Abstract
We investigated the prevalence of Bartonella washoensis in California ground squirrels (Otospermophilus beecheyi) and their fleas from parks and campgrounds located in seven counties of California. Ninety-seven of 140 (69.3%) ground squirrels were culture positive and the infection prevalence by location ranged from 25% to 100%. In fleas, 60 of 194 (30.9%) Oropsylla montana were found to harbor Bartonella spp. when screened using citrate synthase (gltA) specific primers, whereas Bartonella DNA was not found in two other flea species, Hoplopsyllus anomalus (n = 86) and Echidnophaga gallinacea (n = 6). The prevalence of B. washoensis in O. montana by location ranged from 0% to 58.8%. A majority of the gltA sequences (92.0%) recovered from ground squirrels and fleas were closely related (similarity 99.4–100%) to one of two previously described strains isolated from human patients, B. washoensis NVH1 (myocarditis case in Nevada) and B. washoensis 08S-0475 (meningitis case in California). The results from this study support the supposition that O. beecheyi and the flea, O. montana, serve as a vertebrate reservoir and a vector, respectively, of zoonotic B. washoensis in California.
Keywords: Bartonella washoensis, California, ground squirrel, Oropsylla montana, Otospermophilus beecheyi
Introduction
There has been a growing interest in rodent-associated Bartonella spp. due, in part, to the high infection prevalence commonly observed in rodent populations and the potential for disease transmission through contact with rodents or rodent-associated ectoparasites (Breitschwerdt 2014, Gutiérrez et al. 2015).
The genus Bartonella is a relatively large taxonomic group currently containing over 30 species, with more than half of these reported in rodents (Buffet et al. 2013, Gutiérrez et al. 2015). A number of these rodent-associated Bartonella spp. are of known medical and veterinary importance (Chomel et al. 2009, Deng et al. 2012, Breitschwerdt 2014). In humans, symptoms of reported cases associated with rodent-borne Bartonella spp. infections have included endocarditis (B. elizabethae; Daly et al. 1993), fever (B. vinsonii subsp. arupensis; Welch et al. 1999), neuroretinitis (B. grahamii; Kerkhoff et al. 1999), myocarditis (B. washoensis; Kosoy et al. 2003), lymphadenitis (B. alsatica; Angelakkis et al. 2008), meningitis (B. washoensis; Probert et al. 2009), and lymphadenopathy (the Tel-Aviv strain related to both B. elizabethae and B. tribocorum; Kandelaki et al. 2016).
B. washoensis was first isolated in 1995 from a patient with cardiac disease in Washoe County, Nevada (Kosoy et al. 2003). A strain identical to the human B. washoensis isolate was obtained from California ground squirrels, Otospermophilus beecheyi, trapped during follow-up case investigations. The authors concluded that this rodent species was likely to be a reservoir of this bacterium and a source of the infection (Kosoy et al. 2003).
Other documented B. washoensis infections associated with clinical illness include a domestic dog presenting with mitral valve endocarditis (Chomel et al. 2003) and a second human case from northern California diagnosed with meningitis (Probert et al. 2009). A field investigation of the California patient’s property resulted in finding strains of B. washoensis in both O. beecheyi and their fleas (Oropsylla montana) identical to the strain isolated from the patient (Probert et al. 2009), lending further evidence that O. beecheyi and its fleas are the most likely reservoir and vector, respectively, of zoonotic B. washoensis.
The objective of this study was to evaluate the prevalence of B. washoensis in California ground squirrels and their associated fleas at parks and campgrounds across seven counties in north-central and southern California. We were also interested in comparing the Bartonella strains found in ground squirrels with previously documented human strains of B. washoensis. Using multiple gene targets and optical mapping, we intended to better evaluate how circulating strains found in O. beecheyi are related to known zoonotic strains.
Materials and Methods
Study locations and sample collection
Study sites included 13 locations within 7 counties in California: Strawberry Point Campground in El Dorado County; Los Angeles County: Malibu Creek State Park, West Fork Day Use Area, and Mt. Wilson Observatory; Inyo County: Four Jeffery Campground and Mill Pond County Park; Blue Jay Campground in Orange County; Upper Oso Campground in Santa Barbara County; San Bernardino County: Apple White Campground, Hanna Flats Campground, Barton Flats Campground, and San Gorgonio Campground; and Yuba River Ranger District in Yuba County (Table 1).
Table 1.
Sample Type Collected at Each Location
| County | Location | Sample type |
|---|---|---|
| El Dorado | SPC | Blood |
| Los Angeles | MCSP | Blood and fleas |
| WFDA | Blood and fleas | |
| MWO | Blood and fleas | |
| Inyo | FJC | Blood and fleas |
| MPCP | Blood | |
| Orange | BJC | Blood |
| Santa Barbara | UOC | Blood and fleas |
| San Bernardino | AWC | Blood |
| HFC | Blood | |
| BFC | Blood and fleas | |
| SGC | Blood and fleas | |
| Yuba | YRRD | Blood |
AWC, Apple White Campground; BFC, Barton Flats Campground; BJC, Blue Jay Campground; FJC, Four Jeffery Campground; HFC, Hanna Flats Campground; MCSP, Malibu Creek State Park; MPCP, Mill Pond County Park; MWO, Mt. Wilson Observatory; SGC, San Gorgonio Campground; SPC, Strawberry Point Campground; UOC, Upper Oso Campground; WFDA, West Fork Day Use Area; YRRD, Yuba River Ranger District.
