Abstract
Traditionally, somatic mutations are detected by examining DNA sequence. The maturity of sequencing technology has allowed researchers to screen for somatic mutations in the whole genome. Increasingly, researchers have become interested in identifying somatic mutations through RNAseq data. With this motivation, we evaluated the practicability of detecting somatic mutations from RNAseq data. Current somatic mutation calling tools were designed for DNA sequencing data. To increase performance on RNAseq data, we developed a somatic mutation caller GLMVC based on bias reduced generalized linear model for both DNA and RNA sequencing data. Through comparison with MuTect and Varscan we showed that GLMVC performed better for somatic mutation detection using exome sequencing or RNAseq data. GLMVC is freely available for download at the following website: https://github.com/shengqh/GLMVC/wiki.
Keywords: somatic mutation, RNAseq, exome, generalized linear model
INTRODUCTION
Traditionally, somatic mutations are detected using Sanger sequencing or real-time polymerase chain reaction (RT-PCR) by comparing paired tumor and normal samples. One obvious limitation of such methods is that the somatic mutation detection must be limited to a certain genomic region of interest. Now with high-throughput sequencing (HTS), whole exomes or genomes can be screened for somatic mutations at a reasonable cost (Figure S1). There are two major next-generation sequencing (NGS) paradigms: RNA and DNA sequencing. Both RNA and DNA sequencing can be used to answer different sets of scientific questions important for biomedical research. RNAseq refers to the sequencing of the transcriptome. The two most common forms of DNAseq are exome and whole genome sequencing.
Due to the popularity of RNAseq technology for gene expression profiling over microarray technology [1–4], huge amounts of RNAseq data have been accumulated over the past few years. And the majority of these RNAseq data has been only studied for gene expression. More and more researchers have begun to ask the question of whether or not somatic mutations can be detected accurately through RNAseq data. Same as DNAseq, RNAseq is at single nucleotide resolution. Thus, single nucleotide variants (SNVs) can be detected. To date, many tools, such as Varscan [5] and MuTect [6], have been developed for the identification of somatic mutations through DNAseq data. Yet, less effort has been relatively spent on the detection of SNVs using RNAseq data. In contrast to using DNAseq data, identifying mutations using RNAseq data poses stronger challenges for the primary reason of RNAseq data having a much higher false positive rate for SNVs than DNAseq data [7, 8]. The high false positive rate results from several issues, of which include cycle bias [9], strand bias [10] alignment complexity in the transcriptome, RNA editing, and random errors introduced during reverse transcription and PCR. Cycle bias happens in a heterozygous position when one of two alleles in the supporting reads lie heavily at the beginning or end of the reads [11, 12]. Strand bias occurs when alternative allele detection heavily originates from one of the two strands (forward or reverse). Such bias indicates false positive mutation detection in RNAseq data [12]. Most advanced somatic mutation callers[5] [6] [13] have built-in strand bias quality control. Also, the alignment of RNAseq data proves more complicated than DNAseq data [14]. In mRNA, introns are removed by splicing, thus a read is likely to span the splicing junction, causing a higher probability for error. Similarly, processes such as RNA editing and polyadenylation introduce additional mismatches not found in DNAseq alignment. For conducting expression studies, minor mismatches in alignment do not affect expression value because the computation of expression value depends only on the count of reads mapped to a gene’s genomic span and therefore do not require the examination of theRNAseq at single nucleotide resolution for gene expression. However, SNVs are detected by counting the number of mismatches in alignment against a reference. Thus, excessive mismatches due to errors described above will result in a high false positive rate for SNV detection. False positives due to cycle bias may be filtered out through a quality control check that removes all reported mutations at the beginning or end of the reads that are disproportionate. This has been effectively demonstrated by Kleinman et al.[15]. False positives due to splicing locations are more difficult to distinguish from true variants. Thus, SNPs and somatic mutations identified near splicing sites should be removed or flagged for further review. Most RNAseq data specific variant detection tools, such as SNVQ [16] and SNPiR[14], focus on SNV rather than somatic mutation. And none of these tools consider cycle bias.
