Abstract
Objective
Halotolerant bacteria have multiple uses viz. fermentation with lesser sterility control and industrial production of bioplastics. Moreover, it may increase the crop productivity of coastal saline lands in Bangladesh by transferring the salt tolerant genes into the plants. The study focused on the isolation and identification of the halotolerant bacteria from three soil samples, collected from coastal Patenga area. The samples were inoculated in nutrient media containing a wide range of salt concentrations.
Results
All the samples showed 2, 4 and 6% (w/v) salt tolerance. The isolates from Patenga soil (4, 6%) and beach soil (2%) showed catalase activity and all the isolates showed negative results for oxidase activity, indole production, lactose and motility. All the samples provided positive results for dextrose fermentation. Other tests provided mixed results. Based on the morphological characteristics, biochemical tests and ABIS software analysis the isolates fall within the Enterobacteriaceae, Clostridium and Corynebacterium, with a predominance of Vibrios. Overall the isolates can be considered as mild halotolerant, with the best growth observed at lower salinities and no halophilism detected. Among many possibilities, the genes responsible for the salt tolerant trait in these species can be identified, extracted and inserted into the crop plants to form a transgenic plant to result in higher yield for the rest of the year.
Keywords: Halotolerant, Enterobacteriaceae, Clostridium, Corynebacterium, Salinity
Introduction
Salinization is one of the root reasons for the crop destruction in Bangladesh [1, 2]. Almost 20% of the country is covered as the coastal area from which about 53% of lands are affected by very slight to very strong salinity [3]. The upward movement of the saline ground water in the dry season (November–May) is the factor that initiates the development of saline soil. Nevertheless, the intrusion of the seawater also increases the degree of salinity of the coastal drinking water and cause severe health problems viz. hypertension or high blood pressure, stroke, heart diseases, pre-eclampsia.
Bacteria that grow in the absence of salt and in the presence of high salt concentrations are known as halotolerant. Non-halotolerant which can grow in low salt concentration about 1% w/v. Slightly tolerant as pseudomonads, enterobacteria, and vibrios, can survive in up to 2–8%, moderately tolerant 18–20% and extremely tolerant microbes can grow over the whole range of salt concentrations from zero to saturation. The halotolerant organisms maintain a low level of ionic concentrations to synthesize compatible solutes to balance the osmotic level inside the cytoplasm with the outer medium. These maintenance mechanisms of the internal environment and the properties of the cytoplasmic membrane help them to adapt to changes in the saline environment as salt lakes, saline soils, and salted food products [4].
It was reported that amelioration of salt stress inhibitory effect on the canola seed germination was attributed to the inoculation of ACC deaminase-producing halotolerant bacteria modulating ethylene emission and inducing hydrolytic enzymes [5]. A research was executed on plant growth-promoting rhizobacteria (PGPR) containing aminocyclopropane-1-carboxylate (ACC) deaminase, [6] examined their effect on salinity stress tolerance in okra through the induction of ROS-scavenging enzyme activity. PGPR inoculated plants exhibited higher germination percentage, growth parameters, and chlorophyll content than control.
In a salt stress improvement research for red pepper plants by 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase producing halotolerant bacteria was studied [7, 8]. The result showed salt stress ethylene production by increasing enzyme activities of a biosynthetic pathway. It was also reported that the growth promotion in inoculated red pepper plants under inhibitory levels of salt stress is due to ACC deaminase activity present in the halotolerant bacteria [9].
Halophilic and halotolerant bacteria are essential for salty foods production as Thai fish sauce, pickling brines and salt-cured bacon [10]. Isolates from effluents of textile industries also showed the ability to decolorize the utilized azo dyes [11]. Halotolerant bacteria, recovered from the composting process, were able to produce hydrolases, lipases, proteases, amylases, cellulases and biopolymers [12]. Four halotolerant species (Bacillus atrophaeus, Halomonas shengliensis, Halomonas koreensis and Virgibacillus salarius) showed the ability to metabolize hydrocarbons and isolates as V. salarius and Brevibacillus sp. KUMAs1 has the potential to be used for bioremediation [13, 14]. Nevertheless, Corynebacterium xerosis was the potent degraders of hydrocarbons (petrol and diesel) [15]. Halophilic and halotolerant bacteria can be used for the production of enzymes with different immunological properties [16] and also essential for nutrient recycling and for maintaining the soil health in a salty environment [17].
