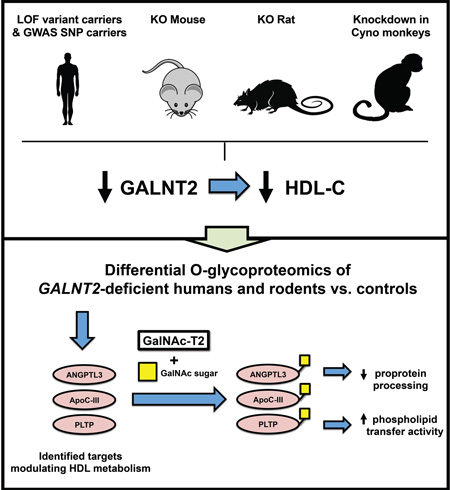
Summary
Human genetics studies have implicated GALNT2, encoding GalNAc-T2, as a novel regulator of high-density lipoprotein cholesterol (HDL-C) metabolism, but the mechanisms relating GALNT2 to HDL-C remain unclear. We investigated the impact of homozygous GALNT2 deficiency on HDL-C in humans and mammalian models. We identified two humans homozygous for loss-of-function mutations in GALNT2 who demonstrated low HDL-C. We also found that GALNT2 loss-of-function in mice, rats, and nonhuman primates decreased HDL-C. O-glycoproteomics studies of a human GALNT2 deficient subject validated ANGPTL3 and ApoC-III as GalNAc-T2 targets. Additional glycoproteomics in rodents identified targets influencing HDL-C, including phospholipid transfer protein (PLTP). GALNT2 deficiency reduced plasma PLTP activity in humans and rodents, and in mice this was rescued by reconstitution of hepatic Galnt2. We also found that GALNT2 GWAS SNPs associated with reduced HDL-C also correlate with lower hepatic GALNT2 expression. These results posit GALNT2 as a direct modulator of HDL metabolism across mammals.
Graphical Abstract
eTOC Paragraph
SNPs in GALNT2 are associated with HDL-C metabolism but whether GALNT2 causes HDL-C to go up or down has been debated. XXX et al show that loss-of-function of GALNT2 reduces HDL-C in humans, rodents and nonhuman primates. They also show species-specific glycosylation targets for GalNAc-T2.
Introduction
Single nucleotide polymorphisms (SNPs) in the first intron of GALNT2 were among the first common variants associated with high-density lipoprotein cholesterol (HDL-C) and triglycerides (TG) through genome-wide association studies (GWAS) for plasma lipids (Kathiresan et al., 2008; Kathiresan et al., 2009; Willer et al., 2008). We previously showed that Galnt2 overexpression reduced and knockdown increased HDL-C in mice, offering further support that GALNT2 was indeed the causal gene at the locus for this trait (Teslovich et al., 2010). GALNT2 encodes the enzyme UDP-N-Acetyl-D-galactosamine:polypeptide N-Acetylgalactosaminyl-transferase 2 (GalNAc-T2), which initiates O-glycosylation of proteins through addition of GalNAc to specific serine or threonine residues (Schjoldager and Clausen, 2012; White et al., 1995). The biological functions of GalNAc-glycosylation are not fully understood but include regulation of protein processing by proprotein convertases (Kato et al., 2006), protein multimerization (Leuenberger et al., 2003) and ectodomain shedding (Goth et al., 2015). Among the proposed targets of GalNAc-T2 that may affect lipoprotein metabolism are angiopoietin-like 3 (ANGPTL3) (Schjoldager et al., 2010) and apolipoprotein C-III (ApoC-III) (Holleboom et al., 2011; Schjoldager et al., 2012), both liver-secreted regulators of TG and HDL-C.
Despite the early association of this locus with plasma lipids from GWAS, the mechanisms underlying how GALNT2 regulates lipids have remained unknown and the directional relationship between GALNT2 expression and HDL-C has been controversial. Our Galnt2 overexpression and knockdown work suggested that GALNT2 is a negative regulator of HDL-C levels since overexpression reduced and knockdown increased HDL-C in mice (Teslovich et al., 2010). This directional relationship was supported by another study identifying heterozygous carriers of a putative loss-of-function (LOF) coding variant in GALNT2, p.Asp314Ala (D314A) (Holleboom et al., 2011). Contrasting these two studies, a recent evaluation of the GWAS SNPs in GALNT2 showed that alleles associated with increased HDL-C correlated with increased hepatic GALNT2 expression (Roman et al., 2015). Given these discordant findings, here we investigated the mechanisms and directionality relating GALNT2 to plasma lipids across humans and mammalian models of GALNT2 loss-of-function.
Results and Discussion
Humans homozygous for LOF mutations in GALNT2 have low HDL-C
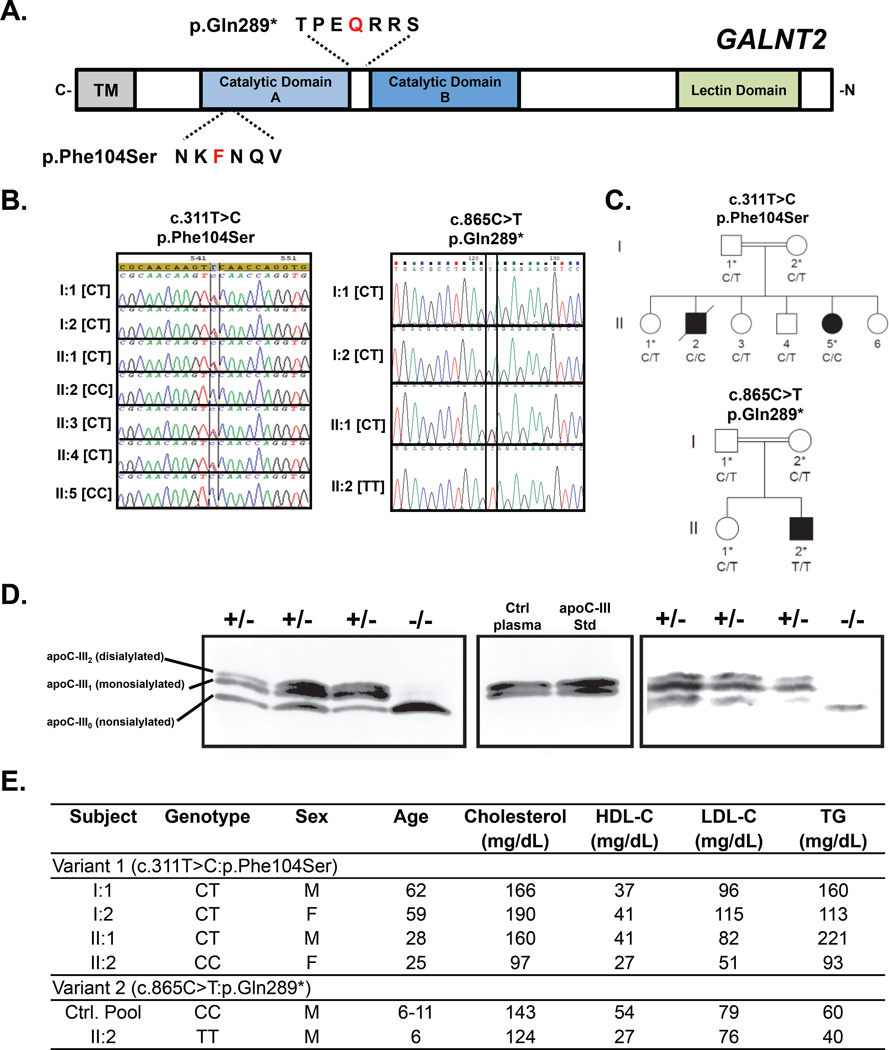
Through exome sequencing and mapping in two independent studies, we identified two unrelated individuals homozygous for different mutations in GALNT2. First, using linkage analysis and whole exome sequencing in families in search of causes of heritable neurological traits, we identified a female of Moroccan origin homozygous for a T>C mutation at position Chr1:230338973 in exon 3 of GALNT2, resulting in a p.Phe104Ser mutation (Figure 1 A–C). This mutation was absent from the 1000 Genomes, NCBI dbSNP, NHLBI Exome Sequencing Project, and Exome Aggregation Consortium (ExAC) variant databases and was predicted deleterious by Align GVGD, SIFT, Polyphen2, MutationTaster and SNP&GO software. We tested recombinant secreted GalNAc-T2 with the p.Phe104Ser variant (mutant) or no variant (WT) for their ability to glycosylate peptide substrates corresponding to two targets of GalNAc-T2, ApoC-III and immunoglobulin A1 (IgA1). While the WT GalNAc-T2 was highly active with both substrates, the p.Phe104Ser GalNAc-T2 mutant was completely inactive with ApoC-III and almost inactive with the IgA1 substrate.