The California Department of Public Health and the Los Angeles County Department of Public Health collected blood and flea samples from April 2013 through July 2014. Ground squirrels were trapped, and blood and fleas were collected according to the protocol presented by Davis et al. (2002), with slight modifications. Live traps (Sherman Traps, Tallahassee, FL and Tomahawk Live Traps, Tomahawk, WI) were set the night before or between 0900 and 1000 h and collected by 1200 the following or same day. Animals were anesthetized using Isoflurane (Butler Schein Animal Health, Dublin, OH), and 200–500 µL of blood was collected using cardiac puncture and placed directly into EDTA tubes (Becton, Dickinson, Co., Franklin Lakes, NJ). Convenience samples during routine plague surveillance of ground squirrels were collected by Los Angeles County Department of Public Health using methods previously described (Billeter et al. 2011, Gundi et al. 2012). Whole blood and fleas were shipped overnight to the Centers for Disease Control and Prevention (CDC) Bartonella and Rodent-Borne Diseases Laboratory in Fort Collins, Colorado for testing and analysis.
Culturing Bartonella from blood
Whole blood samples were tested by culture methods detailed by Bai et al. (2008) to evaluate the presence of viable Bartonella bacteria in ground squirrels from multiple locations. Blood samples were diluted 1:4 with brain heart infusion media containing 5% amphotericin B, and 100 µL of the dilution was plated on brain heart infusion agar containing 10% rabbit blood. The plates were incubated at 35°C with 5% carbon dioxide for a total of 4 weeks. The plates were checked for a growth of bacterial colonies weekly. All Bartonella-like colonies were subcultured from a single colony onto fresh agar. After harvesting in 10%glycerol, bacteria were heated at 95°C for 15 min and used as DNA template for PCR.
The isolates were confirmed to be Bartonella by conventional PCR targeting a 379-bp region of the citrate synthase gene (gltA) using primers 781F (5′-GGG GAC CAG CTC ATG GTG G-3′) and 1137R (5′-AAT GCA AAA AGA ACA GTA AAC A-3′) (Norman et al. 1995). All PCRs were performed in a 25 µL reaction that included DNA template, 12.5 µL of GoTaq® Green Master Mix (Promega, Madison, WI), nuclease-free water, and 1 µL each of 10 µM forward and reverse primer. Nuclease-free water was used as a negative control and B. doshiae DNA (10 pg/µL) as a positive control for each PCR. PCR products were visualized on a 1.5% agarose gel containing ethidium bromide, and samples matching the anticipated amplicon size were confirmed by sequencing.
Detection of Bartonella in fleas
All fleas were identified microscopically using taxonomic keys (Stark et al. 1966, U.S. Public Health Service 1966, Furman and Catts 1982) by either the Los Angeles County Department of Public Health or the CDC. We randomly selected 50 fleas from each location for testing and if fewer than 50 fleas were collected from a site, all fleas were processed.
Individual fleas were placed into a sterile tube containing buffer and 3–5 sterile glass beads. Fleas were then ground in a Mixer Mill MM200 (Retsch GmbH, Haan, Germany) (Rizzo et al. 2015). DNA was extracted from a homogenate using a QIAXtractor automated instrument following manufacturer’s instruction for the tissue protocol (Qiagen, Valencia, CA). The flea DNA was screened by conventional PCR following the procedure described above and using primers targeting a 767-bp region of the gltA gene (CS443f: 5′-GCT ATG TCT GCA TTC TAT CA-3′ and CS1210r: 5′-GAT CYT CAA TCA TTT GTT TCC A-3′) (Billeter et al. 2011).
Sequencing analysis
PCR-positive samples were purified using a QIAquick PCR Purification Kit (Qiagen) and sequenced with a 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). Phylogenetic analysis was performed using the CLUSTAL W alignment with Lasergene 11 software (DNAStar, Madison, WI). Sequences obtained in this study were compared to Bartonella spp. sequences previously deposited in GenBank, including several strains of B. washoensis: B. washoensis AM2-1 (AB444972), B. washoensis AR2-2 (AB444970), B. washoensis AR4-1 (AB444971), B. washoensis CJ 22-1 (AB444956), B. washoensis ER14-3 (AB444974), B. washoensis NVH1 (AF050108), B. washoensis Sb944nv (AF470616), B. washoensis Sb1659nv (AY071858), B. washoensis Sb1859 (AY071859), B. washoensis Sb1865nv (AY071860), B. washoensis SL311nv (AY071861), B. washoensis SR22-1 (AB444968), B. washoensis Tm1794nv (AF451163), B. washoensis 08S-0475 (FJ719016), and B. washoensis subsp. cynomysii (DQ897367).