We want to identify somatic mutations through RNAseq data for several reasons. Due to budget limitations, sample quantities or study goals, researchers often choose RNAseq over DNAseq. However, the potential for RNAseq data should be maximized by performing additional analysis for which RNAseq was not originally designed, such as somatic mutation detection[7]. For somatic mutation, RNAseq has been used primarily in two ways: discovery and validation. Discovery concerns the identification of somatic mutations using RNAseq data alone. For example, Xu et al. identified somatic mutations in prostate cancer using RNAseq data by applying a standard DNA processing pipeline [17]. Validation refers to the use of RNAseq data to validate somatic mutations found within DNAseq data. For example, in the cancer genome atlas (TCGA), RNAseq data has been used to validate somatic mutations found from DNA exome sequencing [18]. We have developed GLMVC which can identify somatic mutations from both DNAseq and RNAseq data. GLMVC uses a bias reduced generalized linear model (brGLM[19]) to identify somatic mutation candidates and to filter out false positives based on several unique characteristics of RNAseq data. GLMVC can take in either BAM files or pileup files as input, and it automatically generates a comprehensive somatic mutation report based on user-defined parameters. Here, we demonstrated GLMVC’s effectiveness using TCGA breast cancer data and compared the results with other popular somatic mutation callers. Through our analyses, we were able to address two important questions: 1) Can somatic mutations be reliably detected through RNAseq data as compared to DNAseq data? 2) What germline reference source (adjacent normal or blood) is better for somatic mutation inference?
METHODS
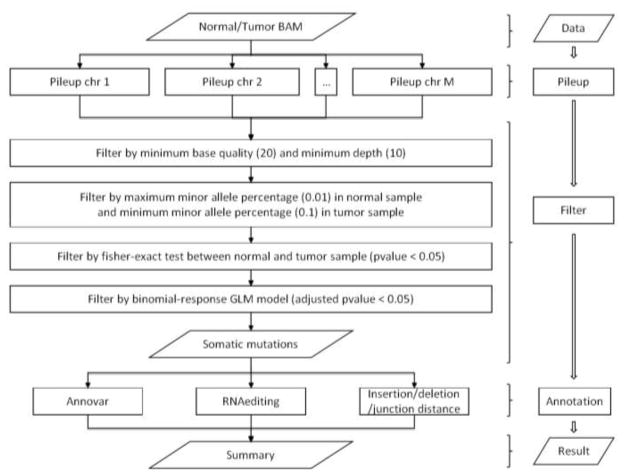
The overall workflow of GLMVC can be seen in Figure 1. GLMVC works in three steps: mutation calling, filtering, and annotaton. Due to the high false positive rate associated with identifying variants using RNAseq data, GLMVC is designed to focus on specificity rather than sensitivity. We employed several filters to eliminate potential false positive SNVs. To identify a somatic mutation in RNAseq data at a particular locus, alignments must be obtained for paired tumor and normal samples. To increase the confidence of somatic mutation calling, a minimum base Phred quality score of 20 [20] is used to filter out low quality reads and bases. A recent report suggests that only >10X coverage is required to ensure 89% accuracy and 92% sensitivity for single nucleotide variations (SNVs) [21]. Thus, a minimum depth of 10 from both tumor data and normal data is required for GLMVC to consider the candidate base. Also, GLMVC requires the percent of observed mutated alleles in the tumor sample to be above a certain threshold (default: 10%), requires a minimum number of observed mutated alleles in the tumor sample (default: 5), and requires the mutated allele frequency in a normal sample to be lower than a determined threshold (default: 2%). The thresholds can be adjusted according to purity of tumor samples and tumor-contamination within normal samples. An initial screening of somatic mutation candidates is done using Fisher’s exact test to significantly reduce the total number of candidate somatic mutations that need to be considered in the filtering steps, which in turn significantly decreases the overall run time of GLMVC.
Figure 1.
GLMVC workflow
Several unique characteristics of RNAseq data were taken into consideration in GLMVC. GLMVC performs a test using brGLM with the bias-reduction method [19] that considers not only the occurance of mutated alleles, but the base quality scores, the strands of the reads, and most importantly, the cycle position at each read. By using a binomial response brGLM model in GLMVC, the probability of somatic mutation signals was adjusted by score, strand and position bias (linear model: Allele ~ tumor/normal + Score + Strand + Position. The somatic mutation candidates passing the brGLM test were sent to the next step for annotation.
During the annotation step, GLMVC uses ANNOVAR [22] to annotate information, such as amino acid change status, dbSNP ID, Polyphen [23], SIFT [24], and other available annotations. Additionally, GLMVC annoates several unique measurements: 1) distance to nearest splicing junction or indel (RNAseq data only); 2)mutation density; 3) RNA editing status (RNAseq data only). If TopHat[25] is used for alignment, BED files generated by TopHat for junctions, insertions and deletions can be used as inputs for GLMVC. If TopHat alignment results are not avaliable, junction information from the GTF format file can be used in its place. Distances to the nearest junction, insertion or deletion are computed for each somatic mutation detected, which may indicate the confidence of the mutation. The mutation density denotes the distance from one mutation to the nearest mutaton. The number of true non-synonymous mutations per tumor ranges from several to around 100 [26]. Thus, it is unlikely to observe two non-nonymous mutations in close proximity (distance smaller than 10 base pairs). It is also difficult to distinguish between a true RNA mutation and an RNA editing event. Thus, we used DARNED, the RNA editing database [27], to flag all somatic mutations observed at known RNA editing locations.