Figure 1. Identification and lipid phenotype of human homozygotes with GALNT2 loss-of-function coding variants.
A. Schematic GALNT2 protein showing position of the two identified variants, p.Phe104Ser and p.Gln289. B. Sanger sequencing of identified variants, c.C311T>C: p.Phe104Ser, and c.C865T: p.Gln289*. C. Pedigree of family members of probands for variants in A-B. Asterisk denotes individuals analyzed by ApoC-III immunoblot. D. Immunoblot of plasma ApoC-III from the probands and family controls for the GALNT2 variants. Migration positions of the 3 major ApoC-III isoforms, nonsialylated ApoC-III (ApoC-III0) lacking any O-glycan modification; monosialylated ApoC-III (ApoC-III1) containing N-acetylgalactosamine, galactose and 1 terminal sialic acid; and disialylated ApoC-III containing N-acetylgalactosamine, galactose, and 2 terminal sialic acids (ApoC-III2), are indicated on the left. E. Plasma lipids of GALNT2 variant carriers.
Another exome sequencing study for recessive heritable causes of intellectual disability (Abou Jamra et al., 2011) identified a nonsense mutation in GALNT2 (NM_004481.4:c.865C>T; p.Gln289*) in a male proband of Pakistani origin. This mutation within the catalytic domain leads to a non-functional protein (Figure 1A) (Milac et al., 2007). The variant was confirmed with Sanger sequencing in the proband and family carriers (Figure 1 B–C). Like the p.Phe104Ser variant, this variant was also absent from existing variant databases.
To further validate the LOF mutations in vivo, we analyzed plasma of the probands and sampled relatives for ApoC-III glycoforms by SDS-PAGE immunoblotting (Figure 1D). While ApoC-III normally circulates as a mono- or di-sialylated protein with a molecular weight of ~9.5 kDa, we found only a band for non-glycosylated ApoC-III in the plasma of the two probands. In plasma from the heterozygous family members, we identified the normal bands with a distribution of mono- and di-sialylated isoforms, suggesting that heterozygosity for both GALNT2 LOF mutations retain sufficient enzyme activity to glycosylate ApoC-III in vivo. Notably, we found that both probands had reduced levels of HDL-C (<5th percentile for age and gender) (Figure 1E). We also found moderately reduced plasma TG in both probands and a moderately reduced LDL-C in the homozygote for the p.Phe104Ser variant (Figure 1E).
Reduced HDL-C in rodent and nonhuman primate models of GALNT2 deficiency
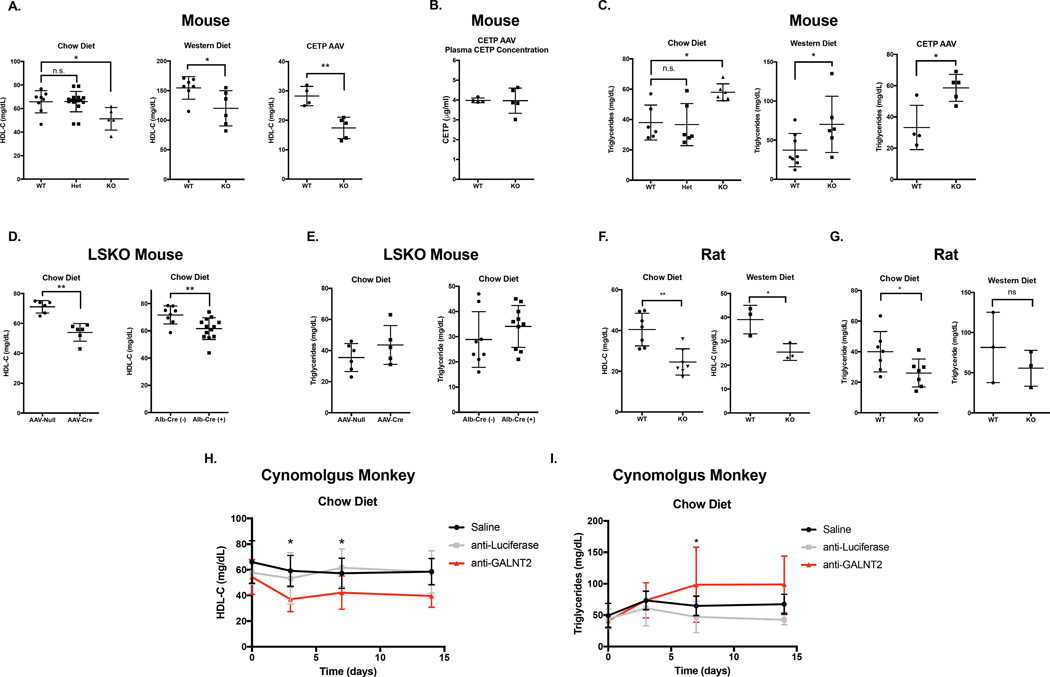
To further address the effect of GalNAc-T2 LOF on plasma lipids, we investigated GALNT2 deficiency in vivo in multiple mammalian models. We generated Galnt2 KO mice and rats, which harbor deletions in exons corresponding to the catalytic domain of the enzyme (Figure S1 A – B). Galnt2-deficient mice had near complete loss of hepatic Galnt2 mRNA levels (Figure S1 C – E), with no observed effects on expression of other GalNAc-transferase genes (Figure S1 G – H). In both chow-fed and Western diet-fed conditions, complete Galnt2 KO mice demonstrated reduced HDL-C relative to WT littermates (20–22% reduction in KOs, P<0.05; Figure 2 A). Because rodents lack cholesteryl ester transfer protein (CETP), a critical regulator of plasma lipids in humans, we tested whether the relationship of Galnt2 to HDL-C was maintained after expression of CETP in Galnt2 KO and WT mice through administering AAV vectors expressing CETP. After 4 weeks of CETP expression, Galnt2 KO mice had 40% lower HDL-C levels compared to WT littermates (P<0.01, Figure 2 A), with similar plasma CETP levels in both groups (Figure 2 B). Galnt2 KO mice also demonstrated a robust elevation in fasting TG (>50% increased TG in Galnt2 KOs on chow diet relative to WT, P<0.05), which was also observed upon 3 weeks of feeding a Western-type diet and after 4 weeks of AAV-mediated CETP expression (P<0.05 for both; Figure 2 C). Importantly, in chow fed mice, we observed no intermediate effect of heterozygous Galnt2 deficiency on HDL-C or TG (Figure 2 A).
Figure 2. Plasma HDL cholesterol in mammalian models of GALNT2 loss-of-function.
A. Plasma HDL-C after a 4 hour fast from Galnt2 WT, heterozygous (Het), and KO mice fed a chow diet (left), a chow diet after 4 weeks or administration of human CETP AAV (center), or a Western-type diet for 3 weeks (right). B. Plasma CETP concentration after 4 weeks in mice from (A, right). C. Plasma triglycerides in mice from (A). D. Plasma HDL-C from Galnt2 WT vs. liver-specific KO mice generated by AAV-Cre delivery in Galnt2fl/fl mice (left) or crossing Galnt2fl/fl with Alb-Cre transgenic mice (right). E. Plasma triglycerides in mice from (D). F. Plasma HDL-C after fasting for 12–16 hours from Galnt2 WT vs. KO rats fed a chow diet (left) or 6 weeks of Western-type diet (right). G. Plasma triglycerides in rats from (F). H. Plasma HDL-C after overnight fasting in cynomolgus monkeys (cyno) treated with saline, anti-luciferase siRNA or anti-GALNT2 siRNA. I. Plasma triglycerides in cyno monkeys from (H). Data is presented as mean values ± S.D.. * P<0.05, ** P<0.01, Student’s unpaired T-test.
Given the importance of the liver in HDL-C metabolism, we also generated liver-specific models of GALNT2 deficiency to test if the association between Galnt2 and plasma lipids is mediated by hepatic expression. Liver-specific Galnt2 KO mice (Galnt2 LSKOs) were generated by injecting Galnt2fl/fl mice with an AAV vector expressing Cre recombinase under the regulation of a liver-specific promoter, as well as by breeding Galnt2fl/fl mice to Albumin-Cre transgenic mice. Galnt2 LSKOs showed near-complete reduction of liver Galnt2 mRNA levels (Figure S1 D). LSKO mice demonstrated 15–22% reduced HDL-C relative to WT controls (P<0.01 for both; Figure 2 D). While the HDL-C reduction in the LSKOs was similar to that of complete KOs, plasma TG in LSKOs were similar to WT controls (Figure 2 E).