Characterization of ground squirrel isolates using additional genetic markers
In addition to gltA, three other housekeeping genes (groEL, ftsZ, and rpoB) and 16S–23S intergenic spacer region (ITS) were used to perform further characterization of 20 selected isolates and reference sequences (Table 2). For each isolate, the sequences of all five loci were combined to form a concatenated sequence of 3684-bp. The concatenated sequences were compared to B. washoensis strains isolated from the two human patients from Nevada (NVH1; Kosoy et al. 2003) and California (08S-0475; Probert et al. 2009). The primer characteristics and cycle conditions for ITS (Diniz et al. 2007), groEL (Zeaiter et al. 2002a), ftsZ (Zeaiter et al. 2002b), and rpoB (Renesto et al. 2001) have been previously described. These sequences were aligned and analyzed using the Lasergene 11 software (DNAStar), using CLUSTAL W alignment.
Table 2.
GenBank Accession Numbers of Reference Strains Used in Concatenated Sequencing Analysis
| GenBank acc. no. | |||||
|---|---|---|---|---|---|
|
|
|||||
| Bartonella spp. | gltA | groEL | ftsZ | ITS | ropB |
| B. henselae | L38987 | AF014829 | AF061746 | L35101 | AF171071 |
| B. elizabethae | Z70009 | AF014834 | AF467760 | L35103 | AF165992 |
| B. tribocorum | AJ005494 | AF304018 | AF467759 | NR025278 | AF165996 |
| B. washoensis NVH1 | AF050108 | AF071193 | This study | This study | This study |
| B. washoensis 08S-0475 | FJ719016 | This study | This study | AB674256 | AB674244 |
ITS, intergenic spacer region.
Whole genome mapping
Five strains of B. washoensis were compared by application of the optical mapping platform; two isolates (B41452, B41511) obtained from O. beecheyi from San Bernardino County, California collected in this study and three additional strains: (1) an O. beecheyi isolate (B10519), B. washoensis strain Sb944nv, from Washoe County, Nevada (Kosoy et al. 2003), (2) an isolate (B42121) obtained from a human patient in Nevada, B. washoensis strain NVH1 (Kosoy et al. 2003), and (3) an isolate (B26896) from a human patient in California, B. washoensis strain 08S-0475 (Probert et al. 2009).
High-molecular weight genomic DNA from these B. washoensis strains was prepared directly from bacteria grown 3 to 4 days (105 colony forming units/µL) using the Argus HMW DNA Isolation Kit (OpGen, Inc., Gaithersburg, MD). In brief, bacteria were washed in no salt buffer, lysed in proteinase K, and diluted in loading/stretching buffer recommended by the manufacturer. To reduce DNA shearing, wide-bore pipette tips were used and DNA samples were gently mixed without vortexing. DNA was examined for quality (only DNA fragments above 150 kilobase pairs [kb] were used) and concentration using the Argus QCards, according to the manufacturer’s instructions.
The software program Enzyme Chooser (OpGen, Inc.) was used to identify BamHI restriction endonuclease cleavage sites in the reference genome that would result in fragments that average 6–12 kb in size and that would not produce any fragments larger than 80 kb. Single genomic DNA molecules were loaded onto a glass surface of a MapCard (OpGen, Inc.) and then digested with BamHI and stained with JOJO-1 with the Argus MapCard Processor (OpGen, Inc.). Map cards then were analyzed by automated fluorescent microscopy using the Argus Whole Genome Mapper (OpGen, Inc.). This software records the size and order of restriction fragments for each DNA molecule.
The single molecule restriction map collections were then tiled according to overlapping fragment patterns to produce a consensus Whole Genome Map. DNA sequence alignment using MapSolver DNA sequence data for B. washoensis strains 085-0475 and Sb944nv from GenBank (acc. no. JH725024, JH725026, JH25025, JH725022, JH725023, and JH725101) in FASTA formatted files was imported into MapSolver software (OpGen, Inc.) and converted into in silico maps using the same restriction enzyme that was used to generate the respective Whole Genome Map.
The contiguous DNA sequence maps were then aligned with the Whole Genome Map using the sequence placement function of MapSolver, and all actual optical maps were scaled according to the size of sequenced fragments. Final alignments were subject for clustering in MapSolver to evaluate a similarity between B. washoensis strains. In addition, the optical maps were evaluated using nearest neighbor chain algorithm.
Results
Prevalence of Bartonella spp. cultured from ground squirrels
Bartonella was successfully cultured from 97 (69.3%) of 140 blood samples collected from O. beecheyi. All isolated bacteria were identified as B. washoensis with some variations in the gltA genotype (Table 3).
Table 3.