The goal of the research was to isolate and identify halotolerant bacteria from natural sources [18]. Multiple uses of these species viz. fermentation with lesser control on sterility and industrial production of bioplastics, can be beneficial for different sectors in Bangladesh. Besides that, it may provide the salt tolerant genes for plants in future. Therefore, plants can uptake and store salt as a nutrient and result in a good yield throughout the year.
Main text
Methods
Sample collection
Three different soil samples (Patenga Beach soil 22°15′06.8″N, 91°45′29.5″E; Land soil 22°14′31.6″N, 91°47′15.9″E and Patenga area soil 22°14′25.4″N, 91°48′59.7″E) were collected from 10 to 12-in. depth at different locations of the coastal Chittagong area; these were kept in a sterile polythene packet at room temperature.
Isolation and screening
5 g of each of the soil samples were taken to prepare a suspension. Then serially diluted (10−3, 10−5 and 10−7, 100 μL) samples were taken and incubated on the nutrient agar plates containing 2% (w/v), 4% (w/v), 6% (w/v), 8% (w/v) and 10% (w/v) NaCl for 24-h at 37 °C. No bacterial growth observed for 8% (w/v) and 10% (w/v) NaCl plates.
Biochemical tests
Total 18 primarily screened isolates (2 each from 2, 4 and 6% plates) were sub-cultured on NA plates without salt with the same dilution factor and nine inocula were tested further. After 24-h of incubation at 37 °C, the plates were observed [19].
Standard gram staining protocols were followed and then slide observed under a microscope. The presence or absence of bubbles or foam was observed to determine the capability of catalase activity [20]. Two drops of oxidase reagent p-aminodimethylaniline oxalate were added to the surface of test organisms’ growth in oxidase test [21]. Nitrate broth (beef extract 3 g/L, peptone 5 g/L, potassium nitrate 5 g/L) [22] and MR-VP broth (peptone, dextrose and potassium phosphate) was prepared and incubated at 24-h at 37 °C for Methyl Red test and VP test [23]. The triple sugar iodine agar was prepared and after inoculation, incubated for about 24-h at 35 °C for TSI test [24]. After inoculation, MIU media (MIU test [25]) and Simmon’s agar slants were also incubated for 24-h at 37 °C [22, 23]. Trypticase 10 g/L, NaCl 5 g/L, phenol red 0.018/L, sugar (glucose, lactose or sucrose) 5 g/L contained broth incubated at 37 °C for 24-h to determine the capability of fermenting carbohydrate substrate with acid and gas production [26]. Autoclaved starch agar (beef extract 3 g/L, soluble starch 10 g/L and agar 15 g/L) plates were incubated for 24-h to indicate the presence of α-amylase [27]. Sterile tubes containing inoculum was transferred in 6.5% NaCl solution and kept in 24-h incubation to assess salt hindrance.
Results
All the samples showed growth in NA media containing 2% (w/v), 4% (w/v) and 6% (w/v) NaCl. But failed to grow in media containing 8% (w/v) and 10% (w/v) NaCl (Table 1).
Table 1.
Results of growth in NA media containing 2% (w/v) to 10% (w/v) NaCl
| Conc. | Patenga soil | Beach soil | Land soil | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2% | 4% | 6% | 8% | 10% | 2% | 4% | 6% | 8% | 10% | 2% | 4% | 6% | 8% | 10% | |
| Growth | ✓ | ✓ | ✓ | x | x | ✓ | ✓ | ✓ | x | x | ✓ | ✓ | ✓ | x | x |
The isolates were all gram positive (Fig. 1; Table 2). Patenga 4, 6% were cocci shaped and rest were rod shaped.