Similarly to the results in mice, Galnt2 KO rats had near complete loss of hepatic Galnt2 mRNA levels (Figure S1 E) and 30% lower plasma HDL-C levels on a chow diet than WT littermates (P<0.01; Figure 2 F). There was a similar reduction in a smaller cohort of WT vs. KO rats (3 per group) fed a Western diet for 6 weeks (P<0.05; Figure 2 F). But unlike the complete KO mice, Galnt2 KO rats demonstrated reduced plasma TG while fed a chow diet. There were no significant differences in plasma TG after 6 weeks of Western diet feeding (3 per group) (Figure 2 G). This may highlight a potentially complex, species-specific regulation of lipoprotein metabolism by GALNT2.
We also explored the impact of GALNT2 deficiency on non-HDL cholesterol (nonHDL-C) levels in our rodent models. Galnt2 KO mice fed a chow diet, a Western diet, or administered CETP AAV all demonstrated no differences in plasma nonHDL-C (Figure S2 A), with similar results when measured by fast-protein liquid chromatography (FPLC) of plasma lipoproteins (Figure S2 B – C). Similarly, liver-specific KO mice also demonstrated no significant differences in nonHDL-C (Figure S2 D – E), while KO rats on chow diet demonstrated a modest reduction in nonHDL-C on a chow diet with no differences in nonHDL-C after 6 weeks of Western diet feeding (Figure S2 F). Collectively, these findings suggest a minimal impact of GALNT2 deficiency on nonHDL-C levels, consistent with the lack of association of this trait with the GALNT2 locus in the prior human GWAS.
Impact of GALNT2 deficiency in mice on HDL-C and TG turnover
Using the LSKO model, we performed clearance studies of HDL radiolabeled with both 3H cholesteryl ether and 125I-ApoA-I. Galnt2 LSKO mice demonstrated a moderate increase in the clearance of 3H cholesteryl ether but not in 125I-ApoA-I relative to controls (Figure S3 A – B), consistent with enhanced HDL cholesterol ester-selective catabolism. Given the previous implication of ApoC-III and ANGPTL3 as putative GalNAc-T2 targets, we also explored postprandial TG clearance in the complete KO and liver-specific KO mice (Figure S3 C – D). We observed delayed TG clearance in the complete KOs but not the liver-specific KO mice. This further supports the notion that the regulation of plasma TG levels by Galnt2 in mice may be extra-hepatic. Despite the observation of delayed lipolysis ex vivo in the complete KO mice, we observed no differences in post-heparin plasma TG or phospholipase activities in these models (Figure S3 E – H)
Knockdown of hepatic GALNT2 in non-human primates reduces HDL-C levels
We also studied the effects of hepatic GALNT2 silencing on plasma lipids in a nonhuman primate model, the cynomolgus monkey (cyno) using siRNA. Hepatic GALNT2 mRNA knockdown mediated by a single dose of siRNA was substantial and persisted for up to 4 weeks (Figure S1 F). GALNT2 knockdown in cyno resulted in >30% reduction of HDL-C relative to controls as early as 3 days after siRNA treatment (Figure 2 G). Knockdown of GALNT2 in cyno resulted in a trend towards elevated fasting TG that was 2-fold higher in the knockdown group 7 days after siRNA treatment (P<0.05; Figure 2 H). Additionally, we found a reduction in nonHDL-C in the anti-GALNT2 siRNA treated monkeys (Figure S2 G), possibly due to the elevation in nonHDL-C in that control group itself; there were no differences in nonHDL-C between the anti-GALNT2 siRNA treated monkeys and saline-treated controls.
Quantitative differential O-glycoproteomics confirm ANGPTL3 and identify PLTP as O-glycoprotein targets controlled by GalNAc-T2
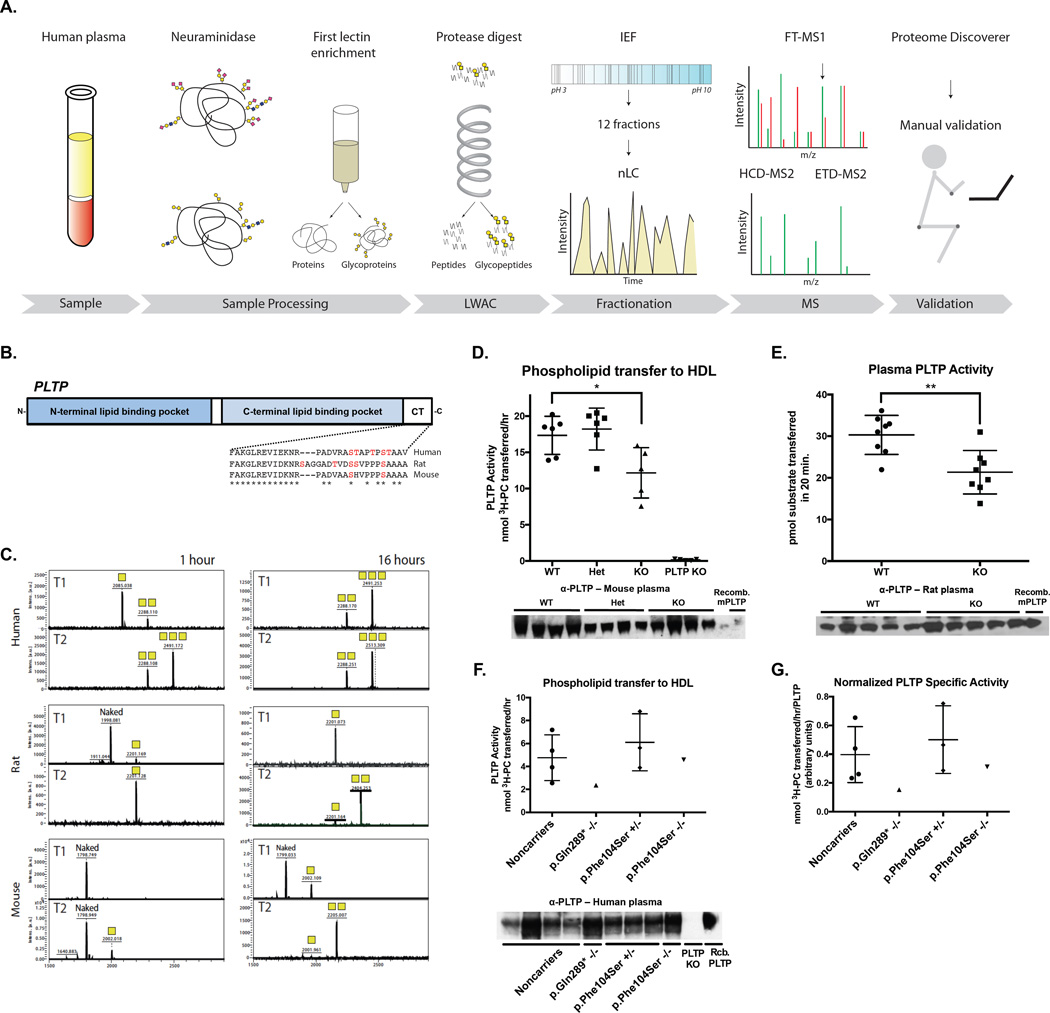
To understand mechanisms underlying the effect of GALNT2 deficiency on plasma lipids we observed in the human subjects and animal models, we employed an unbiased quantitative O-glycoproteomics strategy to identify protein targets with altered glycosylation (Schjoldager et al., 2015). We performed differential quantification of plasma (human, rats and mice) and liver (rats and mice) tissue O-glycopeptides from GALNT2 LOF models (Figure 3 A), revealing multiple glycosites in each model present in WT samples but >10-fold reduced or absent in KO samples (Figure S4 A – C & Table S1).
Figure 3. Differential glycoproteomics of tissues from Galnt2 WT vs. KO rodents identifies PLTP as a candidate target of GalNAc-T2.