To demonstrate the effectiveness of GLMVC, we downloaded sequencing data of 10 breast cancer subjects (TCGA-A7-A0D9, TCGA-BH-A0B3, TCGA-BH-A0B8, TCGA-BH-A0BJ, TCGA-BH-A0BM, TCGA-BH-A0C0, TCGA-BH-A0DK, TCGA-BH-A0DP, TCGA-BH-A0E0, TCGA-BH-A0H7). The sequencing data contains exome sequencing data for tumor, blood, adjacent normal and RNAseq data for tumor and adjacent normal. We performed preprocessing steps on the TCGA BAM files using the GATK’s best practice. For exome sequencing data, we performed mark duplicate, realignment, and recalibaration. For RNAseq data, we added a Split and Trim step prior to the realignment step. Using the released somatic mutation list of these 10 patients by TCGA as the gold standard, we compared somatic mutations identified through both DNAseq and RNAseq data using GLMVC, MuTect (v1.1.7) and Varscan (v2.4.1) for sensitivity, specificity and F-score. The F-score is the combined evaluation of both sensitivity and specifity, and computed as .
RESULTS
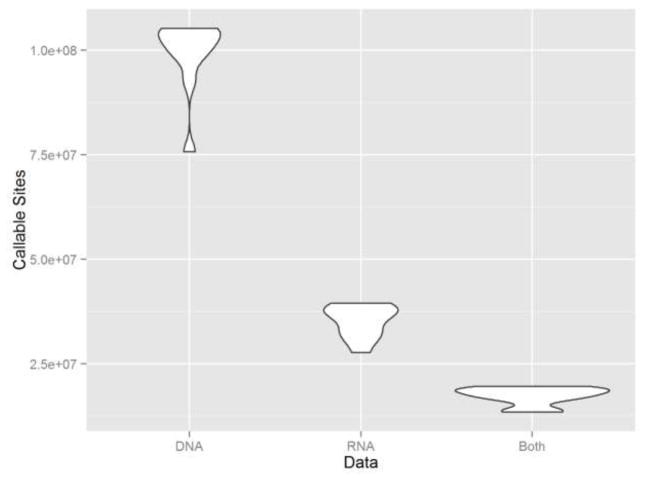
First, we examined the total number of somatic mutation callable sites between DNA and RNA. A somatic mutation callable site is defined as a genomic position with a depth of 10 or greater for both tumor and normal samples at either the DNA or RNA level. We observed more callable sites in exome sequencing data than RNAseq data. On average, exome sequencing data had 103,574,917 callable sites and RNAseq data had 35,309,313 callable sites, with the average overlap between them at 17,610,391 callable sites (Figure 2).
Figure 2.
Callable sites distribution of RNAseq data, exome sequencing data and their overlap. RNAseq data have more callable sites than exome sequencing data.
Of the 10 TCGA breast cancer samples we downloaded, 396 somatic mutations were reported by the TCGA breast cancer study (August, 2015), in which 383 somatic mutations were annotated in exonic region by Annovar. We called somatic mutations using GLMVC, MuTect and Varscan on both DNA (tumor tissue vs. blood and tumor tissue vs. adjacent normal tissue) and RNA (tumor tissue vs. adjacent normal tissue) sequencing data.
