walls and achieved comprehensive spectroscopic information [51]. A very recent study reported that 34 types of pollens underwent Raman spectral measurement and a pollen spectra primary library was built for detecting and identifying pollen in airborne samples, providing an innovative idea and a promising line of investigation for future Raman technology development in the area of aerobiology [52]. As a matter of fact, the method and thought of our study were similar to theirs: we are aiming to build a primary Raman spectral library for corollas of Flos Chrysanthemum. Therefore, in principle, a library including image, Raman spectrum and characteristic peaks distribution pattern of each recorded cultivar could be built to provide potential assistances for recognition and taxonomy related researches. And every new cultivar to be found in the future could be documented in the library by archiving its image, Raman spectrum and characteristic peaks distribution pattern according to the method above. It might have very potential and promising application prospects for plant taxonomy related researches to build, supplement and improve the library in the long run. In addition to the vibration spectral techniques, the hyperspectral imaging was reported as a promising technique for plants identification. The hyperspectral imaging technique was used for nondestructive and fast determination of pigment composition and contents of chrysanthemum. The study revealed the differences inter 160 varieties of chrysanthemum by the hyperspectral measurement of the color phenotypic and pigment contents [53]. The inter-cultivar differences among Flos Chrysanthemum were analyzed in this study by the comprehensive Raman spectra for all Raman-active matters in corollas, which might be also another promising method for plant taxonomy.
The results showed that SERS spectroscopy was potentially capable in the discrimination and identification of the Flos Chrysanthemum by corollas at the cultivar level. The differences in the characteristic peaks distribution patterns at finer scale suggested that there were enough variances among corolla biomolecules that allowed cultivars separation. Plant corollas contained various chemical components, such as chromatoplasm, aromatics, amino acids and polysaccharides. Each of them was distinct in molecular conformations and they interacted with the adjacent molecules in very different ways. The enormous complexity made it nearly impossible to completely assign every spectral band to the individual component. Nevertheless, if it was unnecessary to clearly learn the details of each component they contained, this complexity could be utilized in turn to characterize and discriminate the plants. The plant taxonomy was a much more comprehensive multi-discipline involving morphological, cytological, ecological and genetic characteristics of plants [51]. It was generally recognized that the more characteristics the classification used, the more predictable it would be. Since the Raman spectra could represent not only the type and amount information of the molecules but also the information of molecular conformations and the interacting relationships with the adjacent molecules, it might be a promising method for plant taxonomy.
Potential limitations for this methodology in the study were expected to be analyzed and discussed for further improvement. SERS spectra from other parts and spots of the Flos Chrysanthemum for each cultivar needed to be acquired as supplement information to improve the comprehensiveness of library. A more comprehensive and detail archive could be of benefit to the accuracy for cultivar identification and expanding our knowledge. In addition, more cultivars remained to be measured and documented to enlarge the library. However, this was a pilot study to try charactering the inter-cultivar differences and making classifications for 26 cultivars of Chinese ornamental Flos Chrysanthemum within our reach, and this new methodology might be promising for inducing more valuable findings and innovations in future.
Conclusion
SERS technique is feasible for distinguishing the inter-cultivar differences among Flos Chrysanthemum. A Raman spectral library with images, spectra and characteristic peak distribution patterns was built, and this work presented a new promising method for Flos Chrysanthemum recognition and taxonomy.
Authors’ contributions
SL and TL conceived/designed the experiments. HZ and WX performed the experiments. HZ analyzed the data. ZC and NC contributed reagents/materials/analysis tools. HZ wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank the support of the Key Laboratory of Specialty Fiber Optics and Optical Access Networks (SKLSFO2015-06) and Beijing Advanced Innovation Center for Imaging Technology, Capital Normal University.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The spectral datasets in the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This paper was funded by Natural Science Foundation of China (NSFC) (61422507, 61475095, 61520106014).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- SERS
surface-enhanced Raman scattering
- PAGE
polyacrylamide gel electrophoresis
- POD
peroxidase
- EST
esterase
- AP
analytically pure
- a.u.
arbitrary unit
- ect.
et cetera
Contributor Information
Heng Zhang, Email: zhcn_2010@163.com.
Zhenyi Chen, Email: zychen@shu.edu.cn.
Taihao Li, Email: litaihao2004@yahoo.com.
Na Chen, Email: na.chen@shu.edu.cn.
Wenjie Xu, Email: wenjiexu@163.com.
Shupeng Liu, Email: liusp@shu.edu.cn.