A. Schematic of differential glycoproteomics approach to identify GalNAc-T2 targets from humans and rodent models. B. Phospholipid transfer protein (PLTP) schematic and cross-species alignment showing the C-terminal HDL-binding region. Asterisk shows conversed residues among mice, rats, and humans. Potential O-glycosylated residues (Ser and Thr) are indicated in red. C. In vitro glycosylation assay of PLTP C-terminal peptides by recombinant human GalNAc-T1 and –T2 over 1 hour and 16 hours. Yellow squares denote the number of GalNAc residues on each peptide. D. (Top) Plasma PLTP activity from Galnt2 WT, Galnt2 heterozygous (Het), Galnt2 KO (KO), and PLTP-deficient (PLTP KO) mice. (Bottom) Immunoblot of plasma PLTP from mice from activity assay above. E. (Top) Plasma PLTP activity from Galnt2 WT vs. KO rats. (Bottom) Immunoblot of plasma PLTP from rats from activity assay above. F. (Top) Plasma PLTP activity from human control subjects and homozygotes for GALNT2 nonsynonsymous variants. (Bottom) Immunoblot of plasma PLTP from human participants from PLTP activity assay above. G. Plasma PLTP specific activity from human participants in (F). Specific activity was measured as the plasma activity from (F Top) normalized to the densitometric intensity for the immunoblot from (F bottom). Where applicable, data is presented as mean values ± S.D.. ** P<0.01, ***P<0.001, Student’s unpaired T-test.
From the human O-glycoproteome, we confirmed ApoC-III (Thr74) as a GalNAc-T2 dependent substrate. Notably, ApoC-III was not found in the same analysis of rat and mouse plasma and liver samples (Table S1), in agreement with our finding that the rodent sequences around the glycosites are different and not specifically served by GalNAc-T2 but redundantly by multiple isoforms (Schjoldager et al., 2012). This was confirmed by immunoblotting of ApoC-III from rat and mouse plasma showing no difference in SDS-PAGE migration and levels with only one major band detectable in both WT and KO samples (Figure S4 D – E). We also confirmed that O-glycosylation of ANGPTL3 (Thr226) was selectively lost in the human GALNT2-deficient O-glycoproteome (Table S1), in agreement with our previous in vitro studies (Schjoldager et al., 2012). The sequence around the Thr glycosylation site in ANGPTL3 is conserved in rodents and in vitro analysis of rodent ANGPTL3 confirmed that only GalNAc-T2 was active with this substrate, however we did not detect the corresponding glycopeptide in our glycoproteomics analysis of rodent plasma and liver.
Our differential O-glycoproteomics identified phospholipid transfer protein (PLTP) as O-glycosylated in the plasma and liver of WT mice but absent in KOs (Table S1). The identified O-glycosites were located in the C-terminus of PLTP, a region largely conserved among humans, mice, and rats (Figure 3 B). To validate PLTP as a non-redundant target of GalNAc-T2, we tested the in vitro specificity of the two main GalNAc-Ts expressed in liver, GalNAc-T1 and GalNAc-T2, with peptide substrates corresponding to the C-terminus of human, rat and mouse PLTP (Figure 3 C). We found that GalNAc-T2 was more active than GalNAc-T1 with all three peptides and in particular with the rodent peptides (Figure 3 C). These data support PLTP as a conserved preferential target of GalNAc-T2.
PLTP transfers phospholipids from apoB-containing lipoproteins to HDLs in circulation, and is a known regulator of plasma HDL metabolism (Albers et al., 2012). Mice lacking PLTP activity exhibit reduced HDL-C (Jiang et al., 1999) and increased clearance of HDL-C in vivo (Jiang et al., 2012), a finding resembling that of the Galnt2 KO rodents. We hypothesized that GalNAc-T2 may modulate HDL metabolism at least in part through promoting PLTP function. We compared PLTP activity in plasma from PLTP-deficient mice, rats, and humans relative to controls. Galnt2 KO mice and rats both showed >20% lower plasma PLTP activity relative to WT littermates (P<0.01 for each; Figure 3 D–E), with no apparent difference in total plasma PLTP protein was observed by immunoblotting (Figure 3 D–E), suggesting that the reduction of PLTP activity in the plasma of Galnt2 KO rodents is not due to reduced plasma PLTP protein levels but rather reduced function. This finding is consistent with prior mutagenesis and deletion studies implicating the final 30 residues of PLTP as critical for lipid transfer activity in vitro (Huuskonen et al., 1998). We measured plasma PLTP activity from the p.Gln289* and p.Phe104Ser homozygotes and family carriers and controls (Figure 3 F) and found that plasma from both probands had reduced PLTP activity relative to the mean of the p.Phe104Ser heterozygotes or noncarriers. Furthermore, when we measured a ‘specific activity’ for plasma PLTP, we found a reduction in the two homozygotes relative to controls, with the p.Gln289* homozygous subject’s activity >60% lower and the p.Phe104Ser homozygous subject’s activity >20% lower than the mean of the noncarrier control subjects’ activity (Figure 3 G). These data suggest that GALNT2 is required for facilitating PLTP function and maintaining HDL-C levels in rodents and humans. It has been shown that PLTP deficiency in hyperlipidemic mice models attenuated both VLDL secretion and atherosclerosis (Jiang et al., 2001). Thus we measured VLDL-TG secretion in the Galnt2 WT vs. complete KO mice and found a ~15% reduction in the rate of secretion in the KOs (P<0.05; Figure S3 I). These data further support the notion of reduced PLTP function in the setting of GALNT2 deficiency.
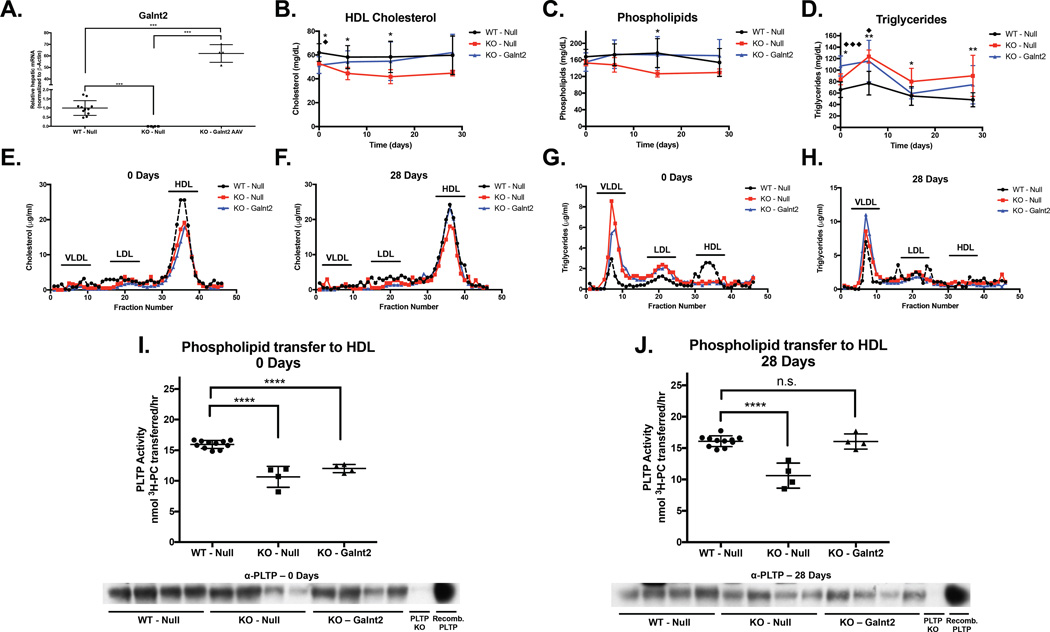
We asked if restoration of Galnt2 expression was sufficient to rescue the HDL-C and PLTP activity reduction in vivo. We reconstituted murine Galnt2 in the liver of Galnt2 KO mice using AAV at a dose of 1×1011 genome copies (GC) of virus per mouse, which was 10-fold lower than the dose used in our prior overexpression study of Galnt2 in WT mice (Teslovich et al., 2010). We compared hepatic Galnt2 restoration at this dose in Galnt2 KO mice to WT mice and Galnt2 KO mice receiving the same dose of Null virus and observed robust restoration of hepatic Galnt2 in the rescue group at 28 days after AAV administration (Figure 4 A). Restoration of hepatic Galnt2 expression markedly raised the reduced levels of HDL-C in the KO mice back to the levels in WT controls receiving Null AAV (Figure 4 B). Similarly, we monitored plasma phospholipid levels, which are low in the setting of PLTP deficiency, and found that while KO mice had lower phospholipids relative to WT initially, the KO mice with hepatic Galnt2 reconstitution showed elevated phospholipids over 4 weeks after dosing (Figure 4 C). We also measured plasma TGs and found that levels were variable across the different timepoints but hepatic Galnt2 rescue in the KO mice caused a moderate reduction in TG levels by 2 weeks post-injection (Figure 4 D). We observed similar trends in cholesterol and triglycerides in the lipoprotein fractions from FPLC separation of pooled plasma of rescued mice as we observed above (Figure 4 E–H).