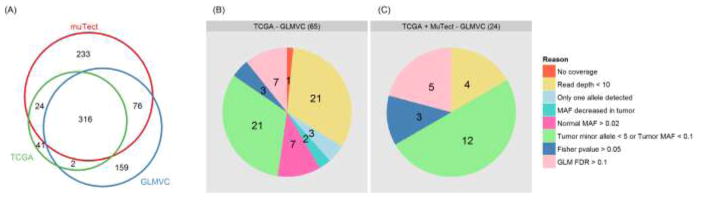
Using the default recommended filters from each caller, MuTect, Varscan, and GLMVC, we found 2533, 3430, 1891 somatic mutations and 649, 539, 553 exonic somatic mutations respectively (Table 1). All three callers found more somatic mutations than reported by TCGA. Using the 383 TCGA reported exonic somatic mutations as the gold standard, MuTect had the highest sensitivity at 88.8%, GLMVC had second best sensitivity at 83%, and Varscan had the lowest sensitivity at 56.7%. However, high sensitivity of MuTect came at the cost of low specificity, with MuTect having 52.4% specificity, Varscan having 40.3% specificity, and GLMVC having the highest specificity at 57.5%. The balance between sensitivity and specificity allowed GLMVC to achieve the highest F-score at 0.679. Out of the 383 TCGA exonic somatic mutations, 260 were non-synonymous. Within the non-synonymous mutations, GLMVC also achieved the highest F-score of 0.766. Figure 3A indicated the overlap of exonic somatic mutations detected by TCGA, MuTect and GLMVC. There were 65 out of 383 TCGA exonic somatic mutations not identified by GLMVC, in which 24 exonic somatic mutations were detected by MuTect. We examined the reasons behind these missing somatic mutations. Of the 65 missing somatic mutations, 22 were due to not being callable sites (read depth < 10); three had no mutated allele after filtering read/base quality in tumor; two had lower mutated allele frequency in tumor than normal; seven passed the upper threshold for mutated allele frequency in normal sample (2%); 21 failed to reach the lower threshold for mutated allele in tumor sample (10% and at least five reads); the remaining 10 failed to reach p-value cutoff for either Fisher’s exact test or brGLM (Figure 3B). Of the 24 missing somatic mutations which were detected by MuTect, four had less than 10 reads in either tumor or normal samples; 12 failed to reach the lower threshold for mutated allele in tumor sample (10% and at least five reads); three failed to pass fisher exact test and another five failed to pass brGLM test (Figure 3C). The results suggest that by default, MuTect inferred somatic mutations from low coverage. For example, in exome sequencing data of sample TCGA-BH-A0B8, at chromosome 11, position 1,268,931, MuTect inferred a somatic mutation with nine reads in blood sample (nine reference, zero mutated) and three reads in tumor sample (one reference, two mutated) were observed. GLMVC can be configured to reach greater sensitivity than MuTect, however, the sacrifice in specificity is too great to ignore. Thus, GLMVC by default focuses more on specificity than sensitivity.
Table 1.
Somatic mutations detected by Mutect, Varscan 2 and GLMVC
| All mutations (396*) | Exonic mutations (383*) | Non-synonymous mutations (260*) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Method | Reference | Detected | TCGA Reported | Detected | TCGA Reported | Sensitivity | Specificity | F-score | Detected | TCGA Reported | Sensitivity | Specificity | F-score |
| MUTECT | DNA NB | 2533 | 349 | 649 | 340 | 0.888 | 0.524 | 0.659 | 403 | 242 | 0.931 | 0.6 | 0.73 |
| VARSCAN 2 | DNA NB | 3430 | 225 | 539 | 217 | 0.567 | 0.403 | 0.471 | 294 | 151 | 0.581 | 0.514 | 0.545 |
| GLMVC | DNA NB | 1891 | 326 | 553 | 318 | 0.83 | 0.575 | 0.679 | 338 | 229 | 0.881 | 0.678 | 0.766 |
|
| |||||||||||||
| MUTECT | DNA NT | 2752 | 308 | 669 | 300 | 0.783 | 0.448 | 0.57 | 388 | 211 | 0.812 | 0.544 | 0.651 |
| VARSCAN 2 | DNA NT | 3851 | 209 | 544 | 202 | 0.527 | 0.371 | 0.436 | 289 | 143 | 0.55 | 0.495 | 0.521 |
| GLMVC | DNA NT | 1840 | 288 | 511 | 282 | 0.736 | 0.552 | 0.631 | 324 | 204 | 0.785 | 0.63 | 0.699 |
|
| |||||||||||||
| MUTECT | RNA NB | 44387 | 88 | 9051 | 87 | 0.227 | 0.01 | 0.018 | 4770 | 71 | 0.273 | 0.015 | 0.028 |
| VARSCAN 2 | RNA NB | 26417 | 88 | 6452 | 87 | 0.227 | 0.013 | 0.025 | 3635 | 73 | 0.281 | 0.02 | 0.037 |
| GLMVC | RNA NB | 8990 | 90 | 1639 | 89 | 0.232 | 0.054 | 0.088 | 755 | 76 | 0.292 | 0.101 | 0.15 |
NB = Normal blood, NT = adjacent normal tissue,
TCGA mutations
Figure 3.
Comparison of exonic somatic mutations from TCGA, MuTect and GLMVC.
Using RNAseq data with a substantially lower number of callable sites, all three callers identified significantly more somatic mutations than in exome sequencing data (Table 1). Using the recommended filters, MuTect, Varscan and GLMVC identified 44387, 26417, 8990 somatic mutations and 79051, 6052, 1639 exonic somatic mutations, respectively. GLMVC achieved the highest sensitivity (23.2%) and the highest specificity (5.4%). MuTect was the lowest specificity (1.8%) and tied sensitivity (22.7%) with Varscan. Varscan was second in sensitivity (1.3%). When combining sensitivity and specificity, GLMVC had the highest F-score for the 383 exonic somatic mutations (0.088) and 260 non-synonymous somatic mutations (0.15). Note that since there were almost three times more callable sites in DNA than RNA sequencing data, a significant portion of the missing sensitivity was caused by not having sequence coverage.