References
- 1.Cheon MS, Yoon T, Lee DY, Choi G, Moon BC, Lee AY, Choo BK, Kim HK. Chtysanthemum indicum linne extract inhibits the inflammatory response by suppressing NF-κb and MAPKs activation in lipopolysaccharide-induced raw 264.7 macrophages. J Ethnopharmacol. 2009;122(3):473–477. doi: 10.1016/j.jep.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 2.Kim C, Kim MC, Kim SM, Nam D, Choi SH, Kim SH, Ahn KS, Lee EH, Jung SH, Ahn KS. Chrysanthemum indicum l. Extract induces apoptosis through suppression of constitutive STAT3 activation in human prostate cancer DU145 cells. Phytother Res. 2013;27(1):30–38. doi: 10.1002/ptr.4689. [DOI] [PubMed] [Google Scholar]
- 3.Kim IS, Ko HM, Koppula S, Kim BW, Choi DK. Protective effect of Chrysanthemum indicum linne against 1-methyl-4-phenylpridinium ion and lipopolysaccharide-induced cytotoxicity in cellular model of parkinson’s disease. Food Chem Toxicol. 2011;49(4):963–973. doi: 10.1016/j.fct.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Lee DY, Choi G, Yoon T, Cheon MS, Choo BK, Kim HK. Anti-inflammatory activity of Chrysanthemum indicum extract in acute and chronic cutaneous inflammation. J Ethnopharmacol. 2009;123(1):149–154. doi: 10.1016/j.jep.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Luyen BT, Tai BH, Thao NP, Cha JY, Lee HY, Lee YM, Kim YH. Anti-inflammatory components of Chrysanthemum indicum flowers. Bioorg Med Chem Lett. 2015;25(2):266–269. doi: 10.1016/j.bmcl.2014.11.054. [DOI] [PubMed] [Google Scholar]
- 6.Shi GB, Zhao MH, Zhao QC, Huang Y, Chen YF. Mechanisms involved in the antinociception of petroleum ether fraction from the EtOH extract of Chrysanthemum indicum in mice. Phytomedicine. 2011;18(7):609–616. doi: 10.1016/j.phymed.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Wang HY, Qi LW, Wang CZ, Li P. Bioactivity enhancement of herbal supplements by intestinal microbiota focusing on ginsenosides. Am J Chin Med. 2011;39(6):1103–1115. doi: 10.1142/S0192415X11009433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang KM, Kim GH. Volatile aroma composition of Chrysanthemum indicum L. flower oil. J Food Sci Nutr. 2008;13:122–127. [Google Scholar]
- 9.HJ Li, Shao J, Shao Investigation, collection and classification of chrysanthemum cultivars in china. J Nanjing Agric Univ. 1990;13(1):30–36. [Google Scholar]
- 10.Luo XY. Taxonomic analysis of morphological characters of large-flowered chrysanthemum cultivars. J Beijing For Univ. 2010;32(3):135–140. [Google Scholar]
- 11.Abbasi S, Mokhtarian F, Kittler J. Reliable classification of chrysanthemum leaves through curvature scale space. In: International conference on scale-space theory in computer vision. 1997.
- 12.Hong Y, Bai X, Sun W, Jia F, Dai S. The numerical classification of chrysanthemum flower colour phenotype. Acta Hortic Sin. 2012;39:1330–1340. [Google Scholar]
- 13.Zhao Y, Zheng L, Li W. Comparision of peroxidase isoenzymes classification and natoral classification in Chinese chrysanthemum. J Yunnan Agric Univ. 1996;11(2):86–90. [Google Scholar]
- 14.Wang C. Study on hybridization technique of Dendranthema × grandiflorum and identification of hybrid f1. Wuhan: Huazhong Agricultural University; 2004. [Google Scholar]
- 15.Ding L, Chen F, Teng N, Fang W. Analysis of genetic diversity of cultivars in Dendranthema × grandiflorum based on pod and est isozyme. Sci Agric Sin. 2008;41(4):1142–1150. [Google Scholar]
- 16.Raman CV. A new radiation. Indian J Phys. 1928;2:387–398. [Google Scholar]
- 17.Fleischmann M, Hendra PJ, McQuillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chem Phys Lett. 1974;26(2):163–166. doi: 10.1016/0009-2614(74)85388-1. [DOI] [Google Scholar]
- 18.Jarvis RM, Goodacre JR. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal Chem. 2004;76(1):40–47. doi: 10.1021/ac034689c. [DOI] [PubMed] [Google Scholar]
- 19.Goeller LJ, Riley MR. Discrimination of bacteria and bacteriophages by Raman spectroscopy and surface-enhanced Raman spectroscopy. Appl Spectrosc. 2007;61(7):679–685. doi: 10.1366/000370207781393217. [DOI] [PubMed] [Google Scholar]