Figure 4. Reconstitution of hepatic Galnt2 expression in Galnt2 KO mice restores HDL-C and PLTP activity.
A. Hepatic Galnt2 expression in WT mice transduced with Null AAV (WT – Null), Galnt2 KO mice transduced with Null AAV (KO – Null), and KO mice transduced with Galnt2 AAV (KO – Galnt2). ***P<0.001, Student’s unpaired T-test. B. Plasma HDL-C in mice from (A). C. Plasma phospholipids from mice in (A). D. Plasma Triglycerides from mice in (A). For B-D, significance was determined at each timepoint. *P<0.05, **P<0.01, Student’s unpaired T-test of comparison of WT – Null to KO – Null. ♦ P<0.05, ♦♦♦ P<0.001, Student’s unpaired T-test of comparison of WT -Null to KO – Galnt2. E. Day 0 cholesterol in fractions after FPLC separation from pooled plasma from each of the groups in (A). F. Day 28 cholesterol from FPLC fractions of pooled plasma from groups in (A). G. Day 0 triglycerides in FPLC fractions from pooled plasma from groups in (A). H. Day 28 triglycerides in FPLC fractions from pooled plasma from groups in (A). I. (Top) Plasma PLTP activity from Day 0 plasma from mice in (A). (Bottom) Immunoblot of plasma PLTP from PLTP activity assay above. J. (Top) Plasma PLTP activity from Day 28 plasma from mice in (A). (Bottom) Immunoblot of plasma PLTP from PLTP activity assay above. For I-J, ****P<0.0001, Student’s unpaired T-test for comparisons between the indicated groups. Data is presented as mean values ± S.D..
Since Galnt2 hepatic reconstitution rescued plasma HDL-C levels in the Galnt2 KO mice, we next assessed the impact of hepatic Galnt2 restoration on plasma PLTP activity. We measured plasma PLTP-mediated transfer of phospholipids to HDL using plasma from the 3 groups of mice described above both before and after AAV administration. Prior to AAV dosing, plasma PLTP activity in the KO groups of mice was approximately 33% lower than that of WT mice (P<0.0001; Figure 4 I). 28 days after Galnt2 reconstitution in KO mice, their plasma PLTP activity had risen to the level of WT mice (Figure 4 J). At both timepoints, total plasma PLTP levels were comparable (Figure 4 I–J, bottom). These data support the notion that hepatic Galnt2 is both necessary and sufficient to potentiate full plasma PLTP activity in mice in vivo.
In addition to PLTP, we identified other specific targets for glycosylation by GalNAc-T2 that may contribute to the differences observed in the rodent phenotypes. These include a non-redundant site in the C-terminus of ApoE (Thr305), a critical ligand for hepatic lipoprotein receptors, in both rat liver and plasma. Additionally, we identified a non-redundant T2-specific glycosite in the N-terminus of hepatic lipase (HL) (Thr38) in mouse liver and plasma. HL is a hepatocyte-expressed TG lipase and phospholipase that hydrolyzes lipids on TG-rich lipoproteins after secretion from liver cells onto the cell surface in a manner dependent on HDL (Chatterjee and Sparks, 2011). Mouse liver glycoproteomics also identified two GalNAc-T2 specific glycosites in the hepatic lipoprotein receptor low-density lipoprotein related receptor 1 (LRP1) (Thr27 and Thr3937). LRP1 is a hepatic receptor mediating the uptake of ApoE-containing TG-rich lipoprotein-remnant particles.
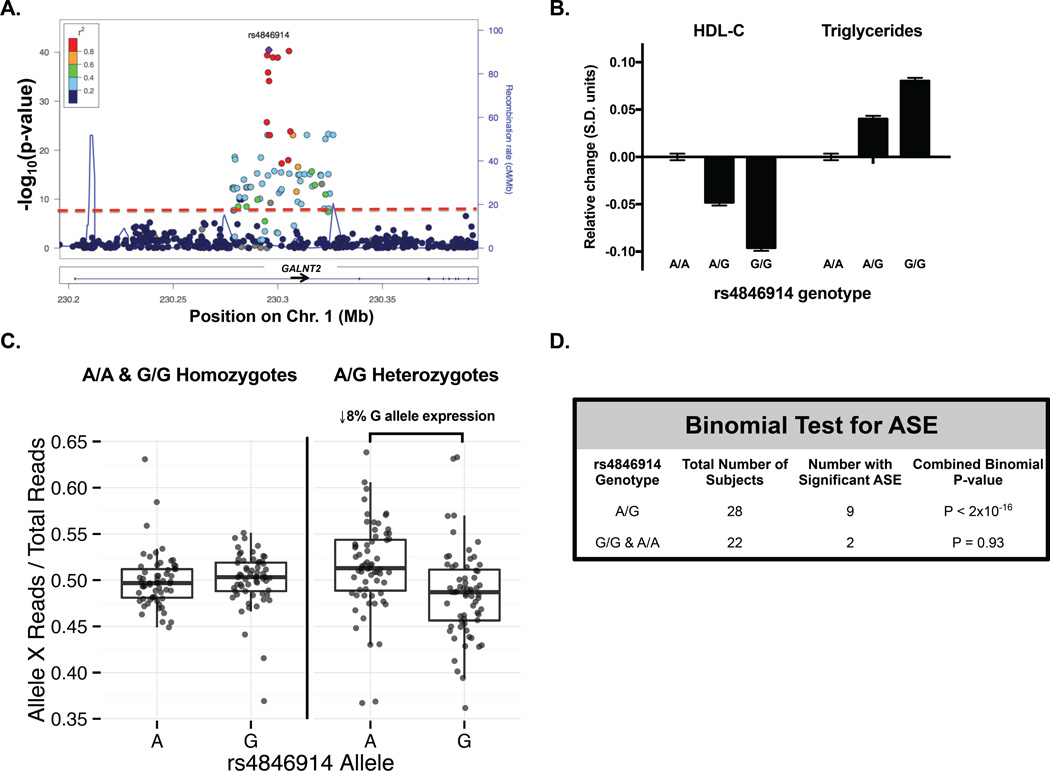
GALNT2 SNPs associated with lower HDL-C are associated with reduced GALNT2 expression in human liver
We next asked if the common SNPs in GALNT2 associated with lipids in GWAS impacted GALNT2 expression in direction consistent with that observed in the LOF models. To answer this, we sequenced the RNA transcriptome from human liver samples and quantified allele-specific GALNT2 expression while accounting for genotype at rs4846914, the lead SNP in GALNT2 associated with HDL-C and TG (Figure 5 A). In a recent GWAS meta-analysis, each copy of the G allele for this SNP was associated with a mean reduction in HDL-C of 0.48 standard deviation (S.D.) units and a 0.40 S.D. unit increase in TG per copy (Figure 5 B). We compared GALNT2 mRNA transcript levels arising from the G allele vs. the A allele for the rs4846914 SNP for each liver RNA transcriptome. From analysis of rs4846914 G/A heterozygotes, we observed an 8% reduction in GALNT2 expression from the G allele relative to the A allele (Figure 5 C). The heterozygotes thus demonstrated a modest but significant allele-specific expression (P < 2 × 10−16; Figure 5 D), with the G allele (associated with lower HDL-C) having lower expression than the A allele (associated with higher HDL-C), consistent with the directionality from LOF variant carriers and animal models. These results are also consistent with a prior eQTL study of plasma lipid GWAS SNP effects on nearest-gene expression levels in human liver (Folkersen et al., 2010), and a recent fine mapping of the GWAS-associated GALNT2 noncoding SNPs (Roman et al., 2015). We note that the directionality found here of reduced GALNT2 expression by alleles associated with higher TG in GWAS contrasts with the finding in the two GALNT2 LOF variant carriers of relatively low TG; this highlights the complexities of assessing plasma TGs in humans and the need to identify additional homozygotes for LOF variants for additional assessment.
Figure 5. Common variants in GALNT2 associated with HDL-C confer allelic imbalance of GALNT2 expression in human liver.