Bartonella washoensis Strains Isolated from Squirrels from Each Location Identified by gltA Sequences
| Tested | |||
|---|---|---|---|
|
|
|||
| County | Location | Pos/n (%) | B. washoensis strain variationa |
| El Dorado | SPC | 12/12 (100) | NVH1 (4/12) |
| 08S-0475 (4/12) | |||
| CJ 22-1 (1/12) | |||
| NVH1 and 08S-0475 (1/12)b | |||
| 08S-0475 and genotype D (1/12)b | |||
| NVH1 and genotype D (1/12)b | |||
| Los Angeles | MCSP | 6/10 (60) | NVH1 (2/6) |
| 08S-0475 (2/6) | |||
| CJ 22-1 (1/6) | |||
| Genotype D (1/6) | |||
| WFDA | 8/8 (100) | NVH1 (7/8) | |
| 08S-0475 (1/8) | |||
| MWO | 4/5 (80) | NVH1 (4/4) | |
| Total | 18/23 (78) | ||
| Inyo | FJC | 5/6 (83) | NVH1 (2/5) |
| NVH1 and 08S-0475 (1/5)b | |||
| Genotype D (2/5) | |||
| MPCP | 2/7 (29) | 08S-0475 (1/2) | |
| Genotype D (1/2) | |||
| Total | 7/13 (54) | ||
| Orange | BJC | 9/13 (69) | 08S-0475 (9/9) |
| Santa Barbara | UOC | 4/16 (25) | NVH1 (2/4) |
| 08S-0475 (1/4) | |||
| Genotype D (1/4) | |||
| San Bernardino | AWC | 11/19 (58) | NVH1 (5/11) |
| 08S-0475 (5/11) | |||
| 08S-0475 and genotype D (1/11)b | |||
| HFC | 8/12 (67) | NVH1 (3/8) | |
| 08S-0475 (5/8) | |||
| BFC | 18/21 (86) | NVH1 (11/18) | |
| 08S-0475 (5/18) | |||
| Genotype D (2/18) | |||
| SGC | 7/8 (88) | NVH1 (2/7) | |
| 08S-0475 (4/7) | |||
| NVH1 and 08S-0475 (1/7)b | |||
| Total | 44/60 (73) | ||
| Yuba | YRRD | 3/3 (100) | NVH1 (2/3) |
| NVH1 and 08S-0475 (1/3)b |
Prevalence of Bartonella spp. detected in fleas
Fleas (n = 286) collected from Los Angeles County, Inyo County, Santa Barbara County, and San Bernardino County belonged to three species: O. montana (194), Hoplopsyllus anomalus (86), and Echidnophaga gallinacea (6). Among these three species, only O. montana was shown to harbor detectable Bartonella DNA (60 of 194 positives; 30.9%), and only B. washoensis was identified (Table 4).
Table 4.
Prevalence of Bartonella washoensis DNA Detected in Fleas from Each Location
| Tested | B. washoensis straina | ||||
|---|---|---|---|---|---|
|
|
|
||||
| County | Location | Species | Pos/n (%) | NVH1 | 08S-0475 |
| Los Angeles | MCSP | O. montana | 0/34 | ||
| H. anomalus | 0/16 | ||||
| MWO | O. montana | 20/34 (59) | 19 | 1 | |
| WFDA | O. montana | 0/10 | |||
| H. anomalus | 0/40 | ||||
| Total | 20/134 (15) | ||||
| Inyo | FJC | O. montana | 0/4 | ||
| H. anomalus | 0/2 | ||||
| Total | 0/6 | ||||
| Santa Barbara | UOC | O. montana | 2/12 (17) | 2 | |
| H. anomalus | 0/28 | ||||
| E. gallinacea | 0/6 | ||||
| Total | 2/46 (4) | ||||
| San Bernardino | BFC | O. montana | 15/50 (30) | 11 | 4 |
| SGC | O. montana | 23/50 (46) | 20 | 3 | |
| Total | 38/100 (38) | ||||
Sequence analysis of B. washoensis identified in ground squirrels and fleas
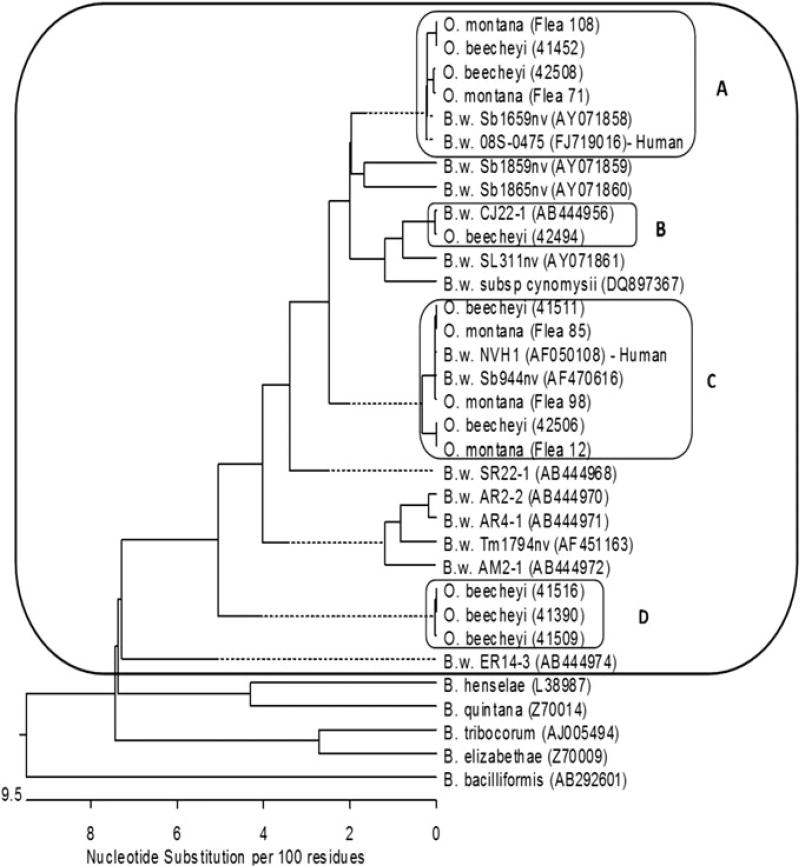
In total, 104 gltA sequences obtained from O. beecheyi were analyzed. The sequences formed four clusters inside the B. washoensis species complex with a sequence similarity of 99.4–100% within each cluster. Three of the clusters aligned with gltA sequences were previously deposited in Gen Bank: AF050108, FJ719016, AF470616, AY071858, and AB444956 (Fig. 1). One genotype was distinguishable from all previously reported sequences of B. washoensis with the closest match (97% similarity) to B. washoensis strain AR4-1 (AB444971) found in an American red squirrel (Tamiasciurus hudsonicus) imported to Japan from the United States (Inoue et al. 2009). This genotype was present in 9.6% of examined sequences and is referred to as B. washoensis genotype D.