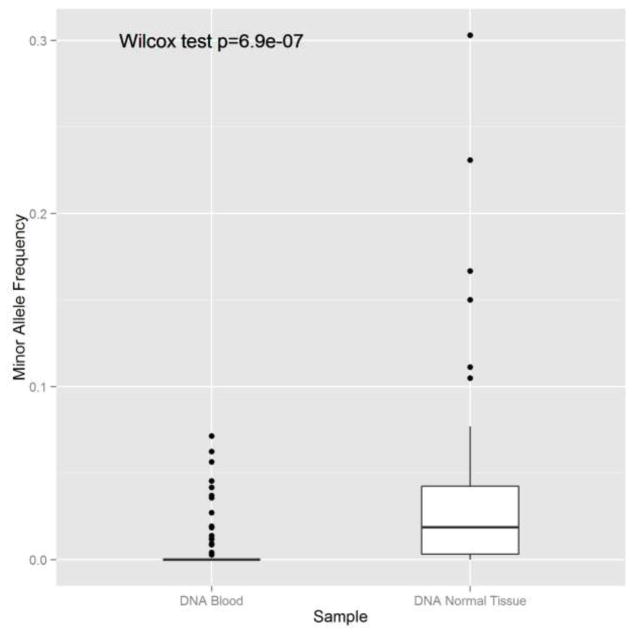
Through our anlaysis, we also showed that blood is better germline reference than adjacent normal tissue for the purpose of somatic mutation inference because adjacent normal tissue is more likely to receive tumor contamination. All three somatic mutation calling tools had higher sensitivity and specificity when using blood as a germline reference as compared to adjacent normal (Table 1). Furthermore, we observed a higher MAF of mutated sites in exome sequencing data generated from adjacent normal tissues than from blood (Figure 4, Wilcox Rank Sum test pvalue < 0.0001). Even for blood, there are some low frequency mutated alleles occasionally, we suspect that these were from mutated circulating cell free DNA or random errors. Overall, the evidence supports the hypothesis that tumor contamination is more prevalant in adjacent normal tissues.
Figure 4.
The MAF of mutated sites is higher in adjacent normal tissue than blood, suggesting that adjacent normal tissues are subjected to tumor contamination.
Similar to other somatic callers, GLMVC can be customized with multiple parameters, such as minimum base quality and minimum read quality. For GLMVC, the two most important parameters are the minimum mutated allele frequency (MMAF) and the minimum mutated allele reads (MMAR) in tumor sample. In Table 2, we presented four combination scenarios for these two parameters. When increasing MMAF or MMAR thresholds, sensitivity increases but specificity decreases, which represents the tradeoff between sensitivity and specificity. The selection of proper parameters entirely depends on the goal of the analysis. If the goal is to identify the most probable somatic mutation candidates, then more stringent parameters are recommended. If the goal is to identify all potential somatic mutation candidates, less stringent parameters should be used. For RNA, due to the high false positive rate in RNAseq data, we strongly recommend stringent parameters, such as MMAF >= 0.1 and MMAR >= 5 in tumor sample.
Table 2.
Comparison of somatic mutations detected using different filter methods
| Source | MMAF1 | MMAR2 | Detected | Exonic mutations | TCGA reported | Sensitivity | Specificity | F-score |
|---|---|---|---|---|---|---|---|---|
| DNA NB | 0.08 | 4 | 2367 | 676 | 325 | 0.849 | 0.481 | 0.614 |
| DNA NB | 0.08 | 5 | 2209 | 652 | 322 | 0.841 | 0.494 | 0.622 |
| DNA NB | 0.1 | 4 | 1996 | 564 | 320 | 0.836 | 0.567 | 0.676 |
| DNA NB | 0.1 | 5 | 1891 | 553 | 318 | 0.83 | 0.575 | 0.679 |
| DNA NT | 0.08 | 4 | 2213 | 609 | 283 | 0.739 | 0.465 | 0.571 |
| DNA NT | 0.08 | 5 | 2113 | 593 | 283 | 0.739 | 0.477 | 0.58 |
| DNA NT | 0.1 | 4 | 1927 | 524 | 280 | 0.731 | 0.534 | 0.617 |
| DNA NT | 0.1 | 5 | 1840 | 511 | 282 | 0.736 | 0.552 | 0.631 |
| RNA NT | 0.08 | 4 | 14258 | 3277 | 92 | 0.24 | 0.028 | 0.05 |
| RNA NT | 0.08 | 5 | 10407 | 1981 | 90 | 0.235 | 0.045 | 0.076 |