A. Manhattan plot of association of GALNT2 SNPs with HDL-C from the Global Lipids Genetics Consortium (GLGC) GWAS. Colors indicate the amount of linkage disequilibrium between plotted SNPs. Purple colored circle indicates lead SNP rs4846914 in GALNT2 intron 1. B. Relative change in HDL-C and TG (in S.E. units) per copy of rs4846914 allele from the GLGC GWAS. C. Allele-specific expression (ASE) of the rs4846914 SNP on GALNT2 expression from 72 human liver samples as measured by RNA-Seq. The fraction of GALNT2 reads arising from either the A or G allele for the rs4846914 SNP relative to total GALNT2 reads is plotted. Triangles indicate samples with significant ASE while circles indicate nonsignificant ASE. D. One-sided binomial test used to assess ASE for the distribution of GALNT2 transcripts arising from rs4846914 A vs. G allele in samples with A/A or A/G genotype for each sample. Data shows mean ± S.D..
Reconciling the directionality of GALNT2 expression to HDL-C
Here we aim to better reconcile the contrasting directionalities from our current work and these previous efforts by further exploring the prior conclusions. We first examined our conclusions from our 2010 report (Teslovich et al., 2010) that showed that Galnt2 overexpression reduced HDL-C levels while shRNA-mediated knockdown of hepatic Galnt2 raised HDL-C in mice. We subsequently found that at the AAV dose of 1×1012 GC/mouse used previously, overexpression of Galnt2 reproducibly decreased HDL-C by 25–30% 14 days after administration (Figure S5 A). This dose also caused a >100,000-fold overexpression above endogenous Galnt2 levels of WT mice treated with Null virus (Figure S5 B). A dose escalation of Galnt2 AAV in WT mice showed that the HDL-C reduction was not observed at AAV doses lower than 3×1011 GC/mouse despite high overexpression of Galnt2. We also questioned whether the HDL-C reduction was due to Galnt2 enzymatic activity. To test this, we engineered an AAV variant that expressed a mutant form of GALNT2 harboring the p.Glu334Gln (E334Q) variant, a highly conserved residue in the catalytic domain critical to glycosyltransferase activity (Fritz et al., 2006). Glycosyltransferase activity assays testing this mutant showed that it conferred no activity relative to WT GalNAc-T2 (Figure S5 C). Comparing WT and E334Q mutant Galnt2 AAVs at the high dose of 1×1012 GC/mouse showed that both equally reduced HDL-C by 30% after 14 days (Figure S5 D). These data support the conclusion that Galnt2 overexpression by AAV at the doses we used previously causes a transgene-specific but glycosyltransferase activity-independent reduction in HDL-C levels.
Our prior work also suggested that AAV shRNA-mediated hepatic Galnt2 knockdown raises HDL-C levels, which contrasts with the findings from the current work in multiple LOF models. Subsequent studies by others have shown that high doses of AAV shRNA expression in the liver can mediate hepatotoxicity due to oversaturation of microRNA (miRNA) export pathways from the nucleus and reduction of essential miRNAs (Grimm et al., 2006). Importantly, these findings were shown to be shRNA sequence-specific but not target gene-specific. Since our 2010 report, we observed that anti-Galnt2 AAV shRNA treatment at the high dose of 1×1012 genome copies/mouse caused an elevation in HDL-C (Figure S5 E) but also hepatic dysfunction as measured by plasma alanine aminotransferase (ALT) levels (Figure S5 F). In our prior AAV shRNA experiments at this dose, we also found reduced levels of miR-122a (Figure SF G), which constitutes a majority of miRNAs in the liver (Lagos-Quintana et al., 2002). As several other hepatic miRNAs have been shown to modulate HDL-C levels (Goedeke et al., 2015; Najafi-Shoushtari et al., 2010; Rayner et al., 2010), we postulate that our results from AAV shRNA experiments may be influenced by additional hepatocellular changes as a result of the degree of shRNA overexpression. Indeed, experiments with other AAV shRNAs targeting Galnt2 at a lower dose of 2.5×1011 GC/mouse caused no elevation in HDL-C and also no increase in plasma aminotransferases (Figure S5 H – K). We posit that the germline Galnt2-deficient models we study here offer a ‘cleaner’ view of the phenotype of heritable LOF, which we demonstrate is consistent across multiple species.
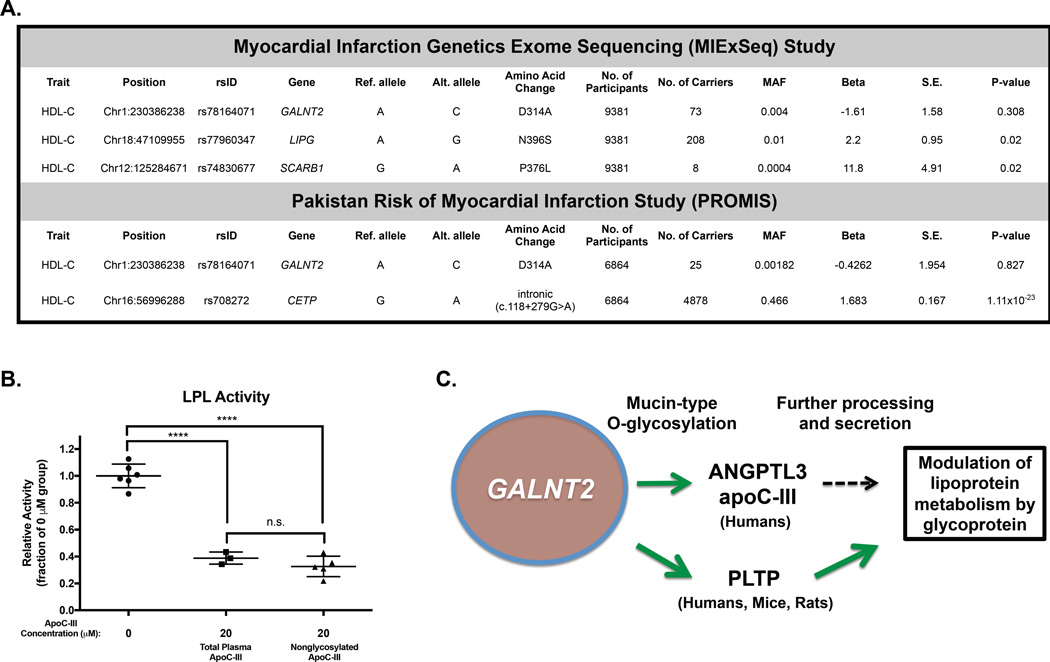
The 2011 study by Holleboom and colleagues (Holleboom et al., 2011) reported two probands with elevated HDL-C who were found to be heterozygotes for a nonsynonymous missense variant in GALNT2, D314A. This report suggested that this variant modestly reduces catalytic activity of GalNAc-T2 as measured by an in vitro enzymatic assay. However, a subsequent identification of this variant in a cohort of 2,000 Danes and functional testing showed the variant to confer normal enzymatic activity (Hansen et al., 2015). Because the 2011 report identified the variant in 2 high HDL-C probands and an additional 6 heterozygotes with normal HDL-C levels, we wondered if identification of additional carriers of this variant and potentially homozygotes would help us better determine the relationship of this variant to HDL-C levels and any allele-dosage effect of the variant on this trait. We searched for carriers of the D314A variant in 2 ethnically distinct exome sequencing cohorts in which HDL-C levels were measured (Figure 6 A). In both cohorts, we identified numerous heterozygous carriers of this GALNT2 variant and one homozygote. We tested the association of this variant with HDL-C levels in both cohorts and found no significant relationship in either cohort; indeed, the D314A homozygote had low HDL-C levels (23 mg/dl). For both cohorts, we identified highly significant associations of known variants at other HDL-C loci, supporting the validity of these findings. These data suggest that in larger populations across ethnicities, the D314A variant in GALNT2 is not associated with higher HDL-C levels and that the initial report was limited by power considerations.
Figure 6. Relationship of the previously reported D314A variant with HDL-C.
A. Association of GALNT2 D314A with HDL-C in the MIExSeq rare variant association study (top) and the PROMIS study (bottom). For each association test, representative variants significantly associated with HDL-C for each respective study are given below the results for the GALNT2 D314A variant. Each study was 100% powered to detect a 1 S.D. change in HDL-C with significance level α=0.05. B. In vitro LPL activity in the presence of 0 µM ApoC-III, 20 µM total plasma ApoC-III, and 20 µM nonglycosylated ApoC-III. Data is shown as mean values ± S.D.. ****P<0.0001, Student’s unpaired T-test. C. Model for proposed targets of GALNT2 that modulate lipoprotein metabolism. Green lines show activating roles of GalNAc-T2 mediated O-glycosylation. Black dotted line indicates yet unclear regulatory effect of the glycosylation of indicated targets.