FIG. 1.
Phylogenetic tree of observed gltA sequences and GenBank reference sequences with accession number in parenthesis. The B. washoensis (B.w.) reference sequences include strain name followed by the GenBank accession number in parenthesis. The sequences obtained from this study are identified by the host organism (O. beecheyi or O. montana) followed by the study ID in parenthesis. Sequence clusters observed from this study are labeled (A, B, C, D). Cluster (A) represents B. washoensis strain 08S-0475. Sequences were obtained from O. beecheyi, O. montana, and California human isolate. Cluster (B) represents B. washoensis strain CJ22-1 with sequences from S. columbinaus and O. beecheyi. Cluster (C) represents B. washoensis strain NVH1, with sequences from O. beecheyi, O. montana, and Nevada human isolate. Cluster (D) represents B. washoensis genotype with sequences from O. beecheyi.
Sequencing analysis from the 60 positive O. montana showed 52 (86.7%) aligned most closely with B. washoensis strain NVH1, with 50 of those sequences identical to this strain and two 99.7% similar. Eight (13.3%) sequences aligned most closely with B. washoensis 08S-0475, with seven 100% identical and one 99.7% similar (Fig. 1).
A comparison of the sequences obtained from ground squirrels with human strains
The majority of the B. washoensis gltA sequences detected in O. beecheyi, 92 of 104 (88.5%), were identical or closely related to the human strains. Forty-three (46.7%) of those sequences were most closely related to B. washoensis 08S-0475 and 17 (39.5%) of those were 100% identical, while 26 (60.5%) sequences had a sequence similarity of 99.4–99.7%. The remaining 49 (48.3%) sequences were most closely related to B. washoensis NVH1, and 46 (93.9%) were 100% identical, while three sequences (6.1%) had a sequence identity of 99.4–99.7%. The gltA sequence homology between B. washoensis NVH1 and B. washoensis 08S-0475 is 98.1%.
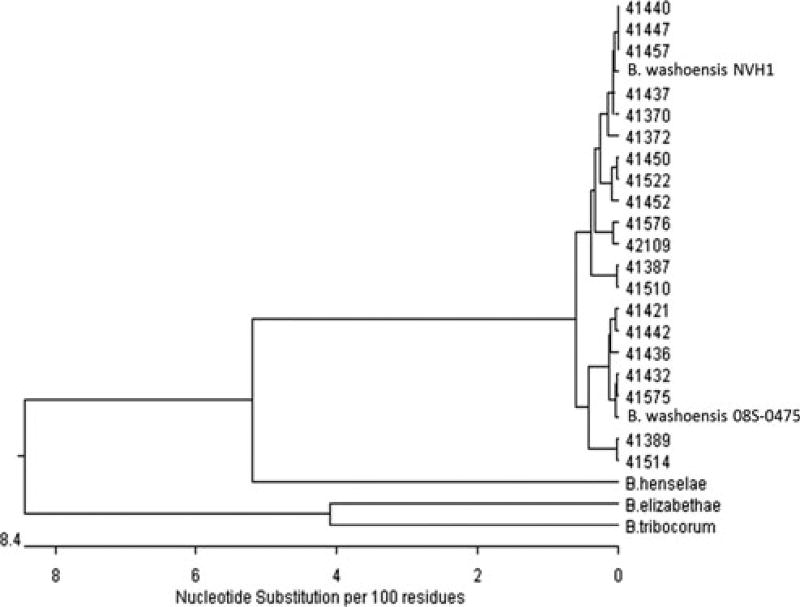
When analyzing concatenated sequences, the 20 selected isolates were most closely related to either one of the two human isolates of B. washoensis NVH1 or 08S-0475, with a sequence similarity of 99.1–99.9% (Fig. 2). The sequence divergence among selected isolates for each genetic marker ranged from 0 to 1.3% for ftsZ, 0 to 2.1% for groEL, 0 to 1.2% for ITS, and 0 to 4.0% for rpoB.
FIG. 2.
