Abstract
Over the past century, X-ray crystallography has been defined by a pursuit for perfection and high resolution. In next Holy Grail of crystallography is to embrace imperfection towards a dynamic picture of enzymes.
Graphical Abstract
Over its hundred-year history, X-ray crystallography has made enormous impacts on diverse fields by providing insight into the structures of countless molecules. Crystallographic structures represent some of the landmark achievements of biology in the past century. From the very first enzyme structure of lysozyme1 in 1965 to the recent high-resolution structure of the oxygen-evolving complex of photosystem II,2 crystallography has given detailed views into the intricate processes by which Nature catalyzes the reactions necessary for life. Indeed, one of the triumphs of crystallography in the last century was the rise of structural enzymology, in which changes in activity are correlated with changes in structure.
Thus far, X-ray crystallography has embodied a quest for perfection. Proteins arranged in a crystal lattice diffract X-rays into Bragg peaks, the sharp reflections from virtual planes of atoms. Subtle imperfections in crystals cause X-rays to scatter away from the peaks, leading to the disappearance of high-resolution peaks and an apparent increase in background scattering. Therefore, the more perfectly ordered the crystal is, the higher resolution diffraction we are able to obtain. In the quest for the best possible resolution, crystallographers screen vast numbers of crystallization conditions and crystals to find the perfect one. Such an enormous effort is needed because in many ways, protein crystals are temperamental. They may grow differently from one batch to another, at the whim of unknown variables. Furthermore, the quality of diffraction can have no correlation with the visual appearance of crystals.
The challenge of obtaining perfectly ordered crystals, however, is not surprising when we consider the fact that, like all molecules, proteins are not static. Rather, they are restless, breathing entities, and this property has consequences for not only how they crystallize but also, and more importantly, how they function. In fact, disorder in a crystal can result from protein motions that are biochemically important, and gaining a dynamic picture of enzymes is a lofty goal that we must strive for. Some of the greatest questions in biochemistry require such a picture: What is the role of dynamics in catalysis? How might inhibitor drugs disrupt such motions? How do multifunctional enzymes coordinate multiple active sites? What are the dynamics involved in protein allostery? The Holy Grail of crystallography in the 21st century is therefore to fully embrace imperfection.
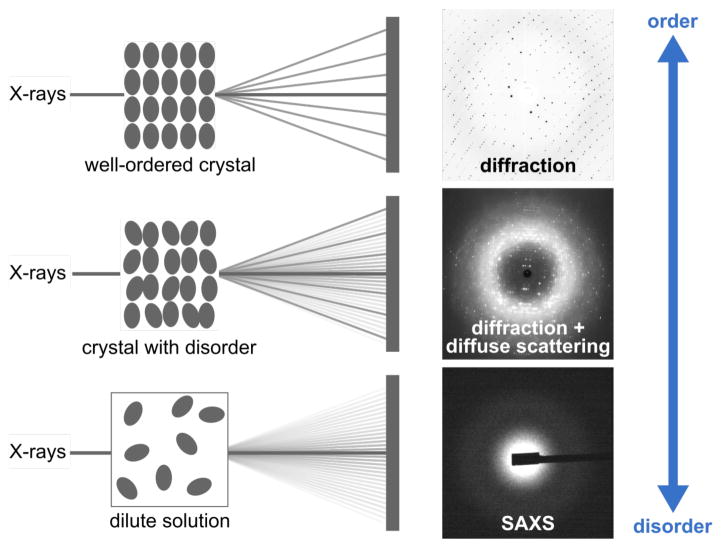
What can we learn when we shift away from our preoccupation with the Bragg peaks in diffraction data? History has shown that we can benefit from thinking outside of the lattice. At a time when protein crystallography was on the rise, André Guinier devoted his 1936 PhD thesis to the study of the peculiar scattering near the beamstop.3 Building the first low-background diffraction instrument for this purpose, he observed that the scattering that arose from silica gel disappeared for vitreous silica, despite having the same chemical composition. What he discovered was that the scattering at low angles provided a wealth of information about disordered materials, information which was not available from Bragg diffraction.3 Today, he is widely regarded as the intellectual father of X-ray scattering and is best known for his contributions to small-angle X-ray scattering (SAXS) (Figure 1, bottom).