- 20.Berger AJ, Koo TW, Itzkan I, Horowitz G, Feld MS. Multicomponent blood analysis by near-infrared Raman spectroscopy. Appl Opt. 1999;38(13):2916. doi: 10.1364/AO.38.002916. [DOI] [PubMed] [Google Scholar]
- 21.Enejder AM, Koo TW, Oh J, Hunter M, Sasic S, Feld MS, Horowitz GL. Blood analysis by Raman spectroscopy. Opt Lett. 2002;27(22):2004–2006. doi: 10.1364/OL.27.002004. [DOI] [PubMed] [Google Scholar]
- 22.Sigurdsson S, Philipsen PA, Hansen LK, Larsen J, Gniadecka M, Wulf HC. Detection of skin cancer by classification of Raman spectra. IEEE Trans Biomed Eng. 2004;51(10):1784–1793. doi: 10.1109/TBME.2004.831538. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z, Mcwilliams A, Lui H, Mclean DI, Lam S, Zeng H. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. Int J Cancer. 2003;107(6):1047–1052. doi: 10.1002/ijc.11500. [DOI] [PubMed] [Google Scholar]
- 24.Shao X, Pan J, Wang Y, Zhu Y, Xu F, Shangguan X, Dong B, Sha J, Chen N, Chen Z. Evaluation of expressed prostatic secretion and serum using surface-enhanced Raman spectroscopy for the noninvasive detection of prostate cancer, a preliminary study. Nanomed Nanotechnol Biol Med. 2016;13:1051–1059. doi: 10.1016/j.nano.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Keidel A, von Stetten D, Rodrigues C, Maguas C, Hildebrandt P. Discrimination of green arabica and robusta coffee beans by Raman spectroscopy. J Agric Food Chem. 2010;58(21):11187–11192. doi: 10.1021/jf101999c. [DOI] [PubMed] [Google Scholar]
- 26.Rubayiza AB, Meurens M. Chemical discrimination of Arabica and Robusta coffees by Fourier transform Raman spectroscopy. J Agric Food Chem. 2005;53(12):4654–4659. doi: 10.1021/jf0478657. [DOI] [PubMed] [Google Scholar]
- 27.Ivleva NP, Niessner R, Panne U. Characterization and discrimination of pollen by Raman microscopy. Anal Bioanal Chem. 2005;381(1):261–267. doi: 10.1007/s00216-004-2942-1. [DOI] [PubMed] [Google Scholar]
- 28.Lee PC, Meisel D. Absorption and surface-enhanced Raman of dyes on silver and gold sols. J Phys Chem. 1982;86(17):3391–3395. doi: 10.1021/j100214a025. [DOI] [Google Scholar]
- 29.Calheiros R, Machado NFL, Fiuza SM, Gaspar A, Garrido J, Milhazes N, Borges F, Marques MPM. Antioxidant phenolic esters with potential anticancer activity: a Raman spectroscopy study. J Raman Spectrosc. 2008;39(1):95–107. doi: 10.1002/jrs.1822. [DOI] [Google Scholar]
- 30.Gamboa J, Munoz R, Quiles MJ. Effects of antimycin a and n-propyl gallate on photosynthesis in sun and shade plants. Plant Sci. 2009;177(6):643–647. doi: 10.1016/j.plantsci.2009.09.004. [DOI] [Google Scholar]
- 31.Ito T, Tada SSS. Aroma constituents of edible chrysanthemum. J Fac Agric Iwate Univ. 1990;20(1):35–42. [Google Scholar]
- 32.Stone N, Kendall C, Smith J, Crow P, Barr H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004;126:141–157. doi: 10.1039/b304992b. [DOI] [PubMed] [Google Scholar]
- 33.Pascal A, Peterman E, Gradinaru C, van Amerongen H, van Grondelle R, Robert B. Structure and interactions of the chlorophyll a molecules in the higher plant Lhcb4 antenna protein. J Phys Chem B. 2000;104(39):9317–9321. doi: 10.1021/jp001504m. [DOI] [Google Scholar]
- 34.Baranski R, Baranska M, Schulz H. Changes in carotenoid content and distribution in living plant tissue can be observed and mapped in situ using NIR–FT–Raman spectroscopy. Planta. 2005;222(3):448–457. doi: 10.1007/s00425-005-1566-9. [DOI] [PubMed] [Google Scholar]
- 35.Smeekens S. Sugar regulation of gene expression in plants. Curr Opin Plant Biol. 1998;1(3):230–234. doi: 10.1016/S1369-5266(98)80109-X. [DOI] [PubMed] [Google Scholar]
- 36.Roitsch T, Ehness R, Goetz M, Hause B, Hofmann M, Sinha AK. Regulation and function of extracellular invertase from higher plants in relation to assimilate partitioning, stress responses and sugar signalling. Aust J Plant Physiol. 2000;27(8–9):815–825. [Google Scholar]