Phylogenetic tree of concatenated sequences, listed by isolate ID, combined from four housekeeping genes (gltA, groEL, ftsZ, and rpoB), and 16S-23S intergenic spacer region. The concatenated sequences of 3684-bp were compared to B. washoensis strains isolated from the two human patients from Nevada (NVH1) and California (08S-0475).
Comparison of genomes of the selected strains of B. washoensis by optical mapping
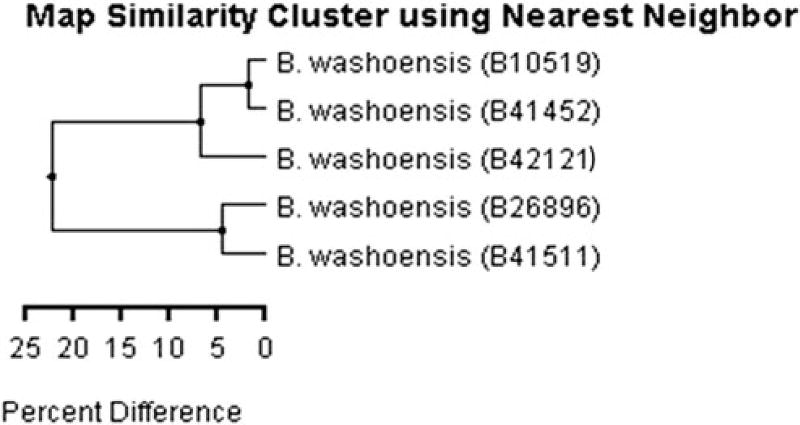
The optical maps scaled by comparison with partial genome sequences of B. washoensis strains 08S-0475 and Sb944nv (Fig. 3) and evaluated by similarity nearest neighbor clustering are shown in Figure 4. Cluster analysis demonstrated the presence of two groups among studied Nevada and California human isolates with ~23% cluster difference on genome architecture and 5% difference inside specific group. In contrast to gltA sequence analysis, O. beecheyi isolate 41,452 clustered with Nevada isolates (human and O. beecheyi) and the isolate 41,511 clustered with California human isolate (Fig. 4).
FIG. 3.
Comparison of optical maps for five isolates of B. washoensis (B41452: isolated from O. beecheyi (this study); B41511: isolated from O. beecheyi (this study); B10519: B. washoensis Sb944nv isolated from O. beecheyi from Nevada (Kosoy et al. 2003); B42121: B. washoensis NVH1 isolated from a human patient in Nevada (Kosoy et al. 2003); B26896: B. washoensis 08S-0475 isolated from a human patient in California (Probert et al. 2009)), as well as partial in silico generated maps for contigs from GenBank of partially sequenced genomes of strain Sb499nv (GenBank acc. no. JH725024, JH725026, JH25025, JH725022, and JH725023) and 08S-0475 (GenBank acc. no. JH725101). Perfect alignment homology of restriction enzyme BamHI maps shown in gray.
FIG. 4.
Evaluation of the optical maps of the five B. washoensis isolates with nearest neighbor clustering. The isolate ID is in parentheses. B41452: isolated from O. beecheyi (this study); B41511: isolated from O. beecheyi (this study); B10519: B. washoensis Sb944nv isolated from O. beecheyi from Nevada (Kosoy et al. 2003); B42121: B. washoensis NVH1 isolated from a human patient in Nevada (Kosoy et al. 2003); and B26896: B. washoensis 08S-0475 isolated from a human patient in California (Probert et al. 2009).
Discussion
In this study, a large percentage of California O. beecheyi and their fleas, O. montana, were shown to harbor B. washoensis strains closely related to two human isolates, strain NVH1 obtained from a Nevada patient diagnosed with cardiac disease and 08S-0475 from a meningitis case from northern California. The usage of multiple molecular markers and optical mapping technology significantly improved our ability to compare B. washoensis strains obtained in this study to previously published results.
Differences in prevalence were observed between certain sites: for example, at MWO in Los Angeles County only B. washoensis NVH1 was cultured and at BJC in Orange County only B. washoensis 08S-0475 was isolated from O. beecheyi. Due to the small number of samples obtained from each site, however, we cannot accurately assess the distribution of these zoonotic B. washoensis strains and future efforts should focus on obtaining a larger number of samples from various locations throughout the state. From a public health perspective, it is important to note that all investigated locations are frequently used by people for recreation purposes; therefore interactions between ground squirrels, fleas, humans, and their pets are likely, increasing the potential of B. washoensis transmission.
Many B. washoensis strain types have been reported in squirrels and other small mammals from Canada (Jardine et al. 2005), China (Inoue et al. 2009), Japan (Sato et al. 2012), Mexico (Rubio et al. 2014), the United Kingdom (Bown et al. 2002), and the United States (Bai et al. 2008). Multilocus sequence analysis conducted by Inoue et al. (2011) demonstrated that B. washoensis strains are likely host specific and that would appear to be the case in most strains detected in this study, as well. Isolates, related to NVH1 and 08S-0475, were the predominant strains found circulating in examined California O. beecheyi. Further testing of additional rodent genera, however, is necessary to confirm whether host specificity occurs with these zoonotic strains.