| RNA NT | 0.1 | 4 | 12087 | 2647 | 91 | 0.238 | 0.034 | 0.06 |
| RNA NT | 0.1 | 5 | 8990 | 1639 | 89 | 0.232 | 0.054 | 0.088 |
minimum minor allele frequency in tumor sample
minimum minor allele reads in tumor sample
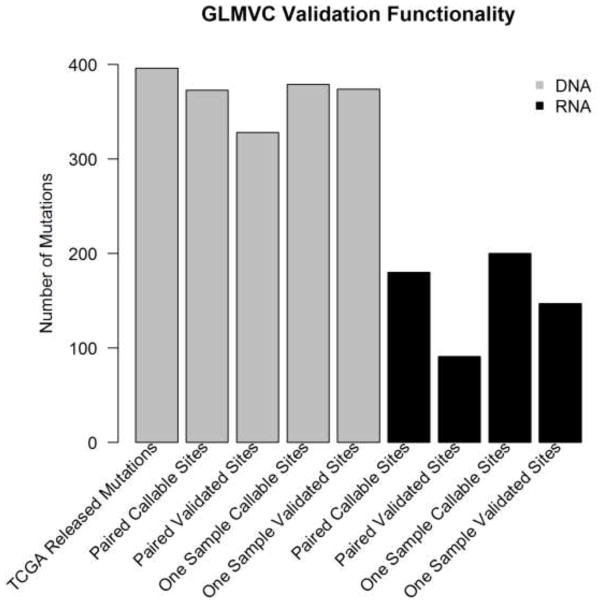
In addition to the somatic mutation detection from all callable sites, GLMVC supports a functionality that validates a list of user predefined somatic mutations using paired normal-tumor or non-paried tumor data. When using normal-tumor paired samples to validate, GLMVC performs somatic mutation detection based on the brGLM model as described earlier. However, raw p-values from brGLM are used instead of adjusted p-values because in validation, only some supporting evidence is required to justify the correctness of the somatic mutations being validated. To demonstrate, we tried to validate all 396 TCGA reported somatic mutations using corresponding paired and non-paired exome sequencing (Table S1) and RNAseq data (Table S2). Exome sequencing data validated more mutations than RNAseq data. This result is not surprising given that the TCGA inferred somatic mutations based on exome sequencing data. Within callable sites, exome sequencing achieves a higher validation rate (validated sites divided by callable sites) as compared to RNAseq (87% vs 49%) for paired data. The complete validation results using tumor-normal paired RNA and exome sequencing data can be found in Table S3 and Table S4.
When using only tumor sample for validation, GLMVC computes the allele frequency of the mutated allele for the callable sites. The user can make his/her own judgment on which percentage of the mutated allele frequency is considered valid based on extracted information. By default, GLMVC considers a somatic mutation valid if the mutated allele frequency is greater than 1%. Using one sample for validation will yield a much higher validation rate due to less filters being used. Using the same 396 somatic mutations as an example, the validation rate increased for both exome sequencing and RNAseq data (99% vs 74%) under default parameters. (Figure 5).
Figure 5.
Validation functionality of GLMVC. RNA can validate around 50% of somatic mutations using tumor-normal paired sample and 80% of somatic mutations using one sample.
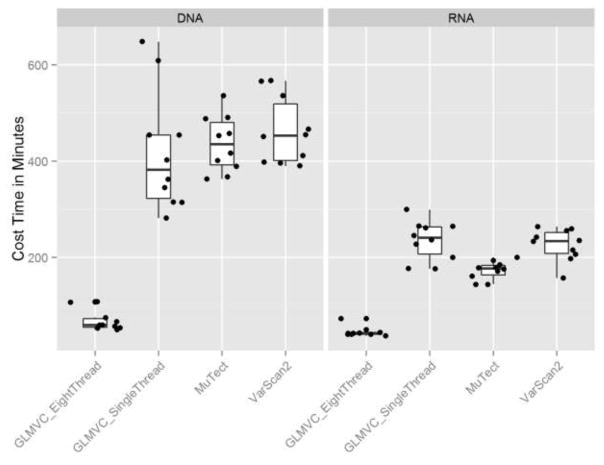
GLMVC is developed using C# in Microsoft .net enviroment. It can be installed and ran on Windows with .net framework 4.0+ or on Linux/Mac systems with mono framework. Because GLMVC was designed to process the sequencing data in parallel on the chromosome level, GLMVC has a better run-time performance than MuTect and Varscan in both single and multi thread scenarioes (Figure 6). For a a pair of tumor-normal bam files of 8GB each, the total runtime for GLMVC is roughly one hour and 40 minutes on an eight core CPU with 2.8 Ghz speed and 16 GB memory. GLMVC is freely available for download at the following website: https://github.com/shengqh/GLMVC/wiki.