Fig. 1.
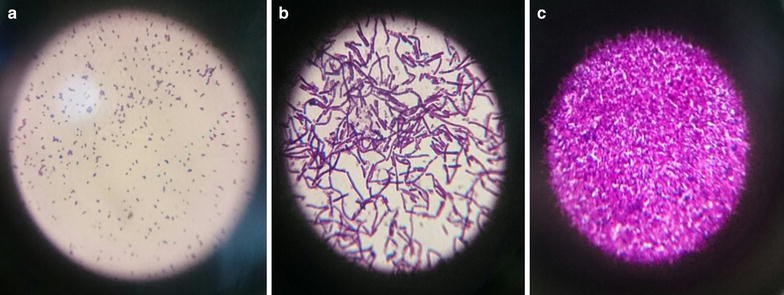
Gram positive bacterial isolates from a Patenga area soil, b Beach soil and c Land soil sample
Table 2.
Results of biochemical and sugar tests of the isolates collected from nutrient agar
| Isolate no | Sample isolate name | Gram stain | TSI | MIU | Catalase | Oxidase | Simmon’s citrate | MR | VP | Nitrate reduction | Starch hydrolysis | Carbohydrate | 6.5% NaCl solution | Presumptive organism | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ± | Shape | BUTT | SLANT | H2S | Gas | Motility | Indole | Urease | Sucrose | Lactose | Dextrose | |||||||||||
| 1. | Patenga area soil 2% | + | Rod | A | A | − | − | − | − | − | + | − | − | + | − | − | − | + | − | + | + | Brevibacillus agri |
| 2. | Patenga area soil 4% | + | Cocci | A | K | − | − | − | − | − | − | − | − | − | + | − | + | + | − | + | + | Pantoea stewartii subsp. stewartii |
| 3. | Patenga area soil 6% | + | Cocci | K | A | − | − | − | − | + | + | − | − | − | + | − | − | + | − | + | − | Corynebacterium xerosis/Corynebacterium minutissimum/Corynebacterium kutscheri |
| 4. | Beach soil 2% | + | Rod | A | A | − | − | − | − | − | + | − | + | + | + | − | + | + | − | + | + | Vibrio metschnikovii |
| 5. | Beach soil 4% | + | Rod | A | A | − | − | − | − | − | − | − | + | + | + | − | + | + | − | + | − | Vibrio metschnikovii |
| 6. | Beach soil 6% | + | Rod | A | A | − | − | − | − | − | − | − | − | + | − | − | + | + | − | + | + | Volucribacter psittacicida |
| 7. | Land soil 2% | + | Rod | K | A | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | + | Aggregatibacter (Haemophilus) segnis |
| 8. | Land soil 4% | + | Rod | K | A | − | − | − | − | − | − | − | + | − | + | + | − | + | − | + | + | Aggregatibacter (Haemophilus) segnis |
| 9. | Land soil 6% | + | Rod | A | K | − | − | − | − | − | − | − | + | − | + | − | − | + | − | + | + | Clostridium innocuum/Clostridium spiroforme |
K alkaline reaction, A acidic reaction, + positive reaction, − negative reaction
In catalase test, hydrogen peroxide was used and broken down to water and oxygen. Patenga 2, 6% and Beach soil 2% were positive. Oxidase test resulted as negative for all (Table 2). Only Land soil 2 and 4% was reduced nitrate by the change of color (red) (Table 2). Most of the samples were responsive to TSI test. Both slant and butt became yellow for Patenga 2%, Beach soil 2, 4 and 6%. The only slant was acidic for Land soil 2 and 4% and Land soil 6% for the butt. Patenga 2%, Beach soil 2, 4 and 6% recorded as MR test positive. Patenga 4, 6%, Beach soil 2, 4%, Land soil 2, 4, and 6% found VP test positive because the color did not change to pink. All negative results recorded at MIU test, except non-motile Patenga 6% sample was urease positive. Simmon’s citrate test showed a positive result for Beach soil 2, 4% and land soil 4, 6%. Land soil 4, 6%, Patenga area 2, 4, 6%, Beach soil 2, 4, and 6% found positive for sucrose metabolism. No positive result observed for lactose fermentation. All the sample result was positive for dextrose fermentation, except Land soil 2%. Patenga area 2% and Beach soil 4% produced gas in fermentation. Patenga area 4% and Beach soil 2, 4, 6% hydrolyzed starch. Patenga 2, 4%, Beach soil 2, 6%, Land soil 2, 4 and 6% showed the ability to grow at 6.5% NaCl solution.