The most common flea species found infesting California ground squirrels are H. anomalus and O. montana and to a lesser extent, E. gallinacea (Metzger and Rust 1999, Hubbart et al. 2011). All three flea species were recovered from O. beecheyi in this study, although a limited number of E. gallinacea was available for testing. Interestingly, only O. montana fleas were found to harbor Bartonella DNA and in contrast to squirrel strains, only sequences identical to homologous sequences of human strains (NVH1 and 08S-0475) were detected in these fleas. These observations provide additional support to the supposition that O. montana is the most likely flea vector for zoonotic B. washoensis. Needless to say, experimental studies utilizing O. montana are needed to verify their role as arthropod vectors of B. washoensis. In addition, the role of other ectoparasites in the transmission of B. washoensis cannot be excluded due to the detection of B. washoensis NVH1 in questing adult Ixodes pacificus ticks in California (Chang et al. 2001).
This study sheds light on the presence and prevalence of B. washoensis in O. beecheyi and their fleas in north-central and southern California. Our results provide evidence to support that O. beecheyi and O. montana are a vertebrate reservoir and vector, respectively, for zoonotic B. washoensis. Future research should focus on understanding the influence of ecological factors and the transmission components of zoonotic B. washoensis, as well as evaluations of potential risks for humans.
Acknowledgments
The authors thank Dr. Marco E. Metzger, CDPH-VBDS, for his assistance with sample collection, John Montenieri, CDC, for assistance with flea identification, and Will Probert for providing B. washoensis strain 08S-0475.
Footnotes
Author Disclosure Statement
No competing financial interests exist.
References
- Angelakkis E, Lepidi H, Canel A, Rispal P, et al. Human case of Bartonella alsatica lymphadenitis. Emerg Infect Dis. 2008;14:1951–1952. doi: 10.3201/eid1412.080757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Kosoy M, Martin A, Ray C, et al. Characterization of Bartonella strains isolated from black-tailed prairie dogs (Cynomys ludovicianus) Vector Borne Zoonotic Dis. 2008;4:1–5. doi: 10.1089/vbz.2007.0136. [DOI] [PubMed] [Google Scholar]
- Billeter S, Gundi VA, Rood MP, Kosoy MY. Molecular detection and identification of Bartonella species in Xenopsylla cheopis fleas (Siphonaptera: Pulicidae) collected from Rattus norvegicus rats in Los Angeles, California. Appl Environ Microbiol. 2011;77:7850–7852. doi: 10.1128/AEM.06012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown KJ, Ellis BA, Birtles RJ, Durden LA, et al. New world origins for haemoparasites infecting United Kingdom grey squirrels (Sciurus carolinensis), as revealed by phylogenetic analysis of Bartonella infecting squirrel populations in England and the United States. Epidemiol Infect. 2002;129:647–653. doi: 10.1017/s0950268802007768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschwerdt EB. Bartonellosis: One health perspective for an emerging infectious disease. ILAR J. 2014;55:46–58. doi: 10.1093/ilar/ilu015. [DOI] [PubMed] [Google Scholar]
- Buffet JP, Kosoy M, Vayssier-Taussat M. Natural history of Bartonella infecting rodents in light of new knowledge on genomics, diversity, and evolution. Future Microbiol. 2013;8:1117–1128. doi: 10.2217/fmb.13.77. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chomel BB, Kasten RW, Romano V, et al. Molecular evidence of Bartonella spp. in questing adult Ixodes pacificus ticks in California. J Clin Microbiol. 2001;39:1221–1226. doi: 10.1128/JCM.39.4.1221-1226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomel BB, Kasten RW, William C, Wey AC, et al. Bartonella endocarditis: A pathology shared by animal reservoirs and Patients. Ann N Y Acad Sci. 2009;1166:120–126. doi: 10.1111/j.1749-6632.2009.04523.x. [DOI] [PubMed] [Google Scholar]
- Chomel BB, Wey AC, Kasten RW. Isolation of Bartonella washoensis from a dog with mitral valve endocarditis. J Clin Microbiol. 2003;41:5327–5332. doi: 10.1128/JCM.41.11.5327-5332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly JS, Worthington MG, Brenner DJ, Moss CW, et al. Rochalimae elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RM, Smith RT, Madon MB, Sitko-Cleugh E. Flea, rodent, and plague ecology at Chuchupate Campground, Ventura County, California. J Vector Ecol. 2002;27:107–125. [PubMed] [Google Scholar]
- Deng H, Le Rhun D, Buffet J, Cotté V, et al. Strategies of exploitation of mammalian reservoirs by Bartonella species. Vet Res. 2012;43:15. doi: 10.1186/1297-9716-43-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz PP, Maggi RG, Schwartz DS, Cadenas MB, et al. Canine bartonellosis: Serological and molecular prevalence in Brazil and evidence of co-infection with Bartonella henselae and Bartonella vinsonii subsp. berkhoffii. Vet Res. 2007;38:697–710. doi: 10.1051/vetres:2007023. [DOI] [PubMed] [Google Scholar]