- 37.Bowie BT, Chase DB, Griffiths PR. Factors affecting the performance of bench-top Raman spectrometers. Part I: instrumental effects. Appl Spectrosc. 2000;54(5):164a–173a. doi: 10.1366/0003702001949924. [DOI] [Google Scholar]
- 38.Bowie BT, Chase DB, Griffiths PR. Factors affecting the performance of bench-top Raman spectrometers. Part II: effect of sample. Appl Spectrosc. 2000;54(6):200a–207a. doi: 10.1366/0003702001950175. [DOI] [Google Scholar]
- 39.Kohler Zimmermann BA. Infrared spectroscopy of pollen identifies plant species and genus as well as environmental conditions. PLoS ONE. 2014;9(4):e95417. doi: 10.1371/journal.pone.0095417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmermann B, Tkalcec Z, Mesic A, Kohler A. Characterizing aeroallergens by infrared spectroscopy of fungal spores and pollen. PLoS ONE. 2015;10(4):e0124240. doi: 10.1371/journal.pone.0124240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Irudayaraj J, Paradkar MM. Discriminant analysis of edible oils and fats by FTIR, FT-NIR and FT-Raman spectroscopy. Food Chem. 2005;93(1):25–32. doi: 10.1016/j.foodchem.2004.08.039. [DOI] [Google Scholar]
- 42.Jone O, Alfredo S, Maitane O, Ibone A, Nestor E. Quantitative analysis of essential oils from rosemary in virgin olive oil using Raman spectroscopy and chemometrics. J Raman Spectrosc. 2012;43(8):1151–1156. doi: 10.1002/jrs.3152. [DOI] [Google Scholar]
- 43.El-Abassy RM, Donfack P, Materny A. Visible Raman spectroscopy for the discrimination of olive oils from different vegetable oils and the detection of adulteration. J Raman Spectrosc. 2010;40(9):1284–1289. doi: 10.1002/jrs.2279. [DOI] [Google Scholar]
- 44.Li-Chan ECY. The applications of Raman spectroscopy in food science. Trends in Food Sci Technol. 1996;7(11):361–370. doi: 10.1016/S0924-2244(96)10037-6. [DOI] [Google Scholar]
- 45.Baranski R, Baranska M, Schulz H, Simon PW, Nothnagel T. Single seed Raman measurements allow taxonomical discrimination of Apiaceae accessions collected in gene banks. Biopolymers. 2006;81(6):497–505. doi: 10.1002/bip.20452. [DOI] [PubMed] [Google Scholar]
- 46.Agarwal UP, Reiner RS. Near-IR surface-enhanced Raman spectrum of lignin. J Raman Spectrosc. 2010;40(11):1527–1534. doi: 10.1002/jrs.2294. [DOI] [Google Scholar]
- 47.Schulz Baranska MH. Determination of alkaloids through infrared and Raman spectroscopy. Alkaloids Chem Biol. 2009;67(09):217. doi: 10.1016/s1099-4831(09)06704-2. [DOI] [PubMed] [Google Scholar]
- 48.Baranska M, Schulz H, Reitzenstein S, Uhlemann U, Strehle MA, Krüger H, Quilitzsch R, Foley W, Popp J. Vibrational spectroscopic studies to acquire a quality control method of eucalyptus essential oils. Biopolymers. 2005;78(5):237–248. doi: 10.1002/bip.20284. [DOI] [PubMed] [Google Scholar]
- 49.Schulz H, Baranska M, Baranski R. Potential of NIR-FT-Raman spectroscopy in natural carotenoid analysis. Biopolymers. 2005;77(4):212–221. doi: 10.1002/bip.20215. [DOI] [PubMed] [Google Scholar]
- 50.Baranska Schulz HM. Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib Spectrosc. 2007;43(1):13–25. doi: 10.1016/j.vibspec.2006.06.001. [DOI] [Google Scholar]
- 51.Sene C, Mccann MC, Wilson RH, Grinter R. Fourier-transform Raman and Fourier-transform infrared-spectroscopy—an investigation of 5 higher-plant cell-walls and their components. Plant Physiol. 1994;106(4):1623–1631. doi: 10.1104/pp.106.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guedes A, Ribeiro H, Fernández-González M, AiraI Abreu MJ. Pollen Raman spectra database: application to the identification of airborne pollen. Talanta. 2014;119:473. doi: 10.1016/j.talanta.2013.11.046. [DOI] [PubMed] [Google Scholar]
- 53.Jing FU, Dai SL. Analysis of color phenotypic and pigment contents of chrysanthemum based on hyperspectral imaging. J Beijing For Univ. 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The spectral datasets in the current study are available from the corresponding author on reasonable request.