Finally, the test results were analyzed in ABIS online software [28], which is used to analyze the genus of the organism. Based on the morphology characteristics, Biochemical test and ABIS software the following results were predicted as Patenga 2%—Brevibacillus agri; Patenga 4%—Pantoea stewartii subsp. stewartii; Patenga 6%—Corynebacterium xerosis/Corynebacterium minutissimum/Corynebacterium kutscheri; Beach soil 2%—Vibrio metschnikovii; Beach soil 4%—Vibrio metschnikovii; Beach soil 6%—Volucribacter psittacicida; Land soil 2%—Aggregatibacter (Haemophilus) segnis; Land soil 4%—Aggregatibacter (Haemophilus) segnis; Land soil 6%—Clostridium innocuum/Clostridium spiroforme.
Discussion
In this study, three soil samples were collected from the coastal area of Chittagong [29–31]. The isolated bacteria from the samples had successfully survived in a limited range of salinities and no halophilism detected. All the isolates showed tolerance to 2% (w/v), 4% (w/v) and 6% (w/v) of NaCl (Table 1). The dilution factor was inversely proportional to the number of colonies. The study divulged the abundance of gram positive bacteria (Table 2). The isolates from Patenga area (2, 6%) and Beach (2%) showed catalase activity and all the isolates showed negative results for oxidase activity, indole production, lactose and motility. Aerobic and facultative aerobes exhibit oxidase activity whereas Enterobacteriaceae are oxidase negative. This provided the evidence of Enterobacteriaceae in the samples. In the MIU test, only the isolates extracted from Patenga area that is 6% (w/v) NaCl tolerant showed urease positive. In addition to this, all the isolates provided positive results for dextrose fermentation. Vibrios were predominating in the investigated soil along with Corynebacterium, Clostridium and Enterobacteriaceae.
Limitations
Extreme halotolerant species as Halomonas elongata CHR63, Thioalkalivibrio versutus or Sporosarcina pasteurii wasn’t detected in the study [32]. The future prospect can be the plantation of transgenic plants in the coastal area for better agricultural use. The research also provides insights for adaption and use of soil microorganisms as natural fertilizers after natural calamity struck the coastal region. Before that, the success of this investigation is not properly accomplished.
Authors’ contributions
NT carried out experimental studies; analyzed, interpreted data and drafted the dissertation. SSR carried out the experimental studies partially and drafted, edited, finalized and revised the article and coordinated to the publisher. RS participated in designing, supervising and coordinating the study. All authors read and approved the final manuscript.
Acknowledgements
Authors are grateful to Prof. Dr. AAZ Ahmad (Chairperson) and Prof. Naiyyum Choudhury of Department of Mathematics and Natural Sciences, BRAC University, Dhaka, for motivation. Their sincere appreciation goes to Dr. M. Mahboob Hossain and Jebunnesa Chowdhury for drawing novel thoughts. And, grateful to Asma Binte Afzal, Nahreen Mirza and Salman Khan Promon for their unremitting help.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data presented within the manuscript.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This study was self-funded by authors and their institution Department of Mathematics and Natural Sciences, BRAC University.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- MIU
motility indole urease test
- et al.
and others
- NA
nutrient agar
- Mg
milligram
- sp.
species
- mL
milliliter
- TSI
triple sugar iodine
- MR
methyl red
- VP
Voges Proskauer
- w/v
weight by volume
- g/L
gram per liter
Contributor Information
Shafkat Shamim Rahman, Email: shafkatshamimrahman@gmail.com.