- Furman D, Catts EP. Manual of Medical Entomology. 4. United Kingdom: Cambridge University Press; 1982. pp. 138–157. [Google Scholar]
- Gundi VA, Billeter S, Rood MP, Kosoy MY. Bartonella spp. in rats and zoonoses, Los Angeles, California, USA. Emerg Infect Dis. 2012;18:631–633. doi: 10.3201/eid1804.110816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez R, Krasnov B, Morick D, Gottlieb Y, et al. Bartonella infection in rodents and their flea ectoparasites: An overview. Vector-Borne Zoonot. 2015;15:27–39. doi: 10.1089/vbz.2014.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbart JA, Jachowski DS, Eads DA. Seasonal and among-site variation in the occurrence and abundance of fleas on California ground squirrels (Otospermophilus beecheyi) J Vector Ecol. 2011;36:117–123. doi: 10.1111/j.1948-7134.2011.00148.x. [DOI] [PubMed] [Google Scholar]
- Inoue K, Kabeya H, Hagiya K, Kosoy MY, et al. Multi-locus sequence analysis reveals host specific association between Bartonella washoensis and squirrels. Vet Microbiol. 2011;148:60–65. doi: 10.1016/j.vetmic.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Inoue K, Maruyama S, Kabeya H, Hagiya K, et al. Exotic small mammals as potential reservoirs of zoonotic Bartonella spp. Emerg Infect Dis. 2009;15:526–532. doi: 10.3201/eid1504.081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine C, Appleyard G, Kosoy MY, McColl D, et al. Rodent-associated Bartonella in Saskatchewan, Canada. Vector-Borne Zoonot. 2005;5:402–409. doi: 10.1089/vbz.2005.5.402. [DOI] [PubMed] [Google Scholar]
- Kandelaki G, Malania L, Bai Y, Chakvetadze N, et al. Human lymphadenopathy caused Ratborne Bartonella Tbilisi, Georgia. Emerg Inf Dis. 2016;22:544–546. doi: 10.3201/eid2203.151823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff FT, Bergmans AM, van Der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluid of a patient with neuroretinitis. J Clin Microbiol. 1999;37:4034–4038. doi: 10.1128/jcm.37.12.4034-4038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy M, Murray M, Gilmore RD, Jr, Bai Y, et al. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J Clin Microbiol. 2003;41:645–650. doi: 10.1128/JCM.41.2.645-650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger ME, Rust ME. Abiotic factors affecting the development of fleas (Siphonaptera) of California Ground Squirrels (Rodentia: Sciuridae) in southern California, USA; Proceedings of the 3rd International Conference on Urban Pests, Prague, Czech Republic; 1999. [Google Scholar]
- Norman AF, Regnery R, Jameson P, Greene C, et al. Differentiation of Bartonella-like isolates at the species level by PCR restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probert W, Louie JK, Tucker JR, Longoria R, et al. Meningitis due to a “Bartonella washoensis”—like human pathogen. J Clin Microbiol. 2009;47:2332–2335. doi: 10.1128/JCM.00511-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renesto P, Gouvernet J, Drancourt M, Roux V, et al. Use of rpo B gene analysis for detection and identification of Bartonella species. J Clin Microbiol. 2001;39:430–437. doi: 10.1128/JCM.39.2.430-437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MF, Billeter SA, Osikowicz L, Luna-Caipo DV, et al. Fleas and flea-associated Bartonella species in dogs and cats from Peru. J Med Entomol. 2015;52:1374–1376. doi: 10.1093/jme/tjv137. [DOI] [PubMed] [Google Scholar]
- Rubio AV, Ávila-Flores R, Osikowicz LM, Bai Y, et al. Prevalence and genetic diversity of Bartonella strains in rodents from northwestern Mexico. Vector-Borne Zoonot. 2014;14:838–845. doi: 10.1089/vbz.2014.1673. [DOI] [PubMed] [Google Scholar]
- Sato S, Kabeya H, Miura T, Suzuki K, et al. Isolation and phylogenetic analysis of Bartonella species from wild carnivores of the suborder Caniformia in Japan. Vet Microbiol. 2012;161:130–136. doi: 10.1016/j.vetmic.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Stark HE, Hudson BW, Pittman B. Plague Epidemiology. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; 1966. [Google Scholar]
- U.S. Department of Health. Pictorial Keys to Arthropods, Reptiles, Birds and Mammals of Public Health Significance. Atlanta, GA: U.S. Department of Health, Education and Welfare Public Health Service Communicable Disease Center; 1966. [Google Scholar]
- Welch DF, Carroll KC, Hofmeister EK, Persing DH, et al. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis from a cattle rancher: Identity with isolates found in conjunction with Borelia burgdoferi and Babesia microti among naturally infected mice. J Clin Microbio. 1999;37:2598–2601. doi: 10.1128/jcm.37.8.2598-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeaiter Z, Fournier PE, Ogata H, Raoult D. Phylogenetic classification of Bartonella species by comparing groEL sequences. Int J Syst Eco Microbiol. 2002a;52:165–171. doi: 10.1099/00207713-52-1-165. [DOI] [PubMed] [Google Scholar]
- Zeaiter Z, Liang Z, Raoult D. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsZ gene sequences. J Clin Microbiol. 2002b;40:3641–3647. doi: 10.1128/JCM.40.10.3641-3647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]