Our data demonstrate that ApoC-III is a human species-specific non-redundant substrate for GalNAc-T2, but the role of O-glycosylation in modulating its lipoprotein modulatory functions remains unclear. Prior work showed that lack of ApoC-III O-glycosylation at Thr74 did not impair protein secretion or binding affinity for lipoproteins in vitro (Roghani and Zannis, 1988). Moreover, human subjects lacking ApoC-III glycosylation due to inheritance of a missense variant at the glycosylation site Thr74 demonstrate normal plasma TG levels and ApoC-III incorporation into lipoproteins (Maeda et al., 1981), suggesting that the glycosylation of ApoC-III alone may not substantially impact plasma lipoprotein levels. Indeed, our own measurement of in vitro LPL activity inhibition by total plasma ApoC-III (containing mostly glycosylated isoforms) and nonglycosylated ApoC-III showed equally potent inhibition of lipolysis of a radiolabeled TG substrate in the presence of recombinant LPL (Figure 6 B).
Integrating our findings, the data presented here support a working model (Figure 6 C) of the regulation of lipoprotein metabolism by GALNT2 by which it regulates at least three substrates, ANGPTL3, ApoC-III, and PLTP, in ways that may have differing impacts on these target proteins with regard to their respective processing and functions. These modifications may be species-specific as in the case of ApoC-III and it is likely that additional targets underlie the complex phenotypic differences among GALNT2 LOF models, which remain our area of continued study.
Conclusions
To date, few of the novel lipid loci identified by GWAS have been shown to physiologically influence plasma lipids in vivo. Strategies for studying function of these novel loci in animal models have largely involved somatic overexpression and knockdown in murine models as well as generation of whole-body or tissue-specific KO mice. These approaches meet with the challenges of 1) the difficulty in pinpointing the causal gene at a newly associated locus harboring multiple genes, 2) the ability to appropriately ascertain the relevant tissue(s) in which expression of the candidate gene regulates the associated phenotype, and 3) the limitations of relating findings in murine models to humans due to the complex physiological differences between rodent and human lipoprotein metabolism. This is an exceptionally difficult task when the candidate gene is involved in partly redundant glycosylation of a multitude of proteins. Through our cross-species studies of GALNT2, we provide an example of how studies of LOF mutations in rare individuals combined with multiple animal model systems might be leveraged to validate and determine the relationship of GWAS candidate genes to complex traits.
Experimental Procedures
Identification of GALNT2 LOF variant carriers
The p.Gln289* variant was identified by linkage analysis by genotyping using the Affymetrix Mapping array 6.0 (Affymetrix) and then exome sequencing using the SureSelect Human All Exon Kit (Agilent). The p.Phe104Ser variant was identified through a genome-wide microarray screen (Illumina 6K panel) and haplotype reconstruction, followed by exome sequencing (Illumina). ApoC-III from plasma of carriers and family members was measured by immunoblotting as described previously (Schjoldager et al., 2012). Plasma lipids were measured by Cobas® Modular analyzer.
Galnt2-deficient animal models
Animal studies were approved by Institutional Animal Care and Use Committees of UPenn (mice), SAGE laboratories (rats), and B.M.S. (cyno). Galnt2−/− rats were created at SAGE labs (Geurts et al., 2009) using zinc finger nuclease (ZFN) targeting. Galnt2 KO mice were generated from intercrossing Galnt2+/− mice (Merck) on a C57Bl/6 background. LSKO mice were generated by either crossing Galnt2fl/fl mice with Cre recombinase transgenic mice (expressed from the albumin promoter) or by administration of AAV8-TBG-Cre. For cyno, the best GALNT2 siRNA from a screen of 20 sequences and luciferase siRNA were chemically modified for stability and formulated into lipid nanoparticles for liver-specific delivery as a single dose. Plasma lipids across models were measured by biochemical autoanalyzer assays. PLTP activity assays from mouse and human plasma samples were performed using a 3H-phosphatidylcholine (PC) substrate as previously described (Jauhiainen et al., 1993; Speijer et al., 1991). PLTP activity from rat plasma samples was measured using the Roar assay (Roar Biochemical, Inc).
Glycoproteomics
For liver O-glycoproteomes, 100 mg rat or mouse liver (N=3 per group) was pooled together by group and further homogenized. For plasma O-glycoproteomes from rats and mice, 200 µl plasma was collected (N=3 per group), neuraminidase treated, and solubilized. Glycoprotein-enriched samples were reduced, alkylated and proteins digested using trypsin (Roche). Peptides were purified using SepPak C18 columns, treated with neuraminidase, and labelled with light (L) or medium (M) isotopomeric dimethyl labels based on group. Labeled Galb1–3GalNAc glycopeptides were separated from non-glycosylated peptides using a long PNA-agarose lectin weak-affinity chromatography (Steentoft et al., 2011). Human plasma O-glycoproteomes were generated as described but the labeling step was omitted. Liquid chromatography-mass spectrometry was performed as described previously (Schjoldager et al., 2015).
Statistics analysis
Data represent mean ± standard deviation, with error bars showing standard deviation. Statistical analysis was performed by two-tailed unpaired Student’s t-test or one-way ANOVA with Tukey’s post-test as appropriate. Statistical significance was defined as P < 0.05 for all analyses.
RNA sequencing of human livers and allelic imbalance
Allele-specific expression (ASE) analysis used data from two cohorts: 40 human liver samples processed at the University of Pennsylvania (UPenn data) and 32 human liver samples processed as a part of the Genotype Tissue Expression (GTEx) consortium (release v4; GTEx data) (GTEx, 2015). UPenn data was genotyped on the Illumina human 610 quad beadchips (GPL8887) at the Northwestern University Center for Genetic Medicine Genomics Core Facility (Innocenti et al., 2011). GTEx data was genotyped on the Illumina Human Omni 2.5 and 5.0 beadchips at the Broad Institute. RNA sequencing was performed using an Illumina HiSeq2500. Sequencing reads were aligned to the human genome (GRCh37/hg19) and heterozygous coding SNPs were retained for analysis if the genotype probabilities estimated by IMPUTE2 were > 0.9. Allele-specific read depth at each coding SNP was extracted, assigning reads to either the ‘A’ or ‘G’ allele of rs4846914 using the phased haplotypes from each individual. Binomial test was used for each individual to test for deviation from null hypothesis of no allelic imbalance.
Data access
Sequencing data for GALNT2 variants has been deposited in GenBank (SRX1888908); additional MIExSeq sequencing data for GALNT2 D314A variant can be accessed by contacting Daniel Rader, rader@mail.med.upenn.edu.
Supplementary Material
Highlights.
Genetic loss-of-function of GALNT2 lowers HDL-C in man, rodents, and nonhuman primates
ANGPTL3 and ApoC-III are non-redundant targets of GalNAc-T2 in humans
PLTP O-glycosylation by GalNAc-T2 potentiates its activity and raises HDL-C
GALNT2 GWAS SNP alleles associated with lower HDL-C reduce hepatic GALNT2 expression
Acknowledgments
We acknowledge Thomas F. Vogt, John S. Mudgett and Merck Research Laboratories for providing Galnt2 KO mice. We thank the UPenn Functional Genomics Core at DRC (P30-DK19525) for RNA sequencing and the University of Pennsylvania Preclinical Vector Core supported by the NHLBI Gene Therapy Resource Program (GTRP), for providing gene vectors used in this study. This work was supported by The Danish Research Councils (Sapere Aude Research Talent Grant to K.T.S.), Kirsten og Freddy Johansen Fonden, A.P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til Almene Formaal, The Novo Nordisk Foundation, a program of excellence from the University of Copenhagen (CDO2016), the Danish National Research Foundation (DNRF107), NIH grants R01 HL111398 and R01 HL089309, and grant 10CVD03 from the Foundation Leducq to D.J.R, the German Research Foundation (DFG) with funding AB393/2-2 to R.A.J and the Mizutani Foundation to S.Y.V.. We also thank Pr. El Fahime El Mostafa for hosting K.T.S in his laboratory. T.G.K., G.F., L.V., P.P., and S.H. are employees and/or stockholders of Bristol-Myers Squibb (Pennington, NJ, USA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information includes five figures, one table, and Supplemental Experimental Procedures.