Romana Siddique, Email: rsiddique@bracu.ac.bd.
Nafisa Tabassum, Email: aayushirahman@gmail.com.
References
- 1.Elaine Colligan. Salinity issues in Bangladesh. Georget J Int Aff. 2011. http://journal.georgetown.edu/guide-to-salinity-issues-in-bangladesh/. Accessed 26 June 2017.
- 2.Rasel HM, Hasan MR, Ahmed B, Miah MS. Investigation of soil and water salinity, its effect on crop production and adaptation strategy. Int J Water Res Environ Eng. 2013;5(8):475–481. [Google Scholar]
- 3.Haque SA. Salinity problems and crop production in coastal regions of Bangladesh. Pak J Bot. 2006;38(5):1359–1365. [Google Scholar]
- 4.Roberts MF. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 2005;1(1):5. doi: 10.1186/1746-1448-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddikee MA, Sundaram S, Chandrasekaran M, Kim K, Selvakumar G, Sa T. Halotolerant bacteria with ACC deaminase activity alleviate salt stress effect in canola seed germination. Appl Biol Chem. 2015;2(58):237–241. [Google Scholar]
- 6.Habib SH, Kausar H, Saud HM. Plant growth-promoting rhizobacteria enhance salinity stress tolerance in okra through ROS-scavenging enzymes. Biomed Res Int. 2016;2016:1–10. doi: 10.1155/2016/6284547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siddikee MA, Glick BR, Chauhan PS, Jong Yim W, Sa T. Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene synthesis with halotolerant bacteria containing 1-aminocyclopropane-1-carboxylic acid deaminase activity. Plant Physiol Biochem. 2011;49(4):427–434. doi: 10.1016/j.plaphy.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Glick BR. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res. 2014;169(1):30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Nabti E, Schmid M, Hartmann A. Application of halotolerant bacteria to restore plant growth under salt stress. In: Halophiles. Berlin: Springer International Publishing; 2015. p. 235–59.
- 10.Margesin R, Schinner F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles. 2001;5(2):73–83. doi: 10.1007/s007920100184. [DOI] [PubMed] [Google Scholar]
- 11.Asad S, Amoozegar MA, Pourbabaee A, Sarbolouki MN, Dastgheib SM. Decolorization of textile azo dyes by newly isolated halophilic and halotolerant bacteria. Bioresour Technol. 2007;98(11):2082–2088. doi: 10.1016/j.biortech.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira LC, Ramos PL, Marem A, Kondo MY, Rocha R, Bertolini T, Silveira MA, Cruz JB, Vasconcellos SP, Juliano L, Okamoto DN. Halotolerant bacteria in the São Paulo Zoo composting process and their hydrolases and bioproducts. Braz J Microbiol. 2015;46(2):347–354. doi: 10.1590/S1517-838246220130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kothari V, Panchal M, Srivastava N. Hydrocarbon degradation potential of halotolerant bacteria. In: Scientific study. 2014. http://www.grin.com/en/e-book/268982/hydrocarbon-degradation-potential-of-halotolerant-bacteria. Accessed 27 June 2017.
- 14.Mallick I, Hossain ST, Sinha S, Mukherjee SK. Use of indigenous bacteria from arsenic contaminated soil for arsenic bioremediation. In: Management of natural resources in a changing environment. Berlin: Springer International Publishing; 2015. p. 155–65.