Figure 6.
Runtime comparison between GLMVC, MuTect and Varscan. Because GLMVC is designed to process sequencing data in parallel by chromosomes, it benefits more from running on a multi-core computer than MuTect and Varscan, thus saves overall runtime substantially.
DISCUSSION
The introduction of HTS technology has made a powerful impact in the biomedical research field. One of the most utilized applications of HTS technology is to screen for somatic mutation candidates. Conventionally, somatic mutations are detected using exome or whole genome sequencing data of DNA. Whether somatic mutations can be accurately inferred from RNAseq data remains unanswered.
Tumor contamination in normal samples can perturb somatic mutation signals, causing the misclassification of somatic mutation into germline variants. The phenomenon of tumor contamination in adjacent normal tissues has been previously documented [28, 29]. To further evaluate the effect of tumor contamination, we examined the mutated allele frequency from three different sources (blood exome sequencing, adjacent normal exome sequencing and adjacent normal RNAseq), and found that the mutated allele frequencies are significantly higher for adjacent normal samples when compared to the blood samples of the same subjects. This suggests that blood is better suited as normal reference when inferring somatic mutation. When adjacent tissue is the only source for normal reference, GLMVC can mitigate the effect of tumor contamination by allowing users to adjust the mutated allele frequency threshold in a normal sample. Tumor purity was another factor we considered during the design of GLMVC. Unlike SNP, where we expect to see alternative alleles near 50%, tumor tissues are most often a mixture of mutated and normal tissues. The mutated allele frequency could vary without pattern. Procedures such as microdissection can help estimate tumor purity in the sample [30]. The expected minimum mutated allele frequency in tumor samples can be adjusted based on the estimated tumor purity.
In this study, we introduced GLMVC, a new somatic mutation caller based on the brGLM model for both high throughput DNA and RNA sequencing data. Because one of the major focuses of GLMVC is RNAseq data somatic mutation identification, GLMVC enforces strong filters to limit false positive rates. When compared with two other popular somatic mutation callers, MuTect and Varscan, GLMVC proved the best in both sensitivity and specificity. In default settings, GLMVC generates a more conservative list of somatic mutations.
We also evaluated the practicality of identifying somatic mutations using RNAseq data. Our results and other reports have shown that high false positive rate is unavoidable when detecting variants from RNAseq data. Thus far, we have given several potential reasons for this, including complexity in the alignment due to splicing, RNA editing, and random errors introduced during reverse transcription or PCR amplification. Another unavoidable drawback of detecting variants from RNAseq data is the limitation that results from RNA expression. RNAseq data’s sequencing depth coverage is highly correlated to RNA expression. For a normal human, as much as 40%–50% of all genes may not be expressed. Thus, RNAseq data does not have enough coverage on the unexpressed genes to infer any variants. Given all of the possible complications with identifying SNVs using RNAseq data, we designed GLMVC to consider these complications while processing RNAseq data, and, by default, GLMVC focuses on specificity rather than sensitivity.
In conclusion, our study made three contributions to the field of somatic mutation research. First, we developed GLMVC, a novel somatic mutation caller with competitive performance for both DNA and RNA sequencing data. The ability to detect somatic mutation in RNAseq data offers new opportunities for reanalyzing and repurposing of accumulated RNAseq data. For example, expression quantitative trait loci (eQTL) are detected by analyzing SNP and gene expression data. Similar study can be conducted with RNAseq data to examine the relationship between gene expression and somatic mutation. It has been shown that somatic mutation resides in or around splice junctions can cause the deletion of exon or partial gene [31]. Second, we demonstrated that in general, somatic mutation detection will be more accurate using DNAseq data than RNAseq data. Due to the high false positive rate in the somatic mutation calls in RNAseq data, we recommend additional validation using different assays, such as RT-PCR or Sanger sequencing. However, RNAseq data can serve as a good validation source for somatic mutations inferred from DNAseq data. Third, we demonstrated that blood is a better germline reference than adjacent normal for somatic mutation inference purposes.
Supplementary Material
Figure S1. Pubmed search results for studies on somatic mutation in cancer, and sequencing. A steady increasing pattern can be observed from 1984 to 2009. When high throughput sequencing technology became popular in around 2010, a steeper slope of increase in somatic mutation studies can be observed.
Research Highlight.
We have developed GLMVC, a freely-available novel mutation caller with improved performance on RNAseq data.
GLMVC has better combination of sensitivity and specificity with faster run-time than comparable programs.
We confirm blood is a better germline reference than adjacent normal tissue.