Author Contributions
Conceptualization: S.A.K., K.T.S., H.C., D.J.R.; Investigation: S.A.K., K.T.S., C.C., A.R., A.C.E., H.M.R., B.A., O.R., G.M.P., C.V., W.Z., A.V., J.S.M., Y.P., G.F., V.L., S.C., E.N., P.P., S.H., C.D.B.; Resources: G.L.G.C., T.G.K., H.H.W., L.H., E.P.B., S.Y.V., D.S., S.K., C.D.B., R.A.J., E.L.; Writing – Original Draft: S.A.K., K.T.S., H.C., D.J.R.; Writing – Reviewing and Editing: S.A.K., K.T.S., C.C., A.C.E., H.M.R., J.S.M., S.H., T.G.K., H.H.W., R.A.J., H.C., D.J.R.; Visualization: S.A.K., K.T.S., H.C., D.J.R.; Supervision: H.C., D.J.R.; Funding Acquisition: K.T.S., S.Y.V., R.A.J., H.C., D.J.R.
References
- Abou Jamra R, Wohlfart S, Zweier M, Uebe S, Priebe L, Ekici A, Giesebrecht S, Abboud A, Al Khateeb MA, Fakher M, et al. Homozygosity mapping in 64 Syrian consanguineous families with non-specific intellectual disability reveals 11 novel loci and high heterogeneity. European journal of human genetics : EJHG. 2011;19:1161–1166. doi: 10.1038/ejhg.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers JJ, Vuletic S, Cheung MC. Role of plasma phospholipid transfer protein in lipid and lipoprotein metabolism. Biochim Biophys Acta. 2012;1821:345–357. doi: 10.1016/j.bbalip.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee C, Sparks DL. Hepatic lipase, high density lipoproteins, and hypertriglyceridemia. Am J Pathol. 2011;178:1429–1433. doi: 10.1016/j.ajpath.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkersen L, van’t Hooft F, Chernogubova E, Agardh HE, Hansson GK, Hedin U, Liska J, Syvanen AC, Paulsson-Berne G, Franco-Cereceda A, et al. Association of genetic risk variants with expression of proximal genes identifies novel susceptibility genes for cardiovascular disease. Circulation. Cardiovascular genetics. 2010;3:365–373. doi: 10.1161/CIRCGENETICS.110.948935. [DOI] [PubMed] [Google Scholar]
- Fritz TA, Raman J, Tabak LA. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-2. The Journal of biological chemistry. 2006;281:8613–8619. doi: 10.1074/jbc.M513590200. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedeke L, Rotllan N, Canfran-Duque A, Aranda JF, Ramirez CM, Araldi E, Lin CS, Anderson NN, Wagschal A, de Cabo R, et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nature medicine. 2015;21:1280–1289. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goth CK, Halim A, Khetarpal SA, Rader DJ, Clausen H, Schjoldager KT. A systematic study of modulation of ADAM-mediated ectodomain shedding by site-specific O-glycosylation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:14623–14628. doi: 10.1073/pnas.1511175112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- GTEx. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L, Lind-Thomsen A, Joshi HJ, Pedersen NB, Have CT, Kong Y, Wang S, Sparso T, Grarup N, Vester-Christensen MB, et al. A glycogene mutation map for discovery of diseases of glycosylation. Glycobiology. 2015;25:211–224. doi: 10.1093/glycob/cwu104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleboom AG, Karlsson H, Lin RS, Beres TM, Sierts JA, Herman DS, Stroes ES, Aerts JM, Kastelein JJ, Motazacker MM, et al. Heterozygosity for a loss-of-function mutation in GALNT2 improves plasma triglyceride clearance in man. Cell Metab. 2011;14:811–818. doi: 10.1016/j.cmet.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huuskonen J, Jauhiainen M, Ehnholm C, Olkkonen VM. Biosynthesis and secretion of human plasma phospholipid transfer protein. Journal of lipid research. 1998;39:2021–2030. [PubMed] [Google Scholar]
- Innocenti F, Cooper GM, Stanaway IB, Gamazon ER, Smith JD, Mirkov S, Ramirez J, Liu W, Lin YS, Moloney C, et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauhiainen M, Metso J, Pahlman R, Blomqvist S, van Tol A, Ehnholm C. Human plasma phospholipid transfer protein causes high density lipoprotein conversion. The Journal of biological chemistry. 1993;268:4032–4036. [PubMed] [Google Scholar]
- Jiang XC, Bruce C, Mar J, Lin M, Ji Y, Francone OL, Tall AR. Targeted mutation of plasma phospholipid transfer protein gene markedly reduces high-density lipoprotein levels. J Clin Invest. 1999;103:907–914. doi: 10.1172/JCI5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang XC, Jin W, Hussain MM. The impact of phospholipid transfer protein (PLTP) on lipoprotein metabolism. Nutr Metab (Lond) 2012;9:75. doi: 10.1186/1743-7075-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang XC, Qin S, Qiao C, Kawano K, Lin M, Skold A, Xiao X, Tall AR. Apolipoprotein B secretion and atherosclerosis are decreased in mice with phospholipid-transfer protein deficiency. Nature medicine. 2001;7:847–852. doi: 10.1038/89977. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Jeanneau C, Tarp MA, Benet-Pages A, Lorenz-Depiereux B, Bennett EP, Mandel U, Strom TM, Clausen H. Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. The Journal of biological chemistry. 2006;281:18370–18377. doi: 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Current biology : CB. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Leuenberger B, Hahn D, Pischitzis A, Hansen MK, Sterchi EE. Human meprin beta: O-linked glycans in the intervening region of the type I membrane protein protect the C-terminal region from proteolytic cleavage and diminish its secretion. Biochem J. 2003;369:659–665. doi: 10.1042/BJ20021398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Uzawa H, Kamei R. Unusual familial lipoprotein C-III associated with apolipoprotein C-III-O preponderance. Biochim Biophys Acta. 1981;665:578–585. doi: 10.1016/0005-2760(81)90273-3. [DOI] [PubMed] [Google Scholar]
- Milac AL, Buchete NV, Fritz TA, Hummer G, Tabak LA. Substrate-induced conformational changes and dynamics of UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferase-2. J Mol Biol. 2007;373:439–451. doi: 10.1016/j.jmb.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghani A, Zannis VI. Mutagenesis of the glycosylation site of human ApoCIII. O-linked glycosylation is not required for ApoCIII secretion and lipid binding. The Journal of biological chemistry. 1988;263:17925–17932. [PubMed] [Google Scholar]
- Roman TS, Marvelle AF, Fogarty MP, Vadlamudi S, Gonzalez AJ, Buchkovich ML, Huyghe JR, Fuchsberger C, Jackson AU, Wu Y, et al. Multiple Hepatic Regulatory Variants at the GALNT2 GWAS Locus Associated with High-Density Lipoprotein Cholesterol. American journal of human genetics. 2015;97:801–815. doi: 10.1016/j.ajhg.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Clausen H. Site-specific protein O-glycosylation modulates proprotein processing – deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim Biophys Acta. 2012;1820:2079–2094. doi: 10.1016/j.bbagen.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Schjoldager KT, Joshi HJ, Kong Y, Goth CK, King SL, Wandall HH, Bennett EP, Vakhrushev SY, Clausen H. Deconstruction of O-glycosylation--GalNAc-T isoforms direct distinct subsets of the O-glycoproteome. EMBO reports. 2015;16:1713–1722. doi: 10.15252/embr.201540796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Vakhrushev SY, Kong Y, Steentoft C, Nudelman AS, Pedersen NB, Wandall HH, Mandel U, Bennett EP, Levery SB, et al. Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9893–9898. doi: 10.1073/pnas.1203563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Vester-Christensen MB, Bennett EP, Levery SB, Schwientek T, Yin W, Blixt O, Clausen H. O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. The Journal of biological chemistry. 2010;285:36293–36303. doi: 10.1074/jbc.M110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speijer H, Groener JE, van Ramshorst E, van Tol A. Different locations of cholesteryl ester transfer protein and phospholipid transfer protein activities in plasma. Atherosclerosis. 1991;90:159–168. doi: 10.1016/0021-9150(91)90110-o. [DOI] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Vester-Christensen MB, Schjoldager KT, Kong Y, Bennett EP, Mandel U, Wandall H, Levery SB, Clausen H. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8:977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Bennett EP, Takio K, Sorensen T, Bonding N, Clausen H. Purification and cDNA cloning of a human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. The Journal of biological chemistry. 1995;270:24156–24165. doi: 10.1074/jbc.270.41.24156. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.