Figure 1.
Diffraction, diffuse scattering, and SAXS result from the elastic scattering of X-rays from electrons. The interference of the scattered waves produces characteristic patterns that reflect the degree of disorder present in the sample. The diffuse scattering image was adapted with permission from ref. 9, Copyright (1997) National Academy of Sciences, U.S.A.
The growth of biological SAXS as a field and indeed, the recent rise of cryo-electron microscopy reflect the fact there is an increasing demand for a dynamic picture of proteins and a perception that crystallography as a technique has already reached its full potential. What is not very widely known is that in fact, dynamic information is not entirely lost in crystal diffraction, and today, we are witnessing the beginnings of a new field known as crystallography beyond Bragg diffraction.
Conventional crystallography is based on measuring the intensity of Bragg peaks, which allows for the average atomic positions to be determined (Figure 1, top). Although Bragg diffraction can suggest mobility in a protein structure, it does not contain information on how motions are correlated. For instance, by Bragg diffraction alone, two protein regions fluctuating in phase cannot be distinguished from the scenario in which identical atomic displacements are made in opposing directions, yet the difference between these two cases can greatly impact our interpretation of enzyme mechanism. Instead, information about the correlation of motions is hidden in the noisy background in between Bragg peaks, the so-called diffuse scattering – the part of diffraction data that we routinely throw away in conventional crystallography (Figure 1, middle). Guinier was perhaps the first to describe this phenomenon with the succinct formula,4
where Idiffuse is the diffuse scattering intensity, F is the Fourier transform of the electron density in the crystal, and brackets denote the ensemble average. From Guinier’s formula, it is clear that Idiffuse is non-zero when the first term, which describes the electron density at any given time or location in the crystal, differs from the average value in the second term. Thus, we reach a surprising conclusion: the types of messy data that we ignore today may help answer long-standing questions about the breathing motions of proteins that are retained within a crystal lattice. By combining Bragg diffraction and diffuse scattering, we can imagine animating crystal structures with biochemically important motions.
In practice, however, interpretation of the diffuse scattering pattern has been exceedingly difficult. Unlike solution scattering, diffuse scattering from crystals is highly complex and spread throughout reciprocal space. The anisotropic component of the diffuse signal appears as small fluctuations on top of a smooth background, coexisting with the intense Bragg peaks. Although diffuse scattering has found utility in materials research,5 macromolecular crystals present unique challenges such as structural complexity, high solvent content, and susceptibility to radiation damage. Not surprisingly, over the past three decades, less than 30 attempts to interpret diffuse scattering from macromolecular crystals have been made.6
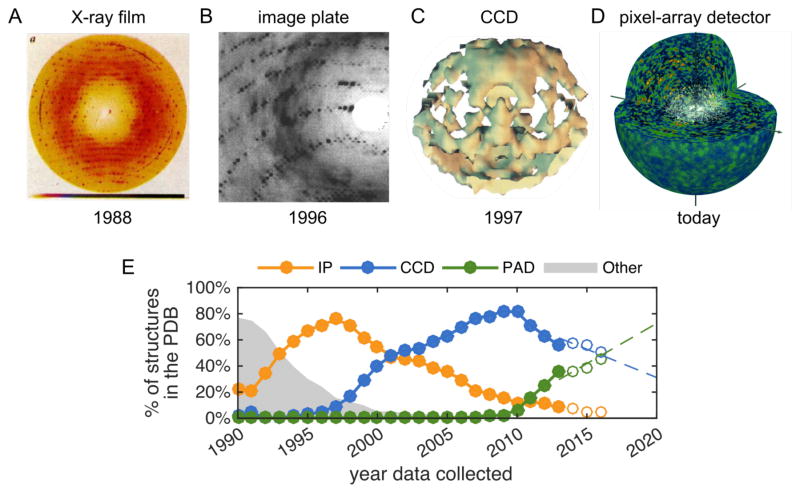