- 15.Jyothi K, Babu KS, Clara NK, Kumar A. Identification and isolation of hydrocarbon degrading bacteria by molecular characterization. Helix. 2012;2:105–111. [Google Scholar]
- 16.Shirazian P, Asad S, Amoozegar MA. The potential of halophilic and halotolerant bacteria for the production of antineoplastic enzymes: l-asparaginase and l-glutaminase. EXCLI J. 2016;15:268. doi: 10.17179/excli2016-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavik G, Stührmann T, Brüchert V, Van der Plas A, Mohrholz V, Lam P, Mußmann M, Fuchs BM, Amann R, Lass U, Kuypers MM. Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature. 2009;457(7229):581–584. doi: 10.1038/nature07588. [DOI] [PubMed] [Google Scholar]
- 18.Grattieri M, Suvira M, Hasan K, Minteer SD. Halotolerant extremophile bacteria from the Great Salt Lake for recycling pollutants in microbial fuel cells. J Power Sources. 2017;356:310–318. doi: 10.1016/j.jpowsour.2016.11.090. [DOI] [Google Scholar]
- 19.Aditi FY, Rahman SS, Hossain MM. A study on the microbiological status of mineral drinking water. Open Microbiol J. 2017;11:31–44. doi: 10.2174/1874285801711010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiner K. Catalase test protocol. Washington: American Society for Microbiology; 2010. http://www.microbelibrary.org/library/laboratorytest/3226-catalase-test-protocol.htm. Accessed 5 July 2017.
- 21.Shields P, Cathcart L. Oxidase test protocol. Washington: American Society for Microbiology; 2010. http://www.microbelibrary.org/library/laboratory-test/3229-oxidase-test-protocol.htm. Accessed 5 July 2017.
- 22.Ferdous TA, Kabir SML, Amin MM, Hossain KMM. Identification and antimicrobial susceptibility of salmonella species isolated from washing and rinsed water of broilers in pluck shops. Int J Anim Vet Adv. 2013;5(1):1–8. [Google Scholar]
- 23.McDevitt S. Methyl red and Voges–Proskauer test protocols 2009 ASM microbe library. 2013. http://www.microbelibrary.org/component/resource/laboratory-test/3204-methyl-red-andvoges-proskauer-test-protocols.htm. Accessed 5 July 2017.
- 24.Cappuccino JG, Sherman N. Microbiology a laboratory manual. Benjamin Cummings: New York; 2005. p. 125–79.
- 25.Acharya T. Tests for bacterial motility: procedure and results 2015. http://microbeonline.com/tests-bacterial-motilityprocedure-results/. Accessed 5 July 2017.
- 26.Rahman SS, Hossain MM, Choudhury N. Effect of various parameters on the growth and ethanol production by yeasts isolated from natural sources. Bangladesh J Microbiol. 2013;30(1–2):49–54. [Google Scholar]
- 27.Starch hydrolysis test. http://www.vumicro.com/vumie/help/VUMICRO/Starch_Hydrolysis_Test.htm. Accessed 5 July 2017.
- 28.Analysis of bacteria identification software. http://www.tgw1916.net/bacteria_logare_desktop.html. Accessed 1 July 2017.
- 29.Arias D, Cisternas LA, Rivas M. Biomineralization of calcium and magnesium crystals from seawater by halotolerant bacteria isolated from Atacama Salar (Chile) Desalination. 2017;405:1–9. doi: 10.1016/j.desal.2016.11.027. [DOI] [Google Scholar]
- 30.Khansha J, Ranjbaran M, Amoozegar MA. Isolation and identification of halophilic and halotolerant bacteria from Badab-e Surt Travertine Spring, Kiasar, Iran, and investigation of calcite biomineralization induction. Geomicrobiol J. 2017:1.
- 31.Laufer K, Niemeyer A, Nikeleit V, Halama M, Byrne JM, Kappler A. Physiological characterization of a halotolerant anoxygenic phototrophic Fe(II)-oxidizing green-sulfur bacterium isolated from a marine sediment. FEMS Microbiol Ecol. 2017;93(5):fix054. doi: 10.1093/femsec/fix054. [DOI] [PubMed] [Google Scholar]
- 32.Aljohny BO. Halophilic bacterium—a review of new studies. Biosci Biotechnol Res Asia. 2015;12(3):2061–2069. doi: 10.13005/bbra/1874. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data presented within the manuscript.