Acknowledgments
QS, SZ, YS, YG were supported by NIH grant P30 CA68485. We would also like to thank Stephanie Page Hoskins for editorial support. The authors declare that there is no conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shendure J. The beginning of the end for microarrays? Nat Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 3.Guo Y, Sheng Q, Li J, Ye F, Samuels DC, Shyr Y. Large Scale Comparison of Gene Expression Levels by Microarrays and RNAseq Using TCGA Data. PLoS One. 2013;8:e71462. doi: 10.1371/journal.pone.0071462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Y, Li CI, Ye F, Shyr Y. Evaluation of read count based RNAseq analysis methods. BMC genomics. 2013;14(Suppl 8):S2. doi: 10.1186/1471-2164-14-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han L, Vickers KC, Samuels DC, Guo Y. Alternative applications for distinct RNA sequencing strategies. Brief Bioinform. 2014 doi: 10.1093/bib/bbu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang P, Samuels DC, Lehmann B, Stricker T, Pietenpol J, Shyr Y, Guo Y. Mitochondria sequence mapping strategies and practicability of mitochondria variant detection from exome and RNA sequencing data. Brief Bioinform. 2015 doi: 10.1093/bib/bbv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Ye F, Sheng Q, Clark T, Samuels DC. Three-stage quality control strategies for DNA re-sequencing data. Briefings in Bioinformatics. 2013 doi: 10.1093/bib/bbt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y, Li J, Li CI, Long J, Samuels DC, Shyr Y. The effect of strand bias in Illumina short-read sequencing data. BMC genomics. 2012;13:666. doi: 10.1186/1471-2164-13-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin W, Piskol R, Tan MH, Li JB. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1210419. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 12.Pickrell JK, Gilad Y, Pritchard JK. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1210484. author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Li J, Li CI, Shyr Y, Samuels DC. MitoSeek: extracting mitochondria information and performing high-throughput mitochondria sequencing analysis. Bioinformatics. 2013;29:1210–1211. doi: 10.1093/bioinformatics/btt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piskol R, Ramaswami G, Li JB. Reliable Identification of Genomic Variants from RNA-Seq Data. Am J Hum Genet. 2013;93:641–651. doi: 10.1016/j.ajhg.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinman CL, Majewski J. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1209658. author reply 1302. [DOI] [PubMed] [Google Scholar]
- 16.Duitama J, Srivastava P, Mandoiu I. Towards accurate detection and genotyping of expressed variants from whole transcriptome sequencing data. BMC genomics. 2012;13:S6. doi: 10.1186/1471-2164-13-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Zhu K, Liu F, Wang Y, Shen J, Jin J, Wang Z, Chen L, Li J, Xu M. Identification of somatic mutations in human prostate cancer by RNA-Seq. Gene. 2013;519:343–347. doi: 10.1016/j.gene.2013.01.046. [DOI] [PubMed] [Google Scholar]
- 18.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosmidis I, Firth D. Bias reduction in exponential family nonlinear models. Biometrika. 2009;96:793–804. [Google Scholar]
- 20.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 21.Quinn EM, Cormican P, Kenny EM, Hill M, Anney R, Gill M, Corvin AP, Morris DW. Development of strategies for SNP detection in RNA-seq data: application to lymphoblastoid cell lines and evaluation using 1000 Genomes data. PLoS One. 2013;8:e58815. doi: 10.1371/journal.pone.0058815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7(Unit7):20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic acids research. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 28.Sadanandam A, Lal A, Benz SC, Eppenberger-Castori S, Scott G, Gray JW, Spellman P, Waldman F, Benz CC. Genomic aberrations in normal tissue adjacent to HER2-amplified breast cancers: field cancerization or contaminating tumor cells? Breast Cancer Res Treat. 2012;136:693–703. doi: 10.1007/s10549-012-2290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elloumi F, Hu ZY, Li Y, Parker JS, Gulley ML, Amos KD, Troester MA. Systematic Bias in Genomic Classification Due to Contaminating Non-neoplastic Tissue in Breast Tumor Samples. Bmc Med Genomics. 2011;4 doi: 10.1186/1755-8794-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otsuka Y, Ichikawa Y, Kunisaki C, Matsuda G, Akiyama H, Nomura M, Togo S, Hayashizaki Y, Shimada H. Correlating purity by microdissection with gene expression in gastric cancer tissue. Scand J Clin Lab Invest. 2007;67:367–379. doi: 10.1080/00365510601046334. [DOI] [PubMed] [Google Scholar]
- 31.Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, Hong D, Park PJ, Lee E. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. 2015;47:1242. doi: 10.1038/ng.3414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Pubmed search results for studies on somatic mutation in cancer, and sequencing. A steady increasing pattern can be observed from 1984 to 2009. When high throughput sequencing technology became popular in around 2010, a steeper slope of increase in somatic mutation studies can be observed.