Pioneering studies of diffuse scattering from protein crystals were reported in the 1980s and 1990s (Figure 2A).6 Notably, Donald Caspar and colleagues observed two types of diffuse scattering from insulin crystals by over-exposing X-ray film: haloes around the Bragg peaks, arising from lattice dynamics, and a faint, textured background, which was attributed to correlated motions within each molecule. Later, image plate detectors made it possible to measure diffuse scattering with reasonable signal to noise without overexposure (Figure 2B).8 A major breakthrough came in 1997 with the measurement of the first complete 3D maps of diffuse scattering by Michael Wall during his PhD with Sol Gruner (Figure 2C), which was made possible by their expertise in detector development.9 For this work, a phosphor-coupled CCD detector was customized to prevent the intense Bragg peaks from bleeding into neighboring pixels, a process known as “blooming”.10 The CCD offered a significant advantage over image plates at the time by reducing the data collection time and quickly replaced image plates as the detector of choice for macromolecular crystallography (Figure 2E). Unfortunately, blooming remained a problem for commercially available CCDs.
Figure 2.
The history of diffuse scattering as a field has tracked developments in detector technology. (A-D) Diffuse scattering from insulin,7 lysozyme,8 Staphylococcal nuclease,9 and photosystem II.13 (E) Fraction of structures in the PDB collected using an image plate (IP), charge-coupled device (CCD), pixel-array detector (PAD), or older recording media (other).16 Statistics from recent years are represented as open circles to emphasize that these are subject to change as more structures are deposited. Dashed lines are a linear extrapolation. Panel A adapted by permission from Macmillan Publishers Ltd: Nature (ref. 7), copyright (1988); panel B reproduced from ref. 8 with permission of the International Union of Crystallography, http://journals.iucr.org/; panel C adapted with permission from ref. 9, Copyright (1997) National Academy of Sciences, U.S.A.; panel D adapted by permission from Macmillan Publishers Ltd: Nature (ref. 13), copyright (2016).
Measuring both Bragg and diffuse signals accurately demands a near-perfect X-ray detector with a uniform response, low noise, a point-spread function that decays rapidly to zero, and high dynamic range. Pixel array detectors (PADs), developed recently, solve many of the issues that affect earlier X-ray detectors. In a PAD, X-rays interact directly with each pixel, and each photon’s interaction is counted. From the perspective of diffuse scattering and conventional crystallography, they are ideal detectors. After two decades of very little activity in the diffuse scattering field, it is heating up again. In three new studies, PADs were used to measure the diffuse scattering from crystals of cyclophilin A,11 trypsin,11 the 70S ribosome,12 and photosystem II.13 Most notably, a PAD was used to perform the first diffuse scattering measurements at an X-ray free electron laser (XFEL).13 Intriguingly, the accumulated diffraction collected from thousands of tiny photosystem-II crystals displayed strong diffuse scattering that extended to higher resolution than the Bragg peaks (Figure 2D). Under the assumption of rigid-body motion of the asymmetric unit, the diffuse scattering was then analyzed using techniques from coherent diffractive imaging with the goal of improving the quality of the crystallographic structure.
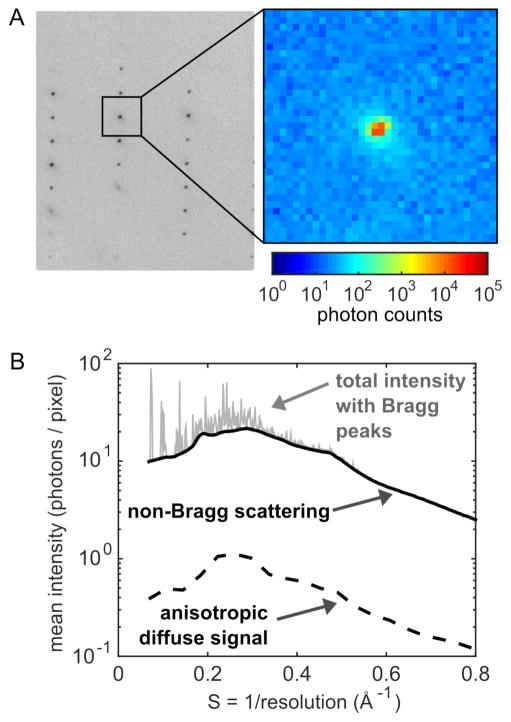
As PADs have begun to replace CCDs, we are now entering an era where perfect X-ray detectors are the norm. According to the most recent statistics from the Protein Data Bank (PDB), PADs account for roughly half of the structural data collected in 2016 (Figure 2E). In this new era, diffuse scattering measurements could become routine. Even with short exposures, we have found that the diffuse scattering can be extracted from PAD images by analyzing the photon count distributions statistically (Figure 3). Thus, as long as steps are taken to reduce or measure the background during a standard crystallography experiment, the diffuse data comes along “for free”. Making use of it, though, requires careful attention to the theory of protein motion in the crystal, as well as software for data analysis and interpretation. Significant progress has been made recently,10–12 and we expect further developments in the near term.
Figure 3.
PAD image of diffraction from a lysozyme crystal (P43212) at ambient temperature. (A) The PAD’s high dynamic range and low noise afford simultaneous recording of weak background scattering (~10 photons/pixel) and intense peaks (~100,000 photons/pixel). (B) Statistical analysis of photon counts within annular rings of constant scattering vector length, S. After correcting for experimental effects, the standard deviation was taken as an estimate of the anisotropic diffuse signal (~0.3 photons/pixel).
What new biological insights will emerge from a renaissance in diffuse scattering? First, diffuse scattering will allow us to validate models of collective motion that are already applied during structure refinement, particularly when those collective motions inform our understanding of biological function. For example, rigid-body motions are often inferred from so-called translation libration screw-axis (TLS) refinement applied to Bragg data,14 and allosteric networks of side-chain interactions have been inferred by fitting multi-conformer ensembles to the electron density.15 Such models imply distinctive diffuse patterns that should be observed in the diffraction images. Second, combining diffuse scattering with molecular dynamics (MD) simulations of crystals will allow us to benchmark force fields and validate simulation results. Diffuse scattering has already proven to be a rich dataset that is straightforward to calculate from MD simulations but very difficult to reproduce accurately,15 suggesting that current MD methods have room for improvement. Finally, it should be possible to refine a general dynamical model to Bragg data and diffuse scattering simultaneously, which would truly revolutionize crystallography. The significant challenge has been finding a model, which captures the relevant degrees of freedom that can be refined to both data sets. Our ability to measure diffuse scattering accurately will go hand-in-hand with the development of new and improved dynamical models. Ultimately, diffuse scattering should allow us to generate novel insights into the dynamics of proteins that are well supported by experimental evidence.
X-ray crystallography has been described as one of the pillars of modern biology. Indeed, it is difficult to overstate how much impact crystallography has had. However, there is a wealth of information obtainable through crystallography that has yet to be fully explored. To achieve some of the most lofty goals of structural enzymology, it is clear that we must venture beyond Bragg diffraction.
Acknowledgments
This work was supported by National Health Institutes (NIH) under award number GM100008 to NA and F32GM117757 to SPM. CHESS is supported by the NSF & NIH/NIGMS via NSF award DMR-1332208, and the MacCHESS resource is supported by NIGMS award GM-103485.
References
- 1.Blake CCF, Koenig DF, Mair GA, North ACT, Phillips DC, Sarma VR. Structure of Hen Egg-White Lysozyme: A Three-Dimensional Fourier Synthesis at 2 Å Resolution. Nature. 1965;206(4986):757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- 2.Suga M, Akita F, Hirata K, Ueno G, Murakami H, Nakajima Y, Shimizu T, Yamashita K, Yamamoto M, Ago H, Shen J-R. Native Structure of Photosystem II at 1.95 Å Resolution Viewed by Femtosecond X-Ray Pulses. Nature. 2015;517(7532):99–103. doi: 10.1038/nature13991. [DOI] [PubMed] [Google Scholar]
- 3.Guinier A. 30 Years of Small-Angle X-Ray Scattering. Phys Today. 1969;22(11):25–30. [Google Scholar]
- 4.Guinier A. X-Ray Diffraction in Crystals, Imperfect Crystals, and Amorphous Bodies. W. H. Freeman and Co; San Francisco: 1963. [Google Scholar]
- 5.Keen DA, Goodwin AL. The Crystallography of Correlated Disorder. Nature. 2015;521(7552):303–309. doi: 10.1038/nature14453. [DOI] [PubMed] [Google Scholar]
- 6.Wall ME, Adams PD, Fraser JS, Sauter NK. Diffuse X-Ray Scattering to Model Protein Motions. Structure. 2014;22(2):182–184. doi: 10.1016/j.str.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caspar DLD, Clarage J, Salunke DM, Clarage M. Liquid-like Movements in Crystalline Insulin. Nature. 1988;332(6165):659–662. doi: 10.1038/332659a0. [DOI] [PubMed] [Google Scholar]
- 8.Pérez J, Faure P, Benoit JP. Molecular Rigid-Body Displacements in a Tetragonal Lysozyme Crystal Confirmed by X-Ray Diffuse Scattering. Acta Crystallogr, Sect D: Biol Crystallogr. 1996;52(4):722–729. doi: 10.1107/S0907444996002594. [DOI] [PubMed] [Google Scholar]
- 9.Wall ME, Ealick SE, Gruner SM. Three-Dimensional Diffuse X-Ray Scattering from Crystals of Staphylococcal Nuclease. Proc Natl Acad Sci U S A. 1997;94(12):6180–6184. doi: 10.1073/pnas.94.12.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wall M. Methods and Software for Diffuse X-Ray Scattering from Protein Crystals. In: Foote RS, Lee JW, editors. Micro and Nano Technologies in Bioanalysis: Methods and Protocols. Vol. 544. Humana Press; Totowa, NJ: 2009. pp. 269–279. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 11.Benschoten AHV, Liu L, Gonzalez A, Brewster AS, Sauter NK, Fraser JS, Wall ME. Measuring and Modeling Diffuse Scattering in Protein X-Ray Crystallography. Proc Natl Acad Sci U S A. 2016;113(15):4069–4074. doi: 10.1073/pnas.1524048113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polikanov YS, Moore PB. Acoustic Vibrations Contribute to the Diffuse Scatter Produced by Ribosome Crystals. Acta Crystallogr, Sect D: Biol Crystallogr. 2015;71(10):2021–2031. doi: 10.1107/S1399004715013838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayyer K, et al. Macromolecular Diffractive Imaging Using Imperfect Crystals. Nature. 2016;530(7589):202–206. doi: 10.1038/nature16949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore PB. On the Relationship between Diffraction Patterns and Motions in Macromolecular Crystals. Structure. 2009;17(10):1307–1315. doi: 10.1016/j.str.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Wilson MA. Visualizing Networks of Mobility in Proteins. Nat Methods. 2013;10(9):835–837. doi: 10.1038/nmeth.2606. [DOI] [PubMed] [Google Scholar]
- 16.Wall ME, Benschoten AHV, Sauter NK, Adams PD, Fraser JS, Terwilliger TC. Conformational Dynamics of a Crystalline Protein from Microsecond-Scale Molecular Dynamics Simulations and Diffuse X-Ray Scattering. Proc Natl Acad Sci U S A. 2014;111(50):17887–17892. doi: 10.1073/pnas.1416744111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.RCSB Protein Data Bank. [accessed Nov 2, 2016]; http://www.rcsb